Abstract
V(D)J recombination involves the stepwise assembly of B and T cell receptor genes as lymphocytes progress through the early stages of development. While the mechanisms that restrict each step in recombination to its appropriate developmental stage are largely unknown, they share many of the components that regulate transcription. For example, enhancer-dependent modifications in histone acetylation and methylation are essential for both germline transcription and rearrangement of antigen receptor genes. Promoters positioned proximal to individual D and J gene segments in Tcra, Tcrb, Tcrd, IgH, and Igk also contribute to antigen receptor gene assembly, though their effects appear more localized than those of enhancers. Tcrb assembly initiates with D-to-J joining at each of two D-J-C gene segment clusters in DN1/2 thymocytes. DJ joints are fused with Vβ elements to complete Tcrb recombination in DN3 cells. We have previously shown that Dβ2 is flanked by upstream and downstream promoters, with the 5′ promoter being held inactive until D-to-J recombination deletes the NFκB-dependent 3′ promoter. We now report that activity of the 5′ promoter reflects a complex interplay between Runx1, GATA-3, and E47 transcription factors. In particular, while multiple E47 and Runx1 binding sites clustered near the Dβ2 5′RS and overlapping inr elements define the core 5′PDβ2, they act in concert with an array of upstream GATA-3 sites to overcome the inhibitory effects of a 110 bp distal polypurine·polypyrimidine (R·Y) tract. The dependence of 5′PDβ2 on E47 is consistent with the reported role of E proteins in post-DN1 thymocyte development and V-to-DJβ recombination.
Keywords: T cell receptor, transcription, V(D)J recombination, thymocytes, double negative, E47, GATA-3, Runx1
1. Introduction
The ability of the immune system to respond to virtually any pathogen depends on developing B and T cells generating a diverse repertoire of antigen receptors. During lymphocyte development, each cell assembles unique antigen receptor genes through stepwise recombination of arrays of variable (V), diversity (D), and joining (J) gene segments (Schatz, 2004). In the thymus, T cell receptor β (Tcrb) gene assembly is initiated by Dβ1-Jβ and Dβ2-Jβ rearrangements in CD4-CD8- double negative (DN) thymocyte subsets including CD44+ CD25-(DN1) and CD44+ CD25+ (DN2) cells, followed by Vβ-DJβ rearrangement in CD44-CD25+ (DN3) thymocytes (Oltz, 2001). Productive Vβ-DJβ rearrangement leads to the cell surface expression of the TCRβ protein, together with preTCRα (pTa) and the CD3 complex as a preTCR. Signaling through the preTCR allows developmental progression to the CD4+CD8+ double positive (DP) stage, rearrangement of Tcra, and exclusion of further Vβ-DJβ rearrangement (Schatz, 2004).
Each V, D, and J gene segment is flanked by recombination signal sequences (RSS), which serve as recognition and cleavage sites for Rag-1 and –2, the lymphocyte-specific components of recombinase (Fugmann et al., 2000). The stepwise nature of antigen receptor gene assembly and its cell type and developmental specificities have long argued that changes in the accessibility of individual RSS targets to RAG proteins must ultimately govern V(D)J recombination. Consistent with the proposition of recombinational accessibility, transcription of unrearranged gene segments, which requires epigenetic chromatin opening (McMurry and Krangel, 2000), correlates with the onset of rearrangement at the same gene segments (Abarrategui and Krangel, 2006). In fact, transcriptional enhancers have been shown to directly influence the recombinational accessibility of each antigen receptor locus (Cobb et al, 2006), coordinating chromatin opening along large segments of the variable region, and initiating germline transcription from promoters positioned proximal to individual D or J segments. The potential impact of promoter activity on recombination appears to be subtler and more localized. Targeted deletion of the T early α promoter (TEA) selectively impairs Vα rearrangement to the most proximal Jα gene segments (Villey et al., 1996), while more distal promoters regulate accessibility of their associated Jα coding segments (Hawwari et al., 2005). Similarly, deletion of the Dβ1-associated promoter, PDβ1, impairs Tcrb Dβ1-to-Jβ recombination (Sikes et al., 1999; Whitehurst et al., 1999) without effecting rearrangement or transcription of the downstream DJβ2 gene segment cluster (Whitehurst et al., 1999).
Whereas both DJβ cassettes possess recombinational accessibility in DN1 cells (McMillan and Sikes, 2008), DJβ2 rearrangements have long been shown to accumulate more slowly than DJβ1 joints (Born et al., 1985; Haars et al., 1986; Lindsten et al., 1987; Uematsu et al., 1988). We have previously shown Dβ2 is flanked by two independently regulated promoters positioned 5′ and 3′ of Dβ2 (McMillan and Sikes, 2008). The 3′Dβ2 promoter is located 400-600 bp downstream of the Dβ2 gene segment and proximal to the Jβ2.1 RSS. Germline DJβ2 transcription during DN thymocyte development is restricted to 3′PDβ2, is dependent on constitutively nuclear P65 RelA-containing NFκB complexes (Sen et al., 1995; Weih et al., 1994), and initiates downstream of the Dβ2 RSS (McMillan and Sikes, 2008). We previously showed that moving PDβ1 to a similar position between Dβ1 and Jβ1.1 impairs its ability to direct recombinational accessibility of Dβ1 transgenes (Sikes et al., 2002).
Transcription from the upstream Dβ2 promoter (5′PDβ2), which passes through the D RSS, was only detected in Tcrb alleles upon Dβ2Jβ2 rearrangement, which deletes 3′PDβ2 and relieves 5′PDβ2 repression (McMillan and Sikes, 2008). Given the coordinated regulation of promoter activity and recombinational accessibility, we wished to define the elements that coordinate 5′PDβ2 activity. In this study, we characterize the regulation of 5′PDβ2 by Runx1, GATA-3, and the E protein, E47. We have previously shown that Dβ1 and Dβ2 are both flanked by multiple GATA-3 binding sites (McMillan and Sikes, 2008; Sikes et al., 1998). We now show that 5′PDβ2 contains 4 distinct GATA-3 binding sites, though GATA-3 binding at endogenous 5′PDβ2 sequences in the Rag1-/-P53-/- DN thymocyte cell line P5424 is modest relative to that at PDβ1. In contrast, endogenous 5′PDβ2 is strongly and preferentially enriched for E47, which has previously been shown to play a critical role in Tcrb assembly (Agata et al., 2007). The minimal sequence necessary for promoter activity localized to a 220 bp region immediately 5′ of Dβ2 that contains both E boxes, as well as a binding site for Runx1 and overlapping RNA polymerase II (RNAP2) initiator (inr) elements. Finally, we show that a 110 bp polypurine·polypyrimidine stretch approximately 700 bp upstream of the minimal promoter acts to antagonize 5′PDβ2 reporter activity. Our data suggest that 5′PDβ2-dependent transcription immediately upstream of the Dβ2 coding sequence is coordinated by a dynamic interplay between positive and negative elements in DN T cell precursors.
2. Materials and Methods
2.1. Cells and antibodies
The Rag1-/-, P53-/- P5424 pro-T cell line has been previously described (Mombaerts et al., 1995). Cells were cultured at 37°C/5% CO2 in RPMI 1640 medium supplemented with 10% fetal calf serum, 2 mM L-glutamine, 0.01% penicillin/streptomycin, and 50 μM β-mercaptoethanol. Antibodies to Runx-1 (sc-28679x), GATA-3 (sc-268x), E47 (sc-763x), Sp1 (sc-59x) and USF-1 (sc-229x) were all purchased from Santa Cruz Biotechnology. Control rabbit IgG (10-4102) was purchased from Rockland Immunochemicals.
