Abstract
MicroRNAs are short non-coding RNAs that regulate the stability and translation of mRNAs. Profiling experiments, using microarray or deep sequencing technology, have identified microRNAs that are preferentially expressed in certain tissues, specific stages of development, or disease states such as cancer. Deep sequencing utilizes massively parallel sequencing, generating millions of small RNA sequence reads from a given sample. Profiling of microRNAs by deep sequencing measures absolute abundance and allows for the discovery of novel microRNAs that have eluded previous cloning and standard sequencing efforts. Public databases provide in silico predictions of microRNA gene targets by various algorithms. To better determine which of these predictions represent true positives, microRNA expression data can be integrated with gene expression data to identify putative microRNA:mRNA functional pairs. Here we discuss tools and methodologies for the analysis of microRNA expression data from deep sequencing.
Keywords: deep sequencing, expression profiling, microRNA
INTRODUCTION
MicroRNAs are ∼22-nt single-stranded RNAs that modulate gene expression in plants and animals [1]. The formation of a double-stranded RNA duplex through the binding of microRNA to mRNA in the RNA-induced silencing complex (RISC) either triggers the degradation of the mRNA transcript or the inhibition of protein translation. The conventional mammalian miRNA–mRNA interaction occurs through complimentary binding sites in the 3′-UTRs of target mRNAs [2]. One of the challenges in the emerging field of microRNA biology is the identification of microRNA targets. A given microRNA may have multiple (up to several hundred) predicted gene targets, and ∼60% of mRNAs have binding sites for several microRNAs in their 3′-UTRs. Decisive roles for microRNAs have been described across mammalian development and human diseases such as cancer. MicroRNAs are generally transcriptionally regulated by transcription factors that act at pol II promoters [3], and they may be preferentially expressed in certain tissues, during specific stages of development, or in disease.
Initially transcribed as a primary microRNA (pri-miRNA) these non-coding RNAs undergo two processing steps. The first step is the generation of stem-loop precursors (pre-miRNAs ∼70 nt) by the enzyme Drosha in a micro-processing complex within the nucleus. The pre-miRNAs are exported into the cytoplasm and processed into double stranded RNAs by the enzyme Dicer to subsequently release the 17–25-nt-long mature miRNA. Dicer plays a role in delivering the double stranded miRNA to the RISC. Argonaute (Ago) is the catalytic component of RISC and participates in the selection of the guide strand of the miRNA while the passenger strand is degraded. The active miRNA-loaded RISC complex binds predominantly to the 3′-UTR of a target mRNA with partial miRNA sequence complementarity where the critical base pairing occurs through the 5′ seed (base pairs 2–8). In vertebrates the RISC complex mediates post-transcriptional gene silencing through deadenylation-mediated mRNA decay and/or translational suppression by mechanisms not yet fully elucidated. In this process, Ago proteins play an important role in sequestering the miRNA–mRNA duplex. Of the four Argonaute proteins expressed in humans Ago-2 has been shown to be central for mRNA cleavage [4,5].
As with conventional protein-coding genes, it is of interest to profile the expression patterns of microRNAs during development and disease. For profiling gene expression, microarrays, consisting of arrayed series of thousands of microscopic spots of DNA oligonucleotides, have been the standard technology. Microarray platforms also exist for profiling microRNA expression. However, the recent introduction of deep sequencing technology, enabling the simultaneous sequencing of up to millions of DNA or RNA molecules, has provided another option for profiling microRNAs. With developing technologies currently trying to meet the goal of generating complete human genome sequences for less than $100 000 [6], using deep sequencing to comprehensively profile mRNA expression remains rather expensive (as compared to microarrays); however, profiling the small RNA fraction that contains microRNAs is much more feasible (though in genomes smaller than human or other eukaryotes, whole transcriptome coverage by sequencing could be feasible as well). Deep sequencing overcomes many of the disadvantages of microarrays, which suffer from background and cross-hybridization problems and measure only the relative abundances of previously discovered microRNAs. In contrast, deep sequencing measures absolute abundance (over a wider dynamic range than possible with microarrays) and is not limited by array content, allowing for the discovery of novel microRNAs or other small RNA species.
