Abstract
Cells respond to stimuli by changes in various processes, including signaling pathways and gene expression. Efforts to identify components of these responses increasingly depend on mRNA profiling and genetic library screens, yet the functional roles of the genes identified by these assays often remain enigmatic. By comparing the results of these two assays across various cellular responses, we found that they are consistently distinct. Moreover, genetic screens tend to identify response regulators, while mRNA profiling frequently detects metabolic responses. We developed an integrative approach that bridges the gap between these data using known molecular interactions, thus highlighting major response pathways. We harnessed this approach to reveal cellular pathways related to alpha-synuclein, a small lipid-binding protein implicated in several neurodegenerative disorders including Parkinson disease. For this we screened an established yeast model for alpha-synuclein toxicity to identify genes that when overexpressed alter cellular survival. Application of our algorithm to these data and data from mRNA profiling provided functional explanations for many of these genes and revealed novel relations between alpha-synuclein toxicity and basic cellular pathways.
Cells live in a dynamic environment in which they confront various perturbations such as sudden environmental changes, toxins, and mutations. The response to such perturbations is typically complex and comprises signaling and metabolic changes, as well as changes in gene expression. Revealing the cellular mechanisms responding to a specific perturbation may unravel its nature, thus illuminating disease mechanisms1 or a drug’s mode of action2 ,3, and identify points of intervention with potential therapeutic value4.
High-throughput experimental techniques including mRNA profiling and genetic screening are commonly used for revealing components of these response pathways because they provide a genome- and proteome-wide view of molecular changes. mRNA profiling experiments rapidly identify genes that are differentially expressed following stimuli. Genetic screening, including deletion, overexpression and RNAi library screens, identify genetic “hits”, genes whose individual manipulation alters the phenotype of stimulated cells. However, each technique has obvious limitations for revealing the full nature of cellular responses. mRNA profiling experiments do not target the series of events that led to the differential expression. Genetic screens provide strong evidence that a gene is functionally related to the response process. Yet, this relationship is often indirect and hard to decipher, especially in high-throughput experiments that typically result in scores of relevant genes with various functions.
It has been noted previously in a few specific instances 2,5–9 that genetic screens do not identify the same genes as mRNA assays conducted in the same conditions. By analyzing the relationship between genetic hits and differentially expressed genes across 179 diverse conditions, we found that this discrepancy is, in fact, a general rule.
Furthermore, we found a striking bias in each technique that led us to a new, more coherent view of cellular responses. To bridge the gap between the two forms of high throughput analysis we developed an algorithm that exploits these experimental biases and that takes advantage of molecular interactions data. This approach simultaneously reveals (i) the functional context of genetic hits, and (ii) additional proteins that participate in the response yet were not detected by either the genetic or the mRNA profiling assays themselves.
Having validated our approach in a wide array of perturbations, we applied it to unravel cellular responses to increased expression of alpha-synuclein. Alpha-synuclein is a small human protein implicated in Parkinson disease whose native function and role in the etiology of the disease remain unclear 10. We screened an established yeast model for alpha-synuclein toxicity 11,12 using an additional set of 3,500 overexpression yeast strains, exposing the multifaceted toxicity of alpha-synuclein. Application of our approach to the high-throughput genetic and transcriptional data of the yeast model illuminated response pathways whose manipulation altered cellular survival, and provided the first cellular map of the proteins and genes responding to alpha-synuclein expression.
The relationship between genetic hits and differentially expressed genes
In order to derive a comprehensive view of the relationship between genetic hits and differentially expressed genes identified in a particular condition, we analyzed published mRNA profiles and genetic hits for 179 distinct perturbations in yeast (Methods). These data included responses to a wide array of chemical and genetic insults affecting a multitude of cellular processes. For 30 of these perturbations complete genetic screens were reported, typically identifying >100 genetic hits; only partial genetic data are available for the remaining perturbations. The number of genetic hits, differentially expressed genes and genes common to both for each perturbation are given in Table 1 and Supplementary Table 1. Intriguingly, in almost all cases the overlap was astonishingly small and statistically insignificant (p>0.05, Methods).
Table 1.
