Abstract
The eukaryotic signaling protein calmodulin (CaM) can bind to more than 300 known target proteins to regulate numerous functions in our body in a calcium-dependent manner. How CaM distinguishes between these various targets is still largely unknown. Here, we investigate fluctuations of the complex formation of CaM and its target peptide sequences using single-molecule force spectroscopy by AFM. By applying mechanical force, we can steer a single CaM molecule through its folding energy landscape from the fully unfolded state to the native target-bound state revealing equilibrium fluctuations between numerous intermediate states. We find that the prototypical CaM target sequence skMLCK, a fragment from skeletal muscle myosin light chain kinase, binds to CaM in a highly cooperative way, while only a lower degree of interdomain binding cooperativity emerges for CaMKK, a target peptide from CaM-dependent kinase kinase. We identify minimal binding motifs for both of these peptides, confirming that affinities of target peptides are not exclusively determined by their pattern of hydrophobic anchor residues. Our results reveal an association mode for CaMKK in which the peptide binds strongly to only partially Ca2+-saturated CaM. This binding mode might allow for a fine-tuning of the intracellular response to changes in Ca2+ concentration.
Keywords: atomic force microscopy, protein engineering, protein–target interactions
Numerous signaling pathways in plants and animals depend on calcium as their second messenger molecule. In eukaryotic cells, the small (148 aa) two-domain protein calmodulin (CaM) is the prevalent Ca2+ signaling protein. Upon binding of four Ca2+ ions to the EF-hand motifs of CaM, the flexible calcium-free apo conformation is converted into an extended holo conformation. In the Ca2+-loaded form, hydrophobic clefts are exposed in both domains of CaM, allowing additional binding to specific recognition sequences in target proteins (Fig. 1A) (1). To date, more than 300 target proteins for CaM have been described, with affinities typically in the nanomolar range (2). Simulations show a high degree of conformational plasticity for CaM, which might be necessary for binding to a large variety of targets (3). Among the targets of CaM, kinases that regulate important cellular processes such as gene transcription, muscle contraction, and neuronal growth take center stage. Reflecting its importance, the signaling pathways of CaM and its ligand binding properties have been studied extensively during the past 30 years using state of the art biophysical techniques (4). However, for most of the CaM-target sequence complexes, structural information is lacking. Furthermore, for a large majority of target peptides, no detailed information about association and dissociation kinetics, as well as about binding modes, is available (4). In particular, association of target peptides to only partially Ca2+-saturated CaM may play a crucial role for fine-tuning the intracellular response to Ca2+ signals, yet dissection of the sequence of peptide binding events remains a challenge.
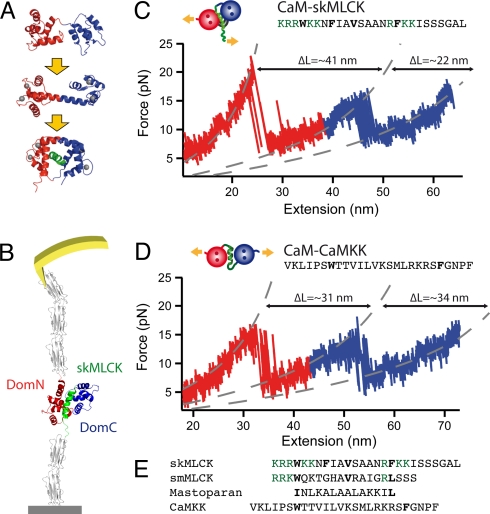
Fig. 1.
(A) Structure of CaM in different ligand binding states. DomN is shown in red, DomC in blue. Upper picture, apo CaM; middle, Ca2+ loaded form, Ca2+ ions are shown in gray; lower picture, Ca2+-CaM bound to target peptide skMLCK (green). (B) Scheme of the experimental setup. CaM-skMLCK is incorporated into filamin domains (gray) that serve as handles for attaching the protein construct to a surface and to an AFM cantilever tip. (C) Typical force vs. extension trace of skMLCK fused to CaM (CaM-skMLCK) at a pulling velocity of vpull = 0.5 nm/s. Unfolding peaks of DomN and DomC of CaM-skMLCK are shown in red and blue, respectively. WLC curves are shown in gray. In the skMLCK amino acid sequence, hydrophobic anchor residues are highlighted in bold type, charged amino acids directly adjacent to these hydrophobic residues are colored in green. (D) Force vs. extension trace of CaMKK inserted between DomN and DomC of CaM (CaM-CaMKK), recorded at a pulling velocity of 1 nm/s. Again, unfolding peaks of DomN and DomC of CaM-skMLCK are shown in red and blue, respectively. (E) Sequence alignment of the target peptides skMLCK, smMLCK, mastoparan, and CaMKK. Hydrophobic anchor residues are shown in bold font, and charged amino acids directly adjacent to hydrophobic anchor residues are colored in green.
