Abstract
Ergot alkaloids are toxins and important pharmaceuticals that are produced biotechnologically on an industrial scale. The first committed step of ergot alkaloid biosynthesis is catalyzed by dimethylallyl tryptophan synthase (DMATS; EC 2.5.1.34). Orthologs of DMATS are found in many fungal genomes. We report here the x-ray structure of DMATS, determined at a resolution of 1.76 Å. A complex of DMATS from Aspergillus fumigatus with its aromatic substrate L-tryptophan and with an analogue of its isoprenoid substrate dimethylallyl diphosphate reveals the structural basis of this enzyme-catalyzed Friedel-Crafts reaction, which shows strict regiospecificity for position 4 of the indole nucleus of tryptophan as well as unusual independence of the presence of Mg2+ ions. The 3D structure of DMATS belongs to a rare β/α barrel fold, called prenyltransferase barrel, that was recently discovered in a small group of bacterial enzymes with no sequence similarity to DMATS. These bacterial enzymes catalyze the prenylation of aromatic substrates in the biosynthesis of secondary metabolites (i.e., a reaction similar to that of DMATS).
Keywords: EC 2.5.1.34, ergot alkaloids, PT barrel, ABBA prenyltransferase
The ergot alkaloids are among the most important natural pharmaceuticals and toxins in human history (1). Ergots are resting structures (sclerotia) of certain fungi that parasitize ears of grain. The medicinal uses of ergots may date back to 600 B.C. in Mesopotamia (now Iraq) (2) and were certainly well described in the 16th century (3). Both the natural ergot alkaloids and their semisynthetic derivatives are in widespread use in modern medicine and exhibit a broad spectrum of pharmacological activities, including uterotonic activity, modulation of blood pressure, control of the secretion of pituitary hormones, migraine prevention, and dopaminergic and neuroleptic activities (4). Ingestion of ergot-infected grass or grains caused severe epidemics in the Middle Ages in Europe, both in humans and in animals, and ergot-infested grasses continue to produce serious epidemics in livestock today (1, 5). The fame of ergot alkaloids is also attributable to the potent hallucinogenic activity of lysergic acid diethylamide, originally reported by Stoll and Hofmann (6).
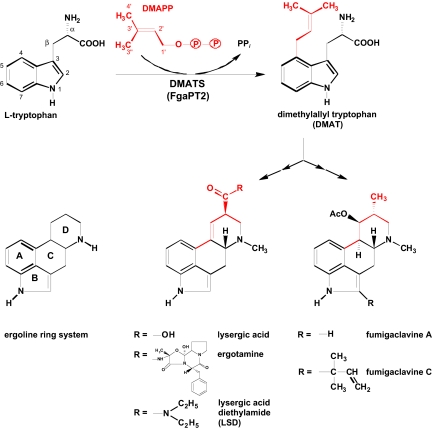
The ergot alkaloids, which are characterized by the ergoline ring system (Fig. 1), can be divided into 2 major classes: the amides of lysergic acid, which are produced by plant-associated fungi of the family Clavicipitaceae, and the clavine alkaloids, which are primarily produced by members of the fungal order Eurotiales such as Aspergillus fumigatus, currently the most common agent of invasive mycosis in humans (7).
Fig. 1.
Reaction catalyzed by DMATS, and structures of ergot alkaloids.
The first committed step in ergot alkaloid biosynthesis is the prenylation of L-tryptophan, catalyzed by the 4-dimethylallyl tryptophan synthase (DMATS; E.C. 2.5.1.34) (Fig. 1). The enzyme was initially described from Claviceps in 1971 (8) and, later on, was purified to apparent homogeneity (9, 10). It is a soluble homodimeric protein with an apparent molecular weight of ≈105 kDa. In sharp contrast to other prenyltransferases (PTs) such as farnesyl diphosphate synthase (11, 12), DMATS does not require magnesium or other divalent cations for its enzymatic activity, although addition of 4 mM CaCl2 moderately increases its reaction velocity (10). The structural gene coding for DMATS in Claviceps, termed dmaW, was identified by Tsai et al. (13). A similar gene, fgaPT2, exists in the biosynthetic gene cluster of fumigaclavine, in the genome sequence of A. fumigatus. Expression of the DMATS sequence from A. fumigatus (hereafter referred to as FgaPT2) as a his-tagged protein yielded the initial enzyme of ergot alkaloid biosynthesis. This enzyme was characterized in homogeneous form after heterologous expression (14). A very similar enzyme was later described from the fungus Malbranchea aurantiaca (15), and orthologs of DMATS have been found in many other fungal genomes.
Recently, several enzymes with sequence similarity to DMATS have been cloned from fungal genomes and were expressed, purified, and biochemically characterized, as reviewed by Steffan et al. (16). These enzymes show different substrate specificities, catalyzing the prenylation of tryptophan or derivatives thereof. In addition, they exhibit different regiospecificities because they can attach the prenyl group to different carbons or to the nitrogen atom of the indole nucleus. Furthermore, the prenyl group can be attached via its C-1 (“regular prenylation”) or its C-3 (“reverse prenylation”).