Measured responses to cellular perturbations.
| Perturbation | Number of differentially expressed genes1 |
Number of genetic hits2 |
Overlap |
|---|---|---|---|
| Growth arrest (HU) 20,79 | 59 | 86 | 0 |
| DNA damage (MMS) 4,13 | 198 | 1448 | 43 |
| ER stress (tunicamycin) 3,20 | 200 | 127 | 5 |
| Fatty acid metabolism (oleate) 9,80 | 269 | 103 | 9 |
| ATP synthesis block (arsenic)2 | 828 | 50 | 9 |
| Protein biosynthesis (cycloheximide) 20,79 | 20 | 164 | 0 |
| Gene inactivation, screen complete (24 data sets 19,20,81,82)3 | 27 | 130 | 0 |
| Gene inactivation, screen incomplete (149 data sets 19,20)3 | 24 | 12 | 0 |
Differentially expressed genes were defined as those showing at least a 2-fold change in expression following the perturbation or as defined in the original papers.
Number of genes whose genetic manipulation affects the phenotype of perturbed cells relative to wild type.
Median results are shown.
One possible explanation for the poor overlap between genetic hits and differentially expressed genes is that each assay may be biased toward distinct aspects of cellular responses. Analysis of Gene Ontology (GO) enrichment confirmed this hypothesis (Methods). The combined hits from all 179 genetic screens were highly enriched for the annotations biological regulation (23.3%, p<10−82), transcription (14%, p<10−44) and signal transduction (6.3%, p<10−30). In contrast, the regulated genes from all perturbations were enriched mostly for various metabolic processes (e.g., organic acid metabolic process 7.1%, p<10−18) and oxidoreductase activities 7.2%, p< 10−34). To ensure these patterns of enrichment do not stem from a handful of data sources but reflect a general tendency, we also analyzed the 30 perturbations for which complete data were available. We found the same enrichment trends, regardless of whether these perturbations were analyzed individually (Supplementary Table 2) or whether all 30 datasets were combined (Supplementary Table 3). Complete enrichment analyses appear in Supplementary Text. Thus, we find that genetic assays tend to probe the regulation of cellular responses, while mRNA profiling assays tend to probe the metabolic aspects of cellular responses.
The striking differences in annotations between genetic hits and differentially expressed genes imply that each gene set alone often provides a limited and biased view of cellular responses. In fact, this hypothesis was often borne out in cases where the pathways are well-studied by other, more classical methods of genetic and molecular biological research. In the yeast DNA damage response pathway, for example, a genetic screen 4 detected proteins that sense DNA damage (Mec3, Ddc1, Rad17 and Rad24), while mRNA profiling detected repair enzymes such as Rnr4 13. Yet core components of this pathway that had been uncovered by other intense investigations over many years, such as the signal transducers Mec1 and Rad53 and the transcription factor Rfx1, remained undetected by either high-throughput assay.
If we are to fully reap the benefits of applying high-throughput methods to new problems and under-explored biological processes, it is essential that we find new routes to connect these data and obtain a true picture of the regulation of cellular responses. Here we provide a novel framework that bridges the gap between genetic and transcriptional data. Based on known pathways such as the response to DNA damage discussed above, we expect that some of the genetic hits, which are enriched for response regulators, will be connected via regulatory pathways to the differentially regulated genes, which are the output of such pathways. Discovering these pathways may uncover additional components of the cellular response to perturbation that are missing from the experimental data (Figure 1).
