Abstract
Background: Patients with high-risk primary breast cancer remain at high risk for relapse. More precise prognostic and predictive tools are needed to improve treatment of such patients.
Patients and methods: Formalin-fixed, paraffin-embedded tumors from 239 high-risk breast cancer patients were examined for expression of human epidermal growth factor receptor 2 (HER2), epidermal growth factor receptor (EGFR), estrogen receptor, progesterone receptor, Ki-67, p16, p21, p27, and p53 by immunohistochemistry. Gene expression of EGFR, HER2, glutathione S-transferase-Pi (GSTP1), excision repair cross complementation1 (ERCC1), p21, β-tubulin-3, multidurg resistance (MDR1), cyclooxygenase2 (COX2), and cyclin-E was measured by RT-PCR.
Results: Eighty percent of patients presented with locally advanced, or ≥10 axillary nodal metastasis, and 20% with inflammatory breast cancer. The median age was 46 years (26–62 years) and the median number of involved axillary lymph nodes was 12 (0–42). At a median follow-up of 86 months, relapse-free survival (RFS) and overall survival for the entire group were 50% (95% CI 43% to 57%) and 62% (95% CI 56% to 69%). Multivariate Cox stepwise analysis resulted in a simple model for RFS consisting only of p21 expression, EGFR expression assessed by RT-PCR, and number of axillary nodal metastases.
Conclusion: A prognostic model on the basis of the expression of a limited number of proteins and genes may help to guide target-specific therapies in patients with high-risk breast cancer.
Keywords: epidermal growth factor receptor, high-risk breast cancer, prognosis, p21
introduction
Early diagnosis and improved therapeutic options are the most likely causes of the recently observed downward trend in breast cancer-related mortality [1]. However, 10%–15% of patients presenting with high-risk primary breast cancer (HRBC) remain at high risk for relapse and death [2–6]. More precise prognostic and predictive tools are needed to improve treatment of such high-risk patients [7].
Characterization of limited numbers of molecular markers has yielded results suitable for practical application for patients with early-stage, mostly low-risk breast cancer [8, 9]. No such methods have been validated in patients with higher stage/high-risk disease.
We have previously reported that patients with HRBC, characterized by mutation of p53, human epidermal growth factor receptor 2 (HER2) overexpression, high grade, high mitotic index, and lack of progesterone receptor (PR) expression, faired poorly if at least three of these features were present [10]. Here, we report our findings in an extended cohort of HRBC patients inclusive of the analysis of a broad spectrum of protein and gene markers associated with cell proliferation, tumor suppression, and resistance to therapeutics, with long-term follow-up.
patients and methods
inclusion criteria
We studied 239 patients with HRBC who were treated with dose-intense chemotherapy (DICT) at the City of Hope Cancer Center (COHCC) from 1989 to 2001 (52% of the eligible population). The study cohort was selected solely on the basis of the availability of formalin-fixed, paraffin-embedded (FFPE) archival samples from the primary tumors. All patients who participated in DICT trials at the COHCC gave their written, voluntary informed consent. Patients received standard neo-adjuvant and/or adjuvant, primarily doxorubicin-containing, chemotherapy; DICT was administered to all patients at the COHCC. Characteristics of patients are shown in Table 1. The study was initiated before the revision of the staging system [11]; therefore, the preceding American Joint Committee on Cancer (AJCC) staging classification was used for this report. Accordingly, patients with stage II disease included those with T1–T2 tumor size and ≥10 axillary nodes involved (and would now be classified as having stage IIIC disease), and those with stage IIIA disease included T3 tumors with any number of lymph axillary nodes involved (this group would now include stages IIIA, IIIB, and IIIC). All but one patient with stage IIIB disease presented with inflammatory breast carcinoma.
Table 1.
Patient characteristics
| Patient population (N = 239) | Median (range) |
|
| Age, in years | 46 (26–62) | |
| Median follow-up (live patients) in months | 86 (20–155) | |
| Number of axillary nodes involved | 12 (0–42) | |
| Time from diagnosis to DICT, in months | 6 (3–24) | |
| N (% with data available) | N (% not available) | |
| Stage | ||
| II | 99 (41) | |
| IIIA | 93 (39) | |
| IIIBa | 47 (20) | |
| <9 nodes (not inflammatory) | 29 (12) | |
| >9 nodes (not inflammatory) | 163 (68) | |
| Inflammatory | 47a (20) | |
| High-grade primary | 122 (53) | 9 (4) |
| Received neo-adjuvant chemotherapy | 47 (20) | |
| Received standard dose adjuvant anthracyclin | 235 (98) | |
| Received standard dose adjuvant taxane | 29 (12) | |
| Received tandem cycle DICT | 15 (6) | |
| Underwent modified radical mastectomy | 203 (85) | |
| Received radiation therapy | 231 (97) | |
| ER/PR+ | 145 (61) | 2 (1) |
DICT, dose-intense chemotherapy; ER, estrogen receptor; PR, progesterone receptor.
Except for one patient, all stage IIIB patients presented with inflammatory breast cancer.