In recent years, single-molecule mechanical methods have opened unique possibilities to study and control biomolecular conformations with unprecedented precision (5–9). In particular, several studies have investigated protein–ligand interactions on the single-molecule level (10–13). We have recently demonstrated that single-molecule force spectroscopy by AFM allows direct observation of equilibrium fluctuations of CaM (9). We compare the CaM binding peptide sequences of two target kinases, skeletal muscle myosin light chain kinase (skMLCK), which phosphorylates the regulatory light chain of myosin II (14), and CaM-dependent kinase kinase (CaMKK), which primarily phosphorylates downstream regulatory kinases that are also activated by CaM (15) (see also sequence alignment in Fig. 1E). skMLCK, for which extensive structural and kinetic data exist, has been regarded as the prototypical CaM target peptide (16, 17). In contrast, the CaMKK peptide exhibits considerable differences in comparison to most other CaM target sequences. The most striking irregularity is the reversed binding orientation relative to other peptides such as skMLCK, i.e., CaMKK binds with its C terminus to the C-terminal lobe of CaM, whereas skMLCK binds with its C terminus to the N-terminal domain of CaM (18). Binding kinetics and interactions with the individual domains of CaM are largely unknown for CaMKK. However, a protein dynamics study using NMR reveals fundamentally different binding interactions of CaMKK as compared to MLCK (19). Here we show that mechanically induced peptide binding/unbinding transitions yield detailed insights into CaM-target peptide interactions and reveal an association mode for CaMKK in which the peptide binds strongly to only partially Ca2+-saturated CaM.
Results
The experimental setup is depicted schematically in Fig. 1B. The 26-aa target sequence of skMLCK is fused to the C-terminal end of CaM via a short peptide linker (CaM-skMLCK, see sketch in Fig. 1C) (20). We incorporated CaM-skMLCK into Ig domains of Dictyostelium discoideum filamin, which serve as handles for attaching the protein construct to a surface and to an AFM cantilever tip, as described previously (9, 21). Because of its reversed binding orientation, CaMKK cannot be attached at the C-terminal end of CaM. We therefore inserted the peptide into the central interdomain linker of CaM (CaM-CaMKK, see sketch in Fig. 1D) (22).
By moving the surface and the cantilever apart, we can now exert force onto the protein chain, which increases the unbinding rate koff of the peptide and, in turn, decreases its rebinding rate kon. Therefore, the binding/unbinding kinetics of CaM target peptides can be moved to readily observable timescales, allowing real-time observation of target peptide binding/unbinding and CaM folding/unfolding transitions in equilibrium (9). A typical force vs. extension trace of CaM-skMLCK is depicted in Fig. 1C. As we have shown previously, the two peaks, colored in red and blue, correspond to the unfolding of the N-terminal (DomN) and the C-terminal domain (DomC) of CaM, respectively (9). At first sight, force vs. extension traces of CaM-CaMKK (Fig. 1D) show the same unfolding pattern, indicating that both domains still unfold separately in this protein construct. For both CaM-skMLCK and CaM-CaMKK, multiple transitions between the folded state (upper level) and the unfolded state (lower level) can be readily detected for DomN and DomC. A first indication of significantly different binding modes of these two target peptides becomes evident when the length gains upon unfolding of DomN and DomC are analyzed. In CaM-skMLCK, the increase in contour length of the first unfolding peak is significantly larger than that of the second peak (≈41 nm as opposed to ≈22 nm). For CaMKK, both length increases have comparable values (≈31 nm and ≈34 nm). The overall length increases are in excellent agreement with the expected values for both CaM-skMLCK and CaM-CaMKK (see SI Text).
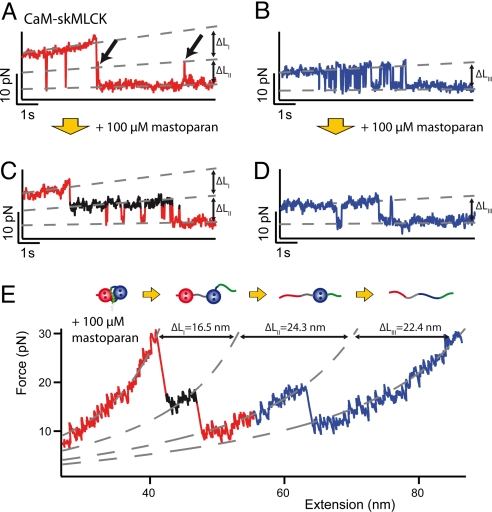
To obtain more insight into peptide binding and unbinding, we zoomed into the transition regions of DomN and DomC for force vs. time traces of CaM-skMLCK (Fig. 2) and of CaM-CaMKK (Fig. 3). Close inspection of the unfolding transitions of DomN in the CaM-skMLCK construct (Fig. 2A) reveals population of a short-lived intermediate (dashed middle level, arrows) between the upper level (fully folded and peptide bound) and the lower level (DomN unfolded). The transitions between the upper and the middle level represent skMLCK binding/unbinding fluctuations, whereas transitions between the middle level and the lower level correspond to unfolding and folding of DomN (9). In contrast, in the time traces of DomC, no additional level can be distinguished (Fig. 2B), suggesting a low affinity of the peptide to isolated DomC (17). Since the intermediate level (arrows in Fig. 2A) is so short-lived that it is barely perceptible in many of the traces, we developed a competitive assay to increase the lifetime of this level and thus study the binding/unbinding pathway in greater detail. To selectively stabilize the intermediate level (CaM completely folded, skMLCK unbound) we added the CaM binding peptide mastoparan to the solution. If skMLCK unbinds under force, mastoparan will bind to DomN and DomC, thereby prolonging the lifetime of the unbound level (middle level) by preventing rebinding of skMLCK. Fig. 2 C and D show typical time traces of CaM-skMLCK in the presence of 100 μM mastoparan in solution. Now, for DomN, the lifetime of the intermediate level is significantly increased (black level in Fig. 2C) and can be readily measured in all traces. This competitive assay now allows a more detailed understanding of the skMLCK unbinding process through precise length measurements of the unbound level. For DomC, no intermediate level is observable, as predicted from the experiments in the absence of mastoparan (Fig. 2D). It is, however, important to note that the frequency of folding/unfolding of DomC is drastically reduced in the presence of mastoparan as becomes evident by comparing Fig. 2 B and D. This effect is due to a stabilization of the folded state by mastoparan binding to DomC (9).
