Abstract
Hydroxyl radical footprinting is a valuable technique for studying protein structure, but care must be taken to ensure that the protein does not unfold during the labeling process due to oxidative damage. Footprinting methods based on sub-microsecond laser photolysis of peroxide that complete the labeling process faster than the protein can unfold have been recently described; however, the mere presence of large amounts of hydrogen peroxide can also cause uncontrolled oxidation and minor conformational changes. We have developed a novel method for sub-microsecond hydroxyl radical protein footprinting using a pulsed electron beam from a 2 MeV Van de Graaff electron accelerator to generate a high concentration of hydroxyl radicals by radiolysis of water. The amount of oxidation can be controlled by buffer composition, pulsewidth, dose, and dissolved nitrous oxide gas in the sample. Our results with ubiquitin and β-lactoglobulin A demonstrate that one sub-microsecond electron beam pulse produces extensive protein surface modifications. Highly reactive residues that are buried within the protein structure are not oxidized, indicating that the protein retains its folded structure during the labeling process. Time-resolved spectroscopy indicates that the major part of protein oxidation is complete in a timescale shorter than that of large scale protein motions.
Introduction
Mass spectrometry is becoming a widely used technique for structural analysis of proteins. Radical-based surface mapping techniques, such as hydroxyl radical protein footprinting, followed by mass spectrometry analysis has become increasingly popular for studying protein structure, protein-protein and protein-ligand interaction interfaces.1 Hydroxyl radicals have gained popularity as a labeling technique because they provide a fast, relatively nonspecific, covalent label that probes a variety of solvent accessible amino acid residues with one experiment.2 Proteins experience structural distortion due to conformational changes, multimerization, ligand binding, and aggregation, and these structural changes expose different solvent accessible surfaces of the protein. By labeling a protein with hydroxyl radicals before and after inducing a structural change, a mass spectrometric comparison can be used to determine protection or exposure of specific residues of each protein conformation.1, 3 However, common methods of production of hydroxyl radicals produce undesired side reactions that complicate analysis. Here a superior method with few uncontrolled side reactions is presented.
Hydroxyl radicals for labeling of proteins have been successfully produced by Fenton chemistry,4, 5 hydrogen peroxide (H2O2) photolysis,6–8 and water radiolysis by either gamma (γ) rays9–13 or exposure to an X-ray synchrotron beam.1, 3, 14–17 Protein oxidation is a labeling process that is dependent on the solvent-accessible surface and the reactivity of the exposed residues,1, 4, 18, 19 although a small dependence on the local sequence has been noted.8 Upon oxidation the native protein structure can be altered due to the oxidative modifications; even a single oxidation event can induce protein unfolding which causes a rapid increase in oxidation as compared to the folded conformation.20–22 In order for hydroxyl radical protein footprinting to be a reliable method for protein structural determination, hydroxyl radicals must react with the protein exclusively in its native conformation. Most hydroxyl radical-generation techniques, including water radiolysis by gamma (γ) rays and X-rays, are performed on a millisecond to minute timescale, making it more likely that the protein will experience unfolding due to modifications.20, 21 Most previous strategies for ensuring that the native conformation is exclusively probed include the use of circular dichroism to determine when conformational changes occur in order to label the protein before structural changes can be detected,23 limiting the amount of oxidation to where the vast majority of protein molecules have one or no oxidations,4, 8 or monitoring of the reaction kinetics to ensure the oxidation of the protein22 or its constituent peptides13, 14, 19 follows an expected pseudo-first order rate law. However, these strategies all require that the overall amount of oxidation be strictly limited to prevent probing oxidatively-unfolded structures, and this limited amount of oxidation results in fewer oxidation sites and lower structural resolution.
A relatively new strategy for ensuring that the native conformation of the protein is probed that still allows for high levels of oxidation is to ensure that all oxidation is completed on a faster timescale than the protein can unfold by using a very short UV laser pulse to photolyse H2O2. This can complete the labeling process on a sub-microsecond timescale.6, 24 The timescale for large scale protein motions, such as helix coiling/uncoiling, is in the long microseconds to milliseconds range;25 therefore, oxidatively labeling a protein at or below this time frame without the use of a precursor oxidant will prevent labeling of the oxidatively unfolded conformation. However, hydrogen peroxide-based methods of oxidation, including Fenton chemistry4, 5, 26 and UV photolysis, 6, 8, 23, 24 all have several problems stemming from the presence of H2O2. The presence of H2O2 in a protein solution can induce uncontrolled Fenton- like chemistry in redox-active metal-binding proteins,27, 28 and cysteine and methionine two-electron oxidation can proceed spontaneously.29 Furthermore, H2O2 has different physical properties than water and may induce protein conformational changes independent of oxidation.
Here we demonstrate the importance of limiting the exposure to hydrogen peroxide using an NMR based assessment of its effects on a model cysteine containing protein (Galectin-3). We then introduce a novel pulsed electron beam water radiolysis technique for hydroxyl radical protein footprinting that does not require hydrogen peroxide. In this method, hydroxyl radicals are generated in less than 1 µs using a 2 MeV Van de Graaff electron accelerator. Using a single electron beam pulse to oxidize the protein sample and subsequently scavenging secondary oxidants using methionine amide, protein oxidation is complete before the protein conformation can change. Ubiquitin, a small protein that ionizes efficiently, was used as a model to determine if controllable protein oxidation could be achieved by this water radiolysis method. Furthermore, β-lactoglobulin A was used as a model protein for method development because of its sensitivity to oxidation-induced conformational changes22 and the availability of a high-resolution X-ray crystal structure.30 In order to account for the motion of the side chains, molecular dynamics (MD) simulations of the protein were performed. These simulations enable the average solvent accessibilities to be computed for each side chain, and thus in principle provide a more complete model for the protein surface, under the conditions of the experiment, than possible from a single static crystal structure.18, 31 We also determined the half-life of the hydroxyl radical label in anoxic solution with the protein by time-resolved UV spectroscopy to ensure that the label is consumed on a timescale consistent with large scale protein motions. The detected sites of oxidation are compared with residues known to be present on the surface of the natively-folded protein to ensure that we are not labeling an oxidatively-unfolded protein.
Materials and Methods
Materials
All solvents were purchased from Sigma-Aldrich (St. Louis, MO), unless otherwise noted, at the highest purity available and used as supplied without further purification. Methionine amide and ammonium phosphate were purchased from Bachem (Torrance, CA) and J.T. Baker (Phillipsburg, NJ), respectively. Deionized water (18 MΩ) was prepared in-house with a Millipore Mill-Q water purification system (Millipore Bedford, MA). Ubiquitin (Sigma-Aldrich) was prepared at 15 µM in 20 mM sodium phosphate buffer. B-lactoglobulin A (Sigma-Aldrich) was prepared at 20 µM in 20 mM sodium phosphate, ammonium bicarbonate, and ammonium phosphate (J.T.Baker, Phillipsburg, NJ) buffers.