Staging/eligibility requirements included tomography of the chest, abdomen, and brain (or magnetic resonance imaging of the brain), bone scan, bilateral bone marrow biopsies [showing no evidence of cancer by routine hematoxylin and eosin (H&E) staining], creatinine clearance of ≥70 ml/min, serum transaminases ≤2 times above the institutional upper limit of normal, and adequate pulmonary function tests. A Karnofsky performance status of ≥80% was required before enrollment on any of the DICT protocols. Patients treated with prior neo-adjuvant/adjuvant doxorubicin exposure of ≤240 mg/m2 and without prior left-sided chest wall radiation received a high-dose doxorubicin-containing DICT regimen; all others received platinum-based regimens.
All DICT protocols have been described earlier and consisted of either doxorubicin- or platinum-based regimens [10, 12]. From 1996 on, patients were preferentially enrolled on tandem cycle DICT trials [12].
post-DICT therapy
Patients received radiation to the primary site/chest wall and draining lymph node areas according to community standards; those with estrogen receptor (ER)- and/or PR-positive breast cancer received antiestrogen (predominantly tamoxifen) therapy for 5 years.
post-treatment follow-up
Following DICT, patients underwent physical examination at least once every 4 months for the first 3 years, and every 6 months thereafter. Yearly mammograms, bone scans, and chest X-rays were carried out for the first 3 years, with yearly mammograms continuing thereafter.
histopathologic and immunohistochemical analysis
Established parameters such as stage, lymph node involvement, grade, and receptor status were assessed. In addition to standard H&E staining, immunohistochemical staining for p16, p21, p27, HER2, epidermal growth factor receptor (EGFR), mitogen-activated protein kinase (MAPK), p53, ER, PR, and Ki-67 was carried out. Representative sections from all primary tumors were generated, reviewed, and analyzed by the same staff member (PC) of the Department of Anatomic Pathology at the COHCC. All tissues were fixed in 10% neutral-buffered formalin and embedded in paraffin. The antibodies used were ER (Clone 1D5, 1:100, Dako, Carpinteria, CA), PR (Clone PR88, 1:50, Biogenex, San Ramon, CA), p16 (Clone G175-405, 1:1000, PharMingen, San Diego, CA), p21 (Clone SX118, 1:200, PharMingen), HER2/neu (Clone CB11, 1:60, Novocastra, Burlingame, CA), p53 (DO7, 1:50, Novocastra), Ki-67 (Clone M7240, 1:200, Dako), EGFR (Clone M3563, 1:200, Dako), p27 (Clone SX53G8, 1:100, Dako), and MAPK (rabbit polyclonal, 1:100, Cell Signaling, Danvers, MA).
Immunohistochemical studies were carried out using the avidin–biotin complex technique, augmented by heat-induced epitope retrieval and/or enzyme digestion. Immunohistochemical staining was carried out on an automated immunohistochemical stainer (TechMate 1000, Ventana Medical System, Tuscon, AZ). Deparaffinized 5-μm sections were rehydrated through a xylene and graded alcohol series, and the slides were rinsed with tap water for 5 min and steamed in 1 mM EDTA buffer (pH 8.0) in a household food steamer (HH90, Black and Decker, Shelton, CT) for 20 min at 100°C. All primary antibody incubations were carried out at room temperature for 30 min. The EnVison universal horseradish peroxidase-labeled polymer detection system was used for antigen localization (Dako). Hematoxylin was used to counterstain the nucleus. Multitissue blocks were used for positive and negative controls. For the nuclear proteins [ER, PR, p16 (both nuclear and cytoplasmic), p21, p27, p53, and Ki-67], the percentage of positively stained nuclei was estimated. HER2, EGFR, and MAPK immunoreactivity was scored as 0+ (negative), 1+ (incomplete membrane staining), 2+ (>10% cells with weak complete cell membrane staining), and 3+ (>10% cells with strong complete cell membrane staining).
RT-PCR analysis
RNA was isolated from FFPE samples according to a proprietary procedure of Response Genetics, Inc. (Los Angeles, CA). After RNA isolation, complementary DNA was prepared from each sample as described previously [13, 14].
RT-PCR was used to create a gene expression profile of genes involved in cell growth, cycle regulation, intracellular drug trafficking/metabolism, and intracellular structure. The following nine genes were tested: EGFR, HER2, glutathione S-transferase (GST)-P1, ERCC1, p21, β-tubulin-3, MDR1, COX2, and cyclin-E. Target genes and an internal reference gene (β-actin) were quantified using a fluorescence-based real-time detection method [ABI PRISM 7900 Sequence detection System (TaqMan®) Perkin-Elmer and Applied Biosystem, Foster City, CA], as previously described [14]. The primers used for RT-PCR testing are listed in Supplementary Table 1, available online.
statistical methods
Outcomes examined included overall survival (OS) and relapse-free survival (RFS). RFS was defined as time to any type of recurrence or death from any cause. Standard Kaplan–Meier and Cox regression methods were applied for survival analysis using the SAS/STAT and S-Plus software [15]. All significance testing was two-sided (log-rank statistics and Wald statistics were used in univariate and multivariate analysis). Univariate and multivariate Cox regression analyses were carried out to assess potential prognostic (inherent to the primary tumors) and predictive indicators (treatment-related variables) of RFS and OS. Disease-specific indicators included age at diagnosis; tumor stage; presence of inflammatory features; grade of the tumor; number of axillary nodes involved with metastasis; ER and PR status; expression of p16, p21, p27, HER2, EGFR, MAPK, p53, and Ki-67 as tested by immunohistochemistry; and expression of EGFR, HER2, GST-P1, ERCC1, p21, β-tubulin-3, MDRI, COX2, and cyclin-E as analyzed by RT-PCR.