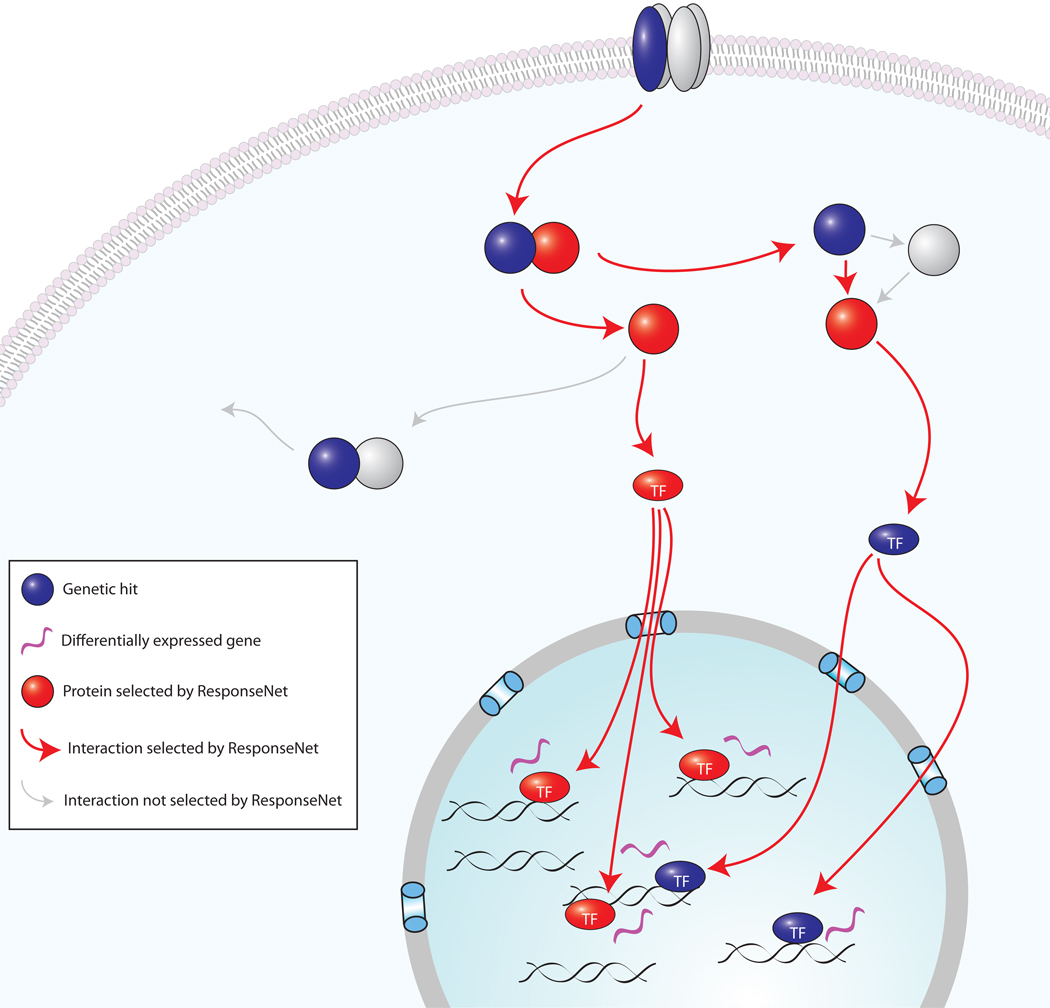
Figure 1. Regulatory relationships between stimulus-specific genetic and transcriptional data.
Cells respond to stimulus with changes in many cellular processes, including signaling and gene expression. The figure shows a general signaling pathway, including receptor binding, transcription factor (TF) translocation into the nucleus and gene expression. Genetic screens and mRNA profiling identify only some of these molecular components and often do not identify the same genes, as shown. We find that the proteins products of genes identified in genetic screens (colored blue) tend to be molecules with regulatory roles. We therefore hypothesize that they may directly or indirectly contribute to the regulation of the observed change in gene expression (colored purple). ResponseNet identifies the likely regulatory pathways, and predicts proteins that are part of these pathways even if they are not identified in either screen (colored red).
ResponseNet algorithm for identification of response networks
The ResponseNet algorithm identifies molecular interaction paths connecting genetic hits and differentially expressed genes that may include hidden components of the cellular response (Figure 1). The yeast Saccharomyces cerevisiae provides a powerful model system for such analysis due to the extensive molecular interactions data now available (Methods and Supplementary Table 4). Taking advantage of these resources we assembled an integrated network model of the yeast interactome that contains protein-protein interactions, metabolic relations and protein-DNA interactions detected by various methods with different levels of reliability14. The resulting interactome relates 5,622 interacting proteins and 5,510 regulated genes, which are represented by network nodes, via 57,955 molecular interactions, which are represented by network edges.
