Abstract
The recently identified RNF125, or TRAC-1, is unique among ubiquitin ligases in being a positive regulator of T cell activation. In addition, TRAC-1 has been shown to down-modulate HIV replication and to inhibit pathogen-induced cytokine production. However, apart from the presence of an N-terminal C3HC4 RING domain, the TRAC-1 protein remains uncharacterized. Here, we report novel interactions and modifications for TRAC-1, and elucidate its domain organisation. Specifically, we determine that TRAC-1 associates with membranes and is excluded from the nucleus through myristoylation. Our data are further consistent with a crucial role for the C-terminus in TRAC-1 function. In this region, novel domains were recognized through the identification of three closely related proteins: RNF114, RNF138 and RNF166. TRAC-1 and its relatives were found to contain, apart from the RING domain, a C2HC and two C2H2 type zinc fingers, as well as an ubiquitin interacting motif (UIM). The UIM of TRAC-1 binds K48-linked poly-ubiquitin chains and is, together with the RING domain, required for auto-ubiquitination. As a consequence of auto-ubiquitination, the half-life of TRAC-1 is shorter than 30 minutes. The identifications of these novel modifications, interactions, domains and relatives, significantly widen the contexts for investigating TRAC-1 activity and regulation.
Keywords: RNF125/TRAC-1, ubiquitin ligase, T cell activation, myristoylation, zinc fingers, ubiquitin interacting motif
INTRODUCTION
The activation of T cells by their ligands, MHC/peptide complexes, is essential for the initiation of an adaptive immune response. The multitude of signaling pathways that are initiated from the T cell receptor (TCR) need careful regulation to ensure that an appropriate response, which can vary from T cell activation through anergy induction to apoptosis, is induced. Recently, it has become clear that ubiquitination, the attachment of the 76 amino acid ubiquitin to target proteins, is crucially involved in the regulation of T cell functions.
Ubiquitination has long been known to target damaged proteins for degradation, but is now also recognized to modulate the function, localization and interactions of target proteins [1-4]. Like phosphorylation, ubiquitination is a common regulatory modification that is rapidly inducible and reversible. The modification of proteins by ubiquitination requires the sequential activity of three classes of proteins [5]. An ubiquitin-activating enzyme, or E1, binds ubiquitin and transfers it to the active site of an ubiquitin-conjugating enzyme, or E2. E2 proteins interact with the ubiquitin ligases, or E3s, and promote the attachment of ubiquitin to substrates of these proteins. Ubiquitin is attached primarily to lysine residues on target proteins and subsequently, poly-ubiquitin chains can be formed. The presence of four or more ubiquitin proteins linked to each other via lysine 48, serves as a signal for degradation by the proteasome. However, a poly-ubiquitin chain linked via alternative lysines, as well as mono-ubiquitination, modulate the activity of substrates in a degradation-independent manner [6-8].
The ubiquitin ligases are central to ubiquitination by selecting substrates and interacting with E2 proteins. These E3s can be recognized by the presence of an E2 interacting domain, of which the RING, HECT and U-box domains are best characterized [9, 10]. In agreement with their role in recruiting selected sets of substrates, the ubiquitin ligases comprise a large and heterogeneous group. It is estimated that >400 are encoded in mammalian genomes [11], the majority of which remain to be characterized.
Given its versatility, ubiquitination can be predicted to impinge on many aspects of T cell function. So far, a role for this modification has been demonstrated in the internalization and degradation of the TCR [12], the activation of NF-κB [13], and the down-modulation of critical enzymes such as ZAP-70, protein kinase C-theta, phospholipase-C-gamma and extracellular signal regulated kinase (ERK)1/2 [14-16]. Furthermore, we, and others have shown the selective ubiquitination of active forms of Lck, a Src-related tyrosine kinase that is essential for T cell activation [17, 18]. The importance of ubiquitination in T cell function is also convincingly illustrated by the critical roles that have emerged for several ubiquitin ligases, such as c-Cbl, Cbl-b, Itch, GRAIL and Roquin (reviewed in [19-22]). For instance, disruptions of the genes for Cbl-b [23, 24] and Roquin [25] in mice leads to an enhanced susceptibility to autoimmune diseases, whereas the absence of c-Cbl results in defects in T cell development [26, 27]. In itchy mice, the gene for Itch is disrupted as the result of a naturally occurring mutation, which causes a wide spectrum of immune and inflammatory disorders [28]. GRAIL is essential for the induction of T cell anergy [29, 30], or unresponsiveness, which is crucial for peripheral tolerance. Although these proteins act via widely differing mechanisms, they share a negative regulatory effect on T cell activation.
The RING domain protein TRAC-1 (T cell RING protein in activation) was identified in a functional screen for T cell regulators [31]. This protein stands out in two major respects from the ubiquitin ligases with functions in T cells described above. First, TRAC-1 is predominantly expressed in lymphoid cells [31], in contrast to the ubiquitous expression of the other proteins. Second, interfering with TRAC-1 expression by anti-sense DNA inhibited T cell activation [32], indicating that unlike the other ligases, TRAC-1 is a positive T cell regulator.
However, recent reports have demonstrated inhibitory activities for TRAC-1 in other processes. TRAC-1 was found to ubiquitinate and down-modulate retinoic acid-inducible gene (RIG-I) [33], a protein that detects viral dsDNA and induces the production of cytokines including type I interferons (IFNs). TRAC-1 was shown to be upregulated in response to IFN and was postulated to provide a negative feedback loop for cytokine production. In another study, TRAC-1 was reported to reduce HIV replication [34]. TRAC-1 was found to be upregulated in IL-4 stimulated CD4+CD38− cells in which HIV replicates with low efficiency relative to CD4+CD38+ cells, and a negative effect of TRAC-1 on HIV transcription was demostrated.
Despite these various important roles, little is known about the TRAC-1 protein and its regulation. Although it is clear that the N-terminal C3HC4 RING domain is important for ubiquitin ligase activity, as it is for other ubiquitin ligases, dominant negative effects of truncated TRAC-1 suggested additional, yet undefined roles for the C-terminal region [32]. This study characterizes the TRAC-1 protein in some detail. Two novel modifications for TRAC-1 are demonstrated, myristoylation and auto-ubiquitination, which are important for membrane-association and stability of TRAC-1, respectively. Furthermore, in the C-terminal region of TRAC-1 the presence of three zinc binding and one ubiquitin-interacting motif are identified. These features permit TRAC-1 to be recognized as belonging to a new subfamily of RING ubiquitin ligases, together with RNF114, RNF138 and RNF166. These findings considerably widen the scope for studying the various functions of TRAC-1 and that of its relatives.
EXPERIMENTAL
Reagents
Chemicals were from Sigma-Aldrich unless otherwise indicated.
cDNA
TRAC-1 cDNA was amplified from cDNA of human primary T cells using primers based on mRNA sequence NM_017831. TRAC-1 cDNA was subcloned into pcDNA3.1A(-) (Invitrogen), pEGFP-N3 (Invitrogen) and PGEX-5X2 (Amersham Pharmacia Biotech) to express, respectively, proteins with C-terminal myc/His, GFP or GST tags. For yeast-two-hybrid experiments, constructs were cloned into pGBKT7 (Clontech) to express GAL4 binding domain fusion proteins, and into a pVP16-derived plasmid [35] to express GAL4 activation domain fusion proteins. The mutants C37,40A-TRAC-1, G2A-TRAC-1 and V217P,S221Q-TRAC-1 were generated by site-directed mutagenesis. The truncated proteins TRAC-1Δ170 and TRAC-1Δ205 were made by PCR using primers that introduce stopcodons at positions 171 and 206, respectively. GST/TRAC-UIM was generated by subcloning a cDNA fragment encoding amino acids 181-232, generated by PCR, in frame with GST in PGEX-5X2. GST/V/S-UIM was made following amplification of a similar fragment from V217P,S221Q-TRAC-1 cDNA. All TRAC-1 constructs were verified by sequencing. The generation of UD-GFP cDNA has been described previously [36]. HA-ubiquitin cDNA was a gift from M. Treier (EMBL, Heidelberg, Germany), and UbcH5a cDNA from M. Malim (King’s College London).