Treatment-related predictive factors included type of conventional dose, neo-adjuvant and/or adjuvant therapy, and radiation treatment to the primary site (yes versus no). The potential effect of the specific DICT regimen [doxorubicin based versus others, taxane-containing regimens versus others, platinum-containing regimens (without taxanes) versus others, and single versus tandem regimen (both by actual delivery and by intent to deliver both treatment cycles)] was also evaluated (data not shown).
In building an RFS and OS model, all factors were included in the initial model build, and a stepwise forward and backward selection Cox regression was employed that accounted for changes in patient numbers due to missing values.
To confirm the final survival models, we verified the model by standard stepwise regression using SAS with entry criteria of 0.2 and stay criteria of 0.05. We also verified that the final model was the same when using S-Plus stepAIC employing Akaike's information criteria. We ran 1000 bootstrap samples with replacement randomly selecting a patient and their survival information and matching this patient with a randomly selected expression profile to verify the correlation structure of the covariates. We employed a leave-one-out validation to search for patients with high leverage and simulated the effects of dropping out 5% (five patients) of patients at random. All variables remained statistically significant in the leave-one-out validation, and the coefficient of variation of the relative risk in the 5% dropout simulation was <8% for EGFR and p21 and <1% for nodes, and the statistical significance was unaltered in 1000 simulations.
For gene expression, before survival analysis, patients were divided into three groups according to the terciles of the gene expression values for all genes except for Cox2, cyclin-E, and EGFR. For those genes, the patients with very low expression were put into one category, and the remaining patients were divided into two groups according to their median. Similarly, for immunohistochemistry, the cut points were selected before survival analysis and were selected on the basis of dividing the range into terciles [16].
results
We analyzed tumor samples from 239 patients. Patient characteristics are described in Table 1. The numbers of samples available for each study, the range of the values obtained, the definition of the cut point used for analysis, and the percent of samples above the cut point are presented in Table 2.
Table 2.
Protein and gene expression profile of primary tumor specimens
| Protein expression by immunohistochemistry | ||||
| Protein | # Patients evaluated | Range | Definition of positive | # (%) Positive |
| Ki-67 | 211 | 0%–95% | ≥60% | 39 (18) |
| p53 | 209 | 0%–100% | >30% | 54 (26) |
| ER/PR | 237 | − or + | + | 145 (61) |
| HER2 | 206 | 0–3 | 2 or 3 | 83 (40) |
| EGFR | 203 | 0–3 | 3 | 44 (22) |
| MAPK | 204 | 0–3 | 3 | 43 (21) |
| p16 | 211 | 0%–100% | >60% | 42 (20) |
| p21 | 212 | 0%–50% | >0% | 109 (51) |
| p27 | 212 | 0%–100% | >0% | 146 (69) |
| Gene expression by RT-PCR | ||||
| Gene | # Patients with interpretable results | Range | Definition of amplified | # (%) Amplified |
| p21 | 171 | 0.63–42.23 | >2.6 | 59 (35%) |
| EGFR | 100 | 0–5.08 | >1.1 | 28 (28%) |
| HER2 | 165 | 0–3.84 | >0.3 | 52 (32%) |
| β-tubulin-3 | 125 | 0–1.54 | >0.7 | 41 (33%) |
| Cox2 | 84 | 0–8.40 | >0.05 | 56 (67%) |
| Cyclin-E | 123 | 0–13.40 | >0.9 | 31 (25%) |
| ERCC1 | 173 | 0.13–6.40 | >1.08 | 114 (66%) |
| GST-P1 | 170 | 0.04–3.77 | >0.65 | 55 (32%) |
| MDR1 | 84 | 0–1.15 | >0.13 | 55 (65%) |
ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; EGFR, epidermal growth factor receptor; MAPK, mitogen-activated protein kinase.
