Abstract
Motivation: Protein–protein interaction (PPI) extraction from published biological articles has attracted much attention because of the importance of protein interactions in biological processes. Despite significant progress, mining PPIs from literatures still rely heavily on time- and resource-consuming manual annotations.
Results: In this study, we developed a novel methodology based on Bayesian networks (BNs) for extracting PPI triplets (a PPI triplet consists of two protein names and the corresponding interaction word) from unstructured text. The method achieved an overall accuracy of 87% on a cross-validation test using manually annotated dataset. We also showed, through extracting PPI triplets from a large number of PubMed abstracts, that our method was able to complement human annotations to extract large number of new PPIs from literature.
Availability: Programs/scripts we developed/used in the study are available at http://stat.fsu.edu/~jinfeng/datasets/Bio-SI-programs-Bayesian-chowdhary-zhang-liu.zip
Contact: jliu@stat.harvard.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Proteins perform their functions by interacting with other molecules, which, in many cases, are also proteins. Protein–protein interactions (PPIs) are of central importance for virtually every cellular process. Information on PPIs is indispensable for our understanding of the mechanisms of biological processes and diseases (Kann, 2007). Several databases, built by painstakingly reading published literatures, provide access to such information (Alfarano et al., 2005; Beuming et al., 2005; Chatraryamontri et al., 2007; Mishra et al., 2006; Pagel et al., 2005; Salwinski et al., 2004; Stark et al., 2006). However, due to the explosive growth of biological publications in recent years, time- and resource-consuming manual annotation has become impractical for systematic extraction of PPIs (Baumgartner et al., 2007). Although author-based PPI annotation has been proposed (Ceol et al., 2008; Leitner and Valencia, 2008), it is still unclear whether they will be adopted by the community in the near future. Consequently, researchers have resorted to automatic methods to address this problem (Jensen et al., 2006). The problem of PPI extraction broadly consists of two components, protein name recognition (protein name tagging) and PPI extraction, both of which are challenging problems. In this study, we have dealt primarily with the second problem, i.e. extracting PPIs given correctly tagged protein names.
Many computational studies have recently attempted to extract PPIs from published literatures, mostly PubMed abstracts due to their easy access (Blaschke et al., 1999; Skusa et al., 2005). All methods detect PPIs based on some rules (or patterns, templates, etc.), which can be generated by two approaches: specifying them either manually (Blaschke et al., 1999; Friedman et al., 2001; Jensen et al., 2006; Leroy and Chen, 2002; Narayanaswamy et al., 2005; Ng and Wong, 1999; Ono et al., 2001; Park et al., 2001; Pustejovsky et al., 2002; Saric et al., 2006; Temkin and Gilder, 2003; Thomas et al., 2000; Wong, 2001; Yakushiji et al., 2001) or computationally inferring/learning them from manually annotated sentences (Huang et al., 2004; Kim et al., 2008b; Malik et al., 2006).
Initial efforts of PPI detection were based on simple rules, such as co-occurrence, which assumes that two proteins likely interact with each other if they co-occur in the same sentence/abstract (Jenssen et al., 2001; Stapley and Benoit, 2000). These approaches tend to produce a large number of false positives, and still require significant manual annotations.
Later studies, aiming to reduce the high false positive rate of earlier methods, used manually specified rules. Although such methods sometimes achieved a higher accuracy than co-occurrence methods by extracting cases satisfying the rules, they have low coverage due to missing cases not covered by the limited number of manually specified rules (Blaschke et al., 1999; Friedman et al., 2001; Jensen et al., 2006; Leroy and Chen, 2002; Narayanaswamy et al., 2005; Ng and Wong, 1999; Ono et al., 2001; Park et al., 2001; Pustejovsky et al., 2002; Saric et al., 2006; Temkin and Gilder, 2003; Thomas et al., 2000; Wong, 2001; Yakushiji et al., 2001).
Recently, machine learning-based methods (Huang et al., 2004; Kim et al., 2008b; Malik et al., 2006; Miwa et al., 2008; Van Landeghem et al., 2008) have achieved better performances than other methods in terms of both decreasing false positive rate and increasing the coverage by automatically learning the language rules using annotated texts. Huang and coworkers (2004) used a dynamic programming algorithm, similar to that used in sequence alignment, to extract patterns in sentences tagged by part-of-speech tagger. Kim and coworkers (2008a, b) used a kernel approach for learning genetic and protein–protein interaction patterns. Van Landeghem et al. (2008) applied rich feature vectors derived from dependency graphs and Miwa et al. (2008) used multiple kernels based on different parsers to extract protein–protein interactions.
Despite extensive studies, current techniques appear to have only achieved partial success on relatively small datasets. Specifically, Park and co-workers (2001) tested their combinatory categorical grammar (CCG) method on 492 sentences and obtained a recall and precision rate of 48% and 80%, respectively. Context-free grammar (CFG) method of Temkin et al. was tested on 100 randomly selected abstracts and obtained a recall and precision of 63.9% and 70.2%, respectively (Temkin and Gilder, 2003). Preposition-based parsing method was tested on 50 abstracts with a precision of 70% (Leroy and Chen, 2002). A relational parsing method for extracting only inhibition relation was tested on 500 abstracts with a precision and recall of 90% and 57%, respectively (Pustejovsky et al., 2002). Ono et al. (2001) manually specified rules for four interaction verbs (interact, bind, complex, associate), which were tested on 1,586 sentences related to yeast and Escherichia coli, and obtained an average recall and precision of 83.6% and 93.2%, respectively. Huang et al. (2004) used a sequence alignment-based dynamic programming approach and obtained a recall rate of 80.0% and precision rate of 80.5% on 1,200 sentences extracted from online articles. However, a closer analysis of Ono's and Huang's datasets showed that they are very biased in terms of the interaction words used [Ono and coworkers' dataset contains just four interaction words, while in Huang's study, although more verbs were mentioned the number of sentences containing interact and bind (and their variants) represents 59.3% of all 1,200 sentences]. In Ono's dataset, there is an unrealistic high proportion of true samples (74.7%), making it much easier to obtain good recall and precision. In Huang's study, an arbitrary number of sentences were chosen from 1,200 sentences as training data and the rest as testing data, while some cross-validation tests should be used. Kim et al. (2008b) developed a Web server, PIE, and tested their method on BioCreative (Krallinger and Valencia, 2007; Krallinger et al., 2008a, b) dataset and achieved very good performance: for PPI article filter task 87.41% precision, 90.53% recall and 88.89% F1-score and for sentence filter task 92.13% precision, 91.78% recall and 91.96% F1-score, making it the best publicly accessible method.
An interaction between two proteins in a sentence is described by at least one and normally only one interaction word, such as interact, bind, phosphorylate and so on. Ideally, one would want to extract not only the names of interacting proteins but also the corresponding interaction words that may describe the type of the interaction (Hatzivassiloglou and Weng, 2002) (we call the two protein names and the interaction word as a PPI triplet). For example, the sentence, ‘We show here that PAHX interacts with FKBP52, but not with FKBP12, suggesting that it is a specific target of FKBP52’, contains four protein names [PAHX, FKBP52, FKBP12 and FKBP52 (the second occurrence of FKBP52 in the sentence)] and one interaction word (interacts). There are totally five triplets [PAHX-interacts-FKBP52, PAHX-interacts-FKBP12, PAHX-interacts-FKBP52 (second FKBP52), interacts-FKBP52-FKBP12 and interacts-FKBP12-FKBP52 (second FKBP52), where FKBP52-interacts-FKBP52 is not counted] with one true interactions (PAHX-interacts-FKBP52). We extract the triplets from sentences and classify them as true or false with probability values based on whether the interaction word correctly describes the interaction relationship between the two protein names.
Our method extracts PPI triplets from text by explicitly quantifying the likelihood of being true for each triplet using Bayesian Network (BN) (Needham et al., 2007) method. This is done by automatically learning the rules that logically connect interacting proteins and their associated interaction words in sentences. Compared to most current methods that filter sentences or abstracts that contain PPIs, our method directly reports PPI triplets. When combined with human annotation, our method models a more realistic situation. Since there are a large number of PPIs in the current PPI databases, a computer programme should recommend only putative new PPIs with their associated text/sentences to human curators. Without explicitly working at PPI triplet (or PPI pair when interaction word is not considered/reported) level, programmes may recommend sentences or abstracts that contain existing PPIs in the databases, thereby increasing the redundancy and the manual annotation labour.
Our PPI triplet extraction system showed an overall accuracy of 87 ± 2% (precision: 76%, recall: 71%, specificity: 92%, F-measure: 74% and area under ROC: 91%) when tested on a set of manually curated sentences using 10 × 10-fold cross-validation. Our method had the same level of accuracy as PIE (Kim et al., 2008b), a state-of-the-art PPI extraction Web server, in PPI sentence extraction, although it was not designed for that task and used a protein name dictionary of fairly small size. In addition, we tested our system on a large dataset of 679,733 samples obtained from PubMed using general interaction terms and showed that our method can be effectively combined with human annotation to discover a substantial number of new protein–protein interactions that were not previously reported in our reference database, BioGRID (Stark et al., 2006). To the best of our knowledge, the current study represents the largest scale general purpose PPI-triplet extraction using automated techniques.
2 METHODS
In our PPI extraction method, we first constructed dictionaries containing keywords that are related to our information extraction task, and then extracted features in sentences that encode the possible rules people use to describe interactions of proteins. An automatic system based on BNs was then built to learn the rules using manually annotated samples. The trained system was tested by cross-validation and then used to discover PPIs from a large collection of biological texts. The workflow of the system is shown in Supplementary Figure S1.
2.1 Dictionaries
We used two types of keyword dictionaries for our task: (i) protein names and (ii) interaction words, which are terms used to describe interaction relationships between two proteins in a sentence. The protein name dictionary, containing totally 68,970 protein names, was constructed based on BioGRID database version 2.0.23 (http://www.thebiogrid.org) (Stark et al., 2006), while the protein interaction word dictionary (shown in Supplementary Material) was constructed by both personal knowledge of the authors and manually reading the sentences from the literature that contained protein interactions. Both the dictionaries are readily extendible for newer terms. In this study, we aim to extract physical protein–protein interactions. The interaction word dictionary we built thus contains only those words that describe physical interactions of proteins.
2.2 PPI triplet features for BN model building
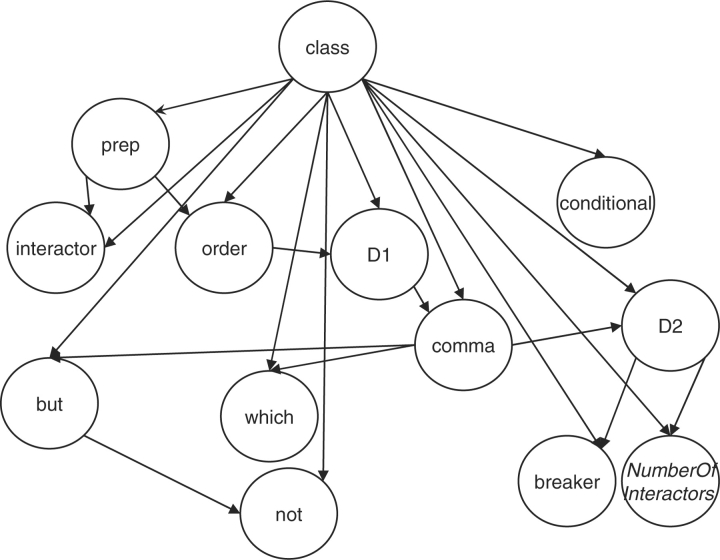
To build BN models, we manually selected the features that we believe are strongly related to the language rules we aim to learn. In the current method, we used 12 features, which are shown along with their possible values in Table S1 of Supplementary Material. Each feature is assumed to capture information/signals associated with certain grammar or language rules that describe PPI relationships. For example, D1 is the distance (number of words in between) of the first two elements of a triplet and D2 is the distance of the last two elements of a triplet. True interactions tend to have smaller values of D1 and D2. The feature order is important because people use certain words in only specific order. For example, when people use the word interacts to describe the interaction of the two proteins it has to be in the order of protein A–interacts–protein B. If the order is interacts–protein A-protein B, then the word interacts must not be used to describe the interaction between protein A and protein B. Similarly, other features we used also affect a triplet being true or false. The overall effect of all the features is modeled by BN (Fig. 1). Using the following sentence as an example, We show here that PAHX interacts with FKBP52, but not with FKBP12, suggesting that it is a specific target of FKBP52. The feature values for PAHX–interacts–FKBP52 (the second FKBP52) are the following: interactor, interacts; D1, 0; D2 16; order, avb; prep, with; conditional, n; comma, ny; but, y; which, n; not, y; breaker, n and NumberOfInteractors, low.
Fig. 1.
Bayesian network structure learned from the cross-validation dataset.
Overall, interactor, NumberOfInteractors, D1, D2, order, comma represent general features that were used to capture more general rules associated with relationships between interacting proteins and the corresponding interaction words; while prep, breaker, not, which, but, conditional represent specific features that were used to capture certain specific rules. For example, while not attempts to capture the negative meaning, but and conditional attempt to capture doubtful and conditional meanings. Likewise, comma, breaker, which and NumberOfInteractors attempt to capture signals/complexities associated with sentences with compound meanings.
2.3 BN model building
Through triplet features, we learn the language rules related to PPIs using a BN. A BN is a graphical model that encodes probabilistic relationships (conditional independencies) among variables of interest, which in the current context are the features that describe PPIs in text. To build/train the BN model, we used the freely available Weka package version 3.4.10 (Witten and Frank, 2005). The model parameters (conditional probability distributions) were calibrated based on the PPI feature set using maximum posterior estimates. The parameters are initialized with default uniform Dirichlet priors with alpha (α)=0.5. Since uniform prior is the only option available in Weka, we modified Weka source code in order to alter the class distribution in the trained model for prediction on the leftover dataset, with different and more realistic class distributions, P(C). To learn the network structure, a local Hill-climbing search algorithm was used with the Bayes scoring metric along with additional settings: structure was initialized as the Naïve Bayes model, the Markov blanket correction was applied to the structure and the maximum number of parents per node was set to two. By limiting the maximum number of parents for a node to two we limited the number of model parameters to be learnt to gain on computational efficiency and reduce the likelihood of over-fitting the training data. With higher and lower number of parent nodes we found that the performance of the model deteriorated (refer to Supplementary Table S2 for results of this analysis).
The learned BN model, including both network structure (as shown in Fig. 1) and parameters, was then used to classify a query feature vector according to the Bayes theorem. That is, given a feature vector E, we compute the posterior probability of the class C being t (true) or f (false), P(C| E), as
| (1) |
where P(C) is the prior probability of C, P(E) is the marginal probability of feature vector E and P(E| C) is the conditional probability of observing the feature vector E in a given class C, which can be simplified by decomposing it as the product of conditional probabilities at each node in the BN based on conditional independencies in the network structure. The value of P(C| E) indicates how likely the interaction (triplet) associated with the feature vector E is in class C. We classify the interaction triplet with feature vector E to be true class if P(C =true| E) is greater than 0.5.
3 RESULTS
3.1 Dataset for training and cross-validation testing
To obtain positive and negative samples to train our system to learn the language rules, we used the BioGRID database to obtain known protein–protein interactions. For each interaction listed in BioGRID, we extracted the corresponding abstract of the article from PubMed and split the abstract into sentences. If two interacting proteins (as reported by BioGRID) and an interaction word were found in a sentence, the triplet was kept in the dataset. Totally, we obtained 17 201 triplets in 6374 sentences, since there could be more than one triplet in a sentence (Dataset 1). We then manually annotated some of the sentences and obtained true and false triplets/interaction cases. We define a triplet as true if the interaction word therein logically associates/directly describes the interaction between the two corresponding protein names. Since the total number of sentences we extracted was large, we used a subset of 1037 sentences (representing 542 abstracts) for training and testing the method through cross-validation. This dataset, called cross-validation dataset or Dataset 2, had 2550 triplets with 668 true cases and 1882 false cases (Supplementary Material).
3.2 Dataset for large-scale PPI extraction
In order to see if our system is capable of extracting new protein–protein interactions from the literature, we collected from PubMed abstracts a large dataset of 679,733 sample sentences containing both true and false PPI triplets. The same protein name dictionary from BioGRID was used. None of the protein pairs in these sentences were present in the BioGRID database, and thus referred as leftover dataset in this article. We then applied the BN model trained using the cross-validation dataset to extract PPIs from this leftover dataset. Unlike the datasets used by some early methods, which were generally interaction rich, the leftover dataset is interaction poor and expectedly contains fewer interactions than a random sample from PubMed. It simulates the real situation for extracting new interactions and presents extra challenges for PPI extraction methods.
3.3 Performance on cross-validation test
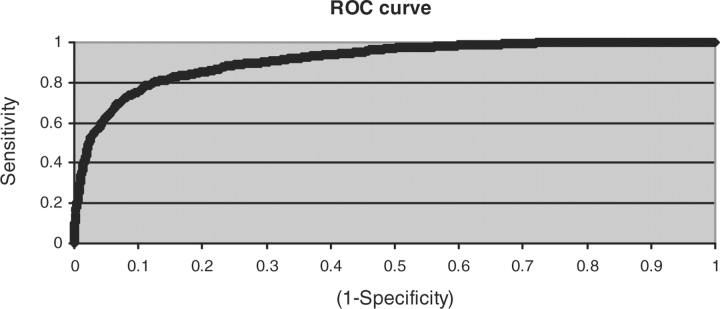
In order to test the performance of our system, we conducted a 10 × 10-fold cross-validation on Dataset 2 containing 2550 triplets with 668 true and 1882 false cases. Here, 10 × 10 means that 10-fold cross validation was repeated 10 times with different random partitionings. Results of this analysis are shown in Table 1. The performance was quite satisfactory with an overall accuracy of 87 ± 2% with area under ROC curve as 0.91 ± 0.02 (shown in Fig. 2) using the default class distribution. The recall (sensitivity), precision (positive predicted value) and F-measure are 71%, 76% and 74%, respectively. We also found that our BN model performed better compared to Naïve Bayes, which gave an overall accuracy of 82 ± 3%. Our method also achieved the same level of accuracy as PIE (Kim et al., 2008b), at PPI sentence extraction, with an F-measure of 72.9 ± 5.4 as against 73.3 for PIE (precision 81.2 and recall 66.8%). We also compared our method with PIE on PPI triplet extraction task and found that PIE performed much worse, which is understandable since PIE is not tuned for extracting PPI triplets.
Table 1.
Results of Bayesian network model on Dataset 2 containing 2550 samples (668 true and 1882 false)
| Dataset | Recall | Precision | Specificity | F-measure | Accuracy |
|---|---|---|---|---|---|
| Dataset 1 | 71 ± 5 | 76 ± 5 | 92 ± 2 | 74 | 87 ± 2 |
| set1 | 83 ± 4 | 83 ± 4 | 82 ± 5 | 83 | 83 ± 3 |
| set2 | 83 ± 4 | 82 ± 4 | 82 ± 4 | 82.5 | 83 ± 3 |
| set3 | 83 ± 4 | 81 ± 4 | 81 ± 5 | 82 | 82 ± 3 |
| set4 | 83 ± 4 | 82 ± 3 | 82 ± 4 | 82.5 | 82 ± 3 |
| set5 | 83 ± 4 | 82 ± 4 | 81 ± 5 | 82.5 | 82 ± 3 |
| set6 | 82 ± 5 | 81 ± 4 | 80 ± 5 | 81.5 | 81 ± 3 |
| set7 | 84 ± 4 | 83 ± 5 | 83 ± 4 | 83.5 | 84 ± 3 |
| set8 | 83 ± 4 | 82 ± 5 | 83 ± 4 | 82.5 | 83 ± 3 |
| set9 | 84 ± 4 | 82 ± 4 | 82 ± 3 | 83 | 83 ± 3 |
| set10 | 82 ± 5 | 80 ± 5 | 80 ± 4 | 81 | 81 ± 4 |
| Average of the 10 sets | 83 | 81.9 | 81.5 | 82.4 | 82.4 |
Datasets, set1 through set10, contained 1336 samples with equal class distribution. Notations are: TP—true positive, FP—false positive, TN—true negative, FN—false negative and recall (sensitivity)=TP/(TP+FN), specificity=TN/(TN+FP), precision (positive predicted value)=TP/(TP+FP), F-measure=2(precision × recall)/(precision+recall), accuracy=(TP+TN)/(TP+TN+FP+FN). The figures shown are all in percent.
Fig. 2.
ROC curve for cross-validation done on cross-validation dataset.
Since the class (true and false) distribution in the cross-validation dataset was unbalanced with more false samples, we conducted an additional analysis to see how our model performance varies on similar datasets with equal class distribution. This was done by generating 10 datasets (set1 through set10 in Table 1), each with 668 true and 668 randomly chosen false cases from cross-validation dataset. We obtained average recall (sensitivity) rate of 83%, precision (positive predicted value) rate of 81.9%, specificity of 81.5%, accuracy of 82.4% and F-measure of 82.4% (Table 1), indicating that our method is quite robust for samples with different portions of true and false cases.
3.4 BN structure
The graphical structure (as shown in Fig. 1) of the BN model learned from cross-validation dataset shows dependencies and conditional independencies among different features that we analyzed. The dependencies for each node are summarized in Table 2 in terms of the neighbours that influence them. Some of these include, e.g. interactor influenced by prep, which makes sense because the preposition should have a bearing on the type of the interactor (interaction term) associated with it in a sentence; which influenced by comma, this again makes sense as which in a sentence is usually preceded by a comma; similarly, presence of but is influenced by comma and not; D2 is influenced by comma, breaker, NumberOfInteractors, which looks reasonable because presence of these three features would normally make a sentence complex and thus should affect length, D2. In order to see the strength of conservation of the directional edges in the BN model shown in Figure 1, we repeated the structure learning task on the training data using 10-folds of cross-validation (thus 9/10th part of the training data was used in each of the 10 runs). The results (Supplementary Table S3) showed that the edges with their directionality (in Fig. 1) remained largely conserved across all these 10 models, indicating the robustness of the learned model structure shown in Figure 1. Overall, these relationships give some insight into the semantics of the sentence structure present in our data, which may be useful for future model enhancements.
Table 2.
Markov blanket of feature nodes in the Bayesian network
| Feature | Neighbourhood of influence |
| Conditional | Class |
| Prep | Interactor, order, class |
| Interactor | Prep, class |
| Order | Prep, D1, class |
| D1 | Order, comma, class |
| Comma | D1, but, which, D2, class |
| Which | Comma, class |
| But | Comma, not, class |
| Not | But, class |
| D2 | Comma, breaker, NumberOfInteractors, class |
| Breaker | D2, class |
| NumberOfInteractors | D2, class |
| Class | All remaining nodes |
In order to see the worthiness of our model features, we conducted a χ2statistic that evaluates and ranks the marginal worth of a feature with respect to the two classes in the cross-validation dataset. The results of this analysis are shown in Table 3. All the features, except conditional, had their χ2 statistic above the critical value of 0.05. We observe that general features, interactor, NumberOfInteractors, D1, D2, order and comma were top-ranking features in their marginal worthiness. We also found that these six general features together contributed significantly towards the performance of our model, giving an overall accuracy of 84.5%, while the remaining specific features when added to the model further improved this accuracy by about 2%. The table also shows loss/gain (Δbaseline) in classification accuracy when each feature was removed individually from the feature set; which also showed that each feature, except conditional, contributed positively towards the classification although the individual ranks did not match with the χ2 rank, which is expected.
Table 3.
Ranking of model features using χ2 statistic on Dataset 2
| Feature | χ2 statistic | DOF | P-value | Rank | Δbaseline |
|---|---|---|---|---|---|
| Interactor | 392.8 | 89 | 0 | 1 | −0.7 |
| NumberOfInteractors | 355.9 | 1 | 0 | 2 | −1.0 |
| D1 | 337.2 | 4 | 0 | 3 | −1.3 |
| D2 | 315.7 | 3 | 0 | 4 | −1.9 |
| Order | 298.8 | 2 | 0 | 5 | −4.6 |
| Comma | 245.4 | 3 | 0 | 6 | −0.5 |
| Prep | 186.2 | 19 | 0 | 7 | −1.8 |
| Breaker | 57.4 | 1 | 3.5E-14 | 8 | −0.3 |
| Not | 49.7 | 1 | 1.8E-12 | 9 | −0.3 |
| Which | 35.0 | 1 | 3.4E-09 | 10 | −0.1 |
| But | 34.3 | 1 | 4.8E-09 | 11 | −0.0 |
| Conditional | 0.3 | 1 | 0.6 | 12 | +0.1 |
3.5 Performance on leftover dataset: a large-scale PPI extraction from 679 733 samples
To test the effectiveness of our method in extracting new PPIs from large collections of unstructured texts, we used the BN model built on cross-validation dataset and applied it on 679 733 PPI triplets of leftover dataset. Since our leftover dataset did not contain any known BioGRID interactions, we expected the dataset to contain much fewer true interactions in percentage terms. We incorporated this fact in our BN model by considering five prior class distributions [P(C=true)=0.05, 0.04, 0.03, 0.02 and 0.01]. By resetting the class distribution in the trained model (see Section 2), i.e. P(C=true), in Equation (1) to 0.05, 97% of these triplets were classified as false, while the remaining 3% (i.e. 18,378 triplets) were classified as true. In order to check if the positive PPI triplets were actually true, we randomly selected 1% of these samples (∼184 triplets, shown in Supplementary Material) for manual verification. Out of these 184 positive predictions, 71 (39%) were found to be true interactions that were not documented in BioGRID. By extrapolating these figures, we estimate that about 7,092 triplet interactions (containing 3,750 non-redundant pairs of protein names) are missed by BioGRID out of all 18,378 (with 9,891 non-redundant pairs of protein names) extracted triplets. Manually verifying them will give us true interactions among those extracted ones, which is a dramatic reduction from manually verifying all 679,733 triplets in the leftover dataset. When we further decreased P(C=true) to 2%, e.g., the number of extracted triplets decreased to 5,679, among which 52% are expected to be true positives. Table 4 shows the results for the five class distributions for which we tested our model. We can see that true positive rate (TP rate=TP/TP+FP) increases and false positive rate (FP/TP+FP) decreases with decreasing P(C=true), as expected. Because of the large number of samples, we were not able to have a good estimate of recall rate for the leftover dataset. In this analysis, we did not carry out gene or any other name disambiguation as we considered this as part of another major challenging problem of Named Entity Recognition (another granularity of PPI problem). While our method is generic and can potentially be integrated with any disambiguation method, the two problems are separate and we plan to work on this problem in the near future.
Table 4.
Performance of the BN model on the leftover dataset based on different prior class distributions, P(C)
| Model class distribution | Total positive predictions from leftover dataset | Based on a random sample of 184 interaction triplets |
||
|---|---|---|---|---|
| TP | FP | TP rate (%) | ||
| True 5%, false 95% | 18 378 | 71 | 113 | 38.59 |
| True 4%, false 96% | 14 947 | 65 | 89 | 42.21 |
| True 3%, false 97% | 10 491 | 53 | 55 | 49.07 |
| True 2%, false 98% | 5679 | 33 | 30 | 52.38 |
| True 1%, false 99% | 1221 | 9 | 4 | 69.23 |
TP—true positive, FP—false positive, TP rate = TP/TP+FP.
4 DISCUSSION
We presented here an accurate, general-purpose method for extracting PPIs from unstructured biological texts. We demonstrated through manually annotated datasets that our method is one of the best performing PPI extraction systems reported so far with a recall of 71%, precision of 76% and accuracy of 87% for extracting PPI triplets. Extracting PPIs from a large number (679 733) of sentences obtained from PubMed using general interaction terms showed that our method can be effectively combined with human annotations to discover a significant number (estimated to be more than 3700) of new PPIs, which were not documented by our reference PPI database BioGRID. The new PPIs can be obtained by manually verifying a much smaller number of triplet samples (18 378 of 679 733). Our study was performed using the same protein name dictionary as BioGRID, indicating the missing PPIs by manual annotation are not due to a poor protein name dictionary. Had we used a more comprehensive protein name dictionary, we could possibly have detected an even larger number of new PPIs. Furthermore, in this study we only used PubMed abstracts. There could be a large number of undiscovered PPIs buried in the full texts. Our study illustrates that human annotation can significantly benefit from robust and accurate computational information extraction methods in building PPI databases from biological texts.
On the basis of the work of Pyysalo and coworkers (2008), we characterized our cross-validation dataset (Supplementary Table S4). The dataset was manually annotated for PPI triplets, whose elements in the dataset were associated with their coordinates and their order of appearance in the sentences. The dataset covered 90 interaction words indicating the type of interaction. The dataset covers multiple species and is limited in its coverage to PPIs present in BioGRID. On an average per sentence, our dataset contained about 29 words, 2.5 entities, 1.5 interaction words and 2.5 triplets. Of the 1037 sentences in our dataset, half of them contained at least one true PPI triplet. Of the 2550 triplets in the dataset, 668 were true triplets. This indicates that the precision of the co-occurrence method on our dataset would be about 26%. Assuming a recall of the co-occurrence method as 1, the F-measure can be calculated as 41%. With respect to these above features, our dataset compares favourably with the datasets presented in Pyysalo (2008) in terms of difficulty. Our method (with F-measure of 74%), therefore, was able to achieve a significant improvement over the co-occurrence method on this dataset.
A common problem in the field is that it is difficult to obtain other researchers' data and programmes to make fair comparisons. This has also been noted as a general problem in computational biology field recently (Veretnik et al., 2008). Without benchmark datasets, it is difficult to compare different approaches in terms of their performances, as there are substantial differences among the used training and testing datasets, which are highly dependent on the selection criteria (Pyysalo et al., 2008). This problem of comparison between programmes gets more aggravated due to their different design objectives and granularity of the PPI problem they attempt. To evaluate the current methods using common standards, BioCreative challenge was created as a community-wide effort for evaluating text mining and information extraction systems applied to the biological domain (Krallinger and Valencia, 2007; Krallinger et al., 2008a, b). We did not test our method on BioCreative datasets because the setup of BioCreative inherently requires also the method to perform name recognition, while our current method focuses on only PPI triplet extraction. In addition, BioCreative does not provide sentences with PPI triplet annotations, which are necessary to test the accuracy of a PPI triplet extraction system like ours in a rigorous and fair way.
In this study, we learned complex language patterns using BN method through manually selected features. The features in our study are defined based on (revolves around) three words that form the concept of the triplets, such as the interaction words, order of triplet elements, the distances D1 and D2, number of interaction words, number of commas and other specific features within the range of triplets. Compared with a method that automatically selects features, our approach is more scientifically meaningful and enables us to learn more complex relationships among the selected features. It was demonstrated in this study that with only 12 features, a rather complex BN can be learned, which gave a quite satisfactory performance on a large and general dataset. Many of the selected features have been used for the first time for the PPI-triplet extraction task. Some similar features (e.g. distances between two protein names, interaction keywords and order) have previously been used in tasks such as identifying interacting protein pairs and sentences that contain interactions. For example, NLP, sequence alignment, and rule-based systems implicitly use the concept of word order. We have adapted this concept implicitly in the formulation of our ‘triplet’ rules. Similarly, some co-occurrence methods and those based on dependency graphs have used the distance between two protein names to improve the extraction accuracy.
One aspect that makes PPI extraction task a non-trivial one is because of the complexity associated with sentences. The complexity arises because there could potentially be numerous unstructured syntactical ways (language patterns) with different contextual meanings to describe the same protein interaction in a sentence. Thus, while a PPI may be present straight-forward in a simple sentence, it may be ambiguously buried in a complex structure in another sentence. Furthermore, many sentences contain multiple protein names and/or interaction words, which are prone to produce false positives. To cope with the sentence complexity, in this study we have analyzed each putative PPI in a sentence separately, rather than all putative PPIs in the sentence together. By dividing a more complex sentence, which may sometimes contain multiple putative PPIs, into individual PPIs with less complex structures, we can extract more specific information related to a PPI and focus on the characteristic language rules describing individual PPIs.
Text-mining methods also need to deal with the ambiguity in inference from extracted information due to missing of other related information. When only part of the information in sentences is used, perfect prediction/extraction cannot be made. Furthermore, the meaning of a sentence can often depend on the contextual meaning of certain words in it. Even given all the rules, without the domain knowledge of certain words, a computer cannot completely understand the meaning of a sentence.
The BN framework can model both the missing data and ambiguity effectively (Needham et al., 2007). The use of Dirichlet parameter priors in the BN framework allowed us to handle missing data, thereby making the method scalable for large-scale PPI extraction. This is evident from the fact that there were 101 distinct interaction words in the leftover dataset that were not present in the cross-validation dataset, which contained only 90 distinct interaction words in all.
Each triplet extracted by our method is associated with a probability value measuring the likelihood of the interaction between two proteins in the triplet. The associated probabilities provide more valuable information on the confidence of the extracted PPIs than those methods giving only binary output (yes or no). The associated probabilities can also be used to rank extracted PPIs to allow a more flexible selection of interactions in terms of their confidence. A weighed or ranked output has also clear advantage for manual revision and assisted curation by human experts.
In this study, we have also tested a few other statistical learning algorithms besides BN, including support vector machine (SVM), logistic regression and decision trees, and found that while all machine-learning methods obtained satisfactory performance, BN is the most accurate and robust method for the PPI extraction task (see Table S2 for a performance comparison of the methods). The BN methodology provides us an intuitive graphical insight into the dependencies between various features that we have used to model interaction relationships. The importance of these dependencies is evident by the superior performance of our BN model compared to the Naïve Bayes model. The insight into these dependencies thus may help us to improve the current feature set.
There is a notable difference in the performance on Dataset 2 and the leftover dataset. One of the reasons is that the proportion of true samples in the leftover dataset is very low (less than 3%), making it more challenging as pointed out in (Pyysalo et al., 2008). The other reason is due to the named entity recognition factor. While our training dataset contains triplets that all were annotated manually for protein names, our leftover dataset is raw and contains triplets with protein names incorrectly tagged by their ambiguous terms that did not properly represent proteins (such as common English words, gene names, part of multiple word protein names)—we annotated such extractions as false in leftover dataset.
Finally, we showed that our method, apart from PPI triplet extraction, performed quite well at PPI sentence extraction with the same accuracy as PIE on our dataset. The comparison between our method and PIE showed that methods that perform well on triplet extraction may likely also perform well on sentence- or abstract-filtering tasks. On the contrary, methods that perform well at sentence or abstract extraction may not perform well at PPI triplet extraction. It is clear, however, that these tasks are quite different, and to extract PPIs including exact interaction words, specific methods need to be developed.
In summary, we have developed a general purpose scalable PPI extraction method that can be used to assist manual annotations. The method is generic and can principally be applied for extracting other types of biological or molecular interactions such as protein–small molecule, gene–gene, drug–drug interactions among others. This can be done by using appropriate domain-specific dictionaries and datasets. The datasets we generated in this study can be used as a PPI extraction benchmark, which will be updated when we annotate more sentences from both Dataset 1 and leftover dataset.
ACKNOWLEDGEMENTS
We sincerely thank Dr Martin Krallinger for his valuable comments on the manuscript. We also thank the anonymous reviewers for many helpful suggestions.
Funding: US NIH grants R01-GM078990, R01-HG02518-02 and US NSF grants DMS-0706989, DMS-0244638, DMS-0204674.
Conflict of Interest: none declared.
REFERENCES
- Alfarano C, et al. The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic Acids Res. 2005;33:D418–D424. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner WA, Jr, et al. Manual curation is not sufficient for annotation of genomic databases. Bioinformatics. 2007;23:i41–i48. doi: 10.1093/bioinformatics/btm229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuming T, et al. PDZBase: a protein-protein interaction database for PDZ-domains. Bioinformatics. 2005;21:827–828. doi: 10.1093/bioinformatics/bti098. [DOI] [PubMed] [Google Scholar]
- Blaschke C, et al. Automatic extraction of biological information from scientific text: protein-protein interactions. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1999:60–67. [PubMed] [Google Scholar]
- Ceol A, et al. Linking entries in protein interaction database to structured text: the FEBS Letters experiment. FEBS Lett. 2008;582:1171–1177. doi: 10.1016/j.febslet.2008.02.071. [DOI] [PubMed] [Google Scholar]
- Chatraryamontri A, et al. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman C, et al. GENIES: a natural-language processing system for the extraction of molecular pathways from journal articles. Bioinformatics. 2001;17:S74–S82. doi: 10.1093/bioinformatics/17.suppl_1.s74. [DOI] [PubMed] [Google Scholar]
- Hatzivassiloglou V, Weng W. Learning anchor verbs for biological interaction patterns from published text articles. Int. J. Med. Inform. 2002;67:19–32. doi: 10.1016/s1386-5056(02)00054-0. [DOI] [PubMed] [Google Scholar]
- Huang M, et al. Discovering patterns to extract protein-protein interactions from full texts. Bioinformatics. 2004:3604–3612. doi: 10.1093/bioinformatics/bth451. [DOI] [PubMed] [Google Scholar]
- Jensen LJ, et al. Literature mining for the biologist: from information retrieval to biological discovery. Nat. Rev. Genet. 2006;7:119–129. doi: 10.1038/nrg1768. [DOI] [PubMed] [Google Scholar]
- Jenssen TK, et al. A literature network of human genes for high-throughput analysis of gene expression. Nat. Genet. 2001;28:21–28. doi: 10.1038/ng0501-21. [DOI] [PubMed] [Google Scholar]
- Kann MG. Protein interactions and disease: computational approaches to uncover the etiology of diseases. Brief Bioinform. 2007;8:333–346. doi: 10.1093/bib/bbm031. [DOI] [PubMed] [Google Scholar]
- Kim S, et al. Kernel approaches for genic interaction extraction. Bioinformatics. 2008a;24:118–126. doi: 10.1093/bioinformatics/btm544. [DOI] [PubMed] [Google Scholar]
- Kim S, et al. PIE: an online prediction system for protein-protein interactions from text. Nucleic Acids Res. 2008b;36:W411–W415. doi: 10.1093/nar/gkn281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krallinger M, Valencia A. Proceedings of the BioCreative Workshop. Madrid, Spain: CNIO; 2007. Assessment of the second BioCreative PPI task: automatic extraction of protein-protein interactions; pp. 41–54. [Google Scholar]
- Krallinger M, et al. Overview of the protein-protein interaction annotation extraction task of BioCreative II. Genome Biol. 2008a;9(Suppl. 2):S4. doi: 10.1186/gb-2008-9-s2-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krallinger M, et al. Evaluation of text-mining systems for biology: overview of the Second BioCreative community challenge. Genome Biol. 2008b;9(Suppl. 2):S1. doi: 10.1186/gb-2008-9-s2-s1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner F, Valencia A. A text-mining perspective on the requirements for electronically annotated abstracts. FEBS Lett. 2008;582:1178–1181. doi: 10.1016/j.febslet.2008.02.072. [DOI] [PubMed] [Google Scholar]
- Leroy G, Chen H. Filling preposition-based templates to capture information from medical abstracts. Pac. Symp. Biocomput. 2002:350–361. [PubMed] [Google Scholar]
- Liu H, et al. BioThesaurus: a web-based thesaurus of protein and gene names. Bioinformatics. 2006;22:103–105. doi: 10.1093/bioinformatics/bti749. [DOI] [PubMed] [Google Scholar]
- Malik R, et al. Combination of text-mining algorithms increases the performance. Bioinformatics. 2006;22:2151–2157. doi: 10.1093/bioinformatics/btl281. [DOI] [PubMed] [Google Scholar]
- Mishra GR, et al. Human protein reference database - 2006 update. Nucleic Acids Res. 2006;34:D411–D414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa M, et al. Combining multiple layers of syntactic information for protein-protein interaction extraction. Proceedings of Third International Symposium on Semantic Mining in Biomedicine (SMBM) 2008:101–108. [Google Scholar]
- Narayanaswamy M, et al. Beyond the clause: extraction of phosphorylation information from medline abstracts. Bioinformatics. 2005;21(Suppl. 1):i319–i327. doi: 10.1093/bioinformatics/bti1011. [DOI] [PubMed] [Google Scholar]
- Needham CJ, et al. A primer on learning in Bayesian networks for computational biology. PLoS Comput. Biol. 2007;3:e129. doi: 10.1371/journal.pcbi.0030129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SK, Wong M. Toward routine automatic pathway discovery from on-line scientific text abstracts. Genome Inform. Ser. Workshop. Genome. Inform. 1999;10:104–112. [PubMed] [Google Scholar]
- Ono T, et al. Automated extraction of information on protein-protein interactions from the biological literature. Bioinformatics. 2001;17:155–161. doi: 10.1093/bioinformatics/17.2.155. [DOI] [PubMed] [Google Scholar]
- Pagel P, et al. The MIPS mammalian protein-protein interaction database. Bioinformatics. 2005;21:832–834. doi: 10.1093/bioinformatics/bti115. [DOI] [PubMed] [Google Scholar]
- Park JC, et al. Bidirectional incremental parsing for automatic pathway identification with combinatory categorial grammar. Pac. Symp. Biocomput. 2001:396–407. [PubMed] [Google Scholar]
- Pustejovsky J, et al. Robust relational parsing over biomedical literature: extracting inhibit relations. Pac. Symp. Biocomput. 2002:362–373. doi: 10.1142/9789812799623_0034. [DOI] [PubMed] [Google Scholar]
- Pyysalo S, et al. Comparative analysis of five protein-protein interaction corpora. BMC Bioinformatics. 2008;9(Suppl. 3):S6. doi: 10.1186/1471-2105-9-S3-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salwinski L, et al. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saric J, et al. Extraction of regulatory gene/protein networks from Medline. Bioinformatics. 2006;22:645–650. doi: 10.1093/bioinformatics/bti597. [DOI] [PubMed] [Google Scholar]
- Skusa A, et al. Extraction of biological interaction networks from scientific literature. Brief Bioinform. 2005;6:263–276. doi: 10.1093/bib/6.3.263. [DOI] [PubMed] [Google Scholar]
- Stapley BJ, Benoit G. Biobibliometrics: information retrieval and visualization from co-occurrences of gene names in Medline abstracts. Pac. Symp. Biocomput. 2000:529–540. doi: 10.1142/9789814447331_0050. [DOI] [PubMed] [Google Scholar]
- Stark C, et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temkin JM, Gilder MR. Extraction of protein interaction information from unstructured text using a context-free grammar. Bioinformatics. 2003;19:2046–2053. doi: 10.1093/bioinformatics/btg279. [DOI] [PubMed] [Google Scholar]
- Thomas J, et al. Automatic extraction of protein interactions from scientific abstracts. Pac. Symp. Biocomput. 2000:541–552. doi: 10.1142/9789814447331_0051. [DOI] [PubMed] [Google Scholar]
- Van Landeghem S, et al. Proceedings of Third International Symposium on Semantic Mining in Biomedicine (SMBM). Turku, Finland: TUCS; 2008. Extracting protein-protein interactions from text using rich feature vectors and feature selection; pp. 77–84. [Google Scholar]
- Veretnik S, et al. Computational biology resources lack persistence and usability. PLoS Comput. Biol. 2008;4:e1000136. doi: 10.1371/journal.pcbi.1000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten IH, Frank E. Data Mining: Practical Machine Learning Tools and Techniques. San Francisco, CA: Morgan Kaufmann; 2005. [Google Scholar]
- Wong L. PIES, a protein interaction extraction system. Pac. Symp. Biocomput. 2001:520–531. [PubMed] [Google Scholar]
- Yakushiji A, et al. Event extraction from biomedical papers using a full parser. Pac. Symp. Biocomput. 2001:408–419. doi: 10.1142/9789814447362_0040. [DOI] [PubMed] [Google Scholar]