The fungal indole PTs are key enzymes in the biosynthesis of a fascinating array of structurally diverse prenylated indole alkaloids in fungi (17). Currently, BLAST searches reveal more than 100 entries with sequence similarity to DMATS in the database. The fungal indole PTs that have been biochemically investigated show remarkable flexibility for their aromatic substrates, and these enzymes may be of considerable interest for chemoenzymatic synthesis of bioactive compounds (16).
Although the crystallization of DMATS from Claviceps was reported in 1981 (18), no structure of an indole PT has been determined. We now report the crystallization and x-ray structural analysis of FgaPT2, the enzyme catalyzing the first committed step of ergot alkaloid biosynthesis in A. fumigatus. Unexpectedly, the structure of this enzyme was found to belong to a very rare ß/α barrel fold, called PT fold (19). This fold has only recently been discovered in a group of bacterial enzymes that also catalyze the prenylation of aromatic substrates but do not show detectable primary sequence similarity to the fungal indole PTs (20, 21).
Results and Discussion
Determination of the Structure of the DMATS FgaPT2.
The crystal structure of unliganded FgaPT2 was determined at 1.76 Å resolution with the use of single-wavelength anomalous dispersion and noncrystallographic symmetry averaging [supporting information (SI), Table S1]. A ternary complex containing both substrates was prepared by incubating an FgaPT2 crystal for 15 min at 4 °C with 3 mM L-tryptophan and with 2 mM dimethylallyl S-thiolodiphosphate (DMSPP). DMSPP serves as an analogue to the natural substrate, dimethylallyl diphosphate (DMAPP). The structure of the ternary complex was then determined to a resolution of 2.2 Å by molecular replacement using rigid body refinement of the unliganded structure. Initial difference electron density maps clearly revealed the presence of both ligands in the complex structure and also showed that a number of side chains in the active site had rearranged in response to the presence of the ligands. Crystallographic refinement of both structures yielded models that possess low free R-factors, excellent stereochemistry, and small rms deviations from ideal values for bond lengths and bond angles (Table S2).
Overall 3D Architecture of DMATS.
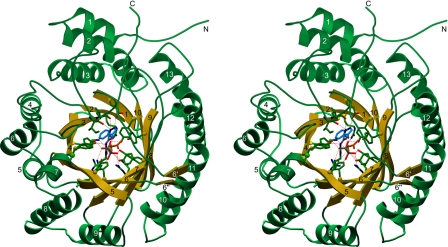
The 3D structure of FgaPT2 (Fig. 2) consists of a single domain, formed by a central barrel of 10 antiparallel β-strands that are surrounded with a ring of solvent-exposed α-helices. The fold is first reminiscent of the TIM barrel, the most common protein fold in the database. However, TIM barrels are constructed from (usually 8) parallel β-strands (Fig. S1), and are therefore not structurally homologous to FgaPT2. A DALI search (22) for structurally related proteins revealed the bacterial enzyme NphB (formerly called Orf2) as the only significant match (z = 16.5). NphB is involved in the biosynthesis of the secondary metabolite naphterpin (19) and, remarkably, catalyzes an aromatic prenylation reaction similar to that of FgaPT2. The comparison of the structures of FgaPT2 and NphB (Fig. S1) clearly demonstrates that both proteins share a common architecture, which has been called PT barrel in the case of NphB (19). However, with 459 residues, FgaPT2 is significantly larger than the 307-residue–containing NphB. The additional residues account for extended loop regions and extra α-helices in FgaPT2 (Fig. S1).
Fig. 2.
Stereo drawing showing the 3D architecture of FgaPT2. The structure is shown as a ribbon drawing, with the 10 β-strands colored yellow and the remaining residues colored green. The ligands L-tryptophan and DMSPP as well as residues that contact them via hydrogen bonds or salt bridges are shown as ball-and-stick models, with the following color code: nitrogen, blue; oxygen, red; phosphate, gray, and sulfur, yellow. Carbon atoms are green (FgaPT2), blue (L-tryptophan), and orange (DMSPP). Hydrogen bonds and salt bridges between protein and ligand atoms are represented with dashed red lines. Secondary structure elements are numbered. N and C denote the N-terminus and C-terminus, respectively.
The FgaPT2 and NphB structures both contain 5 structurally similar repeating units, which can be described as ααββ repeats (Fig. S1). The 2 α-helices pack tightly against the 2 β-strand units in each repeat, forming a hydrophobic core. Superposition shows that the core regions of the 5 repeats agree reasonably well with each other, mostly at positions that contribute to the hydrophobic core in each repeat, whereas the connecting regions exhibit significant diversity. FgaPT2 has an additional N-terminal α-helix, whereas NphB shows an extra C-terminal helix (Fig. S1). A reasonable superposition of the structures of FgaPT2 and NphB, with low rms values, can be achieved based on the β-strands alone. Including the helices drastically increases the rms value.