Recently, deep sequencing technology is becoming more available to researchers studying microRNAs, and the analysis of profiling data by deep sequencing may be carried out using both publicly available and custom-made software. In this review, we discuss tools and methodologies for the analysis of microRNA expression data from deep sequencing, from translating sequencing reads into microRNA abundance levels, to discovering novel microRNAs, to determining lists of differentially expressed microRNAs and their associated gene targets. Key challenges include the requirement for at least 10 µg of total RNA of very high quality; a preparation that includes several gel purifications steps all of which lead to loss of material; lack of a cheap QC step before sequencing; and dominance by a small number of miRNAs of very high copy number, where lower copy sequences may be underrepresented. To date, our own group has sequenced over 300 samples from a wide array of cellular systems (mouse as well as human), including cancer cell lines, stem cells and normal and diseased tissues, including diseases such as cancer, endometriosis and emphysema. While these studies are currently ongoing, they do illustrate the versatility of our deep sequencing methods.
FROM SAMPLE TO microRNA ABUNDANCE LEVELS
For microRNA expression profiling, our own group has chosen Illumina's Genome Analyzer (GA) technology. This platform utilizes massively parallel sequencing of millions of fragments using Illumina's proprietary Clonal Single Molecule Array technology and novel reversible terminator-based sequencing chemistry (www.illumina.com). A number of other next-generation sequencing technologies are currently in widespread use, including 454 and AB SOLiD (all of which are reviewed in [7]). MicroRNA sequencing protocols are available for Illumina (discussed in detail here), 454 [8] and SOLiD [9]. The approaches for analyzing next-generation sequence data described below can readily be applied to data from any of the various platforms.
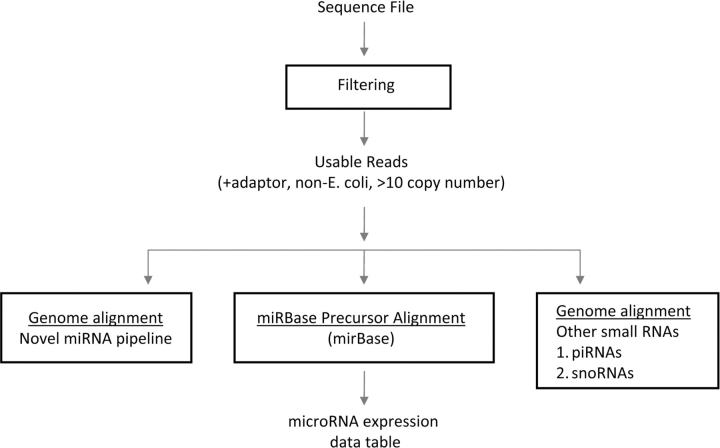
For translating sequence reads into microRNA abundance levels, we have developed our own software in-house, though there are a number of public software tools available for the analysis of sequence data (a number of which are listed in refs [7] and [10], as well as at http://www.sanger.ac.uk/Users/lh3/seq-nt.html), including tools that align sequences to references (which we carry out as described below). Our approach is summarized graphically in Figure 1.
Figure 1:
Analytical steps involved with microRNA profiling by deep sequencing. Raw sequences are first filtered to exclude those most likely to represent sequencing errors. Those sequence-reads that align to known microRNA precursor sequences are compiled into a data table with read counts (a measure of absolute expression level). Sequence reads that do not map to known microRNAs may be searched for potential novel microRNAs. In addition to microRNAs, the sequence reads may be searched for other small RNA species, such as piRNAs or snoRNAs.
The Illumina-ready small RNA template goes through two quality control (QC) steps. The first QC step is really only necessary for the platform development phase. Here, an aliquot of the small RNA template with Illumina adaptors is subjected to PCR-cloning and Sanger sequencing of ∼10–100 colonies. The sequences are analyzed and samples which generate a high fraction of miRNAs (∼25–95%, the typical range of the small RNAome) and other small RNAs with a low fraction of concatenated adaptors are selected for cluster generation on the cluster station of the Illumina GA platform. This insures that the templates to be sequenced are sufficiently enriched for short RNAs and avoids costly sequencing of contaminated and ill-prepared samples. The second QC step is based on the number of clusters generated in the first step of the sequencing pipeline; this becomes the first QC step once the platform has been established. Templates are first captured on a solid matrix and PCR amplified; the resulting cluster of molecules (identical copies of the same template) is called a cluster. Only samples yielding 15 000–35 000 clusters of amplified templates on the first base report are fully sequenced to maximize the yield of sequenced templates. In our experience, failures at the quality control step are almost always due to either the failure to amplify or insufficient starting material, which is indicated by low cluster and read numbers.