NMR Spectroscopy
Heteronuclear single quantum coherence (HSQC) spectra of the 15N-labeled galectin-3 samples were acquired at 600 MHz using a gradient enhanced version of a standard HSQC experiment (gNHSQC from the Varian BioPack). A small number of t1 points (32) and a minimal number of scans were used to reduce acquisition times to three minutes and still allow observation at a reasonable S/N radio. Data were processed using nmrPipe and plotted using nmrDraw.32 Unambiguous resonance assignments were taken from BMRB deposition # 4909.33
Electron Pulse Irradiation
20 µM buffered (pH=7) protein solutions (ubiquitin or β-lactoglobulin A) were subjected to electron pulses from Radiation Laboratory 2 MeV Van de Graaff accelerator. Solutions were saturated prior irradiation either with air or a gas mixture of nitrous oxide/oxygen (N2O/O2; 4/1 v/v Mittler Supply Inc., ultrahigh purity, South Bend, IN). For initial studies with ubiquitin the dose was changed either by varying the beam current, via changes in heater settings of the dispenser cathode, at fixed electron pulsewidth (660 ns) or by varying the electron pulsewidth (480–660 ns) at fixed beam current. Typically to attain high levels of labeled protein, the dose had to be around 300 Gy achieved by a heater setting of 15 and we routinely used a dose in this range. A 250 µl syringe (Hamilton, Reno, NV, custom grinded to minimize the loss of electrons due to scattering in the glass wall) was placed in the front of the electron beam exit window. The beam was focused to a 2 mm diameter and was spatially adjusted to hit the very front volume of the syringe (Supplemental Data). From the glass coloration after irradiation we could determine the spread of the beam in the sample cell and estimate the total irradiated volume to be about 5 µl. After each pulse of electrons the piston was moved to release irradiated solution plus a 0.5 µL volume of unirradiated solution and refill the irradiation volume with the fresh protein solution. The released solution was directed to an empty 1 mL centrifuge vial or a 1 mL centrifuge vial prior filed with 50 µl of buffered (pH=7) 20 mM methionine amide (Bachem) to stop any chain oxidation processes. After 10 subsequent electron pulses the vial containing 100 µl of 10 µM oxidized protein solution was stopped, frozen and stored awaiting labeling analysis. In initial studies the frequency of pulses and corresponding flow in the irradiated volume was varied to establish contribution of secondary irradiation before contact with the quencher. Electron pulse frequency of 1 Hz was used for results presented in this report. The doses applied to the protein solutions were estimated with the Fricke dosimeter.34 Oxygen saturated solutions of Fricke dosimeter were irradiated at the same conditions like the protein samples (beam current, electron pulsewidth, cell position) after protein oxidation process was completed. Over twelve months of studies, the dose varied in the range of 30% at initially established accelerator settings i.e. the beam current and the electron pulsewidth.
Time-Resolved UV Spectroscopy
Pulse radiolysis experiments were performed using 100–1500 ns pulses from the Radiation Laboratory 8 MeV electron linac to obtain the range of doses comparable with doses applied in other protein oxidation experiments. Analyzing light from a pulsed 75 W xenon lamp (Photon Technology International, Birmingham, NJ) was selected using monochromator SPEX-270M. UV kinetics were measured at 250 nm where hydroxyl radicals extinction coefficient35 is ε=535 M−1 cm−1. Samples of protein were prepared by dissolving β-lactoglobulin A to a concentration of 20 µM in ammonium phosphate buffer (pH=7). Buffer solutions were prepared in deionized water (18.2 MΩ-cm, Barnstead Nanopure System, Dubuque, IA). Prior to experiment, samples were bubbled with N2O (Mittler Supply Inc., Ultrahigh Purity) at ambient conditions resulting in a final concentration of N2O of 25 mM. To improve the signal-to-noise ratio, UV signals were averaged over 15 consecutive traces. Due to transient absorption of quartz in the flow sample cell after the pulse of electrons (pronounced mostly at the highest doses) the blank traces for a given dose were collected in the empty cell purged with argon gas (Mittler Supply Inc., Ultrahigh Purity). These blank traces were subtracted from the corresponding sample traces before kinetic analysis. The UV transient absorption traces were fit using a system of first order kinetic differential equations model (fitting code written in Igor Pro 5.00, WaveMetrics Inc., Lake Oswego, OR) that incorporates most important reactions expected after pulse radiolysis of β-lactoglobulin A in N2O saturated solutions. The initial concentration of hydroxyl radicals after the radiation pulse was taken as a sum of initial yields of hydroxyl radicals and hydrated electrons given that hydrated electrons convert to hydroxyl radicals within the duration of the pulse in N2O saturated solutions (see reaction (7)). Extinction coefficients of hydrated electrons and hydroxyl radicals at 250 nm are comparable within the experimental error.36 Thus, their rapid inter-conversion occurring in the first couple hundreds of nanoseconds does not obstruct the overall kinetic analysis extending up to 40 µs after the irradiation pulse. A fitting program was setup to fit all collected UV traces at once fitting the reaction rate of hydroxyl radicals with protein and the resulting product extinction coefficient, while keeping most other (known) parameters of the model fixed. For simplicity the rate constant of the protein radical decay was assumed to be first order since this reaction is many orders of magnitude slower than the initial decay of hydroxyl radicals in the monitored transient absorption time window.
LC-MS of Oxidized Protein
Intact ubiquitin samples were analyzed using a hand pulled fused silica (Technologies, Phoenix, AZ) spray column (75 µm × 10 cm; tip 15 ± 1µm) that was prepared by packing silica C18 resins (Rainin Microsorb MV, 5 µm, 300 Å pore size) from a 50% isopropanol and 50% methanol slurry into the pulled fused silica capillary using a pressurized stainless steel bomb. Prior to reverse-phase HPLC, the column was equilibrated with 0.1% formic acid in water and the intact samples were loaded onto the column using a stainless steel bomb pressurized with nitrogen gas at 1,000 psi for 45 minutes. Liquid chromatography was initiated at a primary flow rate of 4 µL through the Agilent 1100 Series reversed-phase HPLC system (Agilent Technologies, Waldbronn, Germany) that ran through a splitter and resulted in a flow rate of 400 nL/min over the column with a 10 min rinse in 95% Buffer A (0.1% formic acid in water) followed by a 20-min linear gradient of 5 to 95% Buffer B (0.1% formic acid in acetonitrile). The spectra were acquired by nano-electrospray ionization on a Thermo Finnigan LTQ-FT mass spectrometer (San Jose, CA). The capillary temperature was 250°C and the spray voltage was 2.2 kV for ubiquitin.
For all intact β-lactoglobulin A samples, 8 µL were injected using an Agilent autosampler module (Agilent Technologies) over an Agilent ZORBAX 300SB C18 (150 × 0.3 mm, 5 µM particles) reverse-phase column (Agilent Technologies). The HPLC, directly coupled to the LTQ-FT for mass spectrometry, was run using a linear gradient of 95% Buffer A to 95% Buffer B over 20 minutes at a flow rate of 2 µL/min, followed by a 10 minute wash with 95% Buffer B and an 80 minute wash with 95% Buffer A. The capillary temperature was 250°C and the spray voltage was 2.1 kV for β-lactoglobulin A.
Tryptic Digestion of Oxidized β-Lactoglobulin A
One β-lactoglobulin A irradiated with 480 ns electron pulses was selected for tryptic digestion to determine the sites and amounts of oxidation. Ammonium bicarbonate (50 uL, 50 mM) and DTT (25 µL, 25 mM) were added to the β-lactoglobulin A sample (50 µL) and the samples were incubated at 50°C for 3 hours to denature and reduce the protein. Iodoacetamide (25 µL, 90 mM) was added to the solution to carbamidomethylate the reduced disulfide bonds, and incubated in the dark at room temperature for 45 minutes. Sequencing grade modified trypsin (Promega, Madison, WI) was added (25 µL, 0.1 µg/µL) and incubated at 37°C overnight while rotating to digest the protein samples. The samples were analyzed as is in triplicate and the remaining sample was stored at 0°C.
LC-MS/MS of Digested Oxidized β-Lactoglobulin A
8 uL of the oxidized β-lactoglobulin A tryptic digest were injected over an Agilent ZORBAX 300SB C18 (150 × 0.3 mm, 5 µM particles) reverse-phase column (Agilent Technologies). The HPLC, interfaced to an LTQ-FT mass spectrometer, was run using a linear gradient of 95% Buffer A to 60% Buffer B over 47 minutes then to 90% Buffer B over 15 minutes at a flow rate of 2 µL/min, followed by a 5 minute wash with 90% Buffer B and an 55 minute wash with 95% Buffer A. The capillary temperature was 250°C, and the spray voltage was 2.3 kV for β-lactoglobulin A tryptic digest. After the LC-MS/MS experiment, the measured peptides were screened computationally for different modifications using MASCOT in conjunction with ProteIQ,37 and ByOnic.38 Semi-tryptic peptides were included in the search and analysis to obtain better sequence coverage as well as to determine other sites of oxidation. All tandem mass spectra assignments and sites of oxidation verified manually due to sample and search space complexity.