Our representation of the interactome has two important features that facilitate identification of pathways relating genetic hits to transcriptional changes. First, we chose to highlight the role of transcriptional regulatory proteins in determining expression changes by representing differentially expressed genes and their protein products as separate nodes. The only protein nodes that are connected to gene nodes are transcriptional regulatory proteins, and the edges between protein and gene nodes represent observed protein-DNA interactions. Edges between two protein nodes represent protein-protein interaction data. Thus, all pathways connecting genetic hits to the differentially expressed genes must pass through a transcriptional regulatory protein (Supplementary Figure 1). Second, because interactions vary in their reliability, each edge was given a weight that represents the probability that the connected nodes interact in a response pathway. The probabilities were computed using a Bayesian method that considers the types of experimental data supporting the putative interaction and that favors interactions among proteins acting in a common cellular response pathway (Methods).
Due to the vast number of edges, a search for all interaction paths connecting the genetic hits to the differentially expressed genes typically results in “hairball” networks that are very hard to interpret (Figure 2A). One approach to this problem is to identify the highest probability paths. However, pioneering approaches that searched an interactome for high-probability paths had to limit the output path lengths to 3 edges for computational complexity issues15,16. We aimed for a solution that would (i) pick the subset of genetic hits most likely to modulate the differentially expressed genes without limiting it a priori to known regulatory genes, (ii) identify and rank intermediary proteins that are likely to be part of response pathways but escaped detection by high-throughput methods, and (iii) connect the proteins via a high-probability interactome sub-network without restricting its topology.
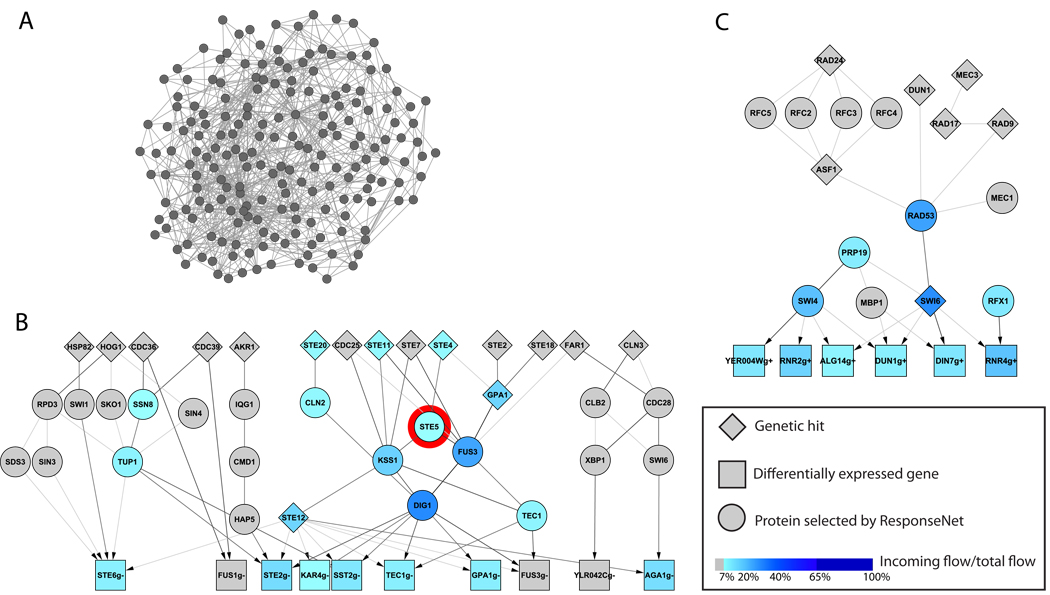
Figure 2. Interactome sub-networks connecting genetic and transcriptional data.
A. A network connecting genetic19 and transcriptional20 data of STE5 deletion strain via paths of length ≤ 3 edges finds 193 nodes and 778 edges.
B. The network created by ResponseNet connects the genetic19 and transcriptional20 data of STE5 deletion strain via 23 intermediary nodes and 96 edges. Higher ranked nodes, as determined by ResponseNet, appear in darker shades of blue and include core components of the pheromone response pathway. Ste5 itself, marked by a red circle, is ranked ninth among the top predicted proteins.
C. The highly-ranked part of the network created by ResponseNet upon connecting genetic hits4,21 to DNA damage signature genes22 identified in yeast treated with the DNA damaging agent MMS. The highest ranking intermediate nodes predicted by ResponseNet include core components of the DNA damage response pathway. The complete network appears in Supplementary Figure 7.
Each node represents either a protein or a gene, and edges represent protein-protein, metabolic, and protein-DNA interactions. The darkness of an edge increases with the amount of flow it carries. Differentially expressed genes are labeled with a suffix of g+ for up- and g- for down-regulation. Networks were visualized using Cytoscape.