In the common TIM barrel fold, the central barrel is completely filled with hydrophobic residues, contributing to the stability of the protein framework. Substrate binding and catalysis are achieved by structurally diverse loop regions without defined secondary structure. In sharp contrast, the structure of FgaPT2, like that of NphB, displays a larger solvent-filled central barrel. The substrates are bound in the center of this barrel (Fig. 2), and nearly all amino acids that interact with the substrates are part of the β-strands (Fig. S2).
Structural Basis for the Reaction Mechanism of DMATS.
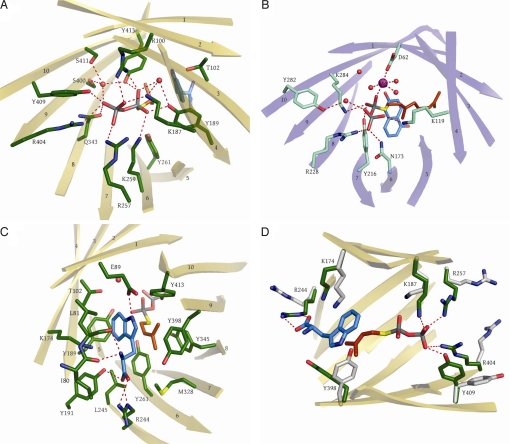
The reaction catalyzed by DMATS is an electrophilic alkylation, or Friedel-Crafts alkylation, of an aromatic ring (23, 24). The structure of FgaPT2, complexed with its natural aromatic substrate L-tryptophan, provides an excellent platform for understanding the mechanism of an enzyme-catalyzed Friedel-Crafts alkylation. In organic synthesis, Friedel-Crafts alkylations are usually carried out with alkyl halides. Friedel-Crafts catalysts like AlCl3 are required to lower the energy barrier for carbocation formation from the alkyl halides. In the DMATS reaction, the energy barrier for carbocation formation from DMAPP is strongly lowered by tight interactions of the diphosphate-leaving group of DMAPP with a large number of amino acid residues (Fig. 3A). In the complex of FgaPT2 with the substrate analogue DMSPP, the positive charges of R100, R257, and R404, as well as those of K187 and K259, negate the negative charges of the diphosphate group, and the oxygen atoms form hydrogen bonds with Y189, Y261, Y345, Y409, Y413, and Q343. The comparison with the unliganded FgaPT2 structure (Fig. 3D) also shows that many of the side chains contacting the substrates, particularly the residues that interact with the pyrophosphate group of DMSPP, are facing away from each other in the unbound FgaPT2 and swing into place only once the ligands are present.
Fig. 3.
Active center of FgaPT2 and comparison with the NphB active site. (A) View of the DMSPP in the active site of FgaPT2. Side chains binding the pyrophosphate are shown. (B) View of the active site of NphB. Side chains binding the pyrophosphate are shown. (C) View of the residues in the active site of FgaPT2 forming the binding pocket for L-tryptophan and the dimethylallyl chain of DMSPP. (D) Active site residues in liganded and unliganded (gray) FgaPT2 undergoing conformational changes upon ligand binding. Dashed red lines show contacts/hydrogen bonds with less than 3.2 Å.
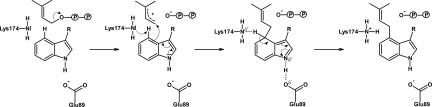
Once the planar allylic carbocation is formed, it is strongly stabilized by cation-π interactions with 2 aromatic rings that are contributed by the substrate tryptophan on one side and the side chain of Y345 on the other side. The reactivity of the indole nucleus toward electrophilic substitution is probably significantly enhanced by a hydrogen bond between the indole N-H and the carboxylate group of E89 (Fig. 3C), which is located in a predominantly hydrophobic environment. As shown in Fig. 4, this hydrogen bond likely serves to reduce the positive charge of the intermediary σ-complex. Notably, the presence of a basic residue that could accept the hydrogen from the indole N-H had been predicted in early biochemical investigations of DMATS (23). The importance of the hydrogen bond with E89 is underlined by the fact that methylation of the ring nitrogen of tryptophan, as well as replacement of the ring nitrogen with sulfur, dramatically reduces or even abolishes the prenylation reaction catalyzed by DMATS. In contrast, methylations in position 5, 6, or 7 interfere much less with the reaction (9, 25).
Fig. 4.
Hypothetical reaction mechanism for DMATS.
The reactive carbocation generated from DMAPP must be prevented from reacting with water or other nucleophiles. In DMATS, this is achieved by the side chains of 5 tyrosine residues that shield the reactive carbocation from the solvent: Y189, Y261, Y345, Y398, and Y413.
The diphosphate group is dissociating from one side of DMAPP, leaving a planar dimethylallyl cation. The indole moiety then approaches from the opposite side, forming the new bond. This orientation of the 2 substrates in the active site readily provides the structural basis for the inversion of the configuration at C-1 of DMAPP in the DMATS reaction, which was proved earlier by studies with isotope-labeled precursors (26).