Fig. 2.
(A and B) Time traces of the unfolding regions of DomN (A) and DomC (B) of CaM-skMLCK, vpull = 0.5 nm/s. A short-lived intermediate level appears for DomN. No such level can be distinguished in DomC. (C and D) Time traces of DomN (C) and DomC (D) of CaM-skMLCK in the presence of 100 μM mastoparan in solution, vpull = 1 nm/s. Under these conditions, the intermediate level of DomN (shown in black) is stabilized and the transition kinetics is slowed down. The intermediate level can be found in all traces, suggesting an obligatory intermediate. (E) Force vs. extension trace of CaM-skMLCK at 100 μM mastoparan, vpull = 10 nm/s, and WLC fit curves (dashed gray lines). The sequence of the structural transitions of CaM-skMLCK under force (scheme above the trace) can be reconstructed from the length gains of the individual unfolding events.
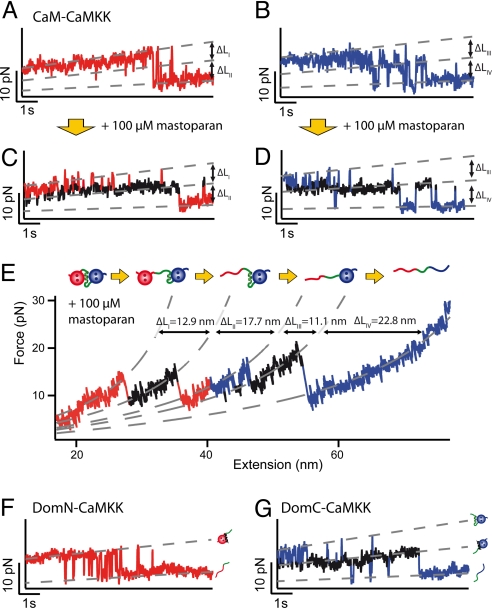
Fig. 3.
(A and B) Time traces of the unfolding regions of DomN (A) and DomC (B) of CaM-CaMKK, vpull = 1 nm/s. For both domains, a short-lived intermediate level appears. (C and D) Time traces of DomN (C) and DomC (D) of CaM-CaMKK at 100 μM mastoparan, vpull = 1 nm/s. The intermediate state (shown in black) is stabilized and can now be clearly distinguished from the two other levels. (E) Force vs. extension trace of CaM-CaMKK at 100 μM mastoparan, vpull = 10 nm/s and WLC fit curves (dashed gray lines). Above the trace, we show a scheme of the sequence of structural transitions as inferred from the increases in contour length. (F) Time trace of isolated DomN fused to CaMKK (DomN-CaMKK) at 10 μM mastoparan. No intermediate level can be detected, hence no sufficiently strong binding of CaMKK to isolated DomN takes place. (G) Time trace of isolated DomC fused to CaMKK (DomC-CaMKK) at 10 μM mastoparan, showing three levels (DomC folded, peptide-bound; DomC folded, peptide-unbound; DomC unfolded).
The assignment of the various transitions to structural events can be carried out by worm-like chain (WLC) fits (23) to force vs. extension traces. To this end, we recorded traces of CaM-skMLCK in the presence of 100 μM mastoparan at a higher pulling velocity (Fig. 2E). Since the skMLCK peptide (26 aa) is shorter than each of the two domains of CaM (ca. 70 aa residues), the lowest increase in contour length upon unfolding must be attributed to peptide unbinding. Thus, the fits clearly show that, in the first step (ΔLI = 16.5 ± 1.4 nm), skMLCK detaches from CaM. Subsequently, DomN (ΔLII = 24.3 ± 1.3 nm) and finally DomC (ΔLIII = 22.4 ± 1.3 nm) unfold (9). A detailed analysis of contour length increases reveals that upon unbinding of skMLCK, also the central interdomain linker (≈15 aa residues) unfolds, leading to a larger increase in contour length than expected for the peptide alone (Table S1 and Fig. S1). Apparently, binding and unbinding of skMLCK happens cooperatively in a single step, i.e., no partial dissociation intermediates of skMLCK can be observed.