MD simulations and solvent accessibility calculations
A crystal structure of β-lactoglobulin (PDB ID code 1BSY)39 was retrieved from the Research Collaboratory for Structural Biology (RCSB) database.40 In order to calculate the time-averaged solvent accessible surface area (<SASA>) of each side chain in β-lactoglobulin, a 10-ns MD simulation of the protein was performed using the all-atom AMBER 841 force field with the PARM99 protein parameters.42 Prior to the MD simulation, all histidine residues were considered as neutral and were protonated at the Nε position. Nine Na+ ions were added to neutralize charge and the protein was solvated by 5,417 TIP3P43 water molecules, using the Protein Builder component of the GLYCAM-Web tool (http://www.glycam.com44), which employs the tLEaP41 module of AMBER. The energy was minimized using the SANDER module41 of AMBER by performing 5000 steps of steepest descent followed by 5000 steps of conjugate gradient minimization, again using GLYCAM-Web. The water molecules were then subjected to a simulated annealing protocol in which they were heated from 5 K to 300 K over a period of 50 ps, held at 300 K for 100 ps, before being cooled to 5 K over a final 50 ps. Following solvent annealing, the entire system was heated from 5 K and to 300 K over 100 ps with no restraints applied to the coordinates. A production MD data set was then collected for 10 ns with the temperature held constant at 300 K. All simulations were performed using the NPT ensemble, at 1 atm using a 2 fs integration time step, with the SHAKE algorithm45 treatment of all hydrogen-containing bonds, and a unit dielectric constant. The atomic co-ordinates were stored every 10 ps for analysis, for a total of 1000 snapshots. SASA values were calculated for individual snapshots employing the NACCESS program46 and averaged over the 10 ns simulation by in-house program to obtain <SASA> values and standard deviations for each of the 162 residues.
Results and Discussion
Rapid NMR Spectroscopy of Proteins in Solution with H2O2
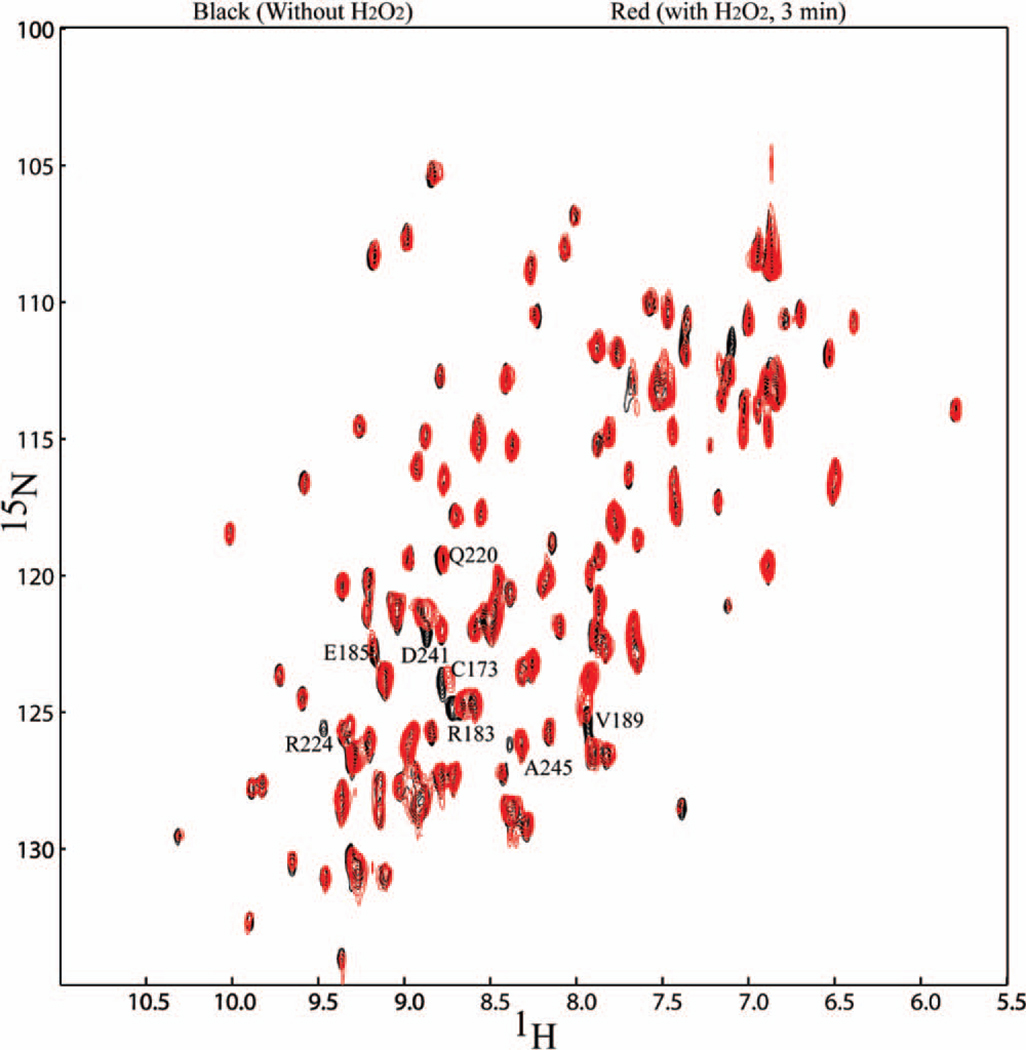
H2O2 photolysis is one of the fastest methods for producing hydroxyl radicals, but there is concern about sensitivity of proteins to the physical and chemical effects that H2O2 can impart. Before questioning the use of methods dependent on H2O2 for transition metal-free protein preparations, we felt it is important to assess the extent to which its presence affected conformation. Using time-resolved NMR spectroscopy we were able to observe changes in the conformation of a model protein, Galectin-3, in the presence of H2O2 versus H2O. Galectin-3 is a small (15 kDa) stable protein that binds galaactose terminated oligosaccharides. It is not suspected to be usually sensitive to hydrogen peroxide. However, it does have a cysteine residue that is not particularly solvent exposed. A three minute HSQC spectrum of galectin-3 was recorded in the presence and absence of H2O2 (Figure 1). The small shifts labeled (Figure 1) in the HSQC spectrum that can be interpreted as a conformational change. Many of the shifted peaks are assigned to resonances near the lactose binding site. Due to the very short time scale (three minutes) and known conformational flexibility of the binding site, direct oxidation by hydrogen peroxide is not thought to be the driving force behind this noted conformation change. The subtle shifts are more likely due to physical association of H2O2 versus water. Some of the perturbed resonances do, however, cluster near C173, the partially buried cysteine that is expected to be most sensitive to oxidative chemical events. At longer times, perturbations of additional resonances more indicative of rapid two-electron oxidation events are seen (data not shown). An LC-MS analysis of galectin-3 after a three minute H2O2 exposure determined the protein is not unusually sensitive to hydrogen peroxide, as no noticeable oxidation products were detected, while a sixty minute H2O2 exposure resulted in detectable protein oxidation (data not shown). Regardless of the origin of the shifts, the data illustrate that proteins are not necessarily probed in their native conformation when using methods that involve H2O2 to produce hydroxyl radicals even in the absence of peroxide-mediated oxidation events, and that these perturbations can occur on short time scales.
Figure 1.