FgaPT2 prenylates the indole moiety with strict regiospecificity for C-4, whereas other fungal PTs show regiospecificity for positions 1, 2, 3, or 7 of the indole moiety (16, 27). In 1976, Lee et al. (9) pointed out that based on their chemical reactivity, positions 2, 5, and 7 of the indole nucleus would be the preferred sites for electrophilic substitution. The architecture of the active site now provides a rationale for the observed regiospecificity of the DMATS reaction for C-4. In addition to the hydrogen bond between the indole N-H and the E89 carboxylate group, the substrate tryptophan is firmly held in place via additional hydrogen bonds to the side chains of residues Y191 and R244 and to the main chain atoms of I80 and L81 (Fig. 3C). The distances from the C-1 atom of the dimethylallyl cation to the carbons 2 and 3 and to the ring nitrogen (position 1) of the indole ring system range from 4.7–5.3 Å. In contrast, the distances to the carbons 4, 5, 6, and 7 range from 3.5–3.9 Å, favoring a reaction at this part of the indole ring system. In comparison, a distance of 3.2 Å (between C-1 of the dimethylallyl group and the C-3–C-4 double bond in isopentenyl diphosphate) has been reported for the active center of farnesyl diphosphate synthase (11). A distance of 4 Å between C-1 of geranyl diphosphate and the aromatic substrate was reported for NphB by Kuzuyama et al. (19). These authors postulated that after cleavage of carbon-oxygen bond to the rigidly fixed pyrophosphate group, the allylic cation rotates toward the aromatic acceptor molecule. A similar movement may occur in the FgaPT2 reaction.
The regiospecificity of DMATS for prenylation at C-4 of the indole nucleus rather than at C-7, which is chemically more reactive and prenylated by another fungal indole PT (28), may be explained by steric constraints that do not allow the formation of a σ-complex with the dimethylallyl group at this position and also not at C-5 or C-6 (Fig. S3). Furthermore, the amino group of K174 appears to be perfectly placed to abstract a proton from the σ-complex at C-4 (Fig. 3D and Fig. S4).
Trans-prenyldiphosphate synthases such as farnesyl diphosphate synthase (11) bind their isoprenyl diphosphate substrates in the form of Mg2+ complexes. The Mg2+ ion is coordinated by oxygens of the pyrophosphate group and by aspartate residues of prototypical (N/D)DxxD motifs of the PTs, lowering the energy barrier for carbocation formation (11). In contrast, neither the bacterial aromatic PT NphB nor FgaPT2 contains the (N/D)DxxD motif. Yet, the reaction catalyzed by NphB is still dependent on Mg2+, and analysis of the NphB crystal structure has revealed that the magnesium ion is indeed located in the active site (Fig. 3B), forming contacts with an oxygen of the diphosphate group, with water molecules, and with 1 aspartic acid carboxylate (19). The reaction of DMATS, in contrast, is independent of Mg2+. The superimposition of the PT barrel structures of NphB and DMATS shows that the diphosphate moiety is localized in a very similar position in both enzymes. However, the positively charged Mg2+ ion in the structure of NphB is replaced by the positively charged side chain of R100, which presumably serves a similar function in FgaPT2.
Because the genuine aromatic substrate of NphB is unknown, this enzyme was structurally characterized using the nonnatural substrate 1,6-dihydroxynaphthalene (19), which is prenylated only with low catalytic efficiency (kcat/km = 7.7 s−1 M−1, in contrast to 46,250 s−1 M−1 calculated for the prenylation of tryptophan by FgaPT2) (14, 29). We note that the aromatic substrates of NphB and FgaPT2 lie in somewhat different positions (Fig. 3). This might explain the lower catalytic efficiency observed for NphB.
Comparison of DMATSs from Different Fungal Species.
FgaPT2 from A. fumigatus exhibits 64%, 62%, and 55% sequence identity to the DMATs MaPT from M. aurantiaca (15), DmaW-Cs from a plant-associated clavicipitacean species (30), and DmaW-Cp from Claviceps purpurea P1 (31), respectively. These 3 enzymes catalyze the same reaction as FgaPT2. Remarkably, all amino acids involved in the coordination of the pyrophosphate moiety in FgaPT2, in the shielding of the allylic cation, in the binding of the aromatic substrate tryptophan, and in the postulated abstraction of the proton from the σ-complex are strictly conserved in all 4 enzymes (Fig. S2). Therefore, the conclusions for the mechanism of the reaction drawn from the structure of FgaPT2 are likely to be valid for these other DMATS orthologs as well. Of particular relevance in this regard is the enzyme DmaW-Cp, from the fungal strain that is used for the industrial production of ergot alkaloids (1, 5).
Mutagenesis of FgaPT2.