We carried out univariate analyses including tumor stage and grade, number of lymph nodes involved, and protein and gene expression. These analyses were carried out with and without adjustment for tumor stage (data not shown), and for inflammatory (stage IIIB) presentation, grade, and the number of axillary nodes involved (>9 versus ≤9, and also as a continuum) (Table 3). In the unadjusted analysis, high tumor grade [RFS: hazard ratio (HR) = 1.47; OS: HR = 1.7, P < 0.05], inflammatory features [OS: HR = 1.84 relative to stage II, P < 0.05], and higher number of nodes (a 3% increase in hazard for each node for RFS and OS, P < 0.05) were associated with adverse outcome. Treatment-related factors did not demonstrate a statistically significant effect (data not shown). In the adjusted analysis, the number of lymph nodes was associated with worse RFS and OS (a 4% increase in hazard for each node, P < 0.05), and inflammatory presentation remained significant (Table 3). Of the molecular markers, low-level protein expression of the tumor suppressor genes p21 and p27, increased expression of EGFR messenger RNA (mRNA), low expression of ERCC1 mRNA, negative hormone receptor status, mutated p53 protein expression, and increased p16 expression had significant adverse associations with the duration of RFS, while the expression of all these parameters and HER2 protein overexpression affected OS in univariate analysis (Table 3). When adjusted for age, inflammatory features, receptor status, nodes (continuous), and grade, only HER2, MAPK, p27 and p21 protein expression, and MDR1 and EGFR gene expression were significant factors in OS, whereas p27, p21, and p16 protein expression and EGFR gene expression were significant factors in RFS (Table 3).
Table 3.
Individual analysis of protein and gene expression profile and survival
| Parameter |
Univariate relapse-free HR (95% CI) | Adjusted relapse-free HR (95% CI) | Univariate overall survival HR (95% CI) | Adjusted overall survival HR (95% CI) | |
| ≤9 nodes, not inflammatory | 1 (baseline) | 1 (baseline) | 1 (baseline) | 1 (baseline) | |
| >9 nodes, not inflammatoryb | 1.52 (0.81–2.85) | 1.65 (0.85–3.22) | 1.44 (0.69–3.03) | 1.55 (0.70–3.44) | |
| Inflammatory breast cancer | 1.78 (0.87–3.63) | 1.68 (0.79–3.56) | 2.07 (0.91–4.67) | 1.88 (0.79–4.49) | |
| Stage II | 1 (baseline) | 1 (baseline) | 1 (baseline) | 1 (baseline) | |
| Stage IIIA | 1.36 (0.92–2.03) | 1.40 (0.91–2.13) | 1.49 (0.93–2.40) | 1.56 (0.94–2.60) | |
| Stage IIIB | 1.44 (0.88–2.35) | 1.43 (0.85–2.40) | 1.84 (1.06–3.20)a | 1.85 (1.03–3.33)a | |
| Inflammatory (all 3B) | 1.24 (0.79–1.93) | 1.23 (0.77–1.96) | 1.51 (0.92–2.45) | 1.47 (0.87–2.43) | |
| Inflammatory (exclude 3B noninflammatory)b | 1.29 (0.82–2.01) | 1.20 (0.76–1.91) | 1.56 (0.96–2.54) | 1.50 (0.89–2.48) | |
| High grade | 1.47 (1.01–2.13)a | 1.23 (0.83–1.83) | 1.70 (1.09–2.63) | 1.31 (0.82–2.09) | |
| Nodes (continuous) | 1.03 (1.01–1.06)a | 1.04 (1.01–1.06)a | 1.03 (1.01–1.06)a | 1.04 (1.01–1.07)a | |
| Immunohistochemistry | |||||
| KI-67 | High | 1.58 (1.00–2.51) | 1.23 (0.73–2.08) | 1.73 (1.03–2.90)a | 1.20 (0.67–2.16) |
| p53 | Low | 0.65 (0.43–0.98)a | 0.80 (0.50–1.26) | 0.54 (0.34–0.86)a | 0.68 (0.41–1.12) |
| ER/PR | + | 0.57 (0.40–0.81)a | 0.59 (0.40–0.87)a | 0.49 (0.32–0.73)a | 0.54 (0.34–0.84)a |
| HER2 | 2/3+ | 1.40 (0.95–2.06) | 1.29 (0.86–1.94) | 1.69 (1.08–2.64)a | 1.77 (1.11–2.83)a |
| EGFR | 3+ | 1.55 (1.00–2.40) | 1.38 (0.87–2.19) | 1.66 (1.02–2.73)a | 1.46 (0.87–2.45) |
| MAPK | 3+ | 0.66 (0.39–1.12) | 0.62 (0.36–1.08) | 0.48 (0.25–0.94)a | 0.44 (0.22–0.89)a |
| p27 | 0 | 1.85 (1.25–2.72)a | 2.04 (1.37–3.04)a | 2.26 (1.45–3.51)a | 2.44 (1.55–3.83)a |
| p21 | Low (0) | 1.77 (1.21–2.61)a | 1.70 (1.15–2.53)a | 1.93 (1.23–3.04)a | 1.96 (1.23–3.10)a |
| p16 | High | 1.99 (1.30–3.05)a | 1.73 (1.06–2.83)a | 1.85 (1.13–3.02)a | 1.44 (0.82–2.52) |
| Gene expression by RT-PCR | |||||
| p21 | Low | 1.66 (1.09–2.54)a | 1.20 (0.76–1.90) | 1.83 (1.13–2.94)a | 1.33 (0.79–2.22) |
| EGFR | High | 2.78 (1.53–5.04)a | 2.73 (1.41–5.26)a | 2.59 (1.36–4.93)a | 2.16 (1.07–4.37)a |
| HER2 | High | 1.31 (0.84–2.04) | 1.14 (0.72–1.81) | 1.33 (0.81–2.17) | 1.15 (0.67–1.96) |
| β-tubulin | High | 1.15 (0.69–1.92) | 1.02 (0.60–1.74) | 1.11 (0.62–1.99) | 1.00 (0.55–1.82) |
| COX2 | High | 0.99 (0.53–1.85) | 0.91 (0.47–1.75) | 0.64 (0.33–1.26) | 0.57 (0.28–1.18) |
| Cyclin-E | High | 1.24 (0.71–2.15) | 0.89 (0.47–1.67) | 1.52 (0.83–2.77) | 1.17 (0.59–2.33) |
| ERCC1 | Low | 1.57 (1.03–2.39)a | 1.27 (0.80–2.01) | 1.53 (0.95–2.47) | 1.18 (0.70–1.99) |
| GST-P1 | High | 1.54 (1.00–2.38) | 1.22 (0.76–1.94) | 1.43 (0.87–2.36) | 1.16 (0.68–1.99) |
| MDR1 | Low | 1.42 (0.77–2.60) | 1.47 (0.77–2.80) | 1.80 (0.90–3.60) | 2.11 (1.00–4.45)a |
P < 0.05. Adjusted for grade, nodes (continuous), ER/PR+, age, and inflammatory.
Excluding single patient with noninflammatory stage IIIB impacted hazard ratio (HR) of genes and proteins by <1% and did not alter statistical significance.
The results of the multivariate analysis on RFS and OS are depicted in Tables 4 and 5. The variables that remained significant were the number of metastatic axillary nodes as a continuum, EGFR gene expression determined by RT-PCR, and p21 protein expression determined by immunohistochemistry. There was a ‘beneficial effect’ of MAPK expression, which was observed mostly in the HER2-positive subset population and was of borderline significance. Of note, co-expression of amplified HER2 and EGFR was not found to be a significant risk factor (data not shown). None of the permuted data sets obtained a P value less than that obtained with the three-variable model (P < 1.6 × 10−6).
Table 4.
Relative risk (RR) of relapse on the basis of a three-variable model
| Parameter | RR | Wald P value |
| Nodes (continuous variable) | 1.06 | 0.002 |
| EGFR (RT-PCR) | 3.98 | 0.00002 |
| p21 (IHC, low) | 2.49 | 0.004 |
EGFR, epidermal growth factor receptor; IHC, immunohistochemistry.
Table 5.
Relative risk (RR) of death on the basis of a four-variable model
| Parameter | RR | Wald P value |
| Nodes (continuous variable) | 1.06 | 0.013 |
| EGFR (RT-PCR, high) | 3.38 | 0.0004 |
| p21 (IHC, low) | 2.89 | 0.004 |
| MAPK (IHC, high) | 0.299 | 0.028 |
EGFR, epidermal growth factor receptor; IHC, immunohistochemistry; MAPK, mitogen-activated protein kinase.
Subset analysis of patients by the type of DICT regimen to establish the potential predictive value of genes involved in mechanisms of resistance against the primary agents in the DICT regimens (ERCC-1, MDR1, GST-P1) yielded no such correlation. There was no statistically significant reduction in the HR (0.589; P = 0.2) when triple negative patients received DICT with a platinum-containing regimen (data not shown).
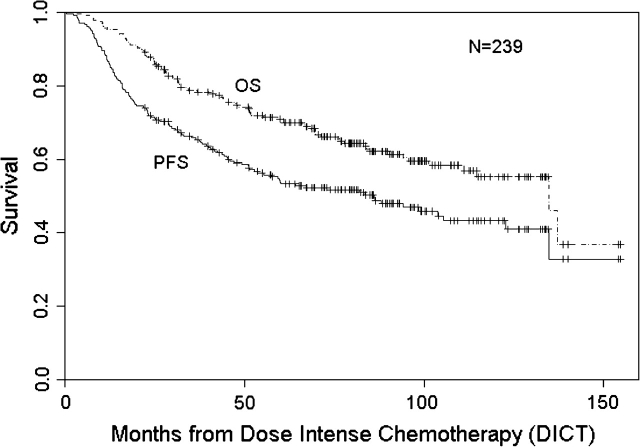
At a median follow-up of 86 months, RFS and OS were 50% (95% CI 43% to 57%) and 62% (95% CI 56% to 69%), respectively (Figure 1). RFS for stages II, IIIA, and IIIB disease was 55% (95% CI 45% to 66%), 47% (95% CI 38% to 59%), and 44% (95% CI 30% to 64%), respectively, following DICT, while OS for stages II, IIIA, and IIIB disease was 70% (95% CI 61% to 80%), 59% (95% CI 49% to 71%), and 51% (95% CI 36% to 72%), respectively (data not shown). When assessing the role of DICT, there was no statistically significant difference between patients treated with any particular regimen (single versus tandem, taxane containing or not).
Figure 1.
Relapse-free and overall survival following dose-intense chemotherapy in patients with high-risk primary breast cancer.
There was a significantly worse outcome in the group characterized by low p21 and high EGFR expression, while patients with tumors characterized by high p21 and low EGFR expression faired much better. Those with tumors expressing only one of the two adverse features fell into an intermediate category (Figure 2).
Figure 2.
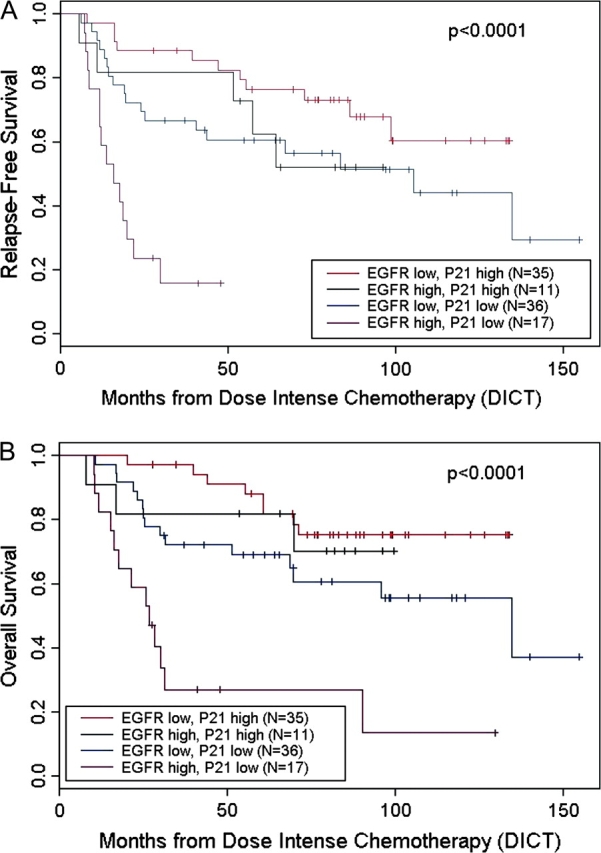
Effect of epidermal growth factor receptor and p21 on survival of patients following dose-intense chemotherapy. (A) Relapse-free survival. (B) Overall survival.
discussion
Current methods including immunohistochemistry and FISH are not easily reproducible or reliable, especially on tumors characterized by low expression of ER/PR and HER2. RT-PCR-based measurement of expression of ER, ER-regulated genes, and HER2 (Oncotype DX™) may provide a quantifiable, more reproducible method that has prognostic and predictive value [17]. Microarray-based molecular classification of breast cancer is still evolving [18]. All RT-PCR and microarray-based assays, such as the MammaPrint™ [19], are still in need of prospective validation, in ongoing trials such as the MINDACT in Europe and the TAYLORx in the United States.
Few studies have been reported on the value of prognostic and predictive factors in patients with HRBC. PR negativity, HER2/neu overexpression, and the presence of p53 protein are associated with poor outcome [10, 20]. RT-PCR analysis of archived FFPE tumor tissue from patients with more than nine axillary nodal metastasis [21] assessed 203 candidate genes: tumor protein p53-binding protein 2, PR, and B-cell CLL/lymphoma2 (Bcl2) were associated with improved distant RFS, while the HER2 adapter GRB7 and CD68 were adverse prognostic indicators. High scores on the 21-gene assay (Oncotype DX™), in addition to increased expression of genes associated with proliferation and immune response, were predictive of pathological complete response in patients with locally advanced breast cancer [22].
The predictive value of gene expression for response to specific chemotherapeutic regimens is the subject of intense investigations [23]. The degree of HER2/HER2 and EGFR expression may influence response to neoadjuvant therapy [25]. In general however, no reliable and reproducable markers of resistance have been established in patients with HRBC, including our current observations. No reliable markers of resistance have been established, so far. Similarly, in our series the expression of drug-resistance genes has not correlated with outcome.
Patients with HRBC remain a high-risk group for relapse, in general. Patients with a basal phenotype face, however, a higher likelihood of early relapse. Such patients trend towards experiencing better outcome when treated with DICT [26]. In a series of 225 patients aimed at better characterizing a HRBC population treated with DICT, EGFR expression, but not phospho-EGFR levels, was identified as a possible independent adverse prognostic indicator of survival, when measured by immunohistochemistry [27]. In our hands, EGFR expression predicted worse prognosis only when measured by RT-PCR, underlying the need for standardization of means of measurement.
We assessed the prognostic and predictive value of a panel of molecular parameters of primary tumors from patients with HRBC, all of whom received treatment with DICT. In the stage- and receptor-adjusted analysis, the combination of low expression of p21 protein and increased expression of the EGFR gene, as determined by RT-PCR, provided a powerful doublet of prognostic indicators. Identification of a few targetable genes of prognostic significance—rather than assessment of entire gene sets—may allow for the rapid incorporation of targeting agents into our current best treatment regimens, whether with or without DICT [28, 29].
Indeed, EGFR- and HER2-directed therapies may be the precursors of improved tumor targeting with drugs such as the Src inhibitor dasatinib, an agent which, in preclinical testing, selectively inhibited basal-like breast cancer cell lines [30]. These and a number of other kinase- or receptor-specific agents may provide the means to help patients with tumor profiles that have the worst prognosis, such as the low p21 and high EGFR expression subset in our patient population. Combinations of an EGFR-inhibiting agent and an inhibitor of AKT (which is associated with reduced signaling through the p21-associated signal transduction/activation pathway) may also be beneficial in this set of patients. Trials incorporating agents that interfere with the phosphatase and tensin homolog (PTEN)/phosphoinositide-3 kinase (PI3K)/AKT pathway are under active development, and tumors with low p21/p27 expression may be particularly good targets for such therapies.
In summary, limited scale, focused, molecular profiling of primary tumors from high-risk breast cancer patients may yield useful information to guide target-specific therapy. Expression of p21 by immunohistochemistry and of EGFR by RT-PCR may be useful for the selection of better treatment strategies for specific subsets of high-risk breast cancer patients. Because standard ‘best’ adjuvant therapy, and even dose-intense therapy, will not help a substantial portion of patients with high-risk disease, the identification of new protein and gene markers is needed to optimize the therapeutic approach to HRBC.
funding
National Cancer Institute (CA 33572 and 62505); National Institutes of Health (M01RR00043).
Supplementary Material
Acknowledgments
The authors thank Judy Brent, RN, Joyce Lawrence, RN, and Melissa Scalia, RN, for coordination of treatment protocols, Eleanor Fermin, Debbie Reardon, Kim Arnold, and Laura Alvarado for data management, and Dawnyetta Reese for secretarial assistance. We would like to thank Keely Walker for editorial assistance. Reported in part in J Clin Oncol; 22 (Suppl 14) (Abstr 9569).
References
- 1.Berry DA, Cronin KA, Plevritis SK, et al. Effective screening and adjuvant therapy on mortality from breast cancer. N Engl J Med. 2005;353(17):1784–1792. doi: 10.1056/NEJMoa050518. [DOI] [PubMed] [Google Scholar]
- 2.Bonadonna G, Zambetti M, Valagussa P. Sequential or alternating doxorubicin and CMF regimens in breast cancer with more than three positive nodes. Ten year results. JAMA. 1995;273(7):542–547. [PubMed] [Google Scholar]
- 3.Citron ML, Berry DA, Cirrincione CT, et al. Randomized trial of dose-dense versus conventionally scheduled and sequential versus concurrent combination chemotherapy as postoperative adjuvant treatment of node-positive primary breast cancer: first Report of Intergroup Trial C9741/Cancer and Leukemia Group B Trial 9741. J Clin Oncol. 2003;21(8):1431–1439. doi: 10.1200/JCO.2003.09.081. [DOI] [PubMed] [Google Scholar]
- 4.Henderson IC, Berry DA, Demetri GD, et al. Improved outcomes from adding sequential paclitaxel but not from escalating doxorubicin dose in an adjuvant chemotherapy regimen for patients with node-positive primary breast cancer. J Clin Oncol. 2003;21(6):976–983. doi: 10.1200/JCO.2003.02.063. [DOI] [PubMed] [Google Scholar]
- 5.Nitz UA, Mohrmann S, Fisher J, et al. Comparison of rapidly cycled tandem high-dose chemotherapy plus peripheral-blood stem-cell support versus dose-dense conventional chemotherapy for adjuvant treatment of high-risk breast cancer: results of a multicentre phase III trial. Lancet. 2005;366(9501):1935–1944. doi: 10.1016/S0140-6736(05)67784-7. [DOI] [PubMed] [Google Scholar]
- 6.Chang S, Parker SL, Pharm T, et al. Inflammatory breast carcinoma incidence and survival: the surveillance, epidemiology, and end results program of the National Cancer Institute. Cancer. 1998;82(12):2366–2372. [PubMed] [Google Scholar]
- 7.Rodenhuis S, Bontenbal M, van Hoesel QG, et al. Efficacy of high-dose alkylating chemotherapy in HER2/neu-negative breast cancer. Ann Oncol. 2006;17(4):588–596. doi: 10.1093/annonc/mdl001. [DOI] [PubMed] [Google Scholar]
- 8.van de Vijver MJ, He YD, van't Veer LJ, et al. A gene expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 9.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 10.Somlo G, Simpson J, Frankel P, et al. Predictors of outcome following high-dose chemotherapy in high-risk primary breast cancer. Br J Cancer. 2002;87(3):281–288. doi: 10.1038/sj.bjc.6600450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singletary SE, Allred C, Ashley P, et al. Staging system for breast cancer: revisions for the 6th edition of the AJCC Cancer Staging Manual. Surg Clin North Am. 2003;83(4):803–819. doi: 10.1016/S0039-6109(03)00034-3. [DOI] [PubMed] [Google Scholar]