In contrast to CaM-skMLCK, three levels appear to be populated in the unfolding time traces of both DomN and DomC of CaM-CaMKK (Fig. 3 A and B). However, similar to CaM-skMLCK, a clear discrimination of the middle level for DomN is not possible due to the fast kinetics. Again, a competitive assay with mastoparan in solution allows stabilizing the short-lived intermediate states, which helps to clearly identify the three distinct levels in the unfolding traces of the two domains (Fig. 3 C and D). For both DomN and DomC, we interpret transitions between the upper and the middle level as peptide binding/unbinding fluctuations and transitions between the middle and the lower level as domain folding/unfolding. The assumption that the middle levels correspond to a folded domain with unbound target peptide is corroborated by the increased lifetime of these two levels in the presence of mastoparan.
As before, the assignment of unfolding transitions to structural events can be analyzed in more detail by WLC fits to traces recorded at 100 μM mastoparan and at a higher pulling velocity (Fig. 3E). As expected, the peptide unbinding events (ΔLI = 12.9 ± 1.3 nm, ΔLIII = 11.1 ± 1.2 nm) exhibit shorter length increases than the domain unfolding transitions (ΔLII = 17.7 ± 1.4 nm, ΔLIV = 22.8 ± 1.2 nm) (see Fig. S2). The fact that ΔLI is smaller than the expected value of ≈20 nm for unbinding of the full CaMKK peptide clearly indicates that not the full peptide, but only a major N-terminal part of CaMKK (CaMKK N) detaches from CaM in the first transition, while the C-terminal part of the target peptide (CaMKK C) unbinds in the third transition (see SI Text for detailed calculations of contour length increases). Midpoint unbinding forces of CaMKK N and CaMKK C are considerably lower than those of skMLCK (see Table 1).
Table 1.
Mean unbinding force of different CaM target peptides at equilibrium conditions (vpull = 1 nm/s, 10 mM CaCl2)
| Target peptides | Mean unbinding force |
|---|---|
| skMLCK(1–26) | 19.3 ± 0.8 pN |
| skMLCK(1–18) | 16.8 ± 0.7 pN |
| smMLCK | 16.6 ± 0.7 pN |
| CaMKK N | 14.5 ± 0.6 pN |
| CaMKK C | 15.4 ± 0.6 pN |
Errors are calculated as SEM + 2% systematic error from force calibration.
Naively, one might expect that the sum of the lengths of the two peptide unbinding transitions ΔLI + ΔLIII = 24.0 ± 1.3 nm should equal the total length of the CaMKK peptide. However, the total length of the CaMKK peptide is only ≈20 nm and hence clearly shorter. At the same time, we measure a length increase of only ΔLII = 17.7 ± 1.4 nm for the unfolding of DomN. For unfolding of DomN, however, we would expect a significantly longer length gain of ≈24 nm (see also corresponding length in Fig. 2E). We can only understand this apparent shortening of DomN and lengthening of CaMKK if we assume that a part of CaMKK N rebinds to DomC after unfolding of DomN (see SI Text, Table S1, and Fig. S1). With ΔLIV = 22.8 ± 1.2 nm, the contour length increase upon unfolding of DomC of CaM-CaMKK is in perfect agreement with the value determined earlier (Fig. 2E). In summary, our analysis allows the following detailed description of the sequence of unfolding events of CaM-CaMKK (see also scheme in Fig. 3E). First, a large part of CaMKK detaches from CaM. In a second step, DomN unfolds, while quasi-simultaneously, several amino acids of CaMKK rebind to DomC. As the force is increased again, the remaining part of the CaMKK peptide unbinds from DomC, which in turn unfolds last.
From the data presented so far, it is unclear whether the observed interaction between CaMKK and DomN necessitates the presence of DomC, i.e., whether folded and Ca2+-loaded DomN is sufficient for CaMKK binding. We therefore deleted DomC from the CaM-CaMKK protein construct (DomN-CaMKK). For this protein construct, we do not detect an intermediate level, suggesting DomN alone cannot bind CaMKK strongly (Fig. 3F). The observation that the N-terminal part of CaMKK can only bind to full-length CaM indicates a cooperative binding mode in which both domains have to interact with the peptide segment to allow formation of a stable bond. As expected, we can still observe CaMKK binding/unbinding transitions in the DomC-CaMKK construct where the complete DomN is removed (Fig. 3G). Contour length increases and unbinding forces of DomC-CaMKK are in excellent agreement with the respective transitions of full-length CaM-CaMKK (Table S2), confirming the above interpretation of the four transitions in Fig. 3E.
Comparing skMLCK and CaMKK experiments, we clearly find different degrees of binding cooperativity. skMLCK binds strongly only to fully folded CaM, but does not form a stable complex with DomC after unfolding of DomN. Also for isolated DomN, no significant affinity to skMLCK has been reported (17). Taken together, these results show a high degree of cooperativity between both domains of CaM for skMLCK binding, since only in the presence of both domains, a strong interaction between skMLCK and CaM is possible. For CaMKK, we observe a lower degree of binding cooperativity. The peptide dissociates in two steps, first CaMKK N unbinds from DomN, then in a second step CaMKK C dissociates from DomC. Folded and Ca2+-loaded DomC is sufficient for stable association of CaMKK. However, binding of CaMKK to DomN is only possible in the presence of DomC, revealing considerable binding cooperativity between both domains of CaM for association of CaMKK N.