Protein Oxidation
High energy electrons passing through the aqueous solution ionize/excite water molecules forming a number of transient species and stable products according to equation (1).
| (1) |
Among all species generated during water radiolysis, hydroxyl radicals (•OH) and hydrated electrons (eaq−) are the most reactive with peptides and proteins.2 When the radiolysis is performed in the presence of oxygen, superoxide radicals and its acidic form of hydroperoxyl radicals are generated in reaction (2) suppressing the effect of hydrated electrons on the proteins.
| (2) |
| (3) |
Since the pKa of HO2• radical is around 4.8 (reaction (3)) and oxidation experiments are performed at buffered pH 7, most of the hydroperoxyl radicals are in the form of a superoxide radical (HO2•/O2•−). HO2•/O2•− radicals are known to react with amino acids at very low rate constants ranging from 10-102 dm3mol−147, 48 therefore one can expect very little effect of HO2•/O2•− radicals with the metal free proteins that are subject of the current studies. HO2•/O2•− radicals undergo comparatively rapid disproportionation to H2O2 and O2 in reactions (4) and (5) via a pH dependent mechanism that also involves reaction (3).
| (4) |
| (5) |
Based on the kinetic equation for this decay mechanism,49, 50 one can estimate the rate constant for disproportionation at pH=7 to be about 2kobs=1.2×106 dm3mol−1. This indicates that if left alone HO•2/O2•− radicals can last milliseconds after the irradiation pulse; however, it is not long enough to cause any significant oxidation of the available protein amino acids. Converting •OH radicals leading to water and recovering initially consumed oxygen in reaction (6) is an additional reaction that consumes HO2•/O2•− radicals before they can react with the protein.
| (6) |
Since the rate constant of reaction (6) is as fast as 1×1010dm3mol−1, it inconveniently consumes some of important •OH radicals for oxidation.
In the initial microseconds following pulse irradiation, the balance of the above reactions leads mostly to the very reactive •OH radicals and relatively nonreactive HO2•/O2•− radicals. •OH radical reaction with amino acids is well established and proceeds with reaction rate constants51 varying from 1010 to 107 dm3mol−1. From a kinetic point of view the majority of free •OH radicals that happen to be close to the protein surface will interact with the fastest reacting amino acids; those include amino acids containing sulfur as well as unsaturated/aromatic side groups. Amino acids with the aliphatic side groups react with •OH radicals relatively slower. In oxygenated solutions protein surface radicals add oxygen forming the corresponding peroxyl radicals. The mechanism of initial sulfur centered radicals is not completely explained yet but leads to well characterized and recognizable products.52, 53 The fate of the relatively fast surface located peroxyl radicals depends entirely on their nature and surroundings. Some of the aromatic peroxyl radicals will eliminate HO•2 radicals leaving behind a stable hydroxyl group52 or a carbonyl group.54 C-centered aliphatic peroxyl radicals can undergo a bimolecular recombination leading to the creation of hydroxyl and/or carbonyl group on the parent carbon atom, or transfer the radical to form a hydroperoxyl group that propagates oxidation to neighboring amino acids.54 Those reactions are well understood and summarized in the number of articles and textbooks.1, 52, 54 In either case in diluted protein solution, sites of oxidation should be anchored to the place where the initial •OH radical attack occurred. Chances are that some of the initially created peroxyl radicals can survive on the surface and do not undergo any of the mentioned transformations. In this case they can react with lengthy present HO•2/O2•− radicals producing hydroxyl group, oxygen and hydrogen peroxide.55 The presence of oxygen is quite essential in the •OH radical induced protein oxidation even though the fastest surface labeling occurs by way of hydroxylation with •OH radicals.1
Our initial experiments were performed with aerated buffered ubiquitin solutions in order to establish optimal doses and concentrations for maximum oxidation. We varied the pulsewidth and current of the electron beam, as well as, the dissolved gas in the sample. In order to double the concentration of hydroxyl radicals in the pulse irradiation we saturated our solutions with nitrous oxide/oxygen combination (N2O/O2 4/1 v/v). In this case the oxygen concentration remained on the same level as in the air saturated solutions, but additionally we benefited from the well known reaction (7) of fast conversion of hydrated electrons to hydroxyl radicals.51 Since the solubility of N2O in water is about 20 times higher than oxygen, most hydrated electrons convert to hydroxyl radicals within the electron pulse, doubling the hydroxyl radical concentration.
| (7) |
The influence of gas composition on ubiquitin oxidation is presented in Figure 2. Due to the large dose applied, we see very substantial oxidation in both air and N2O/O2 saturated solutions. The solutions containing N2O show higher abundance of heavily oxidized ubiquitin. The initial experiments showed that we can easily control any excess of labeled protein by varying the dose and the gas composition. Additionally, we noted that the increase of the applied radiation dose does not increase the level of oxidation proportionally. It is mostly related to the oxygen uptake in the solution after the pulsed irradiation. In aerated or N2O/O2 saturated solutions, oxygen concentrations are on the order of 2.7×10−4 mol/dm3 that corresponds to the concentration of water radiolysis transient species (•OH, H•, eaq−) achievable at roughly 500 Gy of absorbed dose. Considerably increasing the dose above 500 Gy decreases the amount of available oxygen and leads to undesired reactions of hydrated electrons with the protein surface amino acids. Lack of oxygen can distort the formation process of carbon centered peroxyl radicals as precursors of hydroperoxides55 and other oxidation products on the protein surface. The quantitative studies of the dose dependence on the pulse radiolysis •OH protein footprinting were not the subject of the current investigation but will be addressed in the future reports.
Figure 2.
LC-FTMS Analysis of Ubiquitin and β-lactoglobulin A
Sodium phosphate-buffered ubiquitin was used as a model to determine if protein oxidation could be achieved by this method. LC-FTMS of ubiquitin (Figure 2) shows the range of intact protein oxidation obtained with and without N2O, and with and without methionine amide, a scavenger of secondary oxidants. Ubiquitin was also irradiated with various pulsewidths (480 or 660 ns) and electron beam amperages (doses) (Supplemental Data). In the absence of methionine amide (Figure 2b) no unoxidized ubiquitin is detected even at relatively low radiation doses despite the fact that 10% of the solution was unirradiated. However, in the presence of methionine amide (Figure 2c and d), unoxidized ubiquitin from the unirradiated fraction of the solution was still detected despite heavy amounts of oxidation. These results indicate that the methionine amide is sufficient to prevent oxidation of the unirradiated ubiquitin even at high radiation dosages, while secondary oxidation sufficient to consume all of the unirradiated ubiquitin occurs in the absence of the methionine amide even at relatively low radiation dosages (Supplemental Data). Quenching is necessary to ensure the vast majority of labeling occurs on a microsecond time scale and before the protein can unfold, rather than by secondary oxidants (e.g. peroxides in the presence of metal ion traces of UV light) on the second to minute timescale. With electron beam pulsewidths under 700 ns, we detected an extensive amount of oxidation (Figure 2d) using a sub-microsecond electron pulse. By adjusting various parameters of the irradiation (pulsewidth, dissolve N2O and dose), we are able to control the amount of oxidation from very little oxidation to very extensive oxidation (Supplemental Data).
In initial experiments on β-lactoglobulin A, a sodium phosphate buffer was irradiated and we detected sodium adduction (M + n22) in each sample despite multiple attempts to desalt the solution. This interfered with our data analysis (data not shown). Ammonium bicarbonate and ammonium phosphate buffers were used for subsequent experiments. Upon irradiation, the amount of oxidation in the ammonium bicarbonate-buffered samples was significantly lower, probably due to radical scavenging by the bicarbonate buffer. Also, an adduct or modification of unknown origin (observed mass shift of 174) is present in all ammonium bicarbonate-buffered β-lactoglobulin A samples (data not shown). Ammonium phosphate buffer gave the best results for intact β-lactoglobulin A, as it did not scavenge the radical, nor did it result in sodium or other adducts (Supplemental Data).