Cells and transfections
Jurkat T cells were maintained at 37°C, 5% CO2 in RPMI (Cambrex) supplemented with 10% FCS (Helena Biosciences), 100 U/ml penicillin, 0.1 mg/ml streptomycin (pen/strep, Gibco Ltd.). Primary human T cells were enriched from buffy-coats by Lymphoprep (Axis-Shield) gradients and subsequent purification through a nylon wool (Kisker) column. Typically, 90% pure T cell populations were obtained, measured by anti-CD3ε staining and FACS analysis. Isolated T cells were maintained in RPMI, 10% FCS, pen/strep and used for experiments within 24 h of isolation. COS-7, HeLa and 293T human embryonic kidney cells (HEK293T) were grown in DMEM (Cambrex), 10%FCS, pen/strep. For transfection, 107 Jurkat cells were electroporated in 250 μl RPMI, 10% FCS, using a BioRad Gene Pulser at 250 V, 975 μF in the presence of 20 μg DNA. Primary T cells were transfected at 2 × 106 in 100 μl human T cell Nucleofector solution (Amaxa biosystems) with 5 μg DNA, using program U-14 of the Amaxa Nucleofector. HEK293T cells were transfected using the calcium phosphate method with 15 μg DNA per 10 cm tissue culture dish. COS-7 cells were transfected by nucleofection (Amaxa biosystems), and HeLa cells using Fugene (Roche Applied Science). For inhibition of proteasomal degradation, cells were cultured in the presence of 50 μM MG-132 (Calbiochem), added from a 50 mM stock in DMSO. The half-life of TRAC-1 proteins was determined by exposing equal numbers of cells to 100 μg/ml cycloheximide (Sigma) for various times.
Antibodies
To detect tagged TRAC-1 proteins, anti-myc 9E10 (Sigma), anti-GFP (Roche Applied Science) and anti-GST (Amersham Pharmacia Biotech) mAbs were used. Ubiquitination was detected with Covance mAbs P4G7 against Ubiquitin or mAb HA.11 against the haemagglutinin tag. The anti-Transferrin Receptor mAb H68.4 was from Zymed, anti-His rabbit polyclonal from Qiagen, rabbit anti-GM130 (ab40881) from Abcam, rabbit polyclonal C-15 against c-Cbl and H-114 against Hsp90α/β from Santa Cruz Biotechnology. To stimulate T cells, anti-CD3 mAb UCHT1 and anti-CD28 mAb ANC28.1 (Ancell) were used. APC-conjugated anti-CD69 was from BD Pharmingen. The peroxidase-conjugated goat anti-rabbit and rabbit anti-mouse antibodies were from Perbio Science, as was the goat anti-mouse rhodamine-conjugated antibody.
T cell activation and FACS analysis
At 8 - 16 h post-transfection, primary human T cells, or Jurkat T cells, were stimulated with plate-bound anti-CD3 and anti-CD28 antibodies (10 μg/ml each). Cells were activated for 18 h and subsequently stained with APC-conjugated anti-CD69 in 2% FCS/PBS on ice for 30 min. Following fixation in 3% paraformaldehyde, a minimum of 105 cell were acquired by FACS using a FACScalibur (Becton Dickinson), and gated on healthy cells based on forward and side scatter properties (typically 50% cell death after transfection). Instrument settings for acquisition were optimized using unstained, GFP-positive and CD69-APC stained, GFP-negative cells. Analysis was performed using CellQuest (Becton Dickinson) software. Dot blot quadrants were set so that <1% of untransfected controls were present in the GFP+ quadrants and <1% of unactivated controls in the CD69+ quadrants. For each transfection, the percentage of CD69+ cells was calculated for the GFP− and GFP+ cell populations. Thus, %UL/(UL+LL) gave the %CD69+ cells for the GFP− cells, and %UR/(UR+LR) for the GFP+ cells, with UL: upper left, LL: lower left, UR: upper right, LR: lower right quadrant.
IL-2 ELISA
The concentration of IL-2 in the culture supernatants of stimulated and sorted GFP+ Jurkat cells was determined using an IL-2 ELISA kit (eBiosciene) according to the manufacturer’s instructions. Culture medium was used as blank and all samples were analyzed in duplicates. A standard curve was made using purified IL-2.
Immunoprecipitation and Western Blotting
Cells were lysed in NP-40 buffer (1% NonidetP-40, 20 mM Tris pH 7.8, 150 mM NaCl, 2 mM MgCl2), supplemented with the protease inhibitors phenylmethylsulfonylfluoride (PMSF, 1 mM) and CLAP (5 μg/ml each of chymostatin, pepstatin A, antipain hydrochloride and 10 μg/ml leupeptin hemisulphate), and left on ice for 30 min. For the detection of ubiquitinated proteins, the lysis buffer was supplemented with 0.1% SDS and 5 mM N-ethylmaleimide (NEM). Nuclei and cell debris were removed by centrifugation at 13,000 × g for 5 min at 4°C. For direct analysis, lysates were denatured in reducing SDS-sample buffer (5% 2-ME), for 5 min. at 95°C, and loaded on SDS-polyacrylamide gels. Immunoprecipitations, SDS-PAGE and Western blotting were performed as described before [17]. Densitometry of blots was performed using BioRad Quantity One software. The density of TRAC-1 bands was determined following subtraction of background in an equal surface area.
In vitro ubiquitination assays
E.coli strain BL21DE3(pLysS) (Novagen) was transformed with TRAC-1 constructs cloned into PGEX-5X2 to allow production of TRAC-1/GST fusion proteins. Fusion proteins were purified on glutathione-sepharose beads (Amersham Pharmacia Biotech) after lysis of bacteria in the presence of 1% Sarkosyl, according to the manufacturer’s instructions. For the ubiquitination assays, 10 μl GST-beads (~2 μg of protein) were added to 50 μl of ubiquitination buffer (50 mM Tris, pH 7.4; 2.5 mM MgCl2; 0.5 mM dithiothreitol [DTT]; 10 mM ATP) together with 50 ng E1, 2 μg ubiquitin, 1 μg E2 and incubated for 90 min at 30 °C. Ubiquitin, E1 and an E2 sampler pack containing UbcH1/2/3/5a/5b/5c/6/7/9/10 (UW8975) were purchased from Biomol.
Yeast two hybrid experiments
To measure interactions between TRAC-1 proteins and UbcH5a, Y190 cells were cotransformed with combinations of constructs cloned into pGBKT7 and pVP16-derived plasmids. Transformants were selected by growth on media lacking tryptophan and leucine, and β-galactosidase activity was measured in three pools of transformants using Chlorophenol red-β-D-galactopyranoside (CPRG, Roche Applied Science) as substrate.
3H-Myristate labeling
HEK293T cells were labeled 24 h after transfection with [9,10-3H]myristic acid as described before [37]. Briefly, cells were pre-incubated in labeling medium (DMEM, 2% FCS, 5 mM sodium pyruvate [Gibco Ltd.], and 4x non-essential amino acids [Gibco Ltd.]) for 60 min at 37°C, 5% CO2. [9,10-3H]myristic acid in ethanol (10-60 Ci/mmol; DuPont) was dried under nitrogen and taken up in labeling medium at 125 μCi/ml. Cells were labeled with 250 μCi per 10 cm plate for 4 h at 37°C, 5% CO2. After labeling, cells were processed for immunoprecipitation and SDS-PAGE. Proteins were then transferred to nitrocellulose membranes, and visualized by autoradiography using Kodak Biomax MS film and a Biomax Transcreen-LE (Kodak) intensifying screen, or by Western blotting.
Membrane flotation assays
HEK293T or Jurkat cells were harvested 24 h post-transfection, and postnuclear supernatants (PNSs) were generated by sonicating the cells in hypotonic buffer (20 mM Tris pH 7.8, PMSF, CLAP) and pelleting nuclei at 1000 g. PNSs were adjusted to 70% sucrose (w/v) in 2.5 ml, and overlaid with 6 ml of 65% (w/v) and 2.5 ml of 10% (w/v) sucrose according to [38]. Gradients were centrifuged at 120,000 g for 18 h, following which eleven 1 ml fractions were collected. Aliquots of each fraction were analyzed by SDS-PAGE and immunoblotting.
Immunofluorescence
Cells on 13 mm diameter glass coverslips were fixed with 3% paraformaldehyde 24 h after transfection, permeabilized in 0.1% Triton TX-100/PBS and stained as previously described [36]. Coverslips were mounted in Mowiol (Calbiochem-Novachem) and observed by a Leica TCP SP2 AOBSTM confocal microscope. Cross correlation values were determined on an overall pixel scale by thresholding out the area of low intensities (<15%) [39] using TRI2 software version 2.2.2.1 (Gray Cancer Institute, Northwood, UK).