After sequencing, the raw sequence reads are filtered based on quality. Given that the average length of an Illumina sequencer read (∼36 nt) is greater than the average size of a microRNA (∼17–25 nt), we anticipate finding part of the 3′-adaptor at the 3′-end of the sequence (which may not apply to other non-miRNA small RNAs). In the first step, the reads which pass the quality filtering are then passed through an adaptor filter that searches for reads whose 3′-ends align to at least 6 nt of the 3′-adaptor. The adaptor sequences are trimmed and then the adaptor-trimmed reads which have passed both filters are formatted into a non-redundant fasta file where the copy number and sequence is recorded for each unique tag. The unique sequence set then goes through a second filter that searches for potential contamination by Escherichia coli sequence using BLAST against the E. coli sequence database. All sequences that fail to meet the quality cutoff, lack at least a 6-nt subsequence of the 3′-adaptor sequence, and/or match the E. coli sequence database are discarded as unusable reads.
All sequences that pass the above filters are aligned using a local Smith–Waterman alignment [11] of each unique read against each of the precursor microRNA sequences in the reference miRNA precursor set from the latest version of miRBase (http://microrna.sanger.ac.uk/sequences/index.shtml). In order to account for imperfect DICER processing that has been typically reported for mature miRNAs [12], 3-base overhang on the 5′-end and a 6-base overhang on the 3′-end are allowed. We therefore accept any sequence that perfectly matches the mature miRNA precursor in the mature miRNA region ±4 nt at the 5′- and 3′-end as mature miRNAs that are subjected to imperfect Dicer processing. This accounts for tags which match perfectly to known mature miRNAs (allowing for the aforementioned variations at the ends).
In addition, after finding the exact matches to mature miRNAs we also do an alignment of the remaining tags against a list of known miRNA precursors. For this alignment we allow up to three mismatches (loose matches), which allows us to identify miRNAs that carry mutations or that may have undergone RNA editing. The alignments are scored such that a matching or overhanging base counts as 2 points and mismatches as −1. The read counts of all redundantly aligning reads to multiple hairpins generating the same mature miRNAs are equally apportioned to each mature microRNA to which they align. The read counts are then normalized to the number of microRNAs to give the relative abundance of each microRNA. We term this the ‘microRNA expression profile’ of the sample.
We prefer to discard all unique sequence reads of fewer than 10 copies as potential sequence errors. All unique reads of copy number 10 or more are then reported in the output file, where we record a full description of their sequence alignments with known miRNA precursors from miRBase, as well as a tabulation of mismatches in relation to nucleotide position to identify potential sites of RNA editing. The output file also contains a table of copy numbers of exact and loose matches of read sequences to precursor miRNAs, which is used to generate the miRNA expression profile for further analysis.
USING DEEP SEQUENCING DATA TO DISCOVER NOVEL MICRORNAS
One of the key advantages of small RNA sequencing is that it allows for the discovery of novel microRNAs that have eluded previous cloning and standard sequencing efforts. A given sample may generate multiple sequences that are not sufficiently similar to any known microRNA. Below, we outline our analytical approach to evaluate whether these unique sequences represent novel microRNAs, which approach is also summarized graphically in Figure 2.
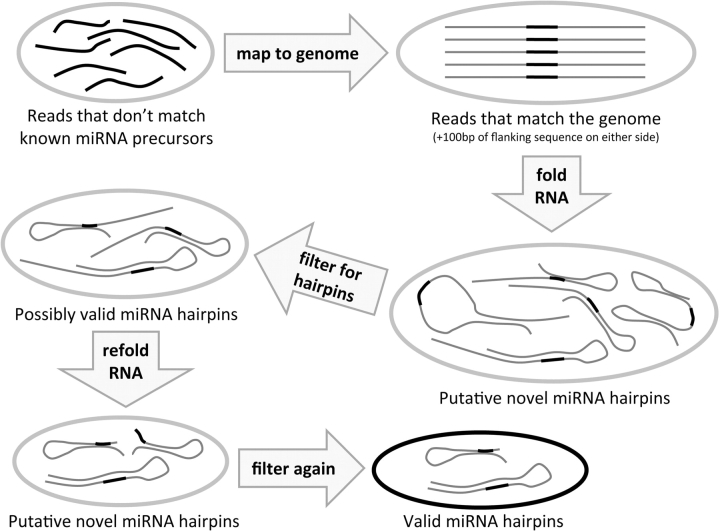
Figure 2:
Novel microRNA discovery pipeline. Taking the sequence reads that do not map to known microRNA precursors, we proceed by mapping them to the whole genome. Those reads that map (exactly) to the genome are taken (plus 100 bp of genomic sequence flanking either side of the read sequence) and folded as RNA using the Vienna package. The putative novel hairpins this produces are filtered for single-loop hairpins with the putative mature microRNA (read sequence) on one side of the hairpin, and those that pass are kept as possibly valid hairpins. These are trimmed down to only the putative hairpin sequence, refolded and filtered again with the Ambros criteria. The remaining hairpins are a valid microRNA hairpin whose putative mature microRNA sequence was found in the short RNA-ome of the sample, and thus are considered putative novel microRNA precursors.