Time-Resolved UV Absorbance Spectra Reveal the Hydroxyl Radical Lifetime
Since proteins can unfold significantly on the order of a few microseconds,56 labeling chemistry should be completed faster than that to prevent labeling of oxidatively unfolded protein. One way to observe the progress of labeling chemistry is to monitor the decay of hydroxyl radicals in the microsecond time scale after their formation during the pulse of electrons. The lifetime of hydroxyl radicals in aqueous solutions after each pulse of electrons can be estimated based on the known reaction rate constants of hydroxyl radicals with solutes.51 In pure buffered water hydroxyl radicals recombine with each other since reactions with buffer components are usually much slower. The second order hydroxyl radical recombination (8) competes with slower first order reactions especially at higher doses when higher concentrations of hydroxyl radicals are produced.
| (8) |
We performed time resolved pulse radiolysis experiments with transient absorption detection at 250 nm to look at the upper limit of hydroxyl radical life time in buffered N2O saturated (no oxygen present) solutions without proteins. The traces from these experiments are provided in Supplemental Data. The initial increase of signal corresponds to formation of hydroxyl radicals and is related to their concentration via an extinction coefficient. Analyzing the transient absorption one can see that the apparent signal decays very quickly and within 8 µs reaches the plateau of H2O2 absorption. Reaction (9) of hydroxyl radicals with H2O2 is relatively slow (2.7×107 mol−1dm3)51 and consumes only about 1% of the formed H2O2 40 microseconds after the irradiation pulse.
| (9) |
However, this minimal concentration of superoxide radical contributes some 20% to the final absorption, since the extinction coefficient of HO2•/O2•− radicals greatly exceeds the extinction coefficient of hydrogen peroxide. Contribution of HO2•/O2•− radical absorption increases, obviously, with the increase of the applied dose since reaction (9) proceeds more efficiently but, the final concentrations of formed HO2•/O2•− are always at a lower fraction than the initial •OH radical concentration. Extension of the pulsewidth from 400 ns to 1500 ns increases the dose almost 4 times but not the peak transient absorption of hydroxyl radicals (Supplemental Data). Reaction (8) effectively decreases the concentration of hydroxyl radicals during the 1500 ns pulse and the rate of hydroxyl radical formation approaches the rate of hydroxyl radical decay giving steady state concentration of hydroxyl radicals just only 50% higher than the peak concentration of hydroxyl radicals after 400 ns pulse. It is important to note that the final absorption of H2O2 and HO2•/O2•− after the completion of reaction (8) is more than 3 times higher for 1500 ns pulse than 400ns (Figure 3), which confirms that reaction (8) with reaction (9) is the main channel of hydroxyl radicals decay in the protein free N2O saturated buffered water. Based on the extinction coefficients at 250 nm of species present, we determined that the time for hydroxyl radicals to decay to 0.2 µM (1% of protein in oxidation experiments) is about 20.6 µs + pulsewidth of electrons. It is very fast but still comparable with the duration of some protein unfolding events.57 Obviously, in the presence of protein, hydroxyl radicals will not stay free for as long because they will react with the protein surface groups. For most amino acids side changes, the reactions are very fast51 and should contribute very effectively to the decrease of hydroxyl radical concentration. To confirm the effect of protein, we performed an experiment in which β-lactoglobulin A was added to the buffered solution and the decay of hydroxyl radicals was monitored. The UV traces in Supplemental Data show that after the addition of 4 µM of protein the apparent UV signal reaches a higher plateau than in buffer alone suggesting that some other absorbing product is being formed in addition to H2O2 and HO2•/O2•−. In fact, a further increase of the protein concentration results in an even higher increase in the amplitude of the formed product. The apparent UV signal results from a sum of contributions from various transient species produced upon hydroxyl radical reaction with the protein surface and can be represented in symbolic reaction (10). The initial step of hydroxyl radical reaction with protein is formation of a protein radical:
| (10) |
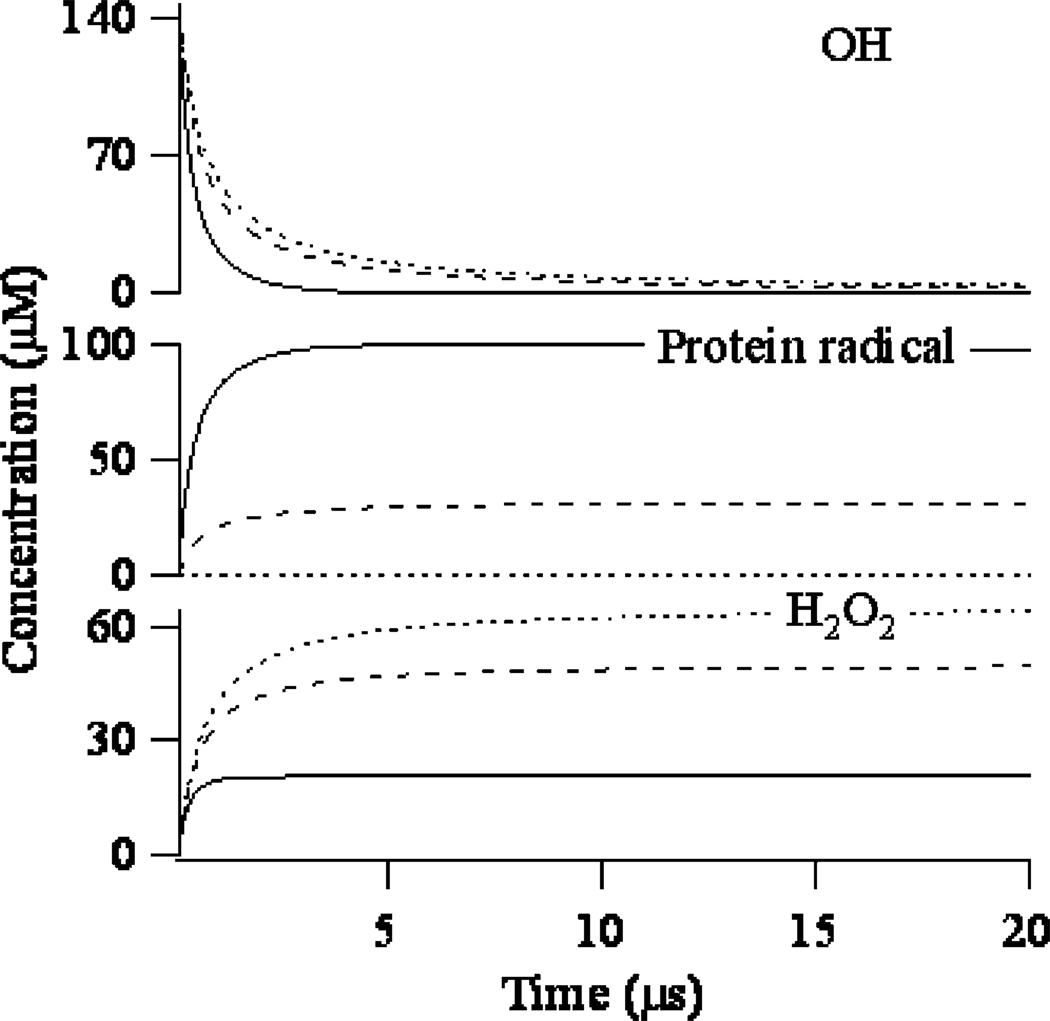
Figure 3.
The protein radical in oxygen-free, N2O saturated solution undergoes further reactions leading to the formation of the final oxidized protein (in case of radicals formed on the sulfur containing or unsaturated/aromatic residues) or decays in the bimolecular processes of disproportionation/dimerization.2, 52 For the time being we are interested in the fate of hydroxyl radicals, and based on the several UV transient signals collected for different protein concentrations, we constructed a simple model to extract the portion of the UV signal that is related to the decay of hydroxyl radicals. The applied global fitting model allowed us to estimate the rate constant of reaction (9) and the overall extinction coefficient of the protein radical for the studied system. All other parameters used to obtain fitting results are tabulated in Supplemental Data. The time profiles of most contributing species absorbing at 250 nm resulting from global fitting are shown on Figure 3. This basic experiment proved that the lifetime of hydroxyl radicals decreases considerably after addition of protein. In addition to the lifetime of hydroxyl radicals, the transient absorption experiment has also shown the importance of properly choosing the concentration of the protein in the pulse irradiation experiment. Lower concentrations of protein may be necessary for some experiments and can increase the extent of protein labeling,56 but at the same time the lower protein concentration prolongs the hydroxyl radical’s lifetime, increasing the chance of oxidizing a previously unfolded residue and necessitating the use of an exogenous quencher to limit the hydroxyl radical lifetime.6
LC-MS/MS Analysis of Oxidized β-lactoglobulin A Tryptic and Semi-Tryptic Fragments
The β-lactoglobulin A sample irradiated with the dose of 260 Gy using a pulse length of 480 ns, was denatured, reduced, carbamidomethylated, and digested using trypsin. The peptide mixture was loaded on a C18 capillary column for LC-MS/MS analysis to locate sites of oxidation. The overall sequence coverage, including all oxidized and unoxidized peptides identified in the LC-MS/MS run, was 100%, with manual verification of all tandem mass spectra assignments. Fourteen oxidation sites were identified for β-lactoglobulin A in these LC-MS/MS experiments; however, one semi-tryptic peptide was detected as completely oxidized and although oxidation could not be mapped to a specific site(s), peptide 108-ENSAEPEQSLACQCLVR-124 contains many reactive residues with large <SASA> values. The identified oxidation sites are shown on the X-ray crystal structure of β-lactoglobulin A30 in Figure 4A.