Ubiqutin pulldown with recombinant GST fusion proteins
GST, GST/TRAC-UIM or GST/V/S-UIM were purified from 10 ml cultures of transformed E. coli BL21DE3(pLysS) with 50 μl glutathione-sepharose beads. 20 μl beads were incubated overnight at 4°C with NP-40 lysates of HEK293T cells transfected with HA-ubiquitin (1/3 of a 10 cm dish per recombinant protein). Beads were washed three times with NP-40 lysis buffer, heated to 65°C in SDS-sample buffer and analysed by SDS-PAGE and Western blotting. To detect binding of purified poly-ubiquitin, 20 μl beads were each incubated with 1 μg K48-linked poly-ubiquitin2-7 (Biomol) in PBS/0.1% BSA and analysed as above.
RESULTS
The C-terminal region of TRAC-1 is important for its function in T cells
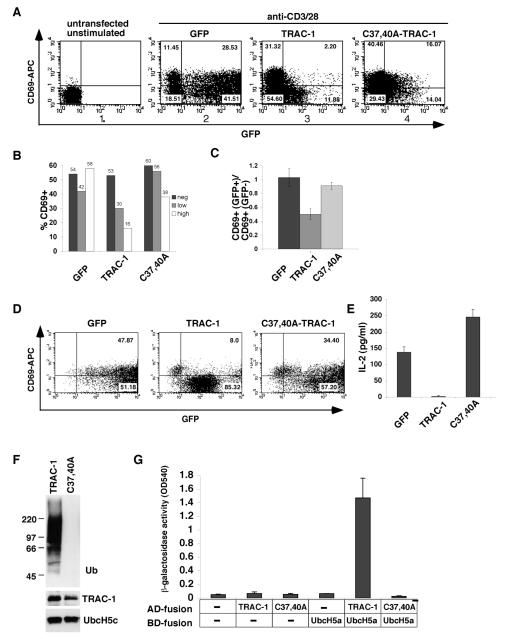
To study the activity of TRAC-1, a construct for a C-terminally GFP-tagged TRAC-1 fusion protein (TRAC-1/GFP) was generated and expressed in primary T cells by nucleofection. The cells were stimulated with anti-CD3 and anti-CD28 and stained for the activation-marker CD69. Analysis of FACS plots showed that TRAC-1/GFP-positive cells to a large extent failed to upregulate CD69, compared to TRAC-1/GFP-negative cells within the same transfection (Fig. 1A, panel 3). Separate analysis of GFP-negative, GFP-low and GFP-high cells from this transfection, showed a dose-dependent effect of TRAC-1/GFP levels on CD69 expression (Fig. 1B), with a 70% decrease observed for GFP-high cells (16% CD69+ cells in the GFP-high versus 53% in the GFP-negative population). On average the expression of TRAC-1/GFP resulted in a ~50% reduction in the induction of CD69 after activation, compared to GFP expressing cells (Fig. 1C). This negative effect was observed in primary as well as in Jurkat T cells (Fig. 1D). TRAC-1/GFP also drastically reduced the induction of another T cell activation marker, the secretion of IL-2 (Fig. 1E). Thus, similar to results previously reported for TRAC-1ΔC, a protein truncated at residue 170, TRAC-1/GFP has a dominant inhibitory effect on T cell activation. This suggests an important role for the C-terminal region of TRAC-1, which is missing in TRAC-1ΔC and may be inaccessible in TRAC-1/GFP owing to the presence of a GFP protein that is similar in size to TRAC-1 itself.
Figure 1. A TRAC-1/GFP fusion protein inhibits T cell activation.
A. Human primary T cells were nucleofected with peGFP-N3 (GFP, panel 2), or with peGFP-N3 containing TRAC-1 (panel 3) or C37,40A-TRAC-1 (panel 4) cDNA to express C-terminally tagged GFP fusion proteins. 16 h after transfection, cells were stimulated with plate-bound anti-CD3/28 for 18 h. Untransfected, unstimulated cells were included as controls (panel 1). Cells were stained with anti-CD69-APC, acquired by FACS and gated on live cells. The percentage of cells in each quadrant is indicated.
B. The percentage of CD69+ cells was calculated separately for GFP-negative (fluorescence intensity 0-10), GFP-low (fluorescence intensity 10-100) and GFP-high (fluorescence intensity >100) populations of the experiment in (A).
C. Six experiments were performed as in A. For each transfection, the percentage of CD69+ cells in the GFP+ population was divided by that in the GFP− population (see materials and methods). The averages of these ratios are represented for each of the transfected constructs.
D. Jurkat T cells were transfected with GFP, TRAC-1/GFP or C37,40A-TRAC-1/GFP expressing constructs. 24 h after transfection, GFP+ cells were sorted by MoFlo® and stimulated with plate-bound anti-CD3/CD28 for 48 h. FACS plots of cells stained with anti-CD69-APC are shown.
E. The culture supernatants of sorted Jurkat cells in (D) were analyzed for the presence of IL-2 by ELISA and represented in a graph (right). A representative of one out of two experiments is shown.
F. Bacterially expressed GST fusion proteins of TRAC-1 and C37,40A-TRAC-1 were captured on glutathione-sepharose beads and incubated for 90 min at 30°C with purified E1, His-tagged E2 UbcH5c, ubiquitin and ATP. After the reaction, beads were analyzed by Western blotting with antibodies against ubiquitin to detect ubiquitin ligase activity (top). The reaction mixture was blotted with anti-GST (middle) and anti-His (bottom) to detect the presence of TRAC-1 proteins and UbcH5c, respectively.
G. Yeast was transformed with GAL4 binding domain (BD) and activation domain (AD) containing vectors, either without insert (-) or with TRAC-1 (wildtype or the C37,40A mutant) or UbcH5a as inserts, as indicated. An interaction between proteins results in β-galactosidase activity and was measured with CPRG as substrate.
For TRAC-1ΔC, mutations in the RING domain were shown to abolish its inhibitory activity [32]. To determine whether this is also the case for TRAC-1/GFP, two of the structurally important Zn2+ chelating cysteines, Cys37 and Cys40, were substituted by alanines (see Fig. 4A for TRAC-1 amino acid sequence). Expression of C37,40A-TRAC-1/GFP in T cells showed that, in contrast to TRAC-1/GFP, this protein did not to any considerable extent inhibit T cell activation as measured by the upregulation of CD69 (Fig. 1A, panel 4; B-D). Similarly, C37,40A-TRAC-1/GFP did not inhibit the production of IL-2 (Fig. 1E). Rather, an increase in IL-2 levels was observed, which may suggest that following T cell activation TRAC-1 has a dampening effect on IL-2 synthesis. The lack of ubiquitin ligase activity for the RING domain mutant C37,40A-TRAC-1 was verified by in vitro auto-ubiquitination experiments (Fig. 1F). Inititial screening of ten different E2 proteins showed that TRAC-1 cooperates with members of the UbcH5 family, similar to the preference reported previously [32]. An inability of the C37,40A-TRAC-1 mutant to associate with UbcH5a was further demonstrated in yeast-two-hybrid experiments (Fig. 1G). Together, these experiments demonstrate that altering the C-terminus of TRAC-1 provokes a RING domain-dependent dominant inhibitory activity. To better understand the normal activity of TRAC-1, we next investigated its modifications, subcellular localisation and the presence of domains other than the RING domain.
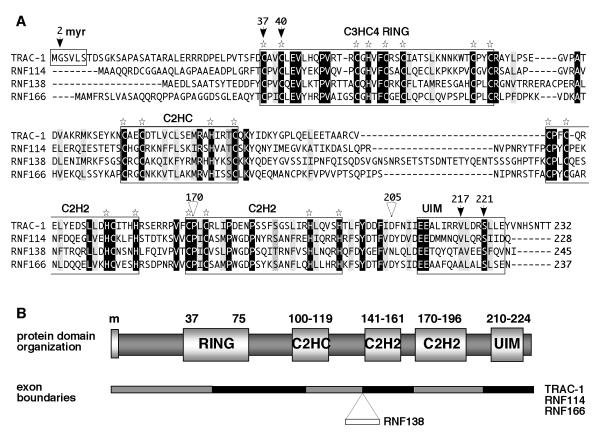
Figure 4. TRAC-1 is homologous to three other small C3HC4 RING proteins.
A. Amino acid alignments of the human proteins TRAC-1 (Q96EQ8), RNF114 (Q9Y508), RNF138 (Q8WVD3) and RNF166 (Q96A37). Alignments were performed using vector NTI software (Invitrogen). Identical residues are highlighted in black, conserved hydrophobic resides in light grey, other conservative changes in darker grey. Star symbols indicate the conserved Cys and His residues in the various domains. The arrows indicate amino acids in TRAC-1 that have been mutated in this study and the triangles show the positions where truncations have been made.