2.2. EMSA
Double-stranded oligonucleotides (Table 2S) were radioactively labeled using Klenow (New England Biolabs) by filling in 3-5 base overhangs with dNTP mixtures containing [α-32P]dCTP and [α-32P]dATP. Nuclear protein extracts were prepared as previously described (Sikes et al., 1998) from either P5424 or thymocytes isolated from 4-8 wk old Rag2-/- mice. Mouse thymus harvests were reviewed and approved by the institutional animal care and use committee at North Carolina State University. Binding reactions (20 μl) were performed at room temperature (30 min.) in a mixture containing nuclear protein extract (20 μg), radiolabeled probe (1 ng), poly(dI-dC) (1 μg), and BSA (10 μg) buffered in 20 mM HEPES (pH 7.9), 5% glycerol, 1 mM EDTA, 1% Nonidet P-40, and 5 mM DTT. Where indicated, unlabeled double-stranded competitor oligonucleotide (100-fold molar excess) or antibody (1 μg) was incubated with the nuclear extract for 30 min. on ice prior to addition of probes. Nucleoprotein complexes were resolved by electrophoresis on a 5% nondenaturing polyacrylamide gel, and visualized by autoradiography.
2.3. Chromatin immunoprecipitation
Chromatin was prepared from formaldehyde-crosslinked P5424 cells as described (Sikes et al., 2009). Prior to ChIP, each antibody (1 μg) was incubated (1 hr) with paramagnetic Dynabeads (Invitrogen) separately coupled to Protein A (10 μl/IP) and Protein G (10 μl/IP) in 50 μl RIPA buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% SDS, 0.1% NaDeoxycholate) supplemented with BSA (50 mg/ml), sheared salmon sperm DNA (0.5 mg/ml), Halt protease inhibitor cocktail (Thermo Scientific), and 1 mM PMSF. Conjugated antibody:bead complexes were washed 2X in supplemented RIPA buffer.
For ChIP, each antibody:bead complex was mixed with chromatin (106 P5424 cell equivalents) in 100 μl supplemented RIPA buffer. Protein-DNA complexes were immunoprecipitated for 2 hours at 4°C with rotation, and then sequentially washed 3X in RIPA buffer and 2X in TE, pH 8.0. Protein-DNA complexes were eluted 15 min. in NaHCO3 (100 mM), and crosslinks were reversed for 15 min. at 95°C after addition of NaCl (200 mM). Samples and matched inputs were incubated with Proteinase K (10 μg/ml) 1 hr @ 45°C, and DNA was purified using Qiaquick nucleotide removal columns (Qiagen) according to the manufacturer's instructions.
For realtime PCR, bound (3 μl) and input (3 μl of 1:100 dilution) samples were amplified using 1X SensiMix Plus (Quantace) and primers specific for PDβ1 (forward: 5′-TCACCTTCCTTATCTTCAACTCCC-3′; reverse: 5′-TCCCATAGAATTGAATCACCGTGG-3′), and 5′PDβ2 (forward: 5′-GTTTCTGAGGCATGTGTCTCTGCG-3′; reverse: 5′TCCTCTTTGTCACAGTGCCCACC-3′). Cycling parameters for 20 μl reactions were 95°C 10 min., followed by 40 cycles of 95°C, 20 sec; 63°C, 30 sec; 72°C, 30 sec. Fold enrichment in the bound fractions relative to input was calculated as previously described (Ciccone et al., 2004), and the average enrichment for triplicate amplifications was reported.
2.4. Plasmids
All luciferase reporter plasmids were generated by cloning individual restriction fragments or PCR amplification products of p5′D2JJ-BS (McMillan and Sikes, 2008) into the SmaI site of pGL3-Eβ (McMillan and Sikes, 2008), in which luciferase expression is driven in a T lineage-dependent manner by the mouse Tcrb enhancer, Eβ. Site-specific mutations were introduced into individual reporter constructs using the Quickchange II site-directed mutagenesis kit (Stratagene) according to the manufacturer's recommendations. Oligonucleotide primers used to introduce each mutation are as shown (Table 1S). The integrity of all promoter constructs was confirmed by restriction digest and sequencing. Plasmid DNA for luciferase reporter assays was prepared using the PureYield Endotoxin-free Midiprep kit (Promega) according to the manufacturer's instructions.
2.5. Luciferase assays
Luciferase reporter assays were performed as previously described (McMillan and Sikes, 2008). Briefly, 107 P5424 cells in log-phase growth were electroporated using a Bio-RAD gene pulser II (260 V, 950 μF, 200 Ω) in 0.4 cm cuvettes containing minimal RPMI 1640 medium (300 μl) 5′PDβ2 reporter construct (10 μg) and pSV-RL (250 ng, Promega). Cells were allowed to recover 10 min. on ice before transfer to complete culture medium. After 24 hr culture, cells were harvested, total protein extracted, and luciferase and renilla luminescence were sequentially determined for 50 μg total protein using the Dual-Luciferase Reporter Plasmid System (Promega) and a Centro LB960 luminometer (Berthold Technologies). All transfections were performed 3 or more times using independent plasmid preparations.
3. Results
3.1. Runx1, GATA-3, and USF1 bind multiple sites 5′ of Dβ2
We previously described two regions of promoter activity flanking the Dβ2 gene segment (McMillan and Sikes, 2008). Before Dβ2-Jβ2 rearrangements, germline transcription in CD44+25- DN1 thymocytes is initiated at multiple sites within a 400 bp region downstream of Dβ2. Upon D-to-J rearrangement, 3′PDβ2 is deleted and transcript initiation is redirected predominantly to overlapping initiator (inr) elements positioned immediately upstream of Dβ2. Consistent with transcription patterns before and after D-to-J joining, we showed in reporter studies that a second Dβ2 promoter is located upstream of the D coding sequence in a position analogous to the Dβ1 promoter, PDβ1 (Sikes et al., 1998), and contains putative binding sites for a number of transcription factors central to hematopoietic development (Fig. 1). Moreover, activity of this promoter in our reporter experiments was only revealed when germline sequence 3′ of Dβ2 was deleted (McMillan and Sikes, 2008), suggesting that 5′PDβ2 activity may play a role in Tcrb assembly after DJβ2 joints are formed.
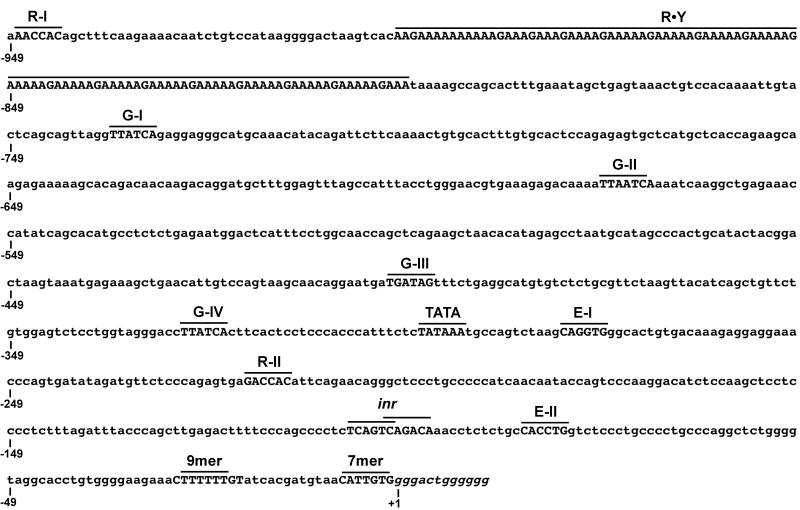
Figure 1.