Figure 4.
The surface average solvent accessibility (<SASA>) value of each amino acid residue for all 162 residues was obtained from 1000 snapshots over a 10 ns MD simulation (Supplemental Data). While 10 ns is too short a period to simulate large-scale protein backbone motions, it is adequate to capture much of the motion of the side chains, and provides a useful approach for achieving statistically significant average SASA values (<SASA>) on a given time scale. In addition, the use of MD simulations on this time scale provides the opportunity to probe the extent to which a model based essentially on the fully folded protein is able to describe the data from an experiment performed on the millisecond timescale. The <SASA> values for each mapped oxidation site on β-lactoglobulin A are listed in Table 1. Using this method we are able to determine that, with the exception of M24, all amino acids determined to be oxidized under our conditions were predicted to be suitably solvent accessible by MD simulations.
Table 1.
Average Solvent Accessibility of Oxidized Residues in β-Lactoglobulin
| Peptide (residues) | Peptide fractional oxidation | site(s) of oxidation | <SASA> [Å2] |
|---|---|---|---|
| 1–8 | 0.3861 ± 0.1686 | L1 | 49.78 |
| M7 | 35.88 | ||
| 23–40 | 0.1184 ± 0.0052 | M24 | 0.21 |
| 41–60 | 0.0127 ± 0.0003a | E51 | 123.54 |
| 0.0227 ± 0.0009 | L57 | 40.63 | |
| 84–91 | 0.0174 ± 0.0103 | L87 | 156.27 |
| E89 | 88.65 | ||
| 92–100 | 0.0091 ± 0.0043 | Y99 | 40.51 |
| 92–101 | 0.0437 ± 0.0218 | ||
| 125–135 | 0.0124 ± 0.0072 | V128 | 28.86 |
| 125–138 | 0.0310 ± 0.0040 | L133 | 58.05 |
| 142–148 | 0.1607b | M145 | 6.7 |
| 149–162 | 0.0590 ± 0.0057 | F151 | 20.35 |
| E158 | 81.89 | ||
| I162 | 83.85 |
Mass Shift of −30 Which is Characteristic of oxidized glutamic acid residue.
Only seen in one replicate.
The sulfur-containing residues (Met and Cys) are the most susceptible to oxidative modifications.29 No cysteine residues were conclusively identified as sites of oxidation; even though they are chemically reactive, they are buried within the protein structure, and therefore should react only slowly with a hydroxyl radical if the protein retains its folded form during the labeling process. Three of the four methionine residues found in β-lactoglobulin A, Met7, Met24 and Met145, were identified as sites of oxidation with a mass shift of +16. Met24 has a very small <SASA> value of 0.21 Å2, but Met7 and Met145 have small to moderate solvent accessibility (<SASA> value = 35.88 and 6.7 Å2 respectively). Met107 also has a large <SASA> value and was not detected as oxidized by our measurements. However, it is present on a large peptide that was determined to be the site of multiple semi-tryptic cleavages and unusual oxidation events; it is quite possible that Met107 was oxidized quite readily, but that we are unable to resolve and identify the oxidized form of the peptide from the MS/MS spectra. All other sites of oxidation identified had large <SASA> values (Table 1), and can be clearly seen on the surface of the protein (Figure 4); importantly, all of the oxidized sites were predicted by MD simulation to be among the most highly accessible residues of each type within the protein (e.g. out of twenty-two total leucine residues, the four leucine residues oxidized were all among the five most solvent accessible according to MD simulations), with the exception of Met24 (Supplemental Data).
This leaves an unresolved question: why was Met24 oxidized under these conditions? One potential cause is incomplete scavenging of secondary oxidants, which have been noted to lead to uncontrolled oxidation of methionine.29 However, these same secondary oxidants would also be capable of oxidizing cysteine, which is not detected. A more likely possibility is that the protein undergoes larger backbone motions, in the region of M24, on the time scale of the experiment, than are captured in the 10 ns MD simulation. In that case, the oxidation of M24 would be larger than predicted by the <SASA> values. It is important to note here that by using average SASA values, rather than those from a single protein structure, the possibility that this discrepancy is related to an anecdotal orientation of the side chain of M24 is greatly reduced.
Because most of the oxidized peptides contain more than one oxidation site, accurate quantitation of the amount of oxidation at individual sites is currently not possible. We were able to quantitate the amount of oxidized versus unoxidized peptide using the triplicate LC-MS/MS runs to determine the peak area and the standard deviation of the MS total ion chromatogram of a given m/z for each peptide and its oxidized form(s). The peak areas were used to calculate peptide fractional oxidation by dividing the peak area of the oxidized peptide by the sum of the peak areas of the oxidized and unoxidized forms of that peptide. The peptide fractional oxidation is shown for each β-lactoglobulin A peptide in Figure 4B and values are listed in Table 1. The sites of oxidation that could be determined by MS/MS analysis are also listed in Table 1; however, while these identified sites are major sites of oxidation for the peptide, they may not be the only substantial sites of oxidation. It is important to note that, as is all LC/MS experiments of complex mixtures, it is quite possible that major oxidation products exist that were not detected or that did not fragment sufficiently well to determine the site(s) of oxidation.
Conclusion
The overall purpose of this study was to demonstrate a method for hydroxyl radical footprinting of proteins in solution that is completed faster than large scale oxidation-induced conformational changes can occur, without requiring a precursor oxidant. Time-resolved NMR spectroscopy illustrates that the presence of hydrogen peroxide can result in uncontrolled oxidation or peroxide-induced conformational changes, causing the labeling of a non-native protein structure. Additionally, the presence of redox-active transition metals in solution with the hydrogen peroxide can lead to uncontrolled metal catalyzed oxidation5, and in many cases it is not possible to completely purify redox-active transition metals from the protein-ligand preparation. The development of a hydrogen peroxide-free method of hydroxyl radical footprinting that is capable of completing the labeling reaction faster than the protein can unfold increases the applicability of hydroxyl radical footprinting technology to peroxide-sensitive proteins, while still allowing for extensive labeling of the native conformation. As shown from the results presented, the electron accelerator pulsed water radiolysis method is suitable for heavy, controlled oxidation of proteins and oxidation of solvent accessible residues. Using an electron beam pulse, controllable protein oxidation can be obtained by adjusting variables such as pulsewidth, dose, and dissolved gas. Time–resolved UV spectroscopy indicates that the most reactive radical species are consumed in less than 2 µs without a quencher, and this rapid timescale allows for extensive oxidation before the protein can unfold due to the modifications. Our ability to heavily oxidize the protein without concern for oxidation-induced unfolding allows us to detect a much greater amount of oxidation than previously-reported results on the same protein,8 allowing for higher resolution hydroxyl radical protein footprinting data. MS/MS analysis and MD simulations indicate that all oxidation sites except one (Met24) identified by this method are moderately to highly solvent accessible. Additionally, as a rapid oxidation method, electron pulse water radiolysis has broad applicability in time-resolved structural studies, most notably UV-based spectroscopy methods where the high concentrations of hydrogen peroxide necessary for flash photolysis of peroxide can confound spectroscopy. Such a pulse labeling method will prove to be extremely useful in designing time-resolved spectroscopic studies of oxidation-induced protein unfolding, which is important for understanding the biophysical basis of oxidative stress-induced protein inactivation.