B. Schematic diagram of the domain composition of TRAC-1 and its family members. Indicated are the myristoylation site (m, present in TRAC-1 only) the RING domain, the newly identified C2HC and C2H2 zinc binding motifs and the UIM domain. The numbers refer to amino acid positions in TRAC-1. The gene composition with the exon boundaries relative to the protein domains is depicted below. RNF138 contains one extra exon compared to the other three proteins.
Myristoylation and localisation of TRAC-1
A myristoylation consensus sequence, (M)GXXT/S, is present at the extreme N-terminus (Fig. 4A), which predicts that TRAC-1 is modified by the attachment of myristic acid to Gly2, following the proteolytic removal of the methionine [40, 41]. Myristoylation is a cotranslational irreversible acylation that has profound effects on the localisation and activity of proteins by enhancing the affinity for membranes and influencing protein-protein interactions.
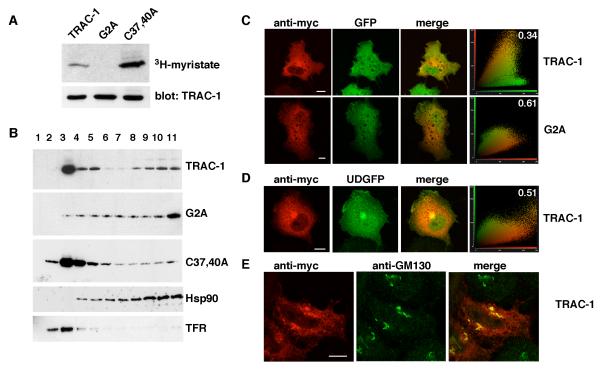
To investigate whether TRAC-1 is myristoylated, HEK293T cells were transfected with TRAC-1/myc constructs and labeled with 3H-myristic acid. Following immunoprecipitation, incorporation of 3H-myristate could be detected for both wildtype TRAC-1 and the RING deficient C37,40A-TRAC-1 (Fig. 2A). Substitution of Gly2 with Ala in the G2A-TRAC-1 mutant prevented this labeling, demonstrating that Gly2 is used as a myristoylation site (Fig. 2A). The higher intensity of 3H-myristic acid labelling for C37,40A-TRAC-1 compared to wildtype TRAC-1 most likely reflects the considerably longer half-life of the mutant (see below), which leads to greater accumulation of protein during the 4 hour labeling.
Figure 2. TRAC-1 is myristoylated and partially membrane-associated.
A. HEK293T cells were transiently transfected with TRAC-1/myc constructs and grown in the presence of 3H-myristic acid for 4 hours. TRAC-1/myc proteins were immunoprecipitated with anti-myc antibodies. One half of the sample was analyzed by SDS-PAGE and autoradiography (top), the other half by immunoblotting with anti-myc antibody (bottom).
B. HEK293T cells transfected with TRAC-1/myc constructs were homogenized in hypotonic buffer and fractionated on discontinuous 70%/65%/10% sucrose gradients. Fractions were collected from the top (fraction 1), and aliquots were analyzed by immunoblotting with anti-myc antibodies to detect TRAC-1 proteins, as well as with antibodies against Hsp90, a soluble protein, and Transferrin Receptor (TFR), a transmembrane protein.
C. COS-7 cells transfected with GFP and either TRAC-1/myc or G2A-TRAC-1/myc cDNAs were grown on 13 mm glass coverslips. 24 hours after transfection, cells were permeabilized, fixed, and stained with anti-myc as primary and rhodamine-conjugated goat-anti-mouse as secondary antibody. Confocal images of single channels as well as overlap images are shown. Scale bar: 10 μm. A cytofluorogram (right panels) is represented to show the intensities of red and green fluorophores per pixel. The relative amount of colocalization was determined using cross correlation analysis and the values (with 0 representing no colocalization, and 1 complete colocalization) are shown in the cytofluorgram.
D. COS-7 cells were transfected with TRAC-1/myc and a plasmid for UD-GFP, a myristoylated and palmitoylated protein that contains the unique domain of Lck fused to the N-terminus of GFP. Cells were stained, observed and analyzed as in (C). Scale bar: 10 μm.
E. HeLa cells were transfected with TRAC-1/myc and stained with anti-myc and anti-GM130, a cis-Golgi marker. Scale bar: 10 μm.
To examine whether TRAC-1 is membrane-associated as a result of myristoylation, membrane flotation experiments were used. HEK293T cells were transfected with TRAC-1/myc constructs and homogenized in isotonic buffer in the absence of detergent. The homogenate was loaded at the bottom of discontinuous 70%/65%/10% sucrose gradients, from which fractions were collected after centrifugation. Membranes and associated proteins are expected to float to the 65%/10% interface at the top of these gradients, which was confirmed by Western blotting for the Transferrin receptor (Fig. 2B). Soluble proteins are excluded from these fractions, as demonstrated with the cytosolic protein Hsp90. For TRAC-1, the majority of the protein was detected in the membrane fractions (Fig. 2B). This membrane-association was dependent on myristoylation, since the myristoylation-deficient G2A-TRAC-1 did not accumulate here (Fig. 2B). Like wildtype TRAC-1, the RING domain mutant C37,40A-TRAC-1 was primarily membrane-associated (Fig. 2B). Small amounts of soluble protein, consistently higher for TRAC-1 than for C37,40A-TRAC-1, were also observed, suggesting that the RING domain influences to some extent myristoylation and/or membrane association.
To study the subcellular localisation of TRAC-1 in more detail, confocal microscopy was used. Staining for TRAC-1/myc constructs after permeabilization showed that, consistent with the presence of a myristic acid, TRAC-1 was excluded from the nucleus (Figs. 2C, D and E). TRAC-1 showed a reticular staining with some concentration in a peri-nuclear region. This is reminiscent of the localisation of other myristoylated proteins [42], and most likely represents the distribution of TRAC-1 at various membrane compartments. In contrast, G2A-TRAC-1 was more homogeneously distributed and able to enter the nucleus. This mutant, but not wildtype TRAC-1, overlapped to a considerable extent with the soluble protein GFP (Fig. 2C).
Localisation of TRAC-1 at the plasma membrane was studied by the co-expression with UD-GFP (Fig. 2D), a myristoylated and palmitoylated protein composed of the unique domain of Lck fused to GFP that localizes to a large extent to the plasma membrane [36]. Very little, if any TRAC-1 co-localized with UD-GFP at the periphery of the cell, showing that TRAC-1 is not present at appreciable levels at the plasma membranee. However, some TRAC-1 colocalized with peri-nuclear UD-GFP, which was previously shown to associate with the Golgi complex [36]. Co-staining with the cis-Golgi marker GM130 confirmed the presence of TRAC-1 in the Golgi region (Fig. 2E). In conclusion, TRAC-1 associates with membranes as a result of myristoylation, and is primarily located at intracellular membrane systems.
TRAC-1 has a short half life as a result of auto-ubiquitination
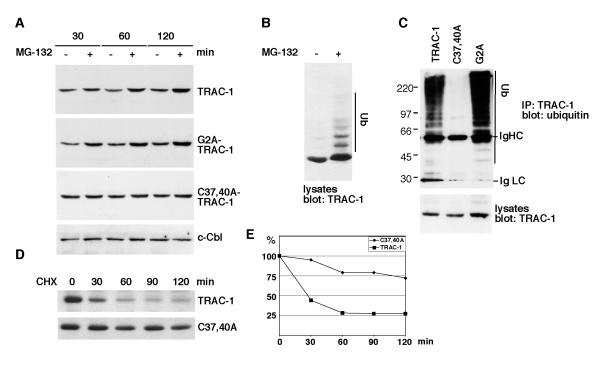
The levels of TRAC-1/GFP in T cells were found to be consistently lower than those of C37,40A-TRAC-1/GFP (see fluorescent intensities in Fig. 1A and D). This suggested that TRAC-1 with an intact RING domain might be targeted for proteasomal degradation. To test whether this is the case, transfected Jurkat T cells were incubated with the proteasome inhibitor MG-132. This treatment had no detectable effect on levels of C37,40A-TRAC-1/GFP, but those of TRAC-1/GFP and G2A-TRAC-1/GFP were increased after an incubation as short as 30 min (Fig. 3A). This effect was not generally observed for ubiquitin ligases with intact RING domains, since levels of c-Cbl were not affected by MG-132. Nor was it specific for TRAC-1/GFP constructs, as levels of TRAC-1/myc were increased in the presence of MG-132 as well. Moreover, higher molecular weight forms of TRAC-1 were detected in the presence of MG-132, indicative of its poly-ubiquitination (Fig. 3B). To investigate TRAC-1 ubiquitination, constructs were cotransfected with HA-ubiquitin, immunoprecipitated and blotted with anti-ubiquitin antibodies. Poly-ubiquitinated forms were detected for TRAC-1 and G2A-TRAC-1 but not for C37,40A-TRAC-1 (Fig. 3C). Therefore, it can be concluded that TRAC-1 is subject to auto-ubiquitination.