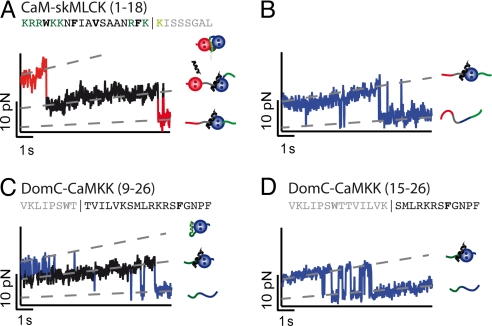
We next investigated the effect of truncation mutants of target peptides on their affinity to determine minimal binding motifs for strong association to CaM. In the 26-residue peptide skMLCK, amino acids 5 to 17 correspond to the canonical CaM binding motif, characterized by a pattern of hydrophobic anchor residues (4) (see also Fig. 1E). In most binding studies, a shortened peptide skMLCK (1–18), which contains the full hydrophobic anchor motif, has been used (24, 25). In Fig. 4 A and B, sample traces of CaM-skMLCK (1–18) in the presence of mastoparan are depicted. Again, an intermediate level appears for DomN (Fig. 4A) but not for DomC (Fig. 4B), so that the protein construct qualitatively shows the same binding/unbinding behavior as full-length CaM-skMLCK (1–26). However, a detailed analysis of the midpoint unfolding forces of the transition region reveals that forces are lower for the truncated peptide (see Fig. 4A and Table 1). This finding suggests that the amino acid residues at the C-terminal end of skMLCK adjacent to the hydrophobic anchor residue Phe-17 are important for optimizing bond strength.
Fig. 4.
(A and B) Time traces of the transition region of DomN (A) and DomC (B) of the CaM-skMLCK (1–18) truncation mutant at 100 μM mastoparan, vpull = 1 nm/s. As in the case of full-length CaM-skMLCK, an intermediate level appears for DomN (shown in black), but not for DomC. In the amino acid sequence of the target peptide, residues that have been removed are colored in gray and light green. (C) Time trace of DomC-CaMKK (9–26) at 10 μM mastoparan, vpull = 1 nm/s. This truncated version of CaMKK is sufficient for strong binding to DomC. (D) Time trace of DomC-CaMKK (15–26) at 10 μM mastoparan, vpull = 1 nm/s. No intermediate level can be resolved, suggesting residues 9–14 of CaMKK are important for high-affinity binding to DomC.
If the skMLCK peptide is shortened even further to amino acid residues 4–17, i.e., to the minimal peptide length compatible with conservation of all hydrophobic anchor residues, a binding/unbinding transition cannot be detected any more (Fig. S2B). Thus, we find an important role of the charged amino acid residues at the N terminus of skMLCK for high-affinity binding (see also Fig. 1E). These N-terminal residues are mostly discussed in the literature as being responsible for the orientation of the peptide in the binding pocket (4, 18). Furthermore, the MLCK variant from smooth muscle, smMLCK, unbinds at similar forces as skMLCK (1–18) (Table 1 and Fig. S2 A and C), whereas no similar cooperative high affinity binding under force takes place for mastoparan (Fig. S2D), corroborating our previous finding of a 2:1 stoichiometry with lower affinities for mastoparan binding to CaM (9).
Next, we set out to determine the minimal CaMKK sequence necessary for strong binding to DomC. To this end, we engineered truncation mutants of DomC-CaMKK by deleting 8 or 14 aa residues at the N-terminal end of CaMKK (Fig. 4 C and D). We find that CaMKK (9–26) still binds strongly to DomC, with midpoint unbinding forces and contour length increases that are indistinguishable from full-length CaMKK (Fig. 4C and Table S2). In particular, this finding suggests that the hydrophobic anchor residue Trp-7 of CaMKK, interacting with DomN in the complex with full-length CaM (18), does not associate with isolated DomC. Removal of the next 6 aa residues [CaM-CaMKK (15–26)], however, completely abolishes binding to DomC (Fig. 4D). Thus, we conclude that the minimal binding motif of CaMKK for binding to DomC starts between residue 9 and 14 of CaMKK (1–26).
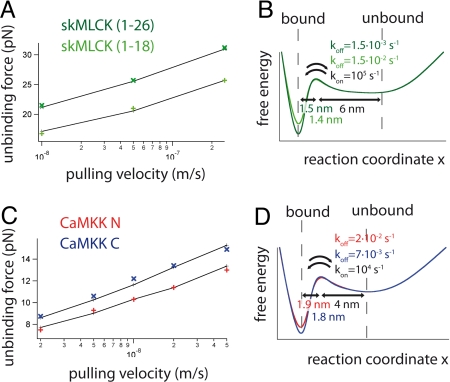
To extrapolate the observed peptide binding/unbinding kinetics to zero force, we measured the pulling speed dependence of unbinding forces under nonequilibrium conditions for both skMLCK and CaMKK. Mean unbinding forces for both full-length skMLCK and the truncated peptide skMLCK (1–18) are plotted in Fig. 5A. As before under equilibrium conditions, the mean unbinding force of skMLCK (1–18) is lower than that of skMLCK (1–26) at all measured pulling velocities. We can now determine the zero-force potential energy landscape for peptide binding and unbinding by reproducing the experimental data with Monte Carlo simulations (see SI Text and Fig. S3) (26). We find a 10-fold higher unbinding rate koff for skMLCK (1–18) as compared to skMLCK (1–26), while binding rate kon and potential widths ΔxN-TS and ΔxU-TS remain roughly the same (Fig. 5B).