Organization of the Dβ2 sequence analyzed for 5′ promoter activity. The positions of putative binding sites for Runx (R), GATA (G), and E proteins (E) are shown, as well as consensus TATA and initiator elements and polypurine·polypyrimidine DNA (R·Y). Dβ2 coding segments (italics) and 5′RSS sequences are also indicated. Numbering is relative to the first base of the Dβ2 coding sequence (+1).
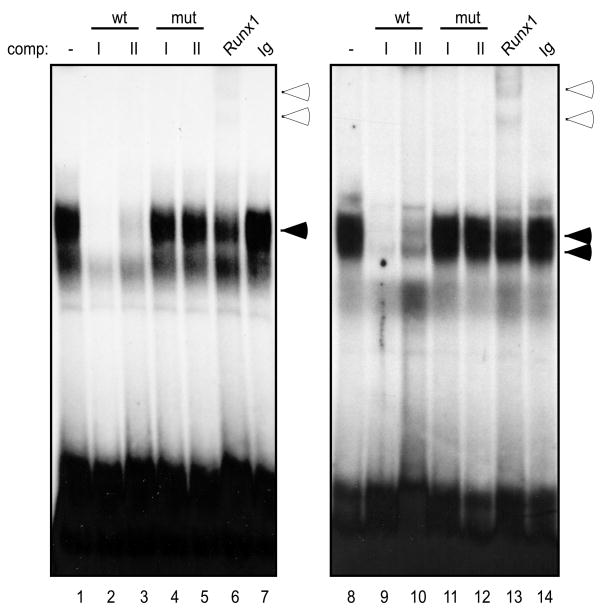
The Runx family of transcription factors (including Runx1, Runx2, and Runx3) are critical for multiple stages of T cell development (Anderson, 2006). Canonical RACCRCA motifs exist in the enhancers of all four TCR loci. Moreover, chromatin immunoprecipitation studies have shown that Runx1 recognizes unidentified sites flanking both Dβ1 and Dβ2 (Oestreich et al., 2006). We identified 2 consensus Runx binding sites in 5′PDβ2 (Fig. 1, R-I and R-II). Radiolabeled probes to both sites detected a complex pattern of binding activities when mixed with nuclear proteins from P5424 cells (Fig. 2, lanes 1 and 8). Unlabeled Runx1-A or Runx1-B oligonucleotides were equally effective at inhibiting all of the binding complexes at both sites (lanes 2-3, 9-10), whereas mutation of core ACC trinucleotide completely abolished each probe's competitive ability (lanes 4-5, 11-12). Nucleoprotein complexes formed between R-I or R-II and proteins from the DN3-staged P5424 cells were supershifted by Runx1 Ab (lanes 6 and 13, respectively), but not by isotype-matched controls (lanes 7 and 14) or by antibodies to either Runx2 or Runx3 (data not shown). Given that expression of the three Runx proteins varies during thymopoiesis (Taghon et al., 2006), we assessed binding site occupancy using nuclear extracts from mature T (BW5147) and B (M12) cell lines. Whereas R-I and R-II were bound by Runx2 in BW5147, both sites were occupied by Runx3 in M12 cells (data not shown).
Figure 2.
Runx1 binds separate sites upstream of Dβ2. Nuclear extracts from the P5424 cell line were incubated with radiolabeled ds oligonucleotide probes to the putative Runx binding sites: R-I (lanes 1-7) and R-II (lanes 8-14). Probes were incubated with nuclear extract alone (lanes 1 and 8), in the presence of 100-fold molar excess of the indicated unlabeled competitors (lanes 2-5 and 9-12), or in the presence of the indicated Abs (lanes 6-7 and 13-14). Specific nucleoprotein (filled arrows) and Ab-supershifted complexes (empty arrows) are indicated.
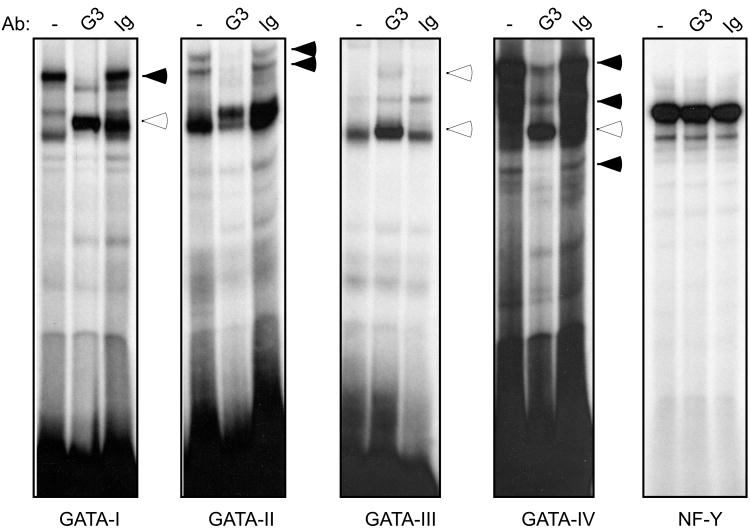
We previously identified 3 canonical GATA binding sites 3′ of the Dβ2 coding sequence, two of which are bound by the T cell specific GATA-3 transcription factor (McMillan and Sikes, 2008). Although these GATA binding sites were dispensable for 3′PDβ2 activity, GATA-3 is essential for activation of the PDβ1 promoter (Sikes et al., 1998) and Eβ (Porritt et al., 2004). The four GATA binding sites upstream of Dβ2 are positioned within a 415 bp stretch. Three sites conform to the canonical GATA motif, WGATAR (Merika and Orkin, 1993), while the fourth (G-II; TGATTA) is mismatched at a single nucleotide. Despite the mismatch in G-II, radiolabeled probes to each of the 4 sites generated a series of complexes with nuclear protein from the P5424 cell line, though binding patterns differed between the separate sites (Fig. 3). Antibodies to GATA-3 either attenuated (G-I, G-II, and G-IV) or supershifted the primary complexes generated with each probe (G-I, G-III, and G-IV). In contrast, GATA-3 Abs had no effect on nucleoprotein complexes assembled with control NF-Y probes (Boothby et al., 1989).
Figure 3.
GATA-3 binds multiple sites within 5′PDβ2. Nuclear extracts from the P5424 cell line were incubated with radiolabeled probes to the 4 putative GATA binding sites in 5′PDβ2 as well as an NF-Y control. In each case, reaction mixtures included probe and nuclear extract alone (-), or in the presence of the indicated Abs. Antibody-attenuated (filled arrows) and supershifted complexes (empty arrows) are indicated.