Figure 3. TRAC-1 is unstable as a result of auto-ubiquitination.
A. Jurkat T cells were transfected with the indicated TRAC-1/GFP constructs. The cells were either left untreated or were incubated with the proteasome inhibitor MG-132 (50 μM) for 30, 60 or 120 min. Equivalent amounts of cell lysates were analysed by Western blotting with anti-GFP to detect TRAC-1/GFP proteins, or with anti-c-Cbl.
B. HEK293T cells transfected with TRAC-1/myc cDNA were either left untreated or incubated with MG-132 (50 μM) for two hours. The cell lysates were blotted with anti-myc antibodies.
C. Plasmids for myc-tagged TRAC-1, C37,40A-TRAC-1 or G2A-TRAC-1 were cotransfected with HA-ubiquitin into HEK293T cells. TRAC-1 proteins were immunoprecipitated with anti-myc and blotted with anti-ubiquitin antibodies to detect ubiquitination (top). Lysates were blotted with anti-myc to detect the transfected TRAC-1 proteins (bottom).
D. COS-7 cells were transfected in suspension with myc-tagged TRAC-1 or C37,40A-TRAC-1 and equal cell numbers were divided over five dishes. Cells were incubated the next day with medium alone (0) or with medium containing cycloheximide (100 μg/ml) for the indicated times. Equal amounts of cell lysates were analyzed by Western blotting with anti-myc antibody.
E. Densitometry of the blots shown in D. For each time point, the density of the TRAC-1 band following subtraction of background levels and equalized for a loading control were determined. TRAC-1 levels are expressed as a percentage of levels in untreated cells.
To examine the effect of auto-ubiquitination on the half life of TRAC-1, cells transfected with TRAC-1 or C37,40A-TRAC-1 were incubated for various times with the protein synthesis inhibitor cycloheximide. These experiments revealed that TRAC-1 has a very short half-life of less than 30 min (Fig. 3D and E). In contrast, C37,40A-TRAC-1 levels were reduced by only 25% after a 2 hour cycloheximide incubation. Thus, TRAC-1 is an unstable protein and its rapid turnover rate is the result of auto-ubiquitination.
TRAC-1 is related to three other RING ubiquitin ligases
Protein domain searches did not recognize any domains C-terminal to the RING domain. However, amino acid alignments of TRAC-1 proteins from different species showed the presence of conserved cysteines and histidines in this region, particularly between residues 141 and 196. Protein blast searches with this part of TRAC-1 revealed that homologous sequences are present in four other human proteins: RNF114 (alternative name Zfp313), RNF138 (or NARF), RNF166 and potassium channel modulatory factor (Kcmf1). In three of these proteins, RNF114, RNF138 and RNF166, the conserved sequence is similarly located close to the C-terminus. Moreover, these three proteins have sizes comparable to the 232 amino acid TRAC-1, ranging from 228 amino acids for RNF114 to 245 amino acids for RNF138. Furthermore, like TRAC-1, these proteins contain a C3HC4 RING domain close to the N-terminus (Fig. 4A). In contrast, Kcmf1 is larger, the homologous sequence is located in the middle of the protein and a C3HC4 RING domain is not present [43, 44]. Therefore, we conclude that TRAC-1 is closely related to RNF138, RNF114 and RNF166 and that these proteins form a novel subfamily of small C3HC4 RING ubiquitin ligases.
The homologous C-terminal region contains two motifs that each correspond to the basic C2H2 zinc finger consensus sequence CX2-4C….HX2-4H [45]. The first “zinc finger like” motif has the conventional 12 amino acid spacing between cysteine and histidine pairs, except in TRAC-1 where only 11 residues are present (Fig. 4A). The consensus sequence for this motif is CPXCX2-3ΦX4ΦΦXHCX3C, with X meaning that any amino acid can be present, and Φ representing a hydrophobic residue. The second motif contains 17 amino acids between the cysteine and histidine pairs and has the consensus sequence CPΦCX2ΦPX3PX3T/SX2ΦX2HΦX3H. These zinc binding motifs diverge from the classical DNA binding zinc fingers by the spacing of the hydrophobic residues and the absence of conserved DNA binding Arg and Asp residues [45]. Thus, the TRAC-1 family motifs should be regarded as novel atypical zinc fingers. The overall similarity between TRAC-1 and its relatives in the region between residues 141 and 196 is 57% for RNF114 and RNF166 (with 29% and 38% identity, respectively) and 54% for RNF138 (36% identity). For comparison, the sequence similarity between C3HC4 RING domains is ~60%.
Further scrutiny of amino acid alignments identified another homologous region with conserved cysteines and histidines. This motif, located between amino acids 100 and 119 in TRAC-1, resembles a C2HC Zinc finger and has the consensus sequence CX2CX3ΦXΦX2ΦRXHX2T/SC (Fig. 4A). Additionally, a ubiquitin interacting motif (UIM) was detected by protein domain searches at the extreme C-terminus of RNF166 and some of the residues important for this domain are conserved in the other TRAC-1 family members (Fig. 4A).
The relationship between the TRAC-1 family members is also evident from the similarities in their gene structure. The genes for TRAC-1, RNF114 and RNF166 contain 6 exons, with exons 1 and 2 together encoding the RING domain, exons 2 and 3 the C2HC-like zinc finger, exon 4 the first C2H2-like zinc finger, exon 5 the second C2H2-like zinc finger, and exon 6 a UIM-type domain. The organisation of the RNF138 gene is the same, except for the presence of an extra exon, encoding 20 amino acids, between exons 3 and 4 (Fig. 4B). The exon boundaries correspond to similar amino acid positions in all four proteins. In summary, TRAC-1 belongs to a new subfamily of small C3HC4 RING ubiquitin ligases. The conservation of three zinc finger like domains and a UIM is consistent with important interactions occurring outside the RING domain.
TRAC-1 binds ubiquitin via a C-terminal UIM
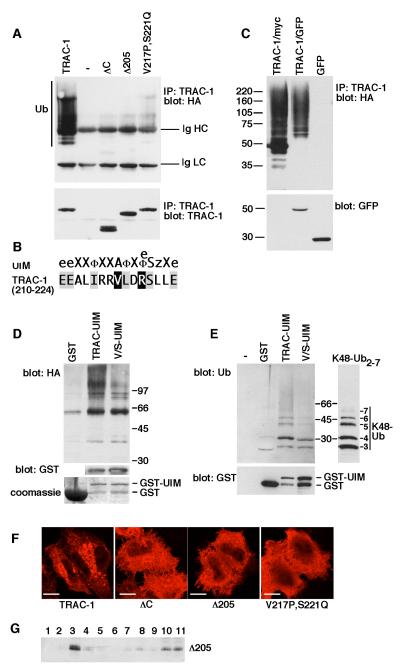
Whilst studying the ubiquitination of TRAC-1, it was observed that the C-terminally truncated TRAC-1ΔC with a deletion of amino acids 171-232 was not subject to auto-ubiquitination (Fig. 5A). This was unexpected since the absent region does not contain any lysines for the attachment of ubiquitin (see Fig. 4A for TRAC-1 amino acid sequence). Since RNF166 is predicted to have a UIM-type ubiquitin-binding domain, it was hypothesized that a similar domain is present in TRAC-1 and cooperates with the RING domain in auto-ubiquitination. The corresponding region of TRAC-1, amino acids 210 – 224, diverts from a consensus UIM motif in two places (Fig. 5B); a valine is present instead of an alanine (position 217), and a positively charged arginine instead of a hydrophobic or acidic amino acid (position 220).
Figure 5. TRAC-1 contains a UIM-type ubiquitin-binding domain that is required for auto-ubiquitination.
A. HEK293T cells were transfected with plasmids expressing HA-ubiquitin and either myc-tagged wildtype TRAC-1, empty vector (-), TRAC-1ΔC, TRAC-1Δ205 or V217P,S221Q-TRAC-1. TRAC-1 proteins were immunoprecipitated with anti-myc and blotted with anti-HA to detect ubiquitination (top). Ub: ubiquitinated TRAC-1; Ig HC and Ig LC: the heavy chain (HC) and the light chain (LC) of the immunoprecipitating anti-myc antibody, respectively. The same blot was re-probed with anti-myc antibody to detect expression of TRAC-1 proteins (bottom).
B. Comparison of the consensus sequence for the UIM domain [according to [47]] with the TRAC-1 sequence of amino acids 210 - 224. The notation is as follows: e, negatively charged; X, helix favouring; Φ, hydrophobic, z, bulky hydrophobic or polar amino acid.
C. HEK293T cells were transfected with plasmids expressing HA-ubiquitin together with either TRAC-1/myc, TRAC-1/GFP or GFP. Proteins were immunoprecipitated with anti-myc (lane 1) or anti-GFP (lanes 2 and 3) and blotted with anti-HA to detect ubiquitination (top). The lysates were blotted for GFP (bottom).
D. GST fusion proteins were isolated on glutathione-sepharose beads and incubated with lysates from HA-ubiquitin transfected HEK293T cells. Beads were analysed by western blotting with anti-HA antibodies to detect the binding of ubiquitinated proteins to the recombinant proteins (top). TRAC-UIM: amino acids 181 – 232 of TRAC-1 fused to the C-terminus of GST; V/S-UIM: amino acids 181 – 232 with point mutations V217P and S221Q fused to the C-terminus of GST. GST itself was similarly purified and used as a control. To detect levels of expression for TRAC-UIM and V/S-UIM the blot was probed with anti-GST antibodies (middle). This is not shown for GST because of the high expression levels of this protein. The expression of GST, relative to the other constructs, is shown on a Coomassie stained gel (bottom).
E. GST, and GST fusion proteins as in (D) were isolated on glutathione beads and incubated with purified K48-linked poly-ubiquitin2-7 chains. Beads alone (-) were included as a control for specific binding. Beads were analysed for binding of ubiquitin with anti-ubiquitin antibodies (top). Input K48 poly-ubiquitin2-7 was analysed on the same polyacrylamide gel but a shorter exposure of the blot is shown. Beads were blotted with anti-GST antibodies to reveal relative levels of the proteins (bottom).
F. HeLa cells were transfected with constructs expressing myc-tagged wildtype TRAC-1, TRAC-1ΔC, TRAC-1Δ205 or V217P,S221Q-TRAC-1. 24 hours after transfection, cells were stained after with anti-myc antibodies and observed by confocal micrsocopy Scale bar: 10 μM.
G. HEK293T cells transfected with TRAC-1Δ205/myc were homogenized in hypotonic buffer and fractionated on discontinuous 70%/65%/10% sucrose gradients as in Fig. 2B. Fractions were collected from the top and aliquots were analyzed by immunoblotting with anti-myc antibodies. Membranes float up to the 65%/10% sucrose interface (fraction 3), whereas soluble proteins remain at the bottom (fractions 10 and 11).
To test whether this region is involved in TRAC-1 auto-ubiquitination, a protein truncated at position 205 was generated (Fig. 4A). Like TRAC-1ΔC, TRAC-1Δ205 was not ubiquitinated (Fig. 5A). To further address the UIM nature of this region, point mutations were introduced at valine 217 and serine 221. These residues correspond to important conserved positions in the UIM consensus sequence [46, 47], with the difference that, as stated before, an alanine is normally present instead of a valine. The resulting mutant, V217P,S221Q-TRAC-1, showed a considerable reduction in ubiquitination (Fig. 5A), strengthening the premise that these residues are part of a ubiquitin binding domain that is important for auto-ubiquitination.
Since both TRAC-1ΔC and TRAC-1/GFP have a dominant inhibitory effect on T cell activation, the auto-ubiquitination of TRAC-1/GFP was determined as well. TRAC-1/GFP ubiquitination was found to occur normally (Fig. 5C) and the UIM of this protein is therefore unlikely to be impaired.
To investigate the ubiqutin-binding properties of the UIM region more directly, amino acids 181 – 232 of TRAC-1 were fused to the C-terminus of GST. The recombinant protein, GST/TRAC-UIM, was purified from bacteria on glutathione-beads and incubated with lysates from HA-ubiquitin transfected HEK293T cells. Indeed, GST/TRAC-UIM, but not GST, was able to pull-down poly-ubiquitinated proteins (Fig. 5D). Moreover, the recognition of ubiquitinated proteins was reduced by the presence of pointmutations V217P and S221Q in GST/V/S-UIM (Fig. 5D). GST/TRAC-UIM was also found to bind purified K48-linked poly-ubiquitin (Fig. 5E), a capacity that was again reduced for GST/V/S-UIM. Thus, TRAC-1 contains a C-terminal UIM-type ubiqutin-binding domain that recognizes K48-linked poly-ubiquitin.
Finally, the effect of C-terminal mutations on the subcellular localisation of TRAC-1 was investigated by immunofluorescence. TRAC-1ΔC, TRAC-1Δ205 and V217P,S221Q-TRAC-1 all showed brighter staining and a more homogeneous distribution than wildtype TRAC-1 (Fig. 5F). Membrane association was not compromised by the absence of a UIM as demonstrated for TRAC-1Δ205 in membrane fractionation experiments (Fig. 5G). The more peripheral staining of these mutants, in contrast to the perinuclear staining of wildtype TRAC-1, suggests that a higher degree of plasma membrane association takes place. Thus, the UIM of TRAC-1 influences its subcellular distribution, which is indicative of important intra- and/or intermolecular interactions taking place via this domain.
DISCUSSION
To provide insights into the function of the ubiquitin ligase TRAC-1, we characterized the protein by determining modifications and identifying novel domains. As a result, a detailed map of TRAC-1 is presented (Fig. 4B). At the N-terminus, TRAC-1 contains a myristoylation site that is modified by the attachment of myristic acid, leading to membrane association. Next, a C3HC4 RING domain is present, essential for ubiquitin ligase activity through the interaction with E2 proteins. C-terminal to this domain follow three atypical zinc fingers, the interactions of which remain to be determined. Finally, at the very C-terminus, a UIM-type domain that binds K48-linked poly-ubiquitin is present. Together with the RING domain, this UIM is required for auto-ubiquitination, which results in a rapid turnover rate of TRAC-1. Thus, a large potential for substrate and regulatory interactions is present in this small ubiquitin ligase. Moreover, identical domain organizations are shown to be present in three other proteins, RNF114, RNF138 and RNF166, which are thereby identified as TRAC-1 relatives.
The results reported here are consistent with a positive regulatory role for TRAC-1 in T cell activation. A potent negative effect was detected for a TRAC-1/GFP fusion protein (Fig 1A-D), similar to the effect of a C-terminally truncated TRAC-1ΔC [32]. The dominant inhibitory effects of TRAC-1/GFP and TRAC-1ΔC are both dependent on an intact RING domain and these proteins are likely to act in similar ways. Their dominant negative activities can be envisioned to result from an ability to compete with endogenous TRAC-1 for protein interactions but an inability to perform normal TRAC-1 function because of the missing (in TRAC-1ΔC), or obstructed (in TRAC-1/GFP) C-terminal region. The C-terminal region could, for instance, influence the ubiquitin ligase activity or the substrate selection of TRAC-1. This hypothesis predicts that important interactions take place on both sides of amino acid 170, the residue where TRAC-1ΔC is truncated. In addition to substrates, TRAC-1 could for instance bind protein-partners that influence its activity.
The RING domain is essential for ubiquitin ligase activity but its role in TRAC-1 function is still confounding. Clearly, mutations in the RING domain of dominant-negative forms negate inhibitory activities (Fig 1A-D, [32]). On the other hand, mutations in the RING domain of the full length protein were reported not to inhibit TRAC-1 function in T cell activation [32]. Moreover, we observed an increase in IL-2 production in the presence of RING-deficient TRAC-1. This suggests that whereas TRAC-1 is required for T cell activation, it may also have a RING domain-dependent inhibitory effect on IL-2 synthesis in activated T cells. RING domain-dependent inhibitory activities have also been reported for TRAC-1 in other processes, such as RIG-I induced cytokine production [33] and HIV transcription [34]. Further insight into the regulation of TRAC-1 and the role of its various domains is needed to clarify these functions.