In a previous study (32), lysine residues K187 and K259 of FgaPT2 were mutated to glutamates. The mutation resulted in a >97% reduction of enzymatic activity, confirming the importance of these residues for catalysis. Analysis of the structure now shows that these 2 residues directly coordinate the pyrophosphate moiety (Fig. 3A), and their replacement with glutamate would therefore interfere with pyrophosphate binding. Mutation of R257, which also contacts the pyrophosphate group (Fig. 3 A and D), to glycine had resulted in a >95% reduction of the enzyme activity (32), probably for the same reason. Additional mutagenesis experiments have been performed previously in the homologous enzyme MaPT of M. aurantiaca. Exchange of T105 and D180 to valine resulted in enzymes that retained only 11% and 7%, respectively, of their original activity (15). The corresponding residues in FgaPT2, T102, and D178, are not directly involved in substrate binding; however, they are located in the active site of the enzyme, in close vicinity to the substrates. Most likely, their exchange to valine leads to structural rearrangements that indirectly affect the enzymatic activity. Both residues are conserved in the 4 orthologs of DMATS (Fig. S2).
The structure of FgaPT2 now suggested that, in addition to K187 and K259 mentioned previously, R100 plays an important role in the binding of the pyrophosphate moiety. We therefore mutated this residue to either aspartate or glutamine, and these mutations indeed reduced the activity by 95% and 99.6%, respectively. Furthermore, our structural investigations suggested that E89 plays a crucial role in the reaction mechanism of FgaPT2 (Fig. 4). When we mutated E89 to alanine, activity was lost completely. These results of mutational studies are consistent with the hypotheses on the reaction mechanisms that we derived from the structure of FgaPT2.
Mutation of K174 to glutamine resulted in a 60% loss of activity (average of 4 independent mutants, range: 50.0–68.8% loss). The glutamine side chain can act as a hydrogen bond acceptor but is not ionizable under physiologic conditions, and it is therefore unlikely to abstract a proton from L-Trp directly. Compared with lysine, the glutamine side chain is also shorter by 1 Å. The observation that the K174Q mutant is still partially active can be explained by a water molecule that is likely to fill the extra space. This water would be polarized as a result of its position next to the negative charge of the L-Trp carboxylate group, and therefore would be able to accept a proton from the σ-complex. Mutation of K174 to glutamate resulted in a 97.7% loss of activity. The K174E mutation introduces a further negative charge and is likely to cause larger structural changes at the active site, explaining the lower activity compared with K174Q.
Comparison of DMATS with Fungal Indole PTs of Different Regiospecificity.
Several enzymes with sequence similarity to DMATS that prenylate tryptophan (or derivatives thereof) with different regiospecificities have been identified and, in some cases, catalyze “reverse” prenylations (i.e., attach the C-3 of the dimethylallyl moiety to the aromatic nucleus). These enzymes include 7-DMATS (catalyzing a regular prenylation at C-7 of the indole moiety), FtmPT1 (regular prenylation at C-2), FtmPT2 (regular prenylation at the ring nitrogen), CdpNPT (reverse prenylation at the ring nitrogen), FgaPT1 (reverse prenylation at C-2), and AnaPT (reverse prenylation at C-3), (16, 27). We have generated structure-based alignments of each of these enzymes with FgaPT2. These confirmed the results of a previously published alignment of these proteins (33) and revealed strict conservation of most residues involved in the coordination of the pyrophosphate moiety [i.e., R100, K187, Y189, R257, K259, Y409, Y413 (numbering according to FgaPT2)] in all the mentioned enzymes. Therefore, the positioning of the pyrophosphate moiety is likely to be identical in all these proteins. Likewise, E89 was strictly conserved in all enzymes, indicating that the interaction of the indole N-H with the carboxylate group of E89 may be essential for the catalytic activity of all these PTs. Finally, 4 tyrosine residues that we suggested to shield the reactive carbocation from water (i.e., Y261 and Y345 as well as Y189 and Y413 mentioned previously) are strictly conserved in each of the named enzymes.
Structural analysis suggested that K174 may be important for the regiospecificity of FgaPT2 for prenylation at C-4 of the indole moiety (Fig. 4). Notably, K174 is conserved in none of the 6 other indole PTs with different regiospecificity than FgaPT2. These enzymes carry valine, leucine, phenylalanine, or threonine residues in the position corresponding to K174 of FgaPT2. Also, the 4 amino acids of FgaPT2, which anchor the polar side chain of the substrate L-tryptophan (I80, L81, Y191 and R244), are not conserved in the 5 other enzymes mentioned.
Therefore, different orientations of the indole moiety in the active center, and possibly different positions of residues that can abstract a proton from the intermediary σ-complex, may be responsible for the different regiospecificities of indole PTs.
A Common Architecture of Aromatic PTs in Bacteria and Fungi.
As described previously (Fig. S1), FgaPT2 and the bacterial enzyme NphB (19) share the same PT barrel fold. This fold is believed to be the common framework of a small family of enzymes called ABBA PTs (20). Currently, 10 bacterial members of this family are found in the database, all from the genus Streptomyces, and 5 of these have been investigated biochemically (19, 29, 34–36). Four additional members of the ABBA family have been identified in fungal genomes, but their function is not yet known (20). The ABBA PTs are soluble enzymes that catalyze aromatic prenylation reactions (i.e., Friedel-Crafts alkylations) but do not show the (N/D)DxxD motifs typical of many other PTs, and with the single exception of NphB, they do not require Mg2+ or other divalent cations to function. These characteristics resemble those of the fungal indole PTs, as noted already when the initial member of the ABBA family was discovered (34). Yet, no significant similarity can be detected between the amino acid sequences of both enzyme groups. A structure-based alignment of FgaPT2 (459 aa) with NphB (307 aa) reveals only 9% identity and 29% similarity between both proteins (Fig. S5). This presents a striking example of the conservation of a 3D structure in proteins of similar function but with highly diverging primary sequence in evolutionary distant taxa.
The PT barrel fold of FgaPT2 and NphB is entirely different from the all–α-helical structure of farnesyl diphosphate synthase (12) and from the structure of cis-polyprenyl diphosphate synthases (37), which, like FgaPT2 and NphB, do not contain an (N/D)DxxD motif. Likewise, this fold bears no resemblance to the recently suggested structures of the membrane-bound aromatic PTs UbiA and LePGT1, which catalyze the 3-prenylation of 4-hydroxybenzoate (38, 39). FgaPT2 and NphB are composed of ααββ structural elements (Fig. S1). A DALI search in the protein structure database did not reveal the presence of this element in other proteins from which both FgaPT2 and NphB might have evolved independently. This favors the hypothesis that the common architecture of FgaPT2 and NphB arises from a common origin of the 2 enzymes rather than from a convergent evolution.
Materials and Methods
Protein Expression, Purification, and Crystallization.
His-tagged FgaPT2 was purified as described by Steffan et al. (25). The N-terminal His-tag was removed by incubating with 0.1 units of thrombin (Sigma) per milligram of FgaPT2 for 15 h at 4 °C. After dialysis, the protein was first passed over a 1-mL Benzamidine Sepharose 4 Fast Flow column (Amersham Biosciences) to remove thrombin and then applied to Ni-NTA agarose (Qiagen) to remove remnants of the tag. The protein was then concentrated to 5 mL using an Amicon Centriprep YM-10 concentrator (Millipore), passed through a 0.2-μm filter, and applied to a Superdex 200 HiLoad 26/60 gelfiltration column (GE Healthcare). The column was eluted with 25 mM Tris-HCl (pH 8.0), 100 mM NaCl, and 2 mM DTT. Fractions containing pure FgaPT2 were pooled, diluted 1:2 with 25 mM Tris-HCl (pH 8.0) and 2 mM DTT and concentrated to 15 mg mL−1 before crystallization. Crystals were grown by vapor diffusion at 4 °C by mixing 2 μL of the protein solution and 1 μL of precipitant [26% (vol/vol) 1,3-butanediol, 50 mM sodium L-lactate, 100 mM sodium 3-morpholino-2-hydroxypropanesulfonic acid (MOPSO) (pH 7.0), 2 mM DTT]. Heavy atom derivative was obtained by soaking crystals for 6 h at 4 °C in 10 mM uranyl nitrate. In all cases, 15–25% (vol/vol) glycerol was used as a cryoprotectant.
Data Collection and Structure Determination.
The FgaPT2 crystals belong to space group P212121 (a = 80.1 Å, b = 98.8 Å, c = 125.6 Å) and contain 2 molecules in the asymmetric unit. Data for the native FgaPT2 crystals were collected at the European Synchrotron Radiation Facility beamline ID14–4 in Grenoble, France, using the Quantum 315r detector (Area Detector Systems Corporation). Data for the uranyl derivative were collected on a rotating anode generator equipped with a Mar345 imaging plate detector, whereas data from the ternary complex resulted from data collection at Swiss Light Source beamline X06SA in Villigen, Switzerland. All diffraction data were recorded at 100 K. The methods for data analysis are described in SI Text.
Mutagenesis.
Mutations were introduced into the fgaPT2 sequence with the QuikChange II Kit (Stratagene) according to the manufacturer's instructions. The proteins were expressed and purified as described previously, and the enzymatic activity was determined as described by Steffan et al. (25).
Supplementary Material
Acknowledgments.
We thank Joseph P. Noel (La Jolla, CA) for plasmid pHis8. We are grateful to the European Synchrotron Radiation Facility and Swiss Light Source beamlines and staff for support. This work was supported by the Deutsche Forschungsgemeinschaft (S.-M.L., L.H., and T.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 3I4X and 3I4Z).
This article contains supporting information online at www.pnas.org/cgi/content/full/0904897106/DCSupplemental.