We reasoned that these requirements could elegantly be met using a “flow algorithm”, a computational method that has been employed previously to analyze signaling or metabolic networks that have already known topology (e.g.,17). In these algorithms flow goes from a source node to a sink node through the graph edges; edges can be associated with a capacity that limits the flow and are also associated with a cost. (As a loose analogy, this resembles water finding the path of least resistance through a complex landscape.) Because we sought to discover the topology of the unknown response pathways connecting genetic hits and differentially expressed genes we required that flow pass from genetic hits through interactome edges to transcriptional regulators of the regulated genes (Supplementary Figure 1). We then formulated our goal as a minimumcost flow optimization problem 18: Cost was defined as the negative log of the probability of an edge. Hence, by minimizing the cost the algorithm gives preference to high-probability paths (Methods).
The solution to the optimization problem is a relatively sparse network that connects many of the genetic hits to many of the regulated genes through known interactions and intermediary proteins (Figure 2B). These intermediary proteins were not identified by either high-throughput genetic analysis or mRNA profiling, but are predicted by the algorithm to play a part in the response. The proteins in the solution are ranked by the amount of flow they carry. The more flow that passes through a protein, the more important it is in connecting the genetic and transcriptional datasets.
Validation of the ResponseNet algorithm
To determine if ResponseNet provides valid biological insights, we used it to connect genetic and transcriptional data from perturbations in well-studied pathways. We then asked if ResponseNet revealed the proteins and pathways that are missing from the genetic and transcriptional data, but that had previously been gleaned from individual analyses. For example, we used ResponseNet to analyze genetic hits19 and transcriptional20 data collected from a strain deleted for the gene encoding Ste5, a scaffold protein that coordinates the MAP kinase cascade activated by pheromone (Figure 2B). The nodes selected by ResponseNet were highly enriched for proteins functioning in the pheromone response pathway (46%, p<10−18), thus revealing the perturbed biological process. The highly ranked intermediary proteins provided biologically meaningful connections between the data sets, as they included key regulators of the pheromone response as well as Ste5, the source of perturbation.
The algorithm also performed well in analyzing the much more complex cellular response to DNA damage4,21,22. The nodes discovered by ResponseNet were highly enriched for the GO categories response to DNA damage stimulus (21%, p<10−14) and DNA repair (19%, p<10−14). Indeed, the most highly ranked part of the network contained core members of the pathway that had previously been uncovered by years of intense investigation but were not detected by high-throughput screens, including the signal transducers Mec1 and Rad53, members of the RFC complex (Rfc2-Rfc5) and the transcription factor Rfx1 (Figure 2C).
To test ResponseNet more broadly, we evaluated its ability to identify hidden components in the cellular response to over one hundred distinct perturbations corresponding to inactivations of well-annotated genes (Methods). For each such perturbation the genetic hits set consisted of the genetic interactors of the inactivated gene (e.g., synthetic lethals), and the differentially expressed genes were based on mRNA profiling of the inactivated strain 20. The identity of the inactivated gene was hidden from the algorithm, and was used to evaluate the predicted network. ResponseNet output was considered successful in revealing the cellular response to the perturbation if the hidden nodes it identified fulfilled one of two criteria: (i) they included the inactivated gene that was the source of perturbation, or (ii) they were significantly enriched for a specific biological process attributed to the inactivated gene. Significance was determined relative to networks generated using randomization techniques (Methods and supplementary text).
ResponseNet success rates are given in Table 2 and Supplementary Table 5. In total, ResponseNet predictions were successful in 63% of the cases. This rate of success is relatively high considering that for the majority of the cases (85%) genetic hits data were rather limited (a median of 14 genetic hits) and no high-throughput genetic screening data are yet available. Notably, ResponseNet typically selected only 1% of the yeast proteins as relevant for the response. Despite the fact that relevant interactions might be missing from our data or have low probability compared with alternative paths, in a third of the cases the inactivated gene was highly ranked among this small fraction.
Table 2.