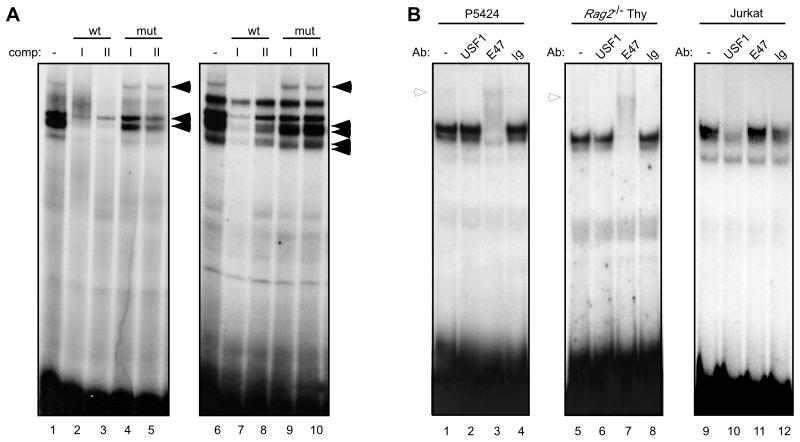
Core 5′PDβ2 activity is confined to a 120 bp region that contains the R-II site and 2 potential E-boxes, E-I and E-II (Fig. 1). Radiolabeled probes of the two E boxes formed at least three nucleoprotein complexes with P5424 extracts that were abolished upon competition with unlabeled probes to either site (Fig. 4A, lanes 1-3 and 6-8). Mutation of the E box 3′ half-site in either probe eliminated its ability to compete for binding (lanes 4-5 and 9-10). ChIP-on-chip analyses showed a strong enrichment of E47 binding across Dβ2 (Agata et al., 2007). Likewise, E47 Abs supershifted complexes formed between P5424 or Rag2-/- thymocyte nuclear extracts and either E-I (Fig. 4B) or E-II probes (data not shown). Abs against upstream stimulatory factor-1 (USF1), a bHLH protein has been shown to bind the Dδ2 promoter (Carabana et al., 2005), failed to supershift E-I complexes in P5424 (lane 2) or Rag2-/- thymocyte nuclear extracts (lane 6). When we assayed nuclear extracts from E47-deficient Jurkat T cells, E-I complexes were now attenuated with USF1 Abs (lanes 9-12), suggesting that loss of E47 during thymocyte maturation may open the 5′PDβ2 E boxes for at least partial USF1 occupancy. At the same time, Abs to other bHLH proteins including c-MYC and MAX failed to alter E-I binding (data not shown).
Figure 4.
Separate E boxes within the minimal 5′PDβ2 bind E47. (A) Nuclear extracts from the P5424 cell line were incubated with radiolabeled probes to the putative E boxes: E-I (lanes 1-5) and E-II (lanes 6-10) alone (lanes 1 and 6), or in the presence of 100-fold molar excess of the indicated unlabeled competitors (lanes 2-5 and 7-10). Specific nucleoprotein complexes are indicated (filled arrows). (B) Probes to the E-I site were incubated with nuclear extracts from P5424 (lanes 1-4), Rag2-/- thymocytes (lanes 5-8), or Jurkat (lanes 9-12) alone (lanes 1, 5, and 9) or in the presence of the indicated Abs (lanes 2-4, 6-8, and 10-12). Ab-supershifted complexes are indicated (empty arrows).
3.2. 5′PDβ2 E boxes are bound by E47
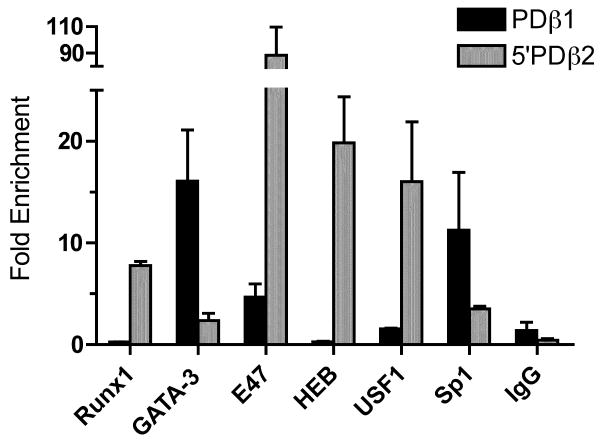
Our EMSA results demonstrate that 5′PDβ2 contains multiple binding sites for a number of different transcription factors. We next used chromatin IP to determine whether GATA-3, Runx1, and E47 bind the promoter region in vivo. To that end, we enzymatically sheared P5424 chromatin that had been crosslinked with formaldehyde, and immunoprecipitated protein-DNA complexes using Abs specific for Runx1, GATA-3, E47, and for Sp1, which binds sites immediately 3′ of Dβ2. As a control, we compared antibody-dependent enrichment of 5′PDβ2 sequence to that of PDβ1. Consistent with the dependence of Dβ1 transcription on GATA-3 and Sp1 (Sikes et al., 1998), both proteins were strongly enriched at PDβ1, relative to a nonspecific IgG control (Fig. 5). Antibodies to Runx1 failed to enrich PDβ1 sequences above isotype control levels. E47 Abs enriched PDβ1 sequences 3.25 fold above IgG levels, though they enriched 5′PDβ2 sequences more than 80 fold over IgG. The HEB and USF1 protiens were also enriched at 5′PDβ2 in P5424, despite their lack of detection by EMSA (Fig. 4). Given the relative levels of enrichment, our ChIP analyses suggest that E47 is the predominant protein at the 5′PDβ2 E boxes in P5424. HEB enrichment may reflect the presence of E47:HEB heterodimers in thymocytes (Sawada and Littman, 1993). There are no reports of E47 heterodimerizing with USF1. Rather, E47 may simply outcompete USF1 for occupancy of the 5′PDβ2 E boxes.
Figure 5.
Transcription factor in vivo association with 5′PDβ2. Chromatin from the P5424 cell line was immunoprecipitated with the indicated antibodies, and analyzed by qPCR for transcription factor association with the 5′PDβ2 (grey) and PDβ1 (black) promoters. Fold enrichment of each promoter sequence following IP with each antibody is shown relative to an untreated input sample. Bars indicate means (± SD) of triplicate qPCRs, and are representative of 2 experiments with independent chromatin preparations.
Consistent with EMSA results, Runx1 and GATA-3 were detected at the Dβ2 upstream promoter (7 and 2 fold over IgG, respectively). Additionally, antibodies to Sp1 enriched 5′PDβ2 sequence approx. 3 fold over IgG. Sequence analysis predicts one potential Sp1 site between R-II and E-II, though it is poorly conserved and failed to bind protein in EMSA studies (data not shown). Although cryptic Sp1 sites may exist in 5′PDβ2, our ChIP results most likely reflect antibody binding to the Sp1 site downstream of Dβ2 (360 bp 3′ of the 5′PDβ2 ChIP amplicon). Similarly, the modest levels of GATA-3 enrichment we detect at 5′PDβ2 may derive from binding to the 2 GATA-3 sites 3′ of Dβ2 (400 and 480 bp 3′ of the 5′PDβ2 amplicon), though EMSA clearly implicated GATA-3 binding 5′ of Dβ2. Alternatively, relative differences between GATA-3 binding at PDβ1 and 5′PDβ2 could reflect the transcriptional activity of the former and inactivity of the latter prior to DJ recombination.