Supplementary Material
Acknowledgments
This research was supported by the National Center for Research Resources of the National Institute of Health (P41RR005351). The Notre Dame Radiation Laboratory is supported by the Office of Basic Energy Sciences at the United States Department of Energy. This is document number NDRL-4795 from the Notre Dame Radiation Laboratory.
References
- 1.Takamoto K, Chance MR. Radiolytic protein footprinting with mass Spectrometry to probe the structure of macromolecular complexes. Annual Review of Biophysics and Biomolecular Structure. 2006;35:251–276. doi: 10.1146/annurev.biophys.35.040405.102050. [DOI] [PubMed] [Google Scholar]
- 2.Garrison WM. Reaction mechanisms in the radiolysis of peptides, polypeptides, and proteins. Chemical Reviews. 1987;87(2):381–398. [Google Scholar]
- 3.Kiselar JG, Maleknia SD, Sullivan M, Downard KM, Chance MR. Hydroxyl radical probe of protein surfaces using synchrotron X-ray radiolysis and mass spectrometry. Int J Radiat Biol. 2002;78(2):101–114. doi: 10.1080/09553000110094805. [DOI] [PubMed] [Google Scholar]
- 4.Sharp JS, Becker JM, Hettich RL. Protein surface mapping by chemical oxidation: structural analysis by mass spectrometry. Anal Biochem. 2003;313(2):216–225. doi: 10.1016/s0003-2697(02)00612-7. [DOI] [PubMed] [Google Scholar]
- 5.Schantz JT, Hutmacher DW, Lam CXF, Brinkmann M, Wong KM, Lim TC, Chou N, Guldberg RE, Teoh SH. Repair of calvarial defects with customised tissue-engineered bone grafts - II. Evaluation of cellular efficiency and efficacy in vivo. Tissue Engineering. 2003;9:S127–S139. doi: 10.1089/10763270360697030. [DOI] [PubMed] [Google Scholar]
- 6.Hambly DM, Gross ML. Laser flash photolysis of hydrogen peroxide to oxidize protein solvent-accessible residues on the microsecond timescale. J Am Soc Mass Spectrom. 2005;16(12):2057–2063. doi: 10.1016/j.jasms.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Sharp JS, Guo JT, Uchiki T, Xu Y, Dealwis C, Hettich RL. Photochemical surface mapping of C14S-Sml1p for constrained computational modeling of protein structure. Anal Biochem. 2005;340(2):201–212. doi: 10.1016/j.ab.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Sharp JS, Becker JM, Hettich RL. Analysis of protein solvent accessible surfaces by photochemical oxidation and mass spectrometry. Anal Chem. 2004;76(3):672–683. doi: 10.1021/ac0302004. [DOI] [PubMed] [Google Scholar]
- 9.Smedley JG, 3rd, Sharp JS, Kuhn JF, Tomer KB. Probing the pH-dependent prepore to pore transition of Bacillus anthracis protective antigen with differential oxidative protein footprinting. Biochemistry. 2008;47(40):10694–10704. doi: 10.1021/bi800533t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franchet-Beuzit J, Spotheim-Maurizot M, Sabattier R, Blazy-Baudras B, Charlier M. Radiolytic footprinting. Beta rays, gamma photons, and fast neutrons probe DNA-protein interactions. Biochemistry. 1993;32(8):2104–2110. doi: 10.1021/bi00059a031. [DOI] [PubMed] [Google Scholar]
- 11.Armstrong RC, Swallow AJ. Pulse-and gamma-radiolysis of aqueous solutions of tryptophan. J Radiat Res. 1969;40(3):563–579. [PubMed] [Google Scholar]
- 12.Kopoldova J, Hrncir S. Gamma-radiolysis of aqueous solution of histidine. Z. Naturforsch. C.: Biosci. 1977;32C(7–8):482–487. [PubMed] [Google Scholar]
- 13.Winchester RV, Lynn KR. X- and gamma-radiolysis of some tryptophan dipeptides. Int J Radiat Biol. 1970;17(6):541–548. doi: 10.1080/09553007014550681. [DOI] [PubMed] [Google Scholar]
- 14.Guan JQ, Vorobiev S, Almo SC, Chance MR. Mapping the G-actin binding surface of cofilin using synchrotron protein footprinting. Biochemistry. 2002;41(18):5765–5775. doi: 10.1021/bi0121104. [DOI] [PubMed] [Google Scholar]
- 15.Liu R, Guan JQ, Zak O, Aisen P, Chance MR. Structural reorganization of the transferrin C-lobe and transferrin receptor upon complex formation: the C-lobe binds to the receptor helical domain. Biochemistry. 2003;42(43):12447–12454. doi: 10.1021/bi0352973. [DOI] [PubMed] [Google Scholar]
- 16.Maleknia SD, Ralston CY, Brenowitz MD, Downard KM, Chance MR. Determination of macromolecular folding and structure by synchrotron x-ray radiolysis techniques. Anal Biochem. 2001;289(2):103–115. doi: 10.1006/abio.2000.4910. [DOI] [PubMed] [Google Scholar]
- 17.Rashidzadeh H, Khrapunov S, Chance MR, Brenowitz M. Solution structure and interdomain interactions of the Saccharomyces cerevisiae "TATA binding protein" (TBP) probed by radiolytic protein footprinting. Biochemistry. 2003;42(13):3655–3665. doi: 10.1021/bi027203f. [DOI] [PubMed] [Google Scholar]
- 18.Charvátová O, Foley B, Bern M, Sharp J, Orlando R, Woods R. Quantifying Protein Interface Footprinting by Hydroxyl Radical Oxidation and Molecular Dynamics Simulation: Application to Galectin-1. Journal of the American Society for Mass Spectrometry. 2008 doi: 10.1016/j.jasms.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldsmith SC, Guan JQ, Almo S, Chance M. Synchrotron protein footprinting: a technique to investigate protein-protein interactions. J Biomol Struct Dyn. 2001;19(3):405–418. doi: 10.1080/07391102.2001.10506750. [DOI] [PubMed] [Google Scholar]
- 20.Sharp JS, Sullivan DM, Cavanagh J, Tomer KB. Measurement of multisite oxidation kinetics reveals an active site conformational change in Spo0F as a result of protein oxidation. Biochemistry. 2006;45(20):6260–6266. doi: 10.1021/bi060470r. [DOI] [PubMed] [Google Scholar]
- 21.Sharp JS, Tomer KB. Analysis of the oxidative damage-induced conformational changes of apo- and holocalmodulin by dose-dependent protein oxidative surface mapping. Biophys J. 2007;92(5):1682–1692. doi: 10.1529/biophysj.106.099093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venkatesh S, Tomer KB, Sharp JS. Rapid identification of oxidation-induced conformational changes by kinetic analysis. Rapid Commun Mass Spectrom. 2007;21(23):3927–3936. doi: 10.1002/rcm.3291. [DOI] [PubMed] [Google Scholar]
- 23.Aye TT, Low TY, Sze SK. Nanosecond laser-induced photochemical oxidation method for protein surface mapping with mass spectrometry. Anal Chem. 2005;77(18):5814–5822. doi: 10.1021/ac050353m. [DOI] [PubMed] [Google Scholar]
- 24.Hambly D, Gross M. Laser flash photochemical oxidation to locate heme binding and conformational changes in myroglobin. International Journal of Mass Spectrometry. 2007;259(1–3):124–129. [Google Scholar]
- 25.Buxton GV, Cattell FCR, Dainton FS. Application of Time-Dependent Rate Constant Theory to Reactions of Solvated Electrons-Reaction Distances, Rate Constants and Diffusion-Coefficients in Concentrated Aqueous-Solutions. Journal of the Chemical Society-Faraday Transactions I. 1975;71(1):115–122. [Google Scholar]
- 26.Heyduk T, Baichoo N, Heyduk E. Hydroxyl radical footprinting of proteins using metal ion complexes. Met Ions Biol Syst. 2001;38:255–287. [PubMed] [Google Scholar]