Assessment of the ResponseNet algorithm using 101 gene inactivation perturbations.
| Source of genetic hits data | Number of genetic data sets | Median number of predicted proteins | Success in predicting inactivated gene | Success in predicting inactivated gene process | Success in predicting inactivated gene or its process | |
|---|---|---|---|---|---|---|
| % predicted | % predicted in top 20 | |||||
| Synthetic genetic arrays (complete screen) | 15 | 102 | 33 | 13 | 67% | 67% |
| Literature (incomplete data) | 86 | 61 | 33 | 27 | 53% | 62% |
| Total | 101 | 64 | 33 | 25 | 56% | 63% |
A map of cellular pathways responding to alpha-synuclein toxicity
Having established the validity of our method to uncover connections between otherwise disparate high-throughput datasets, we applied ResponseNet to investigate the cellular toxicity associated with alpha-synuclein (α-syn). α-syn is a small lipid-binding protein that is natively unfolded when not bound to lipids and prone to forming toxic oligomers 23. It that has been implicated in several neurodegenerative disorders, most particularly Parkinson disease (PD). α-syn is the main component of Lewy bodies, cytoplasmic proteinaceous inclusions that are a hallmark of PD 24; locus duplication or triplication of α-syn lead to familial forms of PD 25,26, and increased expression of α-syn leads to neurodegeneration in several animal models 27. α-syn is linked to alterations in vesicle trafficking 12,28 and mitochondrial function 29, yet despite immense efforts, the cellular pathways by which α-syn leads to cell death are just beginning to be uncovered.
The yeast S. cerevisiae provides a powerful system for studying the molecular basis of α-syn toxicity that result from its intrinsic physical properties. Expression of human α-syn in yeast yields several dosage-dependent defects that are also found in mammalian systems, such as lipid droplet accumulation in the cytosol, the production of reactive oxygen species and impairment of the ubiquitin-proteasome system 11. An initial overexpression screen in yeast for genes that modify α-syn toxicity tested 2,000 strains and identified a class of genes functioning in ER to Golgi vesicle trafficking, leading to the observation that α-syn causes an ER to Golgi vesicle trafficking block. One of these genes, Ypt1/Rab1, a GTPase protein, was tested in neuronal models of PD and was found to rescue dopaminergic neurons from α-syn toxicity 12.
We now report other results from that screen and the results of screening an additional set of 3,500 overexpression yeast strains, thereby covering in total 85% of the yeast proteome. We identified a diverse group of genes including 54 suppressors and 23 enhancers of α-syn toxicity, many with clear human orthologs (Table 3). Major classes of genes that emerged include vesicle-trafficking genes, kinases and phosphatases, ubiquitin-related proteins, transcriptional regulators, manganese transporters, and trehalose biosynthesis genes. Significantly enriched GO categories included ER to Golgi vesicle-mediated transport (12%, p=6.2*10−5), phosphatases (9.1%, p=0.0028) and transcription factors (6.5%, p=0.047). While the identification of additional vesicle trafficking and ubiquitin-related genes is consistent with the defects caused by α-syn expression in yeast, the identification of trehalose biosynthesis genes and manganese transporters was new and intriguing. Trehalose was recently shown to promote the clearance of misfolded mutant α-syn 30, and manganese exposure has been linked with Parkinson-like symptoms albeit with a distinct underlying pathology31. Notably, another suppressor we identified is homologous to the human PD gene PARK9.
Table 3.
Yeast genes that modify α-syn toxicity when overexpressed.