3.3. Promoter activity upstream of Dβ2
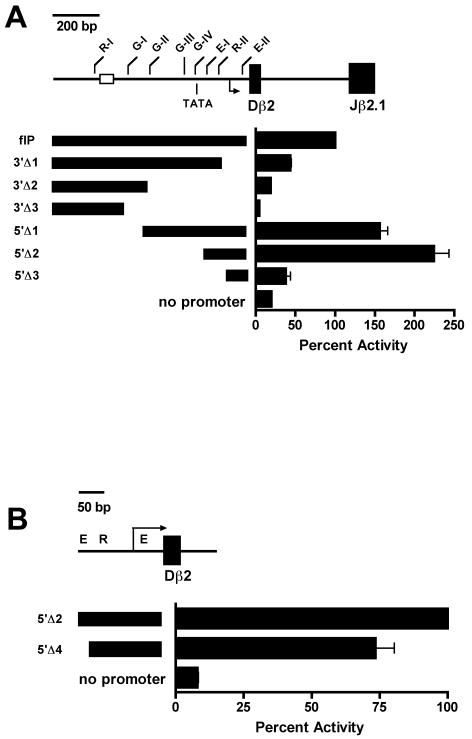
To localize the core 5′PDβ 2 promoter, we generated a series of luciferase reporter constructs in which the Tcrb enhancer, Eβ, was paired with restriction fragments that spanned the 1 kb stretch of DNA immediately upstream of Dβ2. Each construct was transiently transfected into the Rag1-/-P53-/- DN thymocyte cell line P5424, along with control pSV-RL (Promega), and transfectants were assayed for luciferase expression (Fig. 6). Consistent with our previous findings, promoter activity was primarily localized to a 220 bp region immediately 5′ of the Dβ2 5′RS containing the R-II binding site, both E boxes, and 2 overlapping transcription initiator (inr) elements. A 140 bp deletion of the 3′-most portion of this core region that eliminated the inr elements and proximal E box (E-II) reduced promoter activity to 44% of the full-length promoter fragment (Fig. 6A, 3′Δ1 vs flP). Promoter activity was further reduced with subsequent 3′ deletions that eliminated the 4 putative binding sites for the GATA family of transcription factors (3′Δ2 and 3′Δ3). However, 5′ deletions that removed the first 2 (5′Δ1) or all 4 GATA sites (5′Δ2) actually led to modest increases in promoter activity.
Figure 6.
Promoter activity upstream of Dβ2. (A) The indicated DNA fragments were inserted upstream of the luciferase cassette in pGL3-Eβ. Each plasmid was co-transfected with pSV-RL into P5424 cells, and protein extracts were assayed for luciferase activity 24 hours after transfection. Values from 3 or more independent transfections were normalized to renilla controls. Bars represent mean normalized luciferase activity ± SEM, and are expressed as percent activity of a 1 kb StuI/AlwNI full length promoter fragment (flP). (B) pGL3-Eβ constructs containing the indicated DNA fragments were assayed for luciferase activity 24 hrs after co-transfection with pSV-RL into P5424 cells. Bars represent mean normalized luciferase activity ± SEM, and are expressed as percent activity of the 219 bp 5′Δ2 minimal promoter fragment.
Progressive 5′ deletion of E-I and R-II reduced promoter activity to 38% of flP (Fig. 6A, 5′Δ3), suggesting that while GATA binding may or may not enhance 5′PDβ2, E-I and R-II appear to be critical for activity of the core promoter. When normalized to the intact core promoter (5′Δ2), specific deletion of E-I led to a 26% reduction in luciferase activity (Fig. 6B), suggesting that proteins recruited to E-I are required for full activity of the core 5′PDβ2. Similar results were obtained for the 5′ and 3′ deletion studies when constructs were transfected into Jurkat cells or when Eβ was replaced with the SV40 enhancer (data not shown).
3.4. Mutational analysis of 5′PDβ2
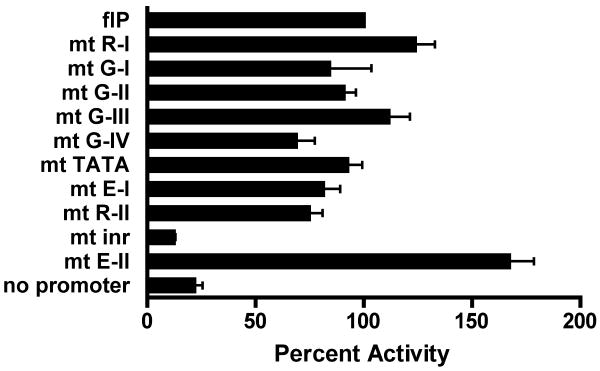
Deletion assays demonstrate that the bulk of promoter activity is retained in the –280/-61 5′Δ2 fragment (Fig. 6A), and suggest only minimal roles for the upstream GATA and R-I sites. To assess individual contributions of transcription factor binding sites to 5′PDβ2 activity, each site in the full-length reporter was selectively mutated (Fig. 7). Consistent with 5′ deletion, disruption of the R-I site had no impact on promoter activity. Likewise, the GATA site at -401 (G-III) appeared dispensable. In contrast, mutation of either G-I or G-II showed a modest reduction in 5′PDβ2 function (16% and 9% below flP, respectively). Our deletion studies suggested that all of the distal sites (R-I and the 4 GATA sites) are dispensable for promoter activity. However, we found that mutation of the most proximal GATA site (G-IV) yielded a 31% reduction in promoter activity, similar to mutation of either the E-I or R-II sites within the core 5′PDβ2 (81% and 75% of flP, respectively). Whereas E-I mutation impaired 5′PDβ2, mutation of E-II failed to reduce promoter activity. This unexpected result for E-II suggests that reduced promoter activity in our minimal 3′ deletion construct, 3′Δ1 (Fig. 6A), may solely derive from loss of the overlapping inr elements.
Figure 7.
Promoter activity of pGL3-Eβ containing the indicated PCR fragments is expressed as percent of PCR-generated full-length promoter control (flP′). Bars represent mean normalized luciferase activity ± SEM of 4 independent transfections. Targeted binding site mutations are indicated as white boxes.
We observed little effect from mutation of the canonical TATAA sequence that lies between G-IV and E-I, 193-197 bp upstream of the inr elements (Fig. 7). Introduction of 4-bp mutation into the paired inr elements resulted in an 88% decrease in promoter activity, consistent with their predominant role in 5′PDβ2 transcript initiation (McMillan and Sikes, 2008). 3′ deletions that removed a 142 bp BanI/AlwNI fragment containing the inr and E-II elements proved somewhat less destructive than selective inr mutation, reducing promoter activity by 56% (Fig. 6A, compare 3′Δ1 to flP). Although the inr elements are not closely flanked by canonical TATA or downstream promoter elements (DPE, RGWCGTG), we did note the presence of a partial DPE sequence (GGTCTCC) 27 bp 3′ of the first inr. Since motif searches and EMSA failed to detect alternative binding moities in the BanI/AlwNI fragment, the specific sensitivity of 5′PDβ2 reporter constructs to targeted inr mutation may indicate activities that normally restrict transcription initiation to the inr sites, and are lost in 3′Δ1.
3.5. Distal elements modulate 5′PDβ2
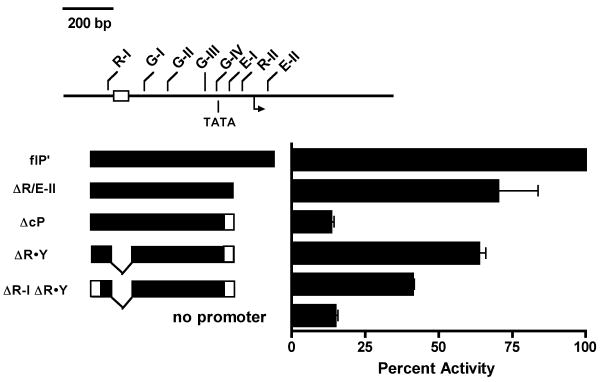
Our deletion analyses suggested that, while the GATA sites appear dispensable for core 5′PDβ2 activity, fragments that span the R-I and GATA sites nonetheless maintain minimal promoter activity. We previously found that a minority of 5′PDβ2 transcripts initiated upstream of the consensus inr elements (McMillan and Sikes, 2008). Indeed, one start site was positioned 26 nucleotides downstream of a consensus TATA box that sits between G-IV and E-I. To more directly test the potential of the GATA sites in contributing to 5′PDβ2 activity, we generated a second panel of pGL3-Eβ luciferase vectors that contained PCR fragments that selectively removed core 5′PDβ2 components (Fig. 8). Relative to the PCR-generated full-length promoter control (flP′), a fragment truncated immediately 3′ of the E-I site (ΔR/E-II) retained 70% promoter activity, while mutagenesis of ΔR/E-II to remove E-I reduced activity to promoterless control levels (ΔcP).
Figure 8.
Mutational analysis of 5′PDβ2 regulation. Individual transcription factor binding sites within the full-length promoter fragment were selectively mutated to assess their contribution to 5′PDβ2 activity. P5424 transfectants of the indicated mutant flP pGL3-Eβ reporters were assayed for promoter activity. Results from 7 independent co-transfections with pSV-RL were normalized to Renilla levels, and expressed as percent wildtype flP.
Polypurine·polypyrimidine sequences have the potential to assemble as triplex or quadruplex DNA (Frank-Kamenetskii and Mirkin, 1995), and have been associated with a diverse array of functions. R·Y stretches are enriched in the upstream and intronic regions of genes (Gaddis et al., 2006). Moreover, R-Y tracts have been linked to mutagenesis, ds breaks, and transcriptional repression (Jain et al., 2008). 5′ deletions that removed the relatively long R-Y tract upstream of the core 5′PDβ2, along with 1 or more GATA-3 sites appeared to enhance promoter activity (Fig. 6A). Given the presence of a canonical TATA box downstream of G-IV, we wished to determine if R·Y deletion might restore promoter activity to the GATA-rich AcP fragment. Indeed, R·Y deletion essentially restored ΔcP activity to parental ΔR/E-II levels (Fig. 8). The upstream Runx binding site, R-I, is located 114 bp 5′ of the polypurine sequence. When we destroyed R-I by site-directed mutagenesis (ΔR-I ΔR·Y), we observed a 36% reduction in promoter activity relative to ΔR·Y, suggesting that R-I has modest activating potential that is masked by the R·Y tract. Taken together, these studies suggest that the R-I and GATA sites may work with downstream Runx and E box elements of the core promoter to overcome the inhibitory effects of the RY tract and drive 5′PDβ2.
4. Discussion
In contrast to the regulatory simplicity of 3′PDβ2, a number of factors appear to contribute to overall 5′PDβ2 activity. The core promoter is localized to a 220 bp sequence that contains binding sites for E47 and Runx1, as well as the predominant transcription start sites positioned over paired inr elements. An array of 4 GATA-3 sites and an additional Runx1 site upstream of the core promoter appear dispensable for core promoter activity, acting instead to counterbalance the inhibitory effects of a 110 bp polypurine repeat. Multiple GATA-3 sites have similarly been found in the other 2 Tcrb germline promoters: PDβ1 and 3′PDβ2. While paired GATA-3 sites upstream of Dβ1 are critical for PDβ1 activity (Sikes et al., 1998), similarly paired sites between Dβ2 and Jβ2.1 failed to augment 3′PDβ2 reporter activity (McMillan and Sikes, 2008). Alternatively, the 6 GATA-3 sites flanking Dβ2 may act to promote a general state of chromatin opening across the Dβ2 regulatory region, rather than directly activating transcription as in PDβ1.
Transcriptional promoters positioned proximal to D and J gene segments play pivotal roles in antigen receptor gene assembly; serving as synaptic targets for downstream enhancers (Oestreich et al., 2006), recruiting chromatin modifying proteins (Osipovich et al., 2007), and driving transcription through germline sequences. We have previously shown that promoter activity 5′ of the Dβ1 element is essential for Dβ1 recombination (Sikes et al., 1999). In contrast, germline transcription of the DJβ2 cassette is dependent on the actions of a promoter positioned between Dβ2 and Jβ2.1 (McMillan and Sikes, 2008), likely leading to the less efficient assembly of DJβ2 joints in early DN thymocytes (Born et al., 1985; Haars et al., 1986; Lindsten et al., 1987; Uematsu et al., 1988). Mutations that diminish germline transcription also reduce V assembly with D reporter targets in Tcrb (Khor et al., 2009) and J segments in Tcra (Abarrategui and Krangel, 2006; Hawwari et al., 2005), indicating a conserved role for germline promoters at each stage of recombination. Whereas downstream positioning of 3′PDβ2 may enhance overall Tcrb diversity by limiting initial Vβ recombination with DJβ2 joints, activation of 5′PDβ2 upon 3′PDβ2 deletion may be necessary to ensure that DJβ2 joints are recombinationally accessible once formed. In that regard, the close proximity of the 5′ promoter to the Dβ2 recombination signal sequences mirrors that of Dβ1 and Dδ2 promoters. Such proximity may be essential for recombinational accessibility, targeting nucleosomal disruption at the D RSS elements that have recently been suggested to initiate both D-to-J and V-to-DJ stages of Tcrb assembly (Franchini et al., 2009).
The class I bHLH proteins (E proteins) have significant and diverse roles in antigen receptor gene rearrangement and lymphocyte development (Murre, 2005). In particular, E47 is required for expression of RAG1 and RAG2 in pro-B cells (Hsu et al., 2003), as well as for regulating Igk recombination (Romanow et al., 2000). Likewise, E47 enforces developmental arrest of DN3 thymocytes until assembly of a functional Tcrb gene (Engel et al., 2001). More recently, Murre and colleagues have shown that E47 binding augments Vβ chromatin accessibility, and that E47 is also strongly localized over DJβ2 and downstream Eβ and Vβ14 elements (Agata et al., 2007). Additionally, histone H3 acetylation and Dβ-to-Jβ recombination are both reduced in DN cells of E47-deficient mice. Our ChIP studies in P5424 suggest that multiple factors including E47 are bound at 5′PDβ2 prior to DJ recombination and promoter activation. As such, E47 binding upstream of Dβ2 may be involved in the recruitment of histone acetyltransferase (HAT) activity to germline D and J sequences, facilitating recombinational accessibility and activation of the 3′PDβ2, before it contributes to 5′PDβ2 activity.
Separate studies confirm that H3Ac levels at the DJβ cassettes remain elevated in DP cells (Morshead et al., 2003; Tripathi, 2002), despite the obligate reduction of E47 DNA binding activity during β selection (Engel et al., 2001). Together, these data suggest a critical but temporally restricted role for E47 in regulating Dβ accessibility during DN development. Since transcripts that initiate at 5′PDβ2 are readily detected in DP cells (McMillan and Sikes, 2008), it is possible that E47 is replaced at the 5′PDβ2 E boxes in DP cells. E47 and E12 can recognize the same sequence, albeit with differing affinities, and HEB binding at 5′PDβ2 is consistent with E47:HEB heterodimers in developing thymocytes (Sawada and Littman, 1993). Alternatively, as suggested by EMSA (Fig. 4), the 5′PDβ2 E boxes could be occupied by other bHLH proteins like USF1 in more mature cells after E47 expression declines, though the significance of DJβ histone acetylation in DP thymocytes remains unclear. Irrespective of function later in development, our data are consistent with a model in which E47 facilitates 5′PDβ2 activation in DN2/DN3 cells.
The RY tract located between the R-I and G-I binding sites was found to inhibit 5′PDβ2 activity. RY elements are disproportionately enriched in the upstream portions of genes, can form stable triplex DNA structures (Jain et al., 2008), and have been shown to repress transcription (Grabczyk and Fishman, 1995; Grabczyk and Usdin, 2000). In particular, the S1 nuclease-sensitive R-Y palindrome upstream of the human c-Myc P1 and P2 promoters has been shown to assemble a short triplex hairpin that partially arrests c-Myc transcription and may underlie c-Myc mutagenicity (Belotserkovskii et al., 2007). The ability of R·Y triplexes to pause RNAP2 has been predicted to have a more significant repressive effect when positioned in the vicinity of other transcription termination elements. Indeed, the Dβ2 R·Y element begins 265 bp downstream of the Cβ1 polyadenylation sequence. Since transcriptional elongation from upstream Tcra promoters has been shown to alter the recombinational accessibility of downstream Jα elements (Abarrategui and Krangel, 2006), we suggest that one role of the Dβ2 R·Y tract may be to isolate the Dβ2 coding and RSS sequences from the effects of readthrough Dβ1 germline transcription.
The partially denatured structure of triplex DNA leaves it as a frequent hotspot for mutagenesis and recombination. The RAG-mediated t(14;18) translocation that juxtaposes bcl-2 with the intronic IgH enhancer and underlies follicular lymphoma has been linked to a triplex-forming RY element within the bcl-2 major breakpoint (Raghavan et al., 2005). If the bcl-2 R·Y repeat can augment RAG recruitment to bcl-2, the Dβ2 R·Y might similarly enhance RAG recruitment to the Eβ regulatory region of Tcrb. In this regard, we note that 2 additional 100 bp R·Y tracts are also found in the DJβ region, one approximately 3.5 kb upstream of Dβ1, and one between Jβ1.4 and Jβ1.5. Future studies will be necessary to determine how these R·Y tracts may affect Tcrb assembly.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institute of Allergy and Infectious Diseases (R56AI070848-01A1). We thank Justin Bradshaw and Jessica Lunsford for critical review of this manuscript.
Abbreviations used in this paper
- RSS
recombination signal sequence
- Tcrb
T cell receptor β locus
- Tcra
T cell receptor α locus
- DN double negative
DP, double positive, Eβ, Tcrb enhancer, PDβ1, Dβ1 promoter
- bHLH
basic helix-loop-helix
- USF1
upstream stimulatory factor 1, ChIP, chomatin immunoprecipitation
- Rag
Recombination activating gene
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abarrategui I, Krangel MS. Regulation of T cell receptor-alpha gene recombination by transcription. Nat Immunol. 2006;7:1109–1115. doi: 10.1038/ni1379. [DOI] [PubMed] [Google Scholar]
- Agata Y, Tamaki N, Sakamoto S, Ikawa T, Masuda K, Kawamoto H, Murre C. Regulation of T cell receptor beta gene rearrangements and allelic exclusion by the helix-loop-helix protein, E47. Immunity. 2007;27:871–884. doi: 10.1016/j.immuni.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Anderson MK. At the crossroads: diverse roles of early thymocyte transcriptional regulators. Immunol Rev. 2006;209:191–211. doi: 10.1111/j.0105-2896.2006.00352.x. [DOI] [PubMed] [Google Scholar]
- Belotserkovskii BP, De Silva E, Tornaletti S, Wang G, Vasquez KM, Hanawalt PC. A triplex-forming sequence from the human c-MYC promoter interferes with DNA transcription. J Biol Chem. 2007;282:32433–32441. doi: 10.1074/jbc.M704618200. [DOI] [PubMed] [Google Scholar]
- Boothby M, Liou HC, Glimcher LH. Differences in DNA sequence specificity among MHC class II X box binding proteins. J Immunol. 1989;142:1005–1014. [PubMed] [Google Scholar]
- Born W, Yague J, Palmer E, Kappler J, Marrack P. Rearrangement of T-cell receptor beta-chain genes during T-cell development. Proc Natl Acad Sci U S A. 1985;82:2925–2929. doi: 10.1073/pnas.82.9.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabana J, Ortigoza E, Krangel MS. Regulation of the murine Ddelta2 promoter by upstream stimulatory factor 1, Runx1, and c-Myb. J Immunol. 2005;174:4144–4152. doi: 10.4049/jimmunol.174.7.4144. [DOI] [PubMed] [Google Scholar]
- Ciccone DN, Morshead KB, Oettinger MA. Chromatin immunoprecipitation in the analysis of large chromatin domains across murine antigen receptor loci. Methods Enzymol. 2004;376:334–348. doi: 10.1016/S0076-6879(03)76022-4. [DOI] [PubMed] [Google Scholar]
- Cobb RM, Oestreich KJ, Osipovich OA, Oltz EM. Accessibility control of V(D)J recombination. Adv Immunol. 2006;91:45–109. doi: 10.1016/S0065-2776(06)91002-5. [DOI] [PubMed] [Google Scholar]
- Engel I, Johns C, Bain G, Rivera RR, Murre C. Early thymocyte development is regulated by modulation of E2A protein activity. J Exp Med. 2001;194:733–745. doi: 10.1084/jem.194.6.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchini DM, Benoukraf T, Jaeger S, Ferrier P, Payet-Bornet D. Initiation of V(D)J recombination by Dbeta-associated recombination signal sequences: a critical control point in TCRbeta gene assembly. PLoS ONE. 2009;4:e4575. doi: 10.1371/journal.pone.0004575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank-Kamenetskii MD, Mirkin SM. Triplex DNA structures. Annu Rev Biochem. 1995;64:65–95. doi: 10.1146/annurev.bi.64.070195.000433. [DOI] [PubMed] [Google Scholar]
- Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annual Review of Immunology. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- Gaddis SS, Wu Q, Thames HD, DiGiovanni J, Walborg EF, MacLeod MC, Vasquez KM. A web-based search engine for triplex-forming oligonucleotide target sequences. Oligonucleotides. 2006;16:196–201. doi: 10.1089/oli.2006.16.196. [DOI] [PubMed] [Google Scholar]
- Grabczyk E, Fishman MC. A long purine-pyrimidine homopolymer acts as a transcriptional diode. J Biol Chem. 1995;270:1791–1797. doi: 10.1074/jbc.270.4.1791. [DOI] [PubMed] [Google Scholar]
- Grabczyk E, Usdin K. The GAA*TTC triplet repeat expanded in Friedreich's ataxia impedes transcription elongation by T7 RNA polymerase in a length and supercoil dependent manner. Nucleic Acids Res. 2000;28:2815–2822. doi: 10.1093/nar/28.14.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haars R, Kronenberg M, Gallatin WM, Weissman IL, Owen FL, Hood L. Rearrangement and expression of T cell antigen receptor and gamma genes during thymic development. J Exp Med. 1986;164:1–24. doi: 10.1084/jem.164.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawwari A, Bock C, Krangel MS. Regulation of T cell receptor alpha gene assembly by a complex hierarchy of germline Jalpha promoters. Nat Immunol. 2005;6:481–489. doi: 10.1038/ni1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu LY, Lauring J, Liang HE, Greenbaum S, Cado D, Zhuang Y, Schlissel MS. A conserved transcriptional enhancer regulates RAG gene expression in developing B cells. Immunity. 2003;19:105–117. doi: 10.1016/s1074-7613(03)00181-x. [DOI] [PubMed] [Google Scholar]