- 12.Somlo G, Frankel P, Chow W, et al. Prognostic indicators and survival in patients with stage IIIB inflammatory breast carcinoma after dose-intense chemotherapy. J Clin Oncol. 2004;22(10):1839–1848. doi: 10.1200/JCO.2004.10.147. [DOI] [PubMed] [Google Scholar]
- 13.Kuramochi H, Hayashi K, Uchida K, et al. Vascular endothelial growth factor messenger RNA expression level is preserved in liver metastases compared with corresponding primary colorectal cancer. Clin Cancer Res. 2006;12(1):29–33. doi: 10.1158/1078-0432.CCR-05-1275. [DOI] [PubMed] [Google Scholar]
- 14.Lord RV, Salonga D, Danenberg KD, et al. Telomerase reverse transcriptase expression is increased early in the Barrett's metaplasia, dysplasia, adenocarcinoma sequence. J Gastrointest Surg. 2000;4(2):135–142. doi: 10.1016/s1091-255x(00)80049-9. [DOI] [PubMed] [Google Scholar]
- 15.SAS Institute. SAS/STAT Software, Release 8.1. Cary, NC: Sas Institute Inc.; 2000. [Google Scholar]
- 16.Nagelkerle N. A note on a general definition of the coefficient of determination. Biometrika. 1991;78(3):691–692. [Google Scholar]
- 17.Paik S, Tang G, Shak S, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24(23):3726–3734. doi: 10.1200/JCO.2005.04.7985. [DOI] [PubMed] [Google Scholar]
- 18.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100(14):8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van't veer L, Paik S, Hayes DF. Gene expression profiling of breast cancer: a new tumor marker. J Clin Oncol. 2005;23(8):1631–1635. doi: 10.1200/JCO.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 20.Nieto Y, Cagnoni PJ, Nawaz S, et al. Evaluation of the predictive value of Her-2/neu overexpression and p53 mutations in high-risk primary breast cancer patients treated with high-dose chemotherapy and autologous stem-cell transplantation. J Clin Oncol. 2000;18(10):2070–2080. doi: 10.1200/JCO.2000.18.10.2070. [DOI] [PubMed] [Google Scholar]
- 21.Cobleigh MA, Tabesh B, Bitterman P, et al. Tumor gene expression and prognosis in breast cancer patients with 10 or more positive lymph nodes. Clin Cancer Res. 2005;11(24 Pt 1):8623–8631. doi: 10.1158/1078-0432.CCR-05-0735. [DOI] [PubMed] [Google Scholar]
- 22.Gianni L, Zambetti M, Clark K, et al. Gene expression profiles in paraffin-embedded core biopsy tissue predict response to chemotherapy in women with locally advanced breast cancer. J Clin Oncol. 2005;23(29):7265–7277. doi: 10.1200/JCO.2005.02.0818. [DOI] [PubMed] [Google Scholar]
- 23.Rouzier R, Perou CM, Symmans WF, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005;11(16):5678–5685. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- 24.Rouzier R, Rajan R, Wagner P, et al. Microtubule-associated protein tau: a marker of paclitaxel sensitivity in breast cancer. Proc Natl Acad Sci USA. 2005;102(23):8315–8320. doi: 10.1073/pnas.0408974102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris LN, You F, Schnitt S, et al. Predictors of resistance to preoperative trastuzumab and vinorelbine for HER-2 positive early phase breast cancer. Clin Cancer Res. 2007;13(4):1198–1207. doi: 10.1158/1078-0432.CCR-06-1304. [DOI] [PubMed] [Google Scholar]
- 26.Diallo-Danebrock R, Ting E, Gluz O, et al. Protein expression profiling in high-risk breast cancer patients treated with high-dose or conventional dose–dense chemotherapy. Clin Cancer Res. 2007;13(2 Pt 1):488–497. doi: 10.1158/1078-0432.CCR-06-1842. [DOI] [PubMed] [Google Scholar]
- 27.Nieto Y, Nawaz F, Jones RB, et al. Prognostic significance of overexpression and phosphorylation of epidermal growth factor receptor (EGFR) and the presence of truncated EGFRvIII in locoregionally advanced breast cancer. J Clin Oncol. 2007;25(28):4405–4413. doi: 10.1200/JCO.2006.09.8822. [DOI] [PubMed] [Google Scholar]
- 28.Fan C, Oh DS, Wessels L, et al. Concordance among gene expression-based predictors for breast cancer. N Engl J Med. 2006;355(6):560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 29.Andre F, Pusztai L. Molecular classification of breast cancer: implications for selection of adjuvant therapy. Nat Clin Pract. 2006;3(11):621–632. doi: 10.1038/ncponc0636. [DOI] [PubMed] [Google Scholar]
- 30.Huang F, Reeves K, Han X, et al. Identification of candidate molecular markers predicting sensitivity in solid tumors to dasatinib: rationale for patient selection. Cancer Res. 2007;67(5):2226–2238. doi: 10.1158/0008-5472.CAN-06-3633. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.