We demonstrate here that TRAC-1 is subject to myristoylation (Fig. 2A), which suggests that additional to protein-protein interactions, protein-membrane interactions may regulate its activity. Myristoylation anchors proteins to membranes, but the weak binding of the 14-carbon myristate to membranes can be reversed by, for instance, phosphorylation or protein-protein interactions. The occurrence of small amounts of TRAC-1 in a soluble form (Fig. 2B) may indicate that TRAC-1 can shuttle between a membrane-bound and a soluble form. The virtual absence of soluble protein for the mutant C37,40A-TRAC-1, suggests that interactions via the RING domain may be involved in this. However, the soluble pool of TRAC-1 may also represent proteins that are associated with the proteasomes but not yet degraded. In addition, a direct influence of the RING domain on the efficiency of myristoylation during translation can not be fully excluded.
The identification of novel domains, zinc finger like motifs and a UIM, offers new possibilities for TRAC-1 interactions. (Fig. 4). The three zinc binding motifs of the TRAC-1 family diverge in various ways from the classical zinc finger, and the exclusion of TRAC-1 from the nucleus (Fig. 2C, D and E) argue against a role in DNA binding. Nevertheless, these motifs represent with high probability structural domains that are involved in interactions with either substrates, obligatory binding-partners, TRAC-1 itself, or perhaps RNA. Zinc fingers often occur in tandem or in triplets. Similarly, the three zinc-binding motifs of the TRAC-1 related proteins might act together as one or two domains. In particular, the two juxtaposed C2H2 zinc fingers may represent one functional unit. Such a motif is also found outside the TRAC-1 family in Kcmf1 (potassium channel modifier) where it is located C-terminal to a variant (C6H2) RING domain. Moreover, this motif is conserved in the Drosophila orthologue of Kcmf1. The dominant inhibitory TRAC-1ΔC lacks both a C2H2 zinc finger and the UIM. Since the inhibitory TRAC-1/GFP has intact UIM function based on its auto-ubiquitination (Fig. 5C), it is more likely that the absence or obstruction of the C2H2 motif explains the inhibitory activity. However, the exact contribution of these domains to TRAC-1 function remains to be established.
The UIM domain is one of over 15 different motifs capable of binding to ubiquitin. UIM domains are mostly found in proteasome subunits, deubiquitinating enzymes and in proteins involved in receptor internalisation, such as components of the ESCRT complexes [46]. To our knowledge, TRAC-1 is the first C3HC4 RING protein described to contain a UIM domain. The UIM of TRAC-1 was found to be essential for auto-ubiquitination (Fig. 5A), which appears to control the stability of this protein (Fig. 3D and E). The very short half life of TRAC-1 (< 30 min, Fig. 3D and E) suggests that the levels of this protein can be very rapidly modulated. Shutting down its transcription would lead to a rapid decrease, and shutting down its ubiquitination to a rapid increase in TRAC-1 levels. Down-modulation may for instance be required after T cell activation, to prevent a negative impact on IL-2 synthesis. The levels of TRAC-1 were shown to be increased upon IFNα or polyI:C exposure [33]. It will be interesting to determine whether the auto-ubiquitination of TRAC-1 is reduced under these conditions. In addition to a role in auto-ubiquitination, the UIM could be involved in intermolecular interactions through binding to K48-linked ubiquitinated proteins (Fig. 5E). Additionally, it may bind to K48-linked poly-ubiquitin on TRAC-1 itself and thereby influence the conformation of this protein. An involvement in important interactions is suggested by the drastic change in subcellular distribution for proteins with a deletion of, or mutations in, this domain (Fig. 5F).
Based on amino acid sequence similarities, protein domain organisation and gene organisation, RNF114, RNF138 and RNF166 were identified as TRAC-1 relatives (Fig. 4). Extensive protein database searches did not identify any other proteins with close similarity. Whereas the TRAC-1 gene is found in mammals only, the genes for the other three proteins is restricted to vertebrates. The restricted expression pattern of TRAC-1 compared to its relatives may point towards a more specialized role. However, tissue expression profiles of RNF114, RNF138 and RNF166 have not yet been determined and redundancy between the TRAC-1 family members proteins can at present not be excluded. The similarities between TRAC-1 and its family members suggest that these proteins may be involved in similar activities. Nothing is yet known about the functions of RNF114 and RNF166, but RNF138 was identified in a yeast two hybrid screen for proteins that interact with Xenopus NLK (Nemo-like kinase) [48]. The protein was named NARF for NLK associated RING finger protein, and was shown to ubiquitinate TCF/LEF proteins leading to their degradation, thereby inhibiting Wnt signalling. Whether TRAC-1 performs a similar role in T cells is currently being addressed in our laboratory.
Thus, the identification of TRAC-1 relatives has considerably widened the field of TRAC-1 investigations. At the same time, the identification of modifications and domains has brought the TRAC-1 protein further into focus and opened new avenues for studies into its regulation.
ACKNOWLEDGEMNTS
We would like to thank colleagues at the Programme in Infection and Immunity for sharing ideas and reagents. In particular we are grateful to Wayne Turnbull for cell sorting, Daniel Pennington and John Pang for help with FACS analysis, Monica Agromayor for advice on yeast-two-hybrid experiments, Adrian Hayday, Susan John and Matt Lovatt for critically reading the manuscript. We also thank Tony Ng (Randall Instititue, King’s College London) for help with cross correlation analysis. This work was supported by a Career Development Award from the Medical Resarch Council and a Project grant from The Wellcome Trust.
Abbreviations
- HEK293T
human embryonic kidney 293T cells
- TCR
T cell receptor
- TRAC-1
T cell RING protein in activation
- UIM
ubiquitin interacting motif.
REFERENCES
- 1.Ciechanover A, Orian A, Schwartz AL. Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays. 2000;22:442–451. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 2.Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 3.Varshavsky A. Regulated protein degradation. Trends Biochem. Sci. 2005;30:283–286. doi: 10.1016/j.tibs.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Weissman AM. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 5.Pickart CM. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 6.Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 7.Hochstrasser M. Lingering mysteries of ubiquitin-chain assembly. Cell. 2006;124:27–34. doi: 10.1016/j.cell.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 8.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 9.Cyr DM, Hohfeld J, Patterson C. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem. Sci. 2002;27:368–375. doi: 10.1016/s0968-0004(02)02125-4. [DOI] [PubMed] [Google Scholar]
- 10.Jackson PK, Eldridge AG, Freed E, Furstenthal L, Hsu JY, Kaiser BK, Reimann JD. The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 2000;10:429–439. doi: 10.1016/s0962-8924(00)01834-1. [DOI] [PubMed] [Google Scholar]
- 11.Semple CA. The comparative proteomics of ubiquitination in mouse. Genome Res. 2003;13:1389–1394. doi: 10.1101/gr.980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cenciarelli C, Hou D, Hsu KC, Rellahan BL, Wiest DL, Smith HT, Fried VA, Weissman AM. Activation-induced ubiquitination of the T cell antigen receptor. Science. 1992;257:795–797. doi: 10.1126/science.1323144. [DOI] [PubMed] [Google Scholar]
- 13.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu. Rev. Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 14.Heissmeyer V, Macian F, Im SH, Varma R, Feske S, Venuprasad K, Gu H, Liu YC, Dustin ML, Rao A. Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins. Nat. Immunol. 2004;5:255–265. doi: 10.1038/ni1047. [DOI] [PubMed] [Google Scholar]
- 15.Lu Z, Xu S, Joazeiro C, Cobb MH, Hunter T. The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol. Cell. 2002;9:945–956. doi: 10.1016/s1097-2765(02)00519-1. [DOI] [PubMed] [Google Scholar]
- 16.Rao N, Lupher ML, Jr., Ota S, Reedquist KA, Druker BJ, Band H. The linker phosphorylation site Tyr292 mediates the negative regulatory effect of Cbl on ZAP-70 in T cells. J. Immunol. 2000;164:4616–4626. doi: 10.4049/jimmunol.164.9.4616. [DOI] [PubMed] [Google Scholar]
- 17.Giannini A, Bijlmakers MJ. Regulation of the Src family kinase Lck by Hsp90 and ubiquitination. Mol. Cell. Biol. 2004;24:5667–5676. doi: 10.1128/MCB.24.13.5667-5676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rao N, Miyake S, Reddi AL, Douillard P, Ghosh AK, Dodge IL, Zhou P, Fernandes ND, Band H. Negative regulation of Lck by Cbl ubiquitin ligase. Proc. Natl. Acad. Sci. U S A. 2002;99:3794–3799. doi: 10.1073/pnas.062055999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heissmeyer V, Rao A. E3 ligases in T cell anergy--turning immune responses into tolerance. Sci. STKE. 2004;2004:pe29. doi: 10.1126/stke.2412004pe29. [DOI] [PubMed] [Google Scholar]
- 20.Liu YC. Ubiquitin ligases and the immune response. Annu. Rev. Immunol. 2004;22:81–127. doi: 10.1146/annurev.immunol.22.012703.104813. [DOI] [PubMed] [Google Scholar]
- 21.Mueller DL. E3 ubiquitin ligases as T cell anergy factors. Nat. Immunol. 2004;5:883–890. doi: 10.1038/ni1106. [DOI] [PubMed] [Google Scholar]
- 22.Schartner JM, Fathman CG, Seroogy CM. Preservation of self: An overview of E3 ubiquitin ligases and T cell tolerance. Semin. Immunol. 2007;19:188–196. doi: 10.1016/j.smim.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 23.Bachmaier K, Krawczyk C, Kozieradzki I, Kong YY, Sasaki T, Oliveira-dos-Santos A, Mariathasan S, Bouchard D, Wakeham A, Itie A, Le J, Ohashi PS, Sarosi I, Nishina H, Lipkowitz S, Penninger JM. Negative regulation of lymphocyte activation and autoimmunity by the molecular adaptor Cbl-b. Nature. 2000;403:211–216. doi: 10.1038/35003228. [DOI] [PubMed] [Google Scholar]
- 24.Chiang YJ, Kole HK, Brown K, Naramura M, Fukuhara S, Hu RJ, Jang IK, Gutkind JS, Shevach E, Gu H. Cbl-b regulates the CD28 dependence of T-cell activation. Nature. 2000;403:216–220. doi: 10.1038/35003235. [DOI] [PubMed] [Google Scholar]
- 25.Vinuesa CG, Cook MC, Angelucci C, Athanasopoulos V, Rui L, Hill KM, Yu D, Domaschenz H, Whittle B, Lambe T, Roberts IS, Copley RR, Bell JI, Cornall RJ, Goodnow CC. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature. 2005;435:452–458. doi: 10.1038/nature03555. [DOI] [PubMed] [Google Scholar]
- 26.Liu YC, Gu H. Cbl and Cbl-b in T-cell regulation. Trends Immunol. 2002;23:140–143. doi: 10.1016/s1471-4906(01)02157-3. [DOI] [PubMed] [Google Scholar]
- 27.Naramura M, Kole HK, Hu RJ, Gu H. Altered thymic positive selection and intracellular signals in Cbl-deficient mice. Proc. Natl. Acad. Sci. U S A. 1998;95:15547–15552. doi: 10.1073/pnas.95.26.15547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perry WL, Hustad CM, Swing DA, O’Sullivan TN, Jenkins NA, Copeland NG. The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nat. Genet. 1998;18:143–146. doi: 10.1038/ng0298-143. [DOI] [PubMed] [Google Scholar]
- 29.Anandasabapathy N, Ford GS, Bloom D, Holness C, Paragas V, Seroogy C, Skrenta H, Hollenhorst M, Fathman CG, Soares L. GRAIL: an E3 ubiquitin ligase that inhibits cytokine gene transcription is expressed in anergic CD4+ T cells. Immunity. 2003;18:535–547. doi: 10.1016/s1074-7613(03)00084-0. [DOI] [PubMed] [Google Scholar]
- 30.Seroogy CM, Soares L, Ranheim EA, Su L, Holness C, Bloom D, Fathman CG. The gene related to anergy in lymphocytes, an E3 ubiquitin ligase, is necessary for anergy induction in CD4 T cells. J. Immunol. 2004;173:79–85. doi: 10.4049/jimmunol.173.1.79. [DOI] [PubMed] [Google Scholar]
- 31.Chu P, Pardo J, Zhao H, Li CC, Pali E, Shen MM, Qu K, Yu SX, Huang BC, Yu P, Masuda ES, Molineaux SM, Kolbinger F, Aversa G, De Vries J, Payan DG, Liao XC. Systematic identification of regulatory proteins critical for T-cell activation. J. Biol. 2003;2:21. doi: 10.1186/1475-4924-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao H, Li CC, Pardo J, Chu PC, Liao CX, Huang J, Dong JG, Zhou X, Huang Q, Huang B, Bennett MK, Molineaux SM, Lu H, Daniel-Issakani S, Payan DG, Masuda ES. A Novel E3 Ubiquitin Ligase TRAC-1 Positively Regulates T Cell Activation. J. Immunol. 2005;174:5288–5297. doi: 10.4049/jimmunol.174.9.5288. [DOI] [PubMed] [Google Scholar]
- 33.Arimoto K, Takahashi H, Hishiki T, Konishi H, Fujita T, Shimotohno K. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc. Natl. Acad. Sci. U S A. 2007;104:7500–7505. doi: 10.1073/pnas.0611551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shoji-Kawata S, Zhong Q, Kameoka M, Iwabu Y, Sapsutthipas S, Luftig RB, Ikuta K. The RING finger ubiquitin ligase RNF125/TRAC-1 down-modulates HIV-1 replication in primary human peripheral blood mononuclear cells. Virology. 2007;368:191–204. doi: 10.1016/j.virol.2007.06.028. [DOI] [PubMed] [Google Scholar]
- 35.Martin-Serrano J, Eastman SW, Chung W, Bieniasz PD. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J. Cell Biol. 2005;168:89–101. doi: 10.1083/jcb.200408155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bijlmakers MJ, Isobe-Nakamura M, Ruddock LJ, Marsh M. Intrinsic signals in the unique domain target p56(lck) to the plasma membrane independently of CD4. J. Cell Biol. 1997;137:1029–1040. doi: 10.1083/jcb.137.5.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bijlmakers MJ, Marsh M. Hsp90 is essential for the synthesis and subsequent membrane association, but not the maintenance, of the Src-kinase p56(lck) Mol. Biol. Cell. 2000;11:1585–1595. doi: 10.1091/mbc.11.5.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swanson CM, Puffer BA, Ahmad KM, Doms RW, Malim MH. Retroviral mRNA nuclear export elements regulate protein function and virion assembly. EMBO J. 2004;23:2632–2640. doi: 10.1038/sj.emboj.7600270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 40.Farazi TA, Waksman G, Gordon JI. The biology and enzymology of protein N-myristoylation. J. Biol. Chem. 2001;276:39501–39504. doi: 10.1074/jbc.R100042200. [DOI] [PubMed] [Google Scholar]
- 41.Resh MD. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta. 1999;1451:1–16. doi: 10.1016/s0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- 42.McCabe JB, Berthiaume LG. Functional roles for fatty acylated amino-terminal domains in subcellular localization. Mol. Biol. Cell. 1999;10:3771–3786. doi: 10.1091/mbc.10.11.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jang JH. FIGC, a novel FGF-induced ubiquitin-protein ligase in gastric cancers. FEBS Lett. 2004;578:21–25. doi: 10.1016/j.febslet.2004.10.071. [DOI] [PubMed] [Google Scholar]
- 44.Kreppel M, Aryee DN, Schaefer KL, Amann G, Kofler R, Poremba C, Kovar H. Suppression of KCMF1 by constitutive high CD99 expression is involved in the migratory ability of Ewing’s sarcoma cells. Oncogene. 2006;25:2795–2800. doi: 10.1038/sj.onc.1209300. [DOI] [PubMed] [Google Scholar]
- 45.Iuchi S. Three classes of C2H2 zinc finger proteins. Cell. Mol. Life Sci. 2001;58:625–635. doi: 10.1007/PL00000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem. J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swanson KA, Kang RS, Stamenova SD, Hicke L, Radhakrishnan I. Solution structure of Vps27 UIM-ubiquitin complex important for endosomal sorting and receptor downregulation. EMBO J. 2003;22:4597–4606. doi: 10.1093/emboj/cdg471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamada M, Ohnishi J, Ohkawara B, Iemura S, Satoh K, Hyodo-Miura J, Kawachi K, Natsume T, Shibuya H. NARF, an nemo-like kinase (NLK)-associated ring finger protein regulates the ubiquitylation and degradation of T cell factor/lymphoid enhancer factor (TCF/LEF) J. Biol. Chem. 2006;281:20749–20760. doi: 10.1074/jbc.M602089200. [DOI] [PubMed] [Google Scholar]