References
- 1.Schardl CL, Panaccione DG, Tudzynski P. Ergot alkaloids—Biology and molecular biology. Alkaloids Chem Biol. 2006;63:45–86. doi: 10.1016/s1099-4831(06)63002-2. [DOI] [PubMed] [Google Scholar]
- 2.van Dongen PW, de Groot AN. History of ergot alkaloids from ergotism to ergometrine. Eur J Obstet Gynecol Reprod Biol. 1995;60:109–116. doi: 10.1016/0028-2243(95)02104-z. [DOI] [PubMed] [Google Scholar]
- 3.Lonicer A. In: Kreuterbuch. Lechler M, editor. Germany: Frankfurt am Main; 1582. [Google Scholar]
- 4.de Groot AN, van Dongen PW, Vree TB, Hekster YA, van Roosmalen J. Ergot alkaloids. Current status and review of clinical pharmacology and therapeutic use compared with other oxytocics in obstetrics and gynaecology. Drugs. 1998;56:523–535. doi: 10.2165/00003495-199856040-00002. [DOI] [PubMed] [Google Scholar]
- 5.Gröger D, Floss HG. Biochemistry of ergot alkaloids—Achievements and challenges. Alkaloids Chem Biol. 1998;50:171–218. [Google Scholar]
- 6.Stoll A, Hofmann A. Die optisch aktiven Hydrazide der Lysergsäure und der Isolysergsäure. (4. Mitteilung über Mutterkornalkaloide) Helv Chim Acta. 1943;26:922–928. [Google Scholar]
- 7.Segal BH, Walsh TJ. Current approaches to diagnosis and treatment of invasive aspergillosis. Am J Respir Crit Care Med. 2006;173:707–717. doi: 10.1164/rccm.200505-727SO. [DOI] [PubMed] [Google Scholar]
- 8.Heinstein PF, Lee SI, Floss HG. Isolation of dimethylallylpyrophosphate: Tryptophan dimethylallyl transferase from the ergot fungus (Claviceps spec. ) Biochem Biophys Res Commun. 1971;44:1244–1251. doi: 10.1016/s0006-291x(71)80219-x. [DOI] [PubMed] [Google Scholar]
- 9.Lee SL, Floss HG, Heinstein P. Purification and properties of dimethylallylpyrophosphate:tryptophan dimethylallyl transferase, the first enzyme of ergot alkaloid biosynthesis in Claviceps. sp. SD 58. Arch Biochem Biophys. 1976;177:84–94. doi: 10.1016/0003-9861(76)90418-5. [DOI] [PubMed] [Google Scholar]
- 10.Gebler JC, Poulter CD. Purification and characterization of dimethylallyl tryptophan synthase from Claviceps purpurea. Arch Biochem Biophys. 1992;296:308–313. doi: 10.1016/0003-9861(92)90577-j. [DOI] [PubMed] [Google Scholar]
- 11.Poulter CD. Farnesyl diphosphate synthase. A paradigm for understanding structure and function relationships in E-polyprenyl diphosphate synthases. Phytochem Rev. 2006;5:17–26. [Google Scholar]
- 12.Liang PH, Ko TP, Wang AH. Structure, mechanism and function of prenyltransferases. Eur J Biochem. 2002;269:3339–3354. doi: 10.1046/j.1432-1033.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- 13.Tsai HF, Wang H, Gebler JC, Poulter CD, Schardl CL. The Claviceps purpurea gene encoding dimethylallyltryptophan synthase, the committed step for ergot alkaloid biosynthesis. Biochem Biophys Res Commun. 1995;216:119–125. doi: 10.1006/bbrc.1995.2599. [DOI] [PubMed] [Google Scholar]
- 14.Unsöld IA, Li SM. Overproduction, purification and characterization of FgaPT2, a dimethylallyltryptophan synthase from Aspergillus fumigatus. Microbiology. 2005;151:1499–1505. doi: 10.1099/mic.0.27759-0. [DOI] [PubMed] [Google Scholar]
- 15.Ding Y, Williams RM, Sherman DH. Molecular analysis of a 4-dimethylallyltryptophan synthase from Malbranchea aurantiaca. J Biol Chem. 2008;283:16068–16076. doi: 10.1074/jbc.M801991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steffan N, Grundmann A, Yin WB, Kremer A, Li S-M. Indole prenyltransferases from fungi: A new enzyme group with high potential for the production of prenylated indole derivatives. Curr Med Chem. 2009;16:218–231. doi: 10.2174/092986709787002772. [DOI] [PubMed] [Google Scholar]
- 17.Williams RM, Stocking EM, Sanz-Cervera JF. Biosynthesis of prenylated alkaloids derived from tryptophan. Top Curr Chem. 2000;209:97–173. [Google Scholar]
- 18.Cress WA, Chayet LT, Rilling HC. Crystallization and partial characterization of dimethylallyl pyrophosphate: L-tryptophan dimethylallyltransferase from Claviceps sp. SD58. J Biol Chem. 1981;256:10917–10923. [PubMed] [Google Scholar]
- 19.Kuzuyama T, Noel JP, Richard SB. Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products. Nature. 2005;435:983–987. doi: 10.1038/nature03668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tello M, Kuzuyama T, Heide L, Noel JP, Richard SB. The ABBA family of aromatic prenyltransferases: Broadening natural product diversity. Cell Mol Life Sci. 2008;65:1459–1463. doi: 10.1007/s00018-008-7579-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heide L. Prenyl transfer to aromatic substrates: Genetics and enzymology. Curr Opin Chem Biol. 2009;13:171–179. doi: 10.1016/j.cbpa.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 22.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gebler JC, Woodside AB, Poulter CD. Dimethylallyltryptophan synthase. An enzyme-catalyzed electrophilic aromatic substitution. J Am Chem Soc. 1992;114:7354–7360. [Google Scholar]
- 24.Silverman RB. The Organic Chemistry of Enzyme-Catalyzed Reactions. San Diego, CA: Academic; 2002. [Google Scholar]
- 25.Steffan N, Unsöld IA, Li SM. Chemoenzymatic synthesis of prenylated indole derivatives by using a 4-dimethylallyltryptophan synthase from Aspergillus fumigatus. ChemBioChem. 2007;8:1298–1307. doi: 10.1002/cbic.200700107. [DOI] [PubMed] [Google Scholar]
- 26.Shibuya M, et al. Stereochemistry of the isoprenylation of tryptophan catalyzed by 4-(γ,γ-dimethylally1)tryptophan synthase from Claviceps, the first pathway-specific enzyme in ergot alkaloid biosynthesis. J Am Chem Soc. 1990;1990:297–304. [Google Scholar]
- 27.Yin WB, Grundmann A, Cheng J, Li SM. Acetylaszonalenin biosynthesis in Neosartorya fischeri: Identification of the biosynthetic gene cluster by genomic mining and functional proof of the genes by biochemical investigation. J Biol Chem. 2009;284:100–109. doi: 10.1074/jbc.M807606200. [DOI] [PubMed] [Google Scholar]
- 28.Kremer A, Westrich L, Li SM. A 7-dimethylallyltryptophan synthase from Aspergillus fumigatus: Overproduction, purification and biochemical characterization. Microbiology. 2007;153:3409–3416. doi: 10.1099/mic.0.2007/009019-0. [DOI] [PubMed] [Google Scholar]
- 29.Kumano T, Richard SB, Noel JP, Nishiyama M, Kuzuyama T. Chemoenzymatic syntheses of prenylated aromatic small molecules using Streptomyces prenyltransferases with relaxed substrate specificities. Bioorg Med Chem. 2008;16:8117–8126. doi: 10.1016/j.bmc.2008.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Markert A, et al. Biosynthesis and accumulation of ergoline alkaloids in a mutualistic association between Ipomoea asarifolia (Convolvulaceae) and a clavicipitalean fungus. Plant Physiol. 2008;147:296–305. doi: 10.1104/pp.108.116699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tudzynski P, et al. Evidence for an ergot alkaloid gene cluster in Claviceps purpurea. Mol Gen Genet. 1999;261:133–141. doi: 10.1007/s004380050950. [DOI] [PubMed] [Google Scholar]
- 32.Stec E, et al. Two lysine residues are responsible for the enzymatic activities of indole prenyltransferases from fungi. ChemBioChem. 2008;9:2055–2058. doi: 10.1002/cbic.200800237. [DOI] [PubMed] [Google Scholar]
- 33.Kremer A, Li SM. Tryptophan aminopeptidase activity of several indole prenyltransferases from Aspergillus fumigatus. Chem Biol. 2008;15:729–738. doi: 10.1016/j.chembiol.2008.05.019. [DOI] [PubMed] [Google Scholar]
- 34.Pojer F, et al. CloQ, a prenyltransferase involved in clorobiocin biosynthesis. Proc Natl Acad Sci USA. 2003;100:2316–2321. doi: 10.1073/pnas.0337708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haagen Y, et al. A soluble, magnesium-independent prenyltransferase catalyzes reverse and regular C-prenylations and O-prenylations of aromatic substrates. FEBS Lett. 2007;581:2889–2893. doi: 10.1016/j.febslet.2007.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saleh O, Gust B, Boll B, Fiedler HP, Heide L. Aromatic prenylation in phenazine biosynthesis: Dihydrophenazine-1-carboxylate dimethylallyltransferase from Streptomyces anulatus. J Biol Chem. 2009;284:14439–14447. doi: 10.1074/jbc.M901312200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo RT, et al. Crystal structures of undecaprenyl pyrophosphate synthase in complex with magnesium, isopentenyl pyrophosphate, and farnesyl thiopyrophosphate: Roles of the metal ion and conserved residues in catalysis. J Biol Chem. 2005;280:20762–20774. doi: 10.1074/jbc.M502121200. [DOI] [PubMed] [Google Scholar]
- 38.Bräuer L, Brandt W, Schulze D, Zakharova S, Wessjohann L. A structural model of the membrane-bound aromatic prenyltransferase UbiA from E. coli. ChemBioChem. 2008;9:982–992. doi: 10.1002/cbic.200700575. [DOI] [PubMed] [Google Scholar]
- 39.Ohara K, Muroya A, Fukushima N, Yazaki K. Functional characterization of LePGT1, a membrane-bound prenyltransferase involved in the geranylation of p-hydroxybenzoic acid. Biochem J. 2009;421:231–241. doi: 10.1042/BJ20081968. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.