Fig. 5.
(A) Pulling speed dependence of mean unbinding force (first peak of the trace) of skMLCK (1–26; dark green) and skMLCK (1–18; light green) at nonequilibrium conditions. Results of Monte Carlo simulations are shown in black. (B) Calculated potential energy landscape at zero force for binding/unbinding of skMLCK (1–26) and skMLCK (1–18) to CaM. (C) Unbinding forces vs. pulling speed are shown for CaMKK N (first peak in Fig. 3E) and CaMKK C (third peak in Fig. 3E) in red and blue, respectively. Since equilibrium conditions prevail at low pulling velocities, forces of the first unbinding transition were analyzed. Results of Monte Carlo simulations are shown in black. (D) Calculated potential energy landscape at zero force for binding/unbinding of CaMKK N and CaMKK C.
In a further set of experiments, we determined the pulling speed dependence of the peptide unbinding transitions of CaM-CaMKK. In Fig. 5C, we plot the average unbinding forces of the two parts of CaMKK that detach from CaM-CaMKK (CaMKK N and CaMKK C) at different pulling velocities. The unbinding forces of CaMKK N lie ≈1 pN below the corresponding values for DomC, highlighting again a relatively strong association with isolated DomC. In the extrapolated potential energy landscape for CaMKK binding/unbinding at zero-force conditions, we find slightly higher potential widths ΔxN-TS as compared to skMLCK (Fig. 5D). The zero-force lifetimes of CaMKK complexed with full-length CaM or DomC are on the order of ≈100 s, similar to the lifetime of skMLCK bound to CaM.
Discussion
Single-molecule force spectroscopy by AFM has allowed us to observe mechanically induced target peptide binding/unbinding transitions in real-time. By applying mechanical force to CaM-target peptide complexes, we were able to slow down the transition kinetics to observable timescales and to clearly separate the individual steps of peptide unbinding. The hierarchy of folding and target peptide binding as well as the sequence of peptide unbinding events could be determined in great detail for skMLCK and CaMKK by steering the protein-target complex through its potential energy landscape from the unfolded state to the fully folded target bound state. Denaturants typically used in bulk experiments like urea or guanidinium hydrochloride act rather unspecifically on the potential energy landscape, which would make acquisition of comparable information difficult.
Our results reveal different degrees of cooperativity for the prototypical CaM-binding sequence skMLCK and the more irregular CaMKK sequence. The skMLCK peptide necessitates fully folded CaM for stable binding and dissociates from CaM in a single step upon force application. This finding is in full agreement with a previous experiment in which we found that full-length CaM unfolds at significantly higher forces than isolated domains if free skMLCK is added to the buffer (9). In contrast, CaMKK already binds strongly to isolated DomC and detaches from CaM in a two-step manner, first from DomN, finally also from DomC. Thus, skMLCK displays a highly cooperative binding behavior, whereas a lower degree of interdomain cooperativity emerges for CaMKK. The affinity of skMLCK to isolated DomC has been reported to be at least three orders of magnitude lower than the affinity to full-length CaM (17). Hence, the lifetime of the DomC-skMLCK complex is too short to allow observation of peptide binding/unbinding transitions under load. In CaMKK, only the binding of the N-terminal part of the peptide requires the cooperative interplay of both domains of CaM. On the spectrum from the high-cooperativity binding in the case of skMLCK to the zero-cooperativity binding mode of mastoparan with a stoichiometry of 2:1 (9), CaMKK is therefore located in the middle, displaying partially cooperative binding to CaM. It is interesting to note, that the true CaM target sequences skMLCK and CaMKK show significantly stronger binding (bond lifetime ≈100 s) to CaM than the wasp venom mastoparan (bond lifetime ≈0.01 s) (9). Low unbinding rates from target sequences might be important for the physiological function of CaM, given that Ca2+ signals often take the form of short-lived spikes (27, 28) and that the number of target sites usually exceeds the concentration of Ca2+-saturated CaM inside the cell (29).
Typically, CaM binding peptides are classified according to their pattern of hydrophobic anchor residues. Our experiments with truncation mutants highlight the importance of other residues for the formation of stable complexes. For skMLCK, we found that deletion of residues 19–26 at the C-terminal end of the peptide increases koff by a factor of ≈10. Additional suppression of residues 1–3 and 18 completely abolishes binding in our assay, an effect suggesting a major role of these charged residues for bond formation (see also Fig. 1E). Also in the case of CaMKK, we find that the pattern of hydrophobic amino acid residues (4) may not be sufficient to characterize a target sequence. Even after removal of the first of only two hydrophobic anchor residues (Trp-7), the peptide still binds strongly to DomC.