Our first step is to map these small RNA sequences to the genome and to fetch each exact sequence match along with 100 bases flanking either side. These ∼220-bp sequences are then tested for microRNA-like hairpin structure. The ∼220-bp putative precursor sequences are then folded with the Vienna package [13], and microRNA hairpin structures which meet the Ambros criteria [14] are then identified. Specifically, the putative microRNA must lie on one arm of a single-loop hairpin with minimum free energy less than −25 kcal/mol; hairpins with overly large or unbalanced loops are rejected. After folding the read plus flanking sequence, the sequences are trimmed down to include only the plausible precursor and then folded again to ensure that the precursor was not artificially stabilized by neighboring sequence. We consider sequence reads that are appropriately placed in these microRNA-like hairpins to be ‘putative mature microRNAs’ (pmms).
After identifying all novel pmms, we separate them into two classes, those which occur frequently in the genome (four or more times), and those which are relatively unique (occur fewer than four times). The former class is likely to have a higher false positive rate, but they may include members of the recently discovered class of repeat-associated microRNAs [15], and so may be kept for future study. We examine the remaining class of pmms for cross-species conservation of the hairpin structure [16]. Specifically, we look for strong conservation of the mature microRNA, significant (but possibly weaker) conservation of the hairpin arm opposite the mature microRNA, and little or no conservation of the hairpin loop. We consider these conservation patterns as definitive confirmation of the microRNA predictions [14].
Publicly available tools for miRNA discovery from deep sequencing of the short RNAome include miRDeep [17], CD-miRNA [18], MiRank [19] and miRCat [20] (miRCat being specific for detecting plant miRNAs). miRDeep relies on deep sequencing and the characteristic pattern of miRNA precursor fragments. Specifically, DICER processing of a miRNA precursor produces three precursor fragments—the mature miRNA, the sequence on the other side of the hairpin which binds to the mature sequence (the so-called mir-star sequence) and the hairpin loop sequence. Novel precursors are identified by the characteristic pattern of higher expression of the mature miRNA over the star and loop sequences. Hairpins which are not processed by DICER will not show this characteristic pattern and so they can be filtered out. As compared to our method, miRDeep is likely more specific for miRNAs, as it relies more on the expression pattern than the structure, and thus it is limited in its ability to discover precursors at low levels of expression where the characteristic miRNA expression signature is not clearly visible.
NORMALIZATION AND COMPARISON OF microRNA EXPRESSION PROFILES
For each microRNA sequence-based profile, the number of small RNA sequence reads can be used to estimate expression level of each microRNA. Given a set of sequence-based profiles, a normalization step is needed for profile-to-profile comparisons. A number of algorithms have been developed for the normalization of array-based gene expression profiles [21]; however, many of these algorithms rely upon the large number (∼40–50k) of data points representing all of the transcripts, where it can be assumed that the vast majority of genes are not changing from sample to sample. In contrast to genes, the number of known microRNAs is much smaller (500–600 for Homo sapiens), and it cannot necessarily be assumed that most microRNAs would not change or that equal numbers would be increasing or decreasing [22,23], and so total intensity or quantile normalization approaches may not be suitable here.
For sequence-based profiles, the total number of valid sequence reads for each profile (be they from microRNA or other RNA species) may be used as the scaling factor for normalization, as this number would give an indication of the total amount of RNA in the given sample. One profile is used as the reference, and the values of each of the other profiles are divided by the scaling factor [(total sequence reads, given)/(total sequence reads, reference)]. The assumption made here is that the total small RNA population remains fairly constant between samples (without necessarily trying to make assumptions on the microRNA sub-population).