- 27.Bridgewater JD, Vachet RW. Metal-catalyzed oxidation reactions and mass spectrometry: The roles of ascorbate and different oxidizing agents in determining Cu–protein-binding sites. Analytical Biochemistry. 2005;341(1):122–130. doi: 10.1016/j.ab.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 28.Bridgewater JD, Lim J, Vachet RW. Using Metal-Catalyzed Oxidation Reactions and Mass Spectrometry to Identify Amino Acid Residues Within 10 L of the Metal in Cu-Binding Proteins. Journal of the American Society for Mass Spectrometry. 2006;17(11):1552–1559. doi: 10.1016/j.jasms.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Xu G, Kiselar J, He Q, Chance MR. Secondary reactions and strategies to improve quantitative protein footprinting. Anal Chem. 2005;77(10):3029–3037. doi: 10.1021/ac048282z. [DOI] [PubMed] [Google Scholar]
- 30.Qin BY, Bewley MC, Creamer LK, Baker HM, Baker EN, Jameson GB. Structural basis of the tanford transition of bovine beta-lactoglobulin. Biochemistry. 1998;37(40):14014–14023. doi: 10.1021/bi981016t. [DOI] [PubMed] [Google Scholar]
- 31.Zheng X, Wintrode P, Chance M. Complementary Structural Mass Spectrometry Techniques Reveal Local Dynamics in Functionally Important Regions of a Metastable Serpin. Structure. 2008;16(1):38–51. doi: 10.1016/j.str.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 32.Zhuang T, Leffler H, Prestegard JH. Enhancement of bound-state residual dipolar couplings: Conformational analysis of lactose bound to Galectin-3. Protein Science. 2006;15(7):1780. doi: 10.1110/ps.051994306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. Journal of Biomolecular NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 34.Klassen NV, Shortt KR, Seuntjens J, Ross CK. Fricke dosimetry: the difference between G (Fe3+) for 60Co gamma-rays and high-energy x-rays. Physics in Medicine and Biology. 1999;44:1609–1624. doi: 10.1088/0031-9155/44/7/303. [DOI] [PubMed] [Google Scholar]
- 35.Janik I, Bartels DM, Jonah CD. Hydroxyl Radical Self-Recombination Reaction and Absorption Spectrum in Water Up to 350 C. J. Phys. Chem. A. 2007;111(10):1835–1843. doi: 10.1021/jp065992v. [DOI] [PubMed] [Google Scholar]
- 36.Nielsen SO, Michael BD, Hart EJ. Ultraviolet Absorption Spectra of eaq-, H, OH, D, and OD from Pulse Radiolysis of Aqueous Solutions. Journal of Physical Chemistry. 1976;80(22):2482–2488. [Google Scholar]
- 37.Weatherly DB, Atwood JA, Minning TA, Cavola C, Tarleton RL, Orlando R. A Heuristic Method for Assigning a False-discovery Rate for Protein Identifications from Mascot Database Search Results*. Molecular & Cellular Proteomics. 2005;4(6):762–772. doi: 10.1074/mcp.M400215-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Bern M, Cai Y, Goldberg D. Lookup peaks: a hybrid of de novo sequencing and database search for protein identification by tandem mass spectrometry. Anal. Chem. 2007;79(4):1393–1400. doi: 10.1021/ac0617013. [DOI] [PubMed] [Google Scholar]
- 39.Qin BY, Bewley MC, Creamer LK, Baker HM, Baker EN, Jameson GB. Structural Basis of the Tanford Transition of Bovine Beta-lactoglobulin. Biochemistry. 1998;37(40):14014–14023. doi: 10.1021/bi981016t. [DOI] [PubMed] [Google Scholar]
- 40.Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, Fagan P, Marvin J, Padilla D, Ravichandran V, Schneider B, Thanki N, Weissig H, Westbrook JD, Zardecki C. The Protein Data Bank. Acta Crystallogr. D. 2002;58(Pt 6 No 1):899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 41.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Wang B, Pearlman DA, Crowley M, Brozell S, Tsui V, Gohlke H, Mongan J, Hornak V, Cui G, Beroza P, Schafmeister C, Caldwell JW, Ross WS, Kollman PA. AMBER 8. San Francisco: University of California; 2004. [Google Scholar]
- 42.Wang J, Cieplak P, Kollman PA. How Well Does a Restrained Electrostatic Potential (RESP) Model Perform in Calculating Conformational Energies of Organic and Biological Molecules? J. Comput. Chem. 2000;21(12):1049–1074. [Google Scholar]
- 43.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Phys. Chem. 1983;79:926–935. [Google Scholar]
- Group W. GLYCAM Web [Google Scholar]
- 45.Ryckaert J-P, Ciccotti G, Berendsen HJ. Numerical Integration of the Cartesian Equations of Motion of a System with Constraints: Molecular Dynamics of n-Alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 46.Hubbard SJ, Thornton JM. NACCESS 2.1.1. London: University College London; 1993. [Google Scholar]
- 47.Cabelli DE. The Reactions of HO2/O2- Radicals in Aqueous Solution. In: Alfassi ZB, editor. Peroxyl Radicals. New York: John Wiley and Sons Ltd; 1997. pp. 407–437. [Google Scholar]
- 48.Bielski BHJ, Shiue GG. Oxygen Free Radicals and Tissue Damage. Amsterdam: Excerpta Medica; 1979. Reacion rate of superoxide radicals with the essential aminoacids; pp. 43–56. [Google Scholar]
- 49.Bielski BHJ, Allen AO. Mechanism of Disproportionation of Superoxide Radicals. Journal of Physical Chemistry. 1977;81(11):1048–1050. [Google Scholar]
- 50.Bielski BHJ, Cabelli DE, Arudi RL, Ross AB. Reactivity Of Ho2/O-2 Radicals In Aqueous-Solution. Journal Of Physical And Chemical Reference Data. 1985;14(4):1041–1100. [Google Scholar]
- 51.Buxton GV, Greenstock CL, Helman WP, Ross AB. Critical-Review of Rate Constants for Reactions of Hydrated Electrons, Hydrogen-Atoms and Hydroxyl Radicals (.Oh/.O-) in Aqueous-Solution. Journal of Physical and Chemical Reference Data. 1988;17(2):513–886. [Google Scholar]
- 52.von Sonntag C. The Chemical Basis of Radiation Biology. London: Tylor & Francis; 1987. Amino acids, oligopeptides and proteins; pp. 394–457. [Google Scholar]
- 53.Bobrowski K, Houee-Levin C, Marciniak B. Stabilization and reactions of sulfur radical cations: Relevance to one-electron oxidation of methionine in peptides and proteins. Chimia. 2008;62(9):728–734. [Google Scholar]
- 54.von Sonntag C, Schuchmann HP. Peroxyl Radicals in Aqueous Solutions. In: Alfassi ZB, editor. Peroxyl Radicals. New York: John Willey & Sons; 1997. pp. 173–234. [Google Scholar]
- 55.Janik I, Ulanski P, Rosiak JM, von Sonntag C. Hydroxyl-radical-induced reactions of the poly(vinyl methyl ether) model 2,4-dimethoxypentane in the absence and presence of dioxygen: a pulse radiolysis and product study. Journal of the Chemical Society-Perkin Transactions 2. 2000;10:2034–2040. [Google Scholar]
- 56.Tong X, Wren JC, Konermann L. Effects of Protein Concentration on the Extent of-Ray-Mediated Oxidative Labeling Studied by Electrospray Mass Spectrometry. Anal. Chem. 2007;79(16):6376–6382. doi: 10.1021/ac070724u. [DOI] [PubMed] [Google Scholar]
- 57.Harvey AH, Gallagher JS, Sengers J. Revised formulation for the refractive index of water and steam as a function of wavelength, temperature and density. Journal of Physical and Chemical Reference Data. 1998;27(4):761–774. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.