Our observation that the CaMKK peptide can bind strongly already to isolated DomC may have significant physiological implications on the Ca2+ dependent regulation of kinases. Given that DomC has a higher affinity for Ca2+ than DomN, at low Ca2+ concentrations only DomC will be in the Ca2+-loaded form (30). Hence, already partially Ca2+-saturated CaM may bind strongly to CaMKK at low Ca2+ concentrations when binding to other sequences requiring full Ca2+ saturation of CaM for strong binding might still be impaired, as is the case for skMLCK. Since CaMKK is located at the top of several CaM-dependent signaling cascades, discrimination between different targets is key to proper functioning of this signaling molecule (15). Activation of CaMKK by partially Ca2+-loaded CaM could provide a mechanism for distinguishing between the different downstream targets of CaMKK. For instance, protein kinase B might be phoshorylated and activated by a complex of partially Ca2+-loaded CaM and CaMKK, while the activity of other CaMKK targets that necessitate additional binding of fully Ca2+-saturated CaM (e.g., CaM-dependent kinase I and IV) would still be blocked.
In summary, we could show that single-molecule force spectroscopy by AFM allows detailed insights into CaM-target peptide interactions, ligand binding modes, and association kinetics on the single-molecule level. In future research, measurements in the physiologically relevant range of low calcium concentrations <1 μM might allow direct investigation of CaMKK binding to only partially Ca2+-saturated CaM. We postulate that this technique will also reveal a wealth of information about other uncommon CaM binding sequences such as IQ repeats (4).
Methods
Cloning and Protein Expression.
To obtain a multidomain protein suitable for mechanical unfolding experiments, human CaM (148 aa residues) was sandwiched between domains three and four of the five rod domains of the actin cross-linker Dictyostelium discoideum filamin (9, 21). Sequences for skMLCK, smMLCK, and mastoparan were inserted between the C-terminal end of CaM and filamin domain four, with a 4-aa spacer at the N-terminal end and a 2-aa spacer at the C-terminal end of the peptide sequence (20). The sequence of the CaMKK peptide was inserted into the α-helical interdomain linker of CaM between residues 79 and 80, with spacers of 4 aa at both the N-terminal and the C-terminal end of the peptide sequence (22). For single-domain experiments, residues 1–79 (DomN) or 80–148 (DomC) of CaM were removed from the protein construct.
All constructs were verified by DNA sequencing. Proteins were purified by nickel-NTA affinity chromatography followed by gel-filtration chromatography. Mastoparan was purchased from Peptanova.
Single-Molecule Force Spectroscopy.
Single-molecule force spectroscopy was performed on a custom-built low-drift AFM, as described previously (9). Gold-coated cantilevers (Biolever Type B; Olympus) with a typical spring constant of 6 pN/nm were used in all experiments. Experiments were carried out at room temperature in a 150 mM KCl, 50 mM Tris buffer solution at pH 8.0, which is a typical buffer for CaM studies, and at 10 mM CaCl2 (17, 30). In a typical experiment, protein solution (final concentration ≈1 μM) was applied to a freshly activated Ni-NTA surface and incubated for 10 min before starting the experiment. Force vs. time and extension vs. time traces were recorded at pulling speeds ranging from 0.5 nm/s to 250 nm/s with a sampling rate of 20 kHz. Piezo stage movement and the corresponding force vs. extension curve are shown in Fig. S4 for a typical trace recorded at 1 nm/s. All data were screened and analyzed in Igor Pro (Wavemetrics) as described in the SI Text. Before analysis, the traces were smoothed by box filtering. The success rate for protein pick-up upon a fast approach-retract pulling cycle was generally ≈5%, which is a typical value for single-molecule force spectroscopy experiments (31). As soon as a correct single-molecule attachment was established, the success rate for a low velocity (≈1 nm/s) pulling cycle was 10%–50%, mostly limited by premature protein desorption from the cantilever. More detailed information on single-molecule force spectroscopy experiments can be found in the SI Text.
Supplementary Material
Acknowledgments.
We thank M. Bertz and F. Ziegler for inspiring discussions and comments on the manuscript. This work was supported by Deutsche Forschungsgemeinschaft Grant RI 990/3-1. J.P.J. was supported by the International Graduate School “Materials Science of Complex Interfaces.”
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904654106/DCSupplemental.
References
- 1.Chin D, Means AR. Calmodulin: A prototypical calcium sensor. Trends Cell Biol. 2000;10:322–328. doi: 10.1016/s0962-8924(00)01800-6. [DOI] [PubMed] [Google Scholar]
- 2.Shen X, Valencia CA, Szostak JW, Dong B, Liu R. Scanning the human proteome for calmodulin-binding proteins. Proc Natl Acad Sci USA. 2005;102:5969–5974. doi: 10.1073/pnas.0407928102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen YG, Hummer G. Slow conformational dynamics and unfolding of the calmodulin C-terminal domain. J Am Chem Soc. 2007;129:2414–2415. doi: 10.1021/ja067791a. [DOI] [PubMed] [Google Scholar]
- 4.Yamniuk AP, Vogel HJ. Calmodulin's flexibility allows for promiscuity in its interactions with target proteins and peptides. Mol Biotechnol. 2004;27:33–57. doi: 10.1385/MB:27:1:33. [DOI] [PubMed] [Google Scholar]
- 5.Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cecconi C, Shank EA, Bustamante C, Marqusee S. Direct observation of the three-state folding of a single protein molecule. Science. 2005;309:2057–2060. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- 7.Wiita AP, et al. Probing the chemistry of thioredoxin catalysis with force. Nature. 2007;450:124–127. doi: 10.1038/nature06231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng SP, et al. Designing an extracellular matrix protein with enhanced mechanical stability. Proc Natl Acad Sci USA. 2007;104:9633–9637. doi: 10.1073/pnas.0609901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Junker JP, Ziegler F, Rief M. Ligand-dependent equilibrium fluctuations of single calmodulin molecules. Science. 2009;323:633–637. doi: 10.1126/science.1166191. [DOI] [PubMed] [Google Scholar]
- 10.Kedrov A, Krieg M, Ziegler C, Kuhlbrandt W, Muller DJ. Locating ligand binding and activation of a single antiporter. EMBO Rep. 2005;6:668–674. doi: 10.1038/sj.embor.7400455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Junker JP, Hell K, Schlierf M, Neupert W, Rief M. Influence of substrate binding on the mechanical stability of mouse dihydrofolate reductase. Biophys J. 2005;89:L46–L48. doi: 10.1529/biophysj.105.072066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao Y, Balamurali MM, Sharma D, Li H. A functional single-molecule binding assay via force spectroscopy. Proc Natl Acad Sci USA. 2007;104:15677–15681. doi: 10.1073/pnas.0705367104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puchner EM, et al. Mechanoenzymatics of titin kinase. Proc Natl Acad Sci USA. 2008;105:13385–13390. doi: 10.1073/pnas.0805034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blumenthal DK, et al. Identification of the calmodulin-binding domain of skeletal muscle myosin light chain kinase. Proc Natl Acad Sci USA. 1985;82:3187–3191. doi: 10.1073/pnas.82.10.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wayman GA, Lee YS, Tokumitsu H, Silva A, Soderling TR. Calmodulin-kinases: Modulators of neuronal development and plasticity. Neuron. 2008;59:914–931. doi: 10.1016/j.neuron.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikura M, et al. Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science. 1992;256:632–638. doi: 10.1126/science.1585175. [DOI] [PubMed] [Google Scholar]
- 17.Bayley PM, Findlay WA, Martin SR. Target recognition by calmodulin: Dissecting the kinetics and affinity of interaction using short peptide sequences. Protein Sci. 1996;5:1215–1228. doi: 10.1002/pro.5560050701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osawa M, et al. A novel target recognition revealed by calmodulin in complex with Ca2+-calmodulin-dependent kinase kinase. Nat Struct Biol. 1999;6:819–824. doi: 10.1038/12271. [DOI] [PubMed] [Google Scholar]
- 19.Marlow MS, Wand AJ. Conformational dynamics of calmodulin in complex with the calmodulin-dependent kinase kinase alpha calmodulin-binding domain. Biochemistry. 2006;45:8732–8741. doi: 10.1021/bi060420m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyawaki A, et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 21.Schwaiger I, Kardinal A, Schleicher M, Noegel AA, Rief M. A mechanical unfolding intermediate in an actin-crosslinking protein. Nat Struct Mol Biol. 2004;11:81–85. doi: 10.1038/nsmb705. [DOI] [PubMed] [Google Scholar]
- 22.Truong K, et al. FRET-based in vivo Ca2+ imaging by a new calmodulin-GFP fusion molecule. Nat Struct Biol. 2001;8:1069–1073. doi: 10.1038/nsb728. [DOI] [PubMed] [Google Scholar]
- 23.Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 24.Findlay WA, Martin SR, Beckingham K, Bayley PM. Recovery of native structure by calcium binding site mutants of calmodulin upon binding of sk-MLCK target peptides. Biochemistry. 1995;34:2087–2094. doi: 10.1021/bi00007a001. [DOI] [PubMed] [Google Scholar]
- 25.Hultschig C, Hecht HJ, Frank R. Systematic delineation of a calmodulin peptide interaction. J Mol Biol. 2004;343:559–568. doi: 10.1016/j.jmb.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 26.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 27.Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 28.Gu X, Spitzer NC. Distinct aspects of neuronal differentiation encoded by frequency of spontaneous Ca2+ transients. Nature. 1995;375:784–787. doi: 10.1038/375784a0. [DOI] [PubMed] [Google Scholar]
- 29.Teruel MN, Chen W, Persechini A, Meyer T. Differential codes for free Ca(2+)-calmodulin signals in nucleus and cytosol. Curr Biol. 2000;10:86–94. doi: 10.1016/s0960-9822(00)00295-5. [DOI] [PubMed] [Google Scholar]
- 30.Linse S, Helmersson A, Forsen S. Calcium binding to calmodulin and its globular domains. J Biol Chem. 1991;266:8050–8054. [PubMed] [Google Scholar]
- 31.Rounsevell RW, Forman JR, Clarke J. Atomic force microscopy: Mechanical unfolding of proteins. Methods. 2004;34:100–111. doi: 10.1016/j.ymeth.2004.03.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.