| Yeast Gene | Type | Strength | Human ortholog(s) | Proposed function |
|---|---|---|---|---|
| Amino Acid Transport | ||||
| AVT4 | suppressor | 3 | SLC36A1 | Vacuolar transporter; exports large neutral amino acids from the vacuole |
| SLC36A2 | ||||
| SLC36A3 | ||||
| SLC36A4 | ||||
| DIP5 | suppressor | 3 | SLC7A1 | Dicarboxylic amino acid permease |
| SLC7A14 | ||||
| SLC7A2 | ||||
| SLC7A3 | ||||
| SLC7A4 | ||||
| SLC7A13 | ||||
| LST8 | suppressor | 3 | GBL | Component of the TOR signaling pathway |
| Autophagy | ||||
| NVJ1 | suppressor | 2 | Nuclear envelope protein; functions during piecemeal microautophagy of the nucleus (PMN) | |
| Cytoskeleton | ||||
| ICY1 | suppressor | 4 | Protein that interacts with the cytoskeleton | |
| ICY2 | suppressor | 4 | Protein that interacts with the cytoskeleton | |
| Manganese transport | ||||
| CCC1 | suppressor | 4 | Putative vacuolar Fe2+/Mn2+ transporter | |
| PMR1 | enhancer | −7 | ATP2C1 | High affinity Ca2+/Mn2+ P-type ATPase required for Ca2+ and Mn2+ transport into Golgi |
| ATP2C2 | ||||
| Protein phosphorylation | ||||
| IME2 | suppressor | 4 | ICK | Serine/threonine protein kinase involved in activation of meiosis |
| PTP2 | suppressor | 3 | PTPRE, PTPRC, PTPN22, PTPRG | Phosphotyrosine-specific protein phosphatase involved in osmolarity sensing |
| GIP2 | suppressor | 3 | PPP1R3A | Putative regulatory subunit of the protein phosphatase Glc7p, involved in glycogen metabolism |
| PPP1R3B | ||||
| PPP1R3C | ||||
| PPP1R3D | ||||
| PPP1R3E | ||||
| YCK3 | suppressor | 3 | CSNK1G1 | Palmitoylated, vacuolar membranelocalized casein kinase I isoform |
| CSNK1G2 | ||||
| CSNK1G3 | ||||
| RCK1 | suppressor | 3 | CAMK1G | Protein kinase involved in the response to oxidative stress |
| CDC5 | suppressor (Cdc5 overexpression is toxic; in presence of a-syn it rescues/rescued) | 3 | PLK2 | Polo-like kinase; found at bud neck, nucleus and SPBs; has multiple functions in mitosis and cytokinesis |
| PTC4 | suppressor | 1 | PPM1G | Cytoplasmic type 2C protein phosphatase |
| SIT4 | enhancer | −2 | PPP6C | Type 2A-related serine-threonine phosphatase. |
| CAX4 | enhancer | −3 | DOLPP1 | Dolichyl pyrophosphate phosphatase, required for Dol-P-P-linked oligosaccharide intermediate synthesis and protein N-glycosylation. |
| PPZ2 | enhancer | −3 | PPP1CC | Serine/threonine protein phosphatase Z |
| PPP1CB | ||||
| PPP1CA | ||||
| PPZ1 | enhancer | −8 | PPP1CA | Serine/threonine protein phosphatase Z |
| PPP1CB | ||||
| PPP1CC | ||||
| Transcription/Translation | ||||
| CUP9 | suppressor | 3 | MEIS1 | Transcriptional repressor involved in copper ion homeostasis |
| MEIS2 | ||||
| MEIS3 | ||||
| NR_002211 | ||||
| .1 PKNOX1 | ||||
| PKNOX2 | ||||
| Q99687-3 | ||||
| TGIF1 | ||||
| TGIF2 | ||||
| TGIF2LX | ||||
| HAP4 | suppressor | 4 | Transcriptional activator and global regulator of respiratory gene expression | |
| FZF1 | suppressor | 3 | KLF15 | Key transcriptional regulator of cellular response to nitrosative stress |
| KLF11 | ||||
| ZNF624 | ||||
| MGA2 | suppressor | 3 | ANKRD1 | ER membrane protein involved in regulation of OLE1 transcription |
| OSBPL1A | ||||
| MKS1 | enhancer | −5 | Pleiotropic negative transcriptional regulator involved in Ras-CAMP and lysine biosynthetic pathways and nitrogen regulation; involved in retrograde (RTG) mitochondria-to-nucleus signaling | |
| VHR1 | suppressor | 3 | Transcriptional activator | |
| JSN1 | suppressor | 2 | PUM1 | Member of the Puf family of RNAbinding proteins, interacts with mRNAs encoding membrane-associated proteins |
| SUT2 | enhancer | −3 | Putative transcription factor; multicopy suppressor of mutations that cause low activity of the cAMP/protein kinase A pathway | |
| TIF4632 | suppressor | 3 | EIF4G1 | Translation initiation factor eIF4G, subunit of the mRNA cap-binding protein complex (eIF4F) |
| EIF4G2 | ||||
| EIF4G3 | ||||
| STB3 | suppressor | 3 | Protein that binds Sin3p in a two-hybrid assay. | |
| MATALPHA1 | enