Abstract
The conventional method of detecting gene-environment interactions, the case-control analysis, suffers from low statistical power. In contrast, the case-only analysis/design can be powerful in certain scenarios, although violation of the assumption of independence between the genetic and environmental factors can greatly bias the results. As an alternative, Bayes model averaging may be used to combine the case-control and case-only analyses. This approach first frames the case-control and case-only analyses as variations of a log-linear model. The weighting between these 2 models is then a function of the data and prior beliefs on the independence of the 2 potentially interacting factors. In this paper, the authors demonstrate via simulations that when there is no prior information on the independence of the genetic and environmental factors, this approach tends to be more powerful than the case-control analysis. Additionally, when the genetic and environmental factors are not independent in the population, bias is substantially reduced, with a corresponding reduction in type I error in comparison with the case-only analysis. Increased power or increased robustness to violations of the independence assumption may be obtained with more appropriate prior specification. The authors use an example data analysis to demonstrate the advantages of this approach.
Keywords: Bayesian estimation, Bayesian model, case-control studies, epidemiologic methods, interaction
There is growing evidence that gene-environment interactions play an important role in complex diseases with a genetic basis (1–8). The conventional method of detecting interactions, the case-control analysis, is known to suffer from low statistical power (9, 10). The case-only analysis (and its analogous log-linear approach) has been proposed for detection of interaction effects with a substantial gain in power (11–15). However, the validity of the case-only design depends greatly upon the assumption that the 2 interacting factors are independent in the underlying population. Previous investigations (16, 17) have shown that the case-only design is highly susceptible to bias arising from nonindependence between the 2 interacting factors in the underlying population.
Generally, prior knowledge of the 2 factors is used to decide the appropriateness of the independence assumption. However, a decision based solely on prior knowledge can lead to uncertainty as to which analytical approach is most appropriate. Alternatively, some investigators have proposed statistically testing the independence of the factors within a representative population and then using the appropriate analysis based on the inference (17–19). This depends greatly on the criteria used to determine evidence for nonindependence and can still lead to increased type I error (20).
Here we propose the use of Bayes model averaging (BMA) to combine case-control and case-only analysis. The data and prior belief in the independence of the 2 potentially interacting factors are used to generate weights between the models. In this paper, we describe the combined approach; perform a simulation experiment to gauge its performance in terms of estimation bias, type I error, and power under various scenarios; and apply the method to a real data example.
MATERIALS AND METHODS
The case-control analysis
Assume we have a case-control sample with a disease outcome Y, a genetic factor G, and a dichotomous environmental factor E. In a case-control analysis, the departure from a multiplicative interaction model can be examined in the following logistic model:
This model can also be viewed as a reduced but equivalent version of the full log-linear model (21), in which the logarithm of the expected numbers of persons in each cell of the 2 × 2 × 2 table of G, E, and Y is modeled as
| (1) |
In both models, β, γ, and σ are exactly the same parameters modeling the main effect of G, E, and the G × E interaction, respectively. In the log-linear model, β0, γ0, and σ0 model the joint distribution of G and E in the controls.
The case-only analysis
With the assumption of G × E independence, the case-only analysis may be used to detect the interaction based on the following model:
Similarly, Umbach et al. (15) proposed a variation of the full log-linear model, in which they demonstrated that the assumption of G × E independence can be specified by setting σ0 in equation 1 equal to 0, resulting in the following model:
| (2) |
When the independence assumption holds, σ* is approximately equivalent to σ (15).
Averaging over the case-only and case-control analyses
BMA may be used to obtain a single estimate of interaction effect and avoid the uncertainty in having to select either the case-control design or the case-only design (22, 23). Specifically, given the observed data D (including Y, G, E, and other potential covariates), the posterior probability of the interaction effect is
To ensure equal sample sizes and comparable likelihoods, the log-linear model for each analysis is employed when calculating the posterior probability of the case-only and case-control models. , the posterior probability of each model, is given by
where is the prior probability for Mk. Priors can be assigned to the case-control model (M1) and the case-only model (M2) by specifying the relative weight of their prior probability W = Pr(M1)/Pr(M2) (i.e., the prior odds). is the integration of the likelihood of Mk over all of the parameters , estimated through a Laplace transformation (24):
is the estimated interaction effect from Mk, and the expectation and the variance of the interaction effect are calculated as
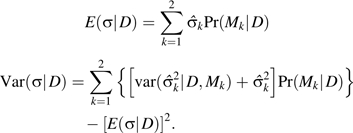 |
Assuming a normal distribution for the interaction estimate, statistical inference on σ is determined via a Wald test,
Simulations
We carried out simulations to illustrate the performance of the BMA approach. The underlying population is generated on the basis of the following logistic regression model:
The baseline disease prevalence rate, p0, is set to 0.05; the odds ratio for E, which equals , is set to 2; the odds ratio for G () is set to 1; and the odds ratio for the interaction effect () is set to either 1 or 1.25. The 2 risk factors, E and G, are binary. Their marginal frequencies, pg and pe, are both set to 0.3. The G × E association in the population is simulated using the following model:
in which is the logarithm of the odds ratio for the population association between G and E.
For each simulation replicate in each scenario, a population of 1,000,000 observations is generated and a 1:1 case-control sample is randomly selected. The empirical power and type I error rate of the conventional case-control analysis, the case-only analysis, and the BMA approach (with W = 1) are compared across various levels of G × E association. To investigate different case-cohort or case-control designs, we perform simulations in which the total sample size is held constant with varying case:control ratios, in addition to scenarios in which the number of cases is fixed and the number of controls is adjusted. Further comparisons of the mean squared error (MSE) and bias for the estimates of interaction effect are also made with varying under the null hypothesis. MSE is defined as the average squared difference between the estimate and the interaction parameter λ, and bias is defined as the mean difference between the estimate and λ. The effect of using different prior information on the results of the BMA approach are also compared across differing values for W.
In all of the analyses, statistical significance is determined by a 2-sided P value at an α level of 0.05, and 1,000 replications are simulated for each scenario. The simulation and analysis are performed in R (25); the specific code is available from the authors upon request.
Application to real data
To illustrate the various approaches in practice, the 3 methods are applied to the Wuhan Smoking Prevention Trial, in which smoking behavior was defined for urban seventh-grade students from Wuhan, China, in 1998 (26). The analysis is limited to 495 male nonsmokers (i.e., controls) and 495 males that have initiated smoking (i.e., cases). A variable number of tandem repeats (VNTR) in the monoamine oxidase A (MAOA) gene, located on chromosome X, and 2 single nucleotide polymorphisms (SNPs) have been genotyped for all participants. The first polymorphism, SNP 1, is unlinked to MAOA and is located in the frequenin homolog (Drosophila) (FREQ) gene on chromosome 9. An interaction analysis between this SNP and the MAOA VNTR represents a scenario in which the 2 factors are independent in the population. The second polymorphism, SNP 2, is located in the MAOA gene region and is linked to the VNTR. An interaction analysis between SNP 2 and the VNTR represents a situation in which the 2 factors are correlated in the population. For the VNTR and the 2 SNPs, the minor allele frequencies are 0.39, 0.16, and 0.27, respectively. Both SNPs are in Hardy-Weinberg equilibrium. Multiplicative interaction between the 2 SNPs and the VNTR with regard to smoking initiation is examined with the case-only, case-control, and BMA approaches. An additive model is used for SNP 1. Since population substructure can lead to correlation of 2 unlinked polymorphisms in the population, 233 ancestry informative markers (27) are used to estimate the coefficient of ancestry for each individual (28). In our analysis, there was very little evidence for substructure, since the mean Asian-specific coefficient of ancestry was 97.5%.
RESULTS
Type I error and power
Figure 1 shows the type I error and power for the case-control analysis, the case-only analysis, and the BMA approach when varies from −0.5 to 0.5 with a sample size of 2,000 cases and 2,000 controls. Figure 1A shows that the case-only analysis has a greatly inflated type I error when is nonzero, while the type I error rate of the case-control analysis remains constant. For the BMA approach, the type I error rate is slightly conservative when there is no G × E association (0.03). When the G × E association is moderate, the type I error rate of the BMA approach increases slightly, albeit at a much lower error rate than in the case-only analysis (the maximum type I error rate is 0.085 when = 0.2 or −0.2). As the G × E association gets stronger, the type I error rate of the BMA analysis approaches the case-control analysis.
Figure 1.

Type I error and power of the case-only, case-control, and Bayes model averaging (BMA) approaches. The odds ratio for the interaction effect is 1.0 (part A) or 1.25 (part B). The prior weight in the BMA approach is set to W = 1:1, with a sample size of 2,000 cases and 2,000 controls in the simulation. represents the association between the genetic factor and the environmental factor in the underlying population.
Figure 1B shows the empirical power of the case-only, case-control, and BMA approaches. While its power is not valid because of the greatly inflated type I error rates when ≠ 0 (as seen in Figure 1A), results of the case-only analysis are displayed for reference. As expected, when the assumption of independence is valid (= 0), the case-only approach is the most powerful approach, and the power of the BMA approach is higher than that of the case-control approach. When the G × E association points in the same direction as the interaction effect (i.e., > 0), the BMA approach is more powerful than the case-control approach. However, if the G × E association points in the opposite direction (< 0), the BMA approach is slightly less powerful than the case-control approach. As the G × E association increases, the power of the BMA analysis approaches that of the case-control analysis.
Specifically, for = 0.2 with 2,000 cases and 2,000 controls, the type I error is 0.085 and the empirical power is 0.49 (Figure 1B). Decreasing the number of controls to 1,000 while keeping the number of cases at 2,000 (total n = 3,000) increases the power of the BMA approach to 0.51 but also increases the type I error to 0.11. Likewise, increasing the number of controls to 4,000 with 2,000 cases (total n = 6,000) results in stabilization of the type I error (0.06) with no noticeable impact on power (0.47). Holding the total sample size constant (n = 4,000), an altered case:control ratio (1:3) greatly reduces power (0.35) with a corresponding reduction in type I error (0.06). In contrast, using a total sample size of 4,000 and a case:control ratio of 3:1 results in an increase in power (0.52) and an increase in type I error (0.12).
Estimates: MSE and bias
In Table 1, the MSE and bias of different approaches are compared across various levels of G × E association under the null hypothesis (λ = 0). With no G × E association, the MSE is smallest for the case-only approach and largest for the case-control approach. The MSE of the BMA (W = 1) lies between these 2 extremes. As the G × E association increases, the MSE and bias of the case-only approach increase sharply while the MSE of the BMA approach remains close to that of the case-control approach, although for modest values of G × E association in the population, there is a slight bias in estimates from the BMA approach.
Table 1.
Mean Squared Error and Bias of the Estimates of the Interaction Effecta
| θge | Case-Control Method | Case-Only Method | Bayes Model Averaging Method | |||
| MSE | Bias | MSE | Bias | MSE | Bias | |
| 0 | 0.023 | 0.003 | 0.012 | 0.004 | 0.015 | 0.003 |
| 0.1 | 0.021 | 0.001 | 0.020 | 0.100 | 0.018 | 0.037 |
| 0.2 | 0.023 | 0.002 | 0.049 | 0.195 | 0.027 | 0.039 |
| 0.3 | 0.020 | 0.001 | 0.099 | 0.297 | 0.027 | 0.026 |
| 0.4 | 0.023 | 0.001 | 0.169 | 0.398 | 0.026 | 0.009 |
| 0.5 | 0.020 | 0.003 | 0.261 | 0.501 | 0.020 | 0.004 |
Abbreviation: MSE, mean squared error.
The odds ratio for the interaction effect was 1.0. The prior weight in the Bayes model averaging approach was set to W = 1:1, with a sample size of 2,000 cases and 2,000 controls in the simulation.
Influence of priors on the BMA approach
Figure 2 shows the effect of different prior weights (W’s ranging from 0.001 to 1,000) in terms of type I error, empirical power, and bias. When there is no G × E association, there is a stable type I error rate across all prior specifications. Likewise, when the prior weight favors the case-only model (i.e., W < 1), there is a corresponding and valid increase in power. Alternatively, when there is substantial G × E association (= 0.5), the type I error increases for extreme priors in favor of the case-only model (W < 0.01), with a corresponding increase in the empirical power. Note that the increase in power appears to be much more substantial than is indicated by the slight increase in type I error. As the prior better reflects the underlying truth of the G × E association (= 0.5) and begins to favor the case-control model (W > 1), there is a reduction in the type I error rate and the power approaches that of the case-control analysis. When the G × E association is modest (= 0.2), there is a much greater increase in the type I error and only extreme priors favoring the case-control model (W > 5) reduce this to a nearly nominal level. Bias in the estimates follows a similar pattern, with the most substantial bias occurring when there is modest G × E association in the population and a prior weight favoring the case-only analysis (W < 1).
Figure 2.

Influence of prior weights on the performance of the Bayes model averaging approach. W represents the relative weights of the case-control and case-only models in the Bayes model averaging approach. Weights to the left on the x-axis (W < 1) favor the case-only model, while weights to the right (W > 1) favor the case-control model. The odds ratio for the interaction effect is 1.0 (parts A and C) or 1.25 (part B). The sample size is 2,000 cases and 2,000 controls in the simulation.
Influence of sample size
Figure 3 shows, under the null hypothesis, the influence of the sample size on the type I error and bias of the BMA approach when varies from 0 to 0.50. No prior information on the G × E association in the underlying population (W = 1:1) is assumed. In general, as the G × E association increases up to moderate levels (e.g., = 0.2 or 0.3), there is an increase in bias and type I error. However, when the G × E association is strong, the bias and type I error for the BMA approach are decreased. While the overall bias and type I error are greatest for small sample sizes, it is the plateau in this general pattern that is greatly influenced by the sample size, with the BMA approach being robust to smaller levels of G × E association with larger sample sizes.
Figure 3.

Influence of sample size on the type I error and bias of the Bayes model averaging (BMA) approach. Bias is defined as the mean difference between the BMA estimate and the true interaction parameter, which is set to be 0 in both part A and part B. represents the association between the genetic factor and the environmental factor in the underlying population. The prior weight in the BMA approach is set to W = 1:1.
Application to real data
Table 2 shows the results of the case-only, case-control, and BMA approaches with different weights when they are applied to the example data. For SNP 1 and the MAOA VNTR (= 0), there are consistent estimates across all analyses (odds ratio = 1.58). The case-only analysis is the most efficient and results in the smallest observed P value (P = 0.019). The case-control analysis yields a P value of 0.098. With the assumption that SNP 1 is independent of the VNTR, the most appropriate prior specification is one that weighs heavily in favor of the case-only analysis (W = 0.01). Here, an estimate and P value similar to those of the case-only analysis are obtained. In contrast, when exploring the interaction between SNP 2 and the VNTR (≠ 0), the case-only analysis yields a substantial effect (odds ratio = 2.06) and a corresponding P value of 0.002. However, in the case-control analysis, the odds ratio for this interaction is closer to 1, with a corresponding P value of 0.312. In the BMA analysis, a nonsignificant statistical test is obtained across all levels of prior information from equal weight (W = 1) to a much greater weight on the case-control analysis (W = 100). Given that SNP 2 and the VNTR are located within the same gene region and may be in linkage disequilibrium, the most appropriate prior weighting may be that favoring the case-control analysis.
Table 2.
Interaction Analyses for 2 Single Nucleotide Polymorphisms With a Variable Number of Tandem Repeats in the Monoamine Oxidase A (MAOA) Genea
| Interaction and Method | Odds Ratio | 95% Confidence Interval | P Value |
| SNP 1 × VNTR interaction | |||
| Case-only | 1.58 | 1.08, 2.32 | 0.019 |
| Case-control | 1.58 | 0.92, 2.72 | 0.098 |
| BMA (W = 1:1) | 1.58 | 1.03, 2.43 | 0.037 |
| BMA (W = 1:10) | 1.58 | 1.07, 2.33 | 0.021 |
| BMA (W = 1:100) | 1.58 | 1.08, 2.32 | 0.019 |
| SNP 2 × VNTR interaction | |||
| Case-only | 2.06 | 1.29, 3.29 | 0.002 |
| Case-control | 1.39 | 0.73, 2.67 | 0.312 |
| BMA (W = 1:1) | 1.63 | 0.82, 3.26 | 0.165 |
| BMA (W = 10:1) | 1.43 | 0.74, 2.78 | 0.290 |
| BMA (W = 100:1) | 1.40 | 0.73, 2.68 | 0.309 |
Abbreviations: BMA, Bayes model averaging; SNP, single nucleotide polymorphism; VNTR, variable number of tandem repeats.
SNP 1, located in the frequenin homolog (Drosophila) (FREQ) gene, is unlinked with the MAOA VNTR. SNP 2 is located in the MAOA gene region and thus is linked and correlated with the VNTR in the population.
DISCUSSION
While the conventional case-control analysis suffers from low efficiency when detecting interaction effects (10), the validity of the case-only analysis/design relies heavily on the assumption of independence of the 2 interacting factors (16, 17). Instead of an all-or-nothing choice between the 2 approaches, BMA can be used to average the estimates of the 2 models based on their posterior probability. Via simulations, we have shown that in comparison with the case-only and case-control approaches, the BMA approach has better overall performance across a variety of scenarios. This holds true even when little prior information is available regarding the independence of the 2 factors in the population. In general, the BMA approach leverages the statistical efficiency of the case-only estimate with the unbiased properties of the case-control estimate. It is more powerful than the case-control approach when = 0 (albeit less powerful than the case-only approach), but it is also robust when there is non-independence of the 2 interacting factors. Although the BMA estimate can be slightly biased and the type I error can be slightly inflated when correlation is modest, as the correlation further increases there is a reduction in bias and a return to the nominal type I error rate. Thus, the BMA approach performs well in situations in which the correlation of the 2 factors can lead to the largest bias and elevated type I error rates for the case-only approach. The flexibility of specifying an appropriate prior can further enhance the performance of the BMA approach.
By appropriately weighting the case-only and case-control models, the BMA approach has increased power and less bias when 2 factors are positively correlated. When there is correlation between the 2 factors, the nonzero σ0 in the log-linear case-control model leads to a higher posterior probability for the case-control model. Consequently, when averaging over the 2 models, the estimate and its standard error are pulled more towards the case-control results. As the correlation increases, the overall result is weighted even more heavily towards the results of the case-control model. When the direction of the correlation for the 2 factors is opposite to that of the interaction effect, the case-only result has less power (Figure 1B). This is due to the combined impact of the interaction effect and the correlation in the population on the co-occurrence of the 2 factors in the cases. The resulting case-only estimate is a balance between the 2 trends and initially results in a decrease in power. As the negative correlation increases, the case-only analysis detects this increase and power tends to increase. Since the BMA approach is dependent upon the case-only estimate, this phenomenon will affect the BMA approach as well. However, because the BMA approach incorporates the case-control estimate, these trends are greatly tempered.
As the sample size increases, the data provide more information on G × E association with which the BMA approach can weight more towards the appropriate model when averaging the estimates of the 2 models, leading to less bias and reduced type I error rates in comparison with analyses with a smaller sample size and similar G × E dependence. As the sample size continues to increase, the influence of the G × E association becomes negligible. This property makes the BMA approach particularly useful as sample sizes increase in order to detect potentially weak interaction effects in genome-wide association studies of complex diseases. When there exists weak G × E association, slight increases in the type I error from a case-only analysis with large sample sizes may have a substantial impact on the overall results across all polymorphisms. In this situation, the large sample sizes will enhance the detection of a G × E association in the BMA approach and result in more robust estimates. In general, an increase in the number of cases (or an increase in the case:control ratio) aids power while also increasing the type I error. An increase in the number of controls (or an increase in the control:case ratio) tends to stabilize the type I error closer to the nominal rate without a substantial impact on power. Ultimately, the final posterior estimate will be a function of the total sample size, the ratio of cases and controls, and the size/direction of both the interaction effect and the correlation in the population.
Several approaches have been proposed as potential solutions to the problems introduced by G × E dependence in the case-only analysis. Some authors have suggested first testing the G × E association in the controls and then choosing the analysis strategy based on the G × E test results (16, 17). Unfortunately, this approach will be very dependent upon the power for declaring significance from a test within the controls. The BMA approach is similar in spirit to this approach, by using the data to provide an estimate of the correlation between the 2 factors. However, instead of an all-or-none choice, the BMA approach weights the 2 models on the basis of evidence for which is more appropriate. An alternative approach is to adjust for a covariate believed to lead to the G × E association in the case-only analysis (20). While applicable in some situations, this approach relies heavily on the presence and identification of the intermediate covariate. Such prior identification of appropriate covariates may be difficult or even impossible in practice.
In an approach similar to that of the BMA analysis presented here, Mukherjee and Chatterjee (29) proposed an empirical Bayes approach derived from a retrospective maximum-likelihood framework that corresponds to a weighted average between the case-only and case-control estimators. In additional simulations mimicking those presented by Mukherjee and Chatterjee (29) (data not shown), we compared the MSE and bias of the empirical Bayes estimate with those of the BMA estimate. Overall, performance was comparable for the 2 methods.
The BMA approach is applied to real data to illustrate the extremes in the level of prior knowledge regarding the 2 interacting factors. In the analysis of an SNP located on a different chromosome with the VNTR, there is an observed consistency in the estimates from the case-only and case-control analyses. In this scenario, the BMA approach has a P value similar to that of the case-only analysis. In contrast, in the analysis in which there exists correlation between SNP 2 and the VNTR, there is a discrepancy between the case-only and case-control estimates. These analyses indicate the feasibility of the BMA approach in practice.
In summary, the BMA approach can combine case-control and case-only estimates in gene-environment or gene-gene interaction analysis. This approach can be more powerful than the case-control approach when there is no correlation in the population and much more robust than the case-only approach when the independence assumption is violated. Increased power or increased robustness to violations of the independence assumption can also be modulated with appropriate prior information on the assumed dependence in the population.
Acknowledgments
Author affiliation: Department of Preventive Medicine and Zilkha Neurogenetic Institute, Keck School of Medicine, University of Southern California, Los Angeles, California (Dalin Li, David V. Conti).
This work was supported by the National Institute on Drug Abuse (grants DA020830 and CA084735 to D. L. and D. V. C.) and the National Institute of Environmental Health Sciences (grants ES015090 and GM069890 to D. V. C.). Genotyping for the example was performed as part of the Pharmacogenetics of Nicotine Addiction Treatment Program (SRI International and University of California, San Francisco), which received funding from the National Institute on Drug Abuse (grant DA020830).
Conflict of interest: none declared.
Glossary
Abbreviations
- BMA
Bayes model averaging
- FREQ
frequenin homolog (Drosophila)
- MAOA
monoamine oxidase A
- MSE
mean squared error
- SNP
single nucleotide polymorphism
- VNTR
variable number of tandem repeats
References
- 1.Stephens JW, Humphries SE. The molecular genetics of cardiovascular disease: clinical implication. J Intern Med. 2003;253(2):120–127. doi: 10.1046/j.1365-2796.2003.01104.x. [DOI] [PubMed] [Google Scholar]
- 2.Lesch KP. Molecular foundation of anxiety disorders. J Neural Transm. 2001;108(6):717–746. doi: 10.1007/s007020170048. [DOI] [PubMed] [Google Scholar]
- 3.Clément K. Genetics of human obesity. Proc Nutr Soc. 2005;64(2):133–142. doi: 10.1079/pns2005416. [DOI] [PubMed] [Google Scholar]
- 4.Grarup N, Andersen G. Gene-environment interactions in the pathogenesis of type 2 diabetes and metabolism. Curr Opin Clin Nutr Metab Care. 2007;10(4):420–426. doi: 10.1097/MCO.0b013e3281e2c9ab. [DOI] [PubMed] [Google Scholar]
- 5.Malats N. Gene-environment interactions in pancreatic cancer. Pancreatology. 2001;1(5):472–476. doi: 10.1159/000055850. [DOI] [PubMed] [Google Scholar]
- 6.Enoch MA. Genetic and environmental influences on the development of alcoholism: resilience vs. risk. Ann N Y Acad Sci. 2006;109(4):193–201. doi: 10.1196/annals.1376.019. [DOI] [PubMed] [Google Scholar]
- 7.Meyers DA, Larj MJ, Lange L. Genetics of asthma and COPD. Similar results for different phenotypes. Chest. 2004;126(2 suppl):105S–110S. doi: 10.1378/chest.126.2_suppl_1.105S. [DOI] [PubMed] [Google Scholar]
- 8.Klareskog L, Padyukov L, Lorentzen J, et al. Mechanisms of disease: genetic susceptibility and environmental triggers in the development of rheumatoid arthritis. Nat Clin Pract Rheumatol. 2006;2(8):425–433. doi: 10.1038/ncprheum0249. [DOI] [PubMed] [Google Scholar]
- 9.Hwang SJ, Beaty TH, Liang KY, et al. Minimum sample size estimation to detect gene-environment interaction in case-control designs. Am J Epidemiol. 1994;140(11):1029–1037. doi: 10.1093/oxfordjournals.aje.a117193. [DOI] [PubMed] [Google Scholar]
- 10.García-Closas M, Lubin JH. Power and sample size calculations in case-control studies of gene-environment interactions: comments on different approaches. Am J Epidemiol. 1999;149(8):689–692. doi: 10.1093/oxfordjournals.aje.a009876. [DOI] [PubMed] [Google Scholar]
- 11.Piegorsch WW, Weinberg CR, Taylor JA. Non-hierarchical logistic models and case-only designs for assessing susceptibility in population-based case-control studies. Stat Med. 1994;13(2):153–162. doi: 10.1002/sim.4780130206. [DOI] [PubMed] [Google Scholar]
- 12.Khoury MJ, Flanders WD. Nontraditional epidemiologic approaches in the analysis of gene-environment interaction: case-control studies with no controls! Am J Epidemiol. 1996;144(3):207–213. doi: 10.1093/oxfordjournals.aje.a008915. [DOI] [PubMed] [Google Scholar]
- 13.Yang Q, Khoury MJ, Flanders WD. Sample size requirements in case-only designs to detect gene-environment interaction. Am J Epidemiol. 1997;146(9):713–720. doi: 10.1093/oxfordjournals.aje.a009346. [DOI] [PubMed] [Google Scholar]
- 14.Weinberg CR, Umbach DM. Choosing a retrospective design to assess joint genetic and environmental contributions to risk. Am J Epidemiol. 2000;152(3):197–203. doi: 10.1093/aje/152.3.197. [DOI] [PubMed] [Google Scholar]
- 15.Umbach DM, Weinberg CR. Designing and analysing case-control studies to exploit independence of genotype and exposure. Stat Med. 1997;16(15):1731–1743. doi: 10.1002/(sici)1097-0258(19970815)16:15<1731::aid-sim595>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 16.Albert PS, Ratnasinghe D, Tangrea J, et al. Limitations of the case-only design for identifying gene-environment interactions. Am J Epidemiol. 2001;154(8):687–693. doi: 10.1093/aje/154.8.687. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt S, Schaid DJ. Potential misinterpretation of the case-only study to assess gene-environment interaction. Am J Epidemiol. 1999;150(8):878–885. doi: 10.1093/oxfordjournals.aje.a010093. [DOI] [PubMed] [Google Scholar]
- 18.Yang Q, Khoury MJ. Evolving methods in genetic epidemiology. III. Gene-environment interaction in epidemiologic research. Epidemiol Rev. 1997;19(1):33–43. doi: 10.1093/oxfordjournals.epirev.a017944. [DOI] [PubMed] [Google Scholar]
- 19.Goldstein AM, Andrieu N. Detection of interaction involving identified genes: available study designs. J Natl Cancer Inst Monogr. 1999;26(1):49–54. doi: 10.1093/oxfordjournals.jncimonographs.a024226. [DOI] [PubMed] [Google Scholar]
- 20.Gatto NM, Campbell UB, Rundle AG, et al. Further development of the case-only design for assessing gene-environment interaction: evaluation of and adjustment for bias. Int J Epidemiol. 2004;33(5):1014–1024. doi: 10.1093/ije/dyh306. [DOI] [PubMed] [Google Scholar]
- 21.Bishop YMM, Fienberg SE, Holland PW. Discrete Multivariate Analysis: Theory and Practice. Cambridge, MA: The MIT Press; 1975. [Google Scholar]
- 22.Viallefont V, Raftery AE, Richardson S. Variable selection and Bayesian model averaging in case-control studies. Stat Med. 2001;20(21):3215–3230. doi: 10.1002/sim.976. [DOI] [PubMed] [Google Scholar]
- 23.Wasserman L. Bayesian model selection and model averaging. J Math Psychol. 2000;44(1):92–107. doi: 10.1006/jmps.1999.1278. [DOI] [PubMed] [Google Scholar]
- 24.Bernardo JM, DeGroot MH, Lindley DV, et al. Bayesian Statistics. Valencia, Spain: University Press; 1980. [Google Scholar]
- 25.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. ( http://www.R-project.org). (Accessed April 5, 2008) [Google Scholar]
- 26.Unger JB, Yan L, Chen X, et al. Adolescent smoking in Wuhan, China: baseline data from the Wuhan Smoking Prevention Trial. Am J Prev Med. 2001;21(3):162–169. doi: 10.1016/s0749-3797(01)00346-4. [DOI] [PubMed] [Google Scholar]
- 27.Smith MW, Patterson N, Lautenberger JA, et al. A high-density admixture map for disease gene discovery in African Americans. Am J Hum Genet. 2004;74(5):1001–1013. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukherjee B, Chatterjee N. Exploiting gene-environment independence for analysis of case-control studies: an empirical Bayes-type shrinkage estimator to trade off between bias and efficiency. Biometrics. 2008;64(3):685–694. doi: 10.1111/j.1541-0420.2007.00953.x. [DOI] [PubMed] [Google Scholar]