The absolute number of sequence reads for a particular microRNA represents a measure of its relative abundance. When comparing two groups of profile differences (such as disease versus control), microRNA levels may be determined by a two-sided t-test or another analogous statistic (log-transforming the sequence count values is recommended for the t-test, as it stabilizing the variance). The ‘fold change’ in microRNA expression between groups (the ratio of the group averages) may also be computed. When comparing sample groups, one may wish to consider the absolute microRNA levels as well as the relative levels; a change from 1000 to 2000 units is arguably more biologically relevant than a change from 5 to 10 units. In order to filter out from the analysis those microRNAs expressed at very low levels, one option is to add a given low number (e.g. 40 units) to each microRNA value [in the preceding example, (2000 + 40)/(1000 + 40) = 1.96 ∼ 2, while (10 + 40)/(5 + 40) = 1.1]; alternatively, one could require a microRNA to be present at a minimum number of units in a given number of samples.
PREDICTING GENE TARGETS OF DIFFERENTIALLY EXPRESSED microRNAs
Once a set of microRNAs has been identified as being differentially expressed between two cellular states (e.g. disease versus normal), the next logical question is what specific genes are being targeted by these microRNAs. A number of computational algorithms have been developed to help predict microRNA-to-mRNA (microRNA:mRNA) relationships, based on sequence analysis. Some of the more well-known prediction algorithms include PicTar (http://pictar.mdc-berlin.de/) [24], TargetScan (www.targetscan.org) [25,26] and miRanda (www.microrna.org) [27]. Different algorithms may yield different microRNA:mRNA predictions, though most employ the same general criteria [28]: (i) complementarity between the microRNA sequence and the 3′-UTR mRNA sequence (whether a perfect is required or one or more base pair mismatches are allowed) and (ii) degree of conservation of the microRNA-binding site across species. Any given prediction algorithm will produce both false positives (pairs that are statistically significant but cannot be verified) and false negatives (true pairs that are missing from the results), and the balance of lowering false positives versus lowering false negatives would be a balance between using stricter versus looser criteria, respectively.
Published gene targeting prediction databases are often made available via a web interface, where the user can look up predicted microRNA:mRNA functional pairs for a specific microRNA or gene of interest. Depending on the prediction algorithm, up to 60% of genes in mammalian genomes may be targeted by microRNAs [26], and a single microRNA could target as many as hundreds of genes. Determining which predicted microRNA:mRNA pairs to focus on for functional studies can be somewhat challenging. One way to potentially cut down on the number of false positive pairs would be to carry out gene expression profiling of the same samples profiled for microRNAs. If a given microRNA is regulating a putative gene target by mRNA destabilization, this should show up in the gene expression data: where the microRNA is high, the gene should be low, and vice versa. If, however, the microRNA targets a gene at the protein translation level rather than the mRNA level, then this would not show up in mRNA profiling, but only in proteomic data. At the same time, recent studies have found that most targets substantially repressed translationally by microRNAs also show mRNA destabilization [29,30], and so mRNA data might be considered a good surrogate for protein data in most cases.
Gene expression profiling can often reveal hundreds of gene expression changes for a biological system of interest, while the current web interfaces for TargetScan or miRanda facilitate a gene-by-gene search, making integration of target predictions with gene expression data on a large-scale somewhat cumbersome and time consuming for many researchers. To address this, we have developed a desktop software application, called ‘SigTerms’ (http://sigterms.sourceforge.net/), which, for a given target prediction database, retrieves all microRNA:mRNA functional pairs represented by an input set of genes [31]. From the retrieved pairs, the user can quickly filter for those that involve those microRNAs that are moving in an opposite direction from the genes. Predictions from other algorithms other than PicTar, TargetScan and miRanda can also be integrated into SigTerms, so long as these predictions are pre-compiled into a file in the required format. Furthermore, for each microRNA, SigTerms computes an enrichment statistic for over-representation of predicted targets within the input gene set; in rationale similar to that of enrichment testing of gene annotation (GO) terms, a microRNA showing high enrichment of gene targets may be further implicated as having an important role in the system under study. Correlating miRNA expression with mRNA profiles can also be done with other methods of microRNA profiling, as reported previously [32].
CONCLUSIONS
Deep sequencing of the small RNA fraction within cells yields an incredibly rich amount of data, from which we can determine not only the expression levels of microRNAs but also the levels of other small RNA species, such as piRNAs or snoRNAs, as well as discover novel small RNAs. In a time when gene expression profiling experiments alone often yield more genes than researchers know what to do with, characterization of the small RNA fraction in addition to the genes can only further complicate our understanding of molecular systems. As we profile cells at various levels of molecular complexity (transcriptome, small RNAome, genome, epigenome, etc.), one challenge will be to effectively integrate these various types of data with each other, in order to help identify key drivers of cellular processes or disease.
Key Points.
Expression profiling of microRNAs by deep sequencing offers distinct advantages over conventional expression arrays, including the ability discover novel microRNAs or other small RNA species.
Expression profile data from deep sequencing may be analyzed much in the same way that gene array data is analyzed, in order to define differentially expressed genes/microRNAs.
While public databases provide in silico predictions of microRNA gene targets, gene expression data can be leveraged with microRNA expression data, in order to better identify putative microRNA:mRNA functional pairs.
FUNDING
Program Project Development Grant from the Ovarian Cancer Research Fund (in part); National Institutes of Health (grant P30 CA125123); the Dan L. Duncan Cancer Center at Baylor College of Medicine.
Biographies
Chad J. Creighton, PhD, is an Assistant Professor. His work focuses on bioinformatics analysis of molecular profiling data, particularly data from cancer, including integration of different data types and sources.
Jeffrey G. Reid, PhD, is a Research Assistant Professor. His primary research focus is on the development of computational analysis tools and techniques for processing high-throughput sequencing data.
Preethi H. Gunaratne, PhD, is an Assistant Professor. Her work focuses on the application of next generation sequencing to dissect the small RNAome in the context of stem cells and human diseases such as cancer and asthma.
References
- Du T, Zamore P. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–52. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell R, et al. Principles of MicroRNA-Target Recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine S, Cole M, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–33. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–97. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell M, Rivas F, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–41. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Schloss J. How to get genomes at one ten-thousandth the cost. Nat Biotechnol. 2008;26:1113–5. doi: 10.1038/nbt1008-1113. [DOI] [PubMed] [Google Scholar]
- Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–45. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Ruby J, Jan C, Player C, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- SOLiD_sequencing_protocol. [20 March 2009, date last accessed]; http://www3.appliedbiosystems.com/AB_Home/applicationstechnologies/ SOLiDSystemSequencing/SmallRNAAnalysis/index.htm.
- Berninger P, Gaidatzis D, van Nimwegen E, et al. Computational analysis of small RNA cloning data. Methods. 2008;44:13–21. doi: 10.1016/j.ymeth.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Smith T, Waterman M. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Morin R, O’Connor M, Griffith M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–21. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker I, Fontana W, Stadler P, et al. Fast folding and comparison of RNA secondary structures. Chem Month. 1994;125:167–88. [Google Scholar]
- Ambros V, Bartel B, Bartel D, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–9. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese J, Seila A, Yeo G, et al. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2007;104:18097–102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N, Lim L, Weinstein E, et al. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–62. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Friedländer M, Chen W, Adamidi C, et al. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26:407–15. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- Tyagi S, Vaz C, Gupta V, et al. CID-miRNA: a web server for prediction of novel miRNA precursors in human genome. Biochem Biophys Res Commun. 2008;372:831–4. doi: 10.1016/j.bbrc.2008.05.134. [DOI] [PubMed] [Google Scholar]
- Xu Y, Zhou X, Zhang W. MicroRNA prediction with a novel ranking algorithm based on random walks. Bioinformatics. 2008;24:i50–8. doi: 10.1093/bioinformatics/btn175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon S, Schwach F, Dalmay T, et al. A toolkit for analysing large-scale plant small RNA datasets. Bioinformatics. 2008;24:2252–3. doi: 10.1093/bioinformatics/btn428. [DOI] [PubMed] [Google Scholar]
- Quackenbush J. Computational analysis of microarray data. Nat Rev Genet. 2001;2:418–27. doi: 10.1038/35076576. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska E, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Ozen M, Creighton C, Ozdemir M, et al. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–93. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- Krek A, Grün D, Poy M, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Friedman R, Farh K, Burge C, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Enright A, Aravin A, et al. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethupathy P, Megraw M, Hatzigeorgiou A. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat Methods. 2006;3:881–6. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhaüsser B, Thierfelder N, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Baek D, Ville J, Shin C, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton C, Nagaraja A, Hanash S, et al. A bioinformatics tool for linking gene expression profiling results with public databases of microRNA target predictions. RNA. 2008;14:2290–6. doi: 10.1261/rna.1188208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Babak T, Corson T, et al. Using expression profiling data to identify human microRNA targets. Nat Methods. 2007;4:1045–9. doi: 10.1038/nmeth1130. [DOI] [PubMed] [Google Scholar]