- Jain A, Wang G, Vasquez KM. DNA triple helices: biological consequences and therapeutic potential. Biochimie. 2008;90:1117–1130. doi: 10.1016/j.biochi.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor B, Mahowald GK, Khor K, Sleckman BP. Functional overlap in the cis-acting regulation of the V(D)J recombination at the TCRbeta locus. Mol Immunol. 2009;46:321–326. doi: 10.1016/j.molimm.2008.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsten T, Fowlkes BJ, Samelson LE, Davis MM, Chien YH. Transient rearrangements of the T cell antigen receptor alpha locus in early thymocytes. J Exp Med. 1987;166:761–775. doi: 10.1084/jem.166.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan RE, Sikes ML. Differential activation of dual promoters alters Dbeta2 germline transcription during thymocyte development. J Immunol. 2008;180:3218–3228. doi: 10.4049/jimmunol.180.5.3218. [DOI] [PubMed] [Google Scholar]
- McMurry MT, Krangel MS. A role for histone acetylation in the developmental regulation of VDJ recombination. Science. 2000;287:495–498. doi: 10.1126/science.287.5452.495. [DOI] [PubMed] [Google Scholar]
- Merika M, Orkin SH. DNA-binding specificity of GATA family transcription factors. Mol Cell Biol. 1993;13:3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P, Terhorst C, Jacks T, Tonegawa S, Sancho J. Characterization of immature thymocyte lines derived from T-cell receptor or recombination activating gene 1 and p53 double mutant mice. Proc Natl Acad Sci U S A. 1995;92:7420–7424. doi: 10.1073/pnas.92.16.7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morshead KB, Ciccone DN, Taverna SD, Allis CD, Oettinger MA. Antigen receptor loci poised for V(D)J rearrangement are broadly associated with BRG1 and flanked by peaks of histone H3 dimethylated at lysine 4. Proc Natl Acad Sci U S A. 2003;100:11577–11582. doi: 10.1073/pnas.1932643100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C. Helix-loop-helix proteins and lymphocyte development. Nat Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- Oestreich KJ, Cobb RM, Pierce S, Chen J, Ferrier P, Oltz EM. Regulation of TCRbeta gene assembly by a promoter/enhancer holocomplex. Immunity. 2006;24:381–391. doi: 10.1016/j.immuni.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Oltz EM. Regulation of antigen receptor gene assembly in lymphocytes. Immunol Res. 2001;23:121–133. doi: 10.1385/IR:23:2-3:121. [DOI] [PubMed] [Google Scholar]
- Osipovich O, Cobb RM, Oestreich KJ, Pierce S, Ferrier P, Oltz EM. Essential function for SWI-SNF chromatin-remodeling complexes in the promoter-directed assembly of Tcrb genes. Nat Immunol. 2007;8:809–816. doi: 10.1038/ni1481. [DOI] [PubMed] [Google Scholar]
- Porritt HE, Rumfelt LL, Tabrizifard S, Schmitt TM, Zuniga-Pflucker JC, Petrie HT. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity. 2004;20:735–745. doi: 10.1016/j.immuni.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Raghavan SC, Chastain P, Lee JS, Hegde BG, Houston S, Langen R, Hsieh CL, Haworth IS, Lieber MR. Evidence for a triplex DNA conformation at the bcl-2 major breakpoint region of the t(14;18) translocation. J Biol Chem. 2005;280:22749–22760. doi: 10.1074/jbc.M502952200. [DOI] [PubMed] [Google Scholar]
- Romanow WJ, Langerak AW, Goebel P, Wolvers-Tettero IL, van Dongen JJ, Feeney AJ, Murre C. E2A and EBF act in synergy with the V(D)J recombinase to generate a diverse immunoglobulin repertoire in nonlymphoid cells. Mol Cell. 2000;5:343–353. doi: 10.1016/s1097-2765(00)80429-3. [DOI] [PubMed] [Google Scholar]
- Sawada S, Littman DR. A heterodimer of HEB and an E12-related protein interacts with the CD4 enhancer and regulates its activity in T-cell lines. Mol Cell Biol. 1993;13:5620–5628. doi: 10.1128/mcb.13.9.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz DG. V(D)J recombination. Immunol Rev. 2004;200:5–11. doi: 10.1111/j.0105-2896.2004.00173.x. [DOI] [PubMed] [Google Scholar]
- Sen J, Venkataraman L, Shinkai Y, Pierce JW, Alt FW, Burakoff SJ, Sen R. Expression and induction of nuclear factor-kappa B-related proteins in thymocytes. J Immunol. 1995;154:3213–3221. [PubMed] [Google Scholar]
- Sikes ML, Bradshaw JM, Ivory WT, Lunsford JL, McMillan RE, Morrison CR. A streamlined method for rapid and sensitive chromatin immunoprecipitation. J Immunol Methods. 2009 doi: 10.1016/j.jim.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikes ML, Gomez RJ, Song J, Oltz EM. A developmental stage-specific promoter directs germline transcription of D beta J beta gene segments in precursor T lymphocytes. J Immunol. 1998;161:1399–1405. [PubMed] [Google Scholar]
- Sikes ML, Meade A, Tripathi R, Krangel MS, Oltz EM. Regulation of V(D)J recombination: A dominant role for promoter positioning in gene segment accessibility. Proc Natl Acad Sci U S A. 2002;99:12309–12314. doi: 10.1073/pnas.182166699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikes ML, Suarez CC, Oltz EM. Regulation of V(D)J recombination by transcriptional promoters. Mol Cell Biol. 1999;19:2773–2781. doi: 10.1128/mcb.19.4.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV. Developmental and molecular characterization of emerging beta- and gammadelta-selected pre-T cells in the adult mouse thymus. Immunity. 2006;24:53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Tripathi R, Jackson A, Krangel MS. A change in the structure of Vβ chromatin associated with TCR β allelic exclusion. Journal of Immunology. 2002;168:2316–2324. doi: 10.4049/jimmunol.168.5.2316. [DOI] [PubMed] [Google Scholar]
- Uematsu Y, Ryser S, Dembic Z, Borgulya P, Krimpenfort P, Berns A, von Boehmer H, Steinmetz M. In transgenic mice the introduced functional T cell receptor beta gene prevents expression of endogenous beta genes. Cell. 1988;52:831–841. doi: 10.1016/0092-8674(88)90425-4. [DOI] [PubMed] [Google Scholar]
- Villey I, Caillol D, Selz F, Ferrier P, de Villartay JP. Defect in rearrangement of the most 5′ TCR-J alpha following targeted deletion of T early alpha (TEA): implications for TCR alpha locus accessibility. Immunity. 1996;5:331–342. doi: 10.1016/s1074-7613(00)80259-9. [DOI] [PubMed] [Google Scholar]
- Weih F, Carrasco D, Bravo R. Constitutive and inducible Rel/NF-kappa B activities in mouse thymus and spleen. Oncogene. 1994;9:3289–3297. [PubMed] [Google Scholar]
- Whitehurst CE, Chattopadhyay S, Chen J. Control of V(D)J recombinational accessibility of the D beta 1 gene segment at the TCR beta locus by a germline promoter. Immunity. 1999;10:313–322. doi: 10.1016/s1074-7613(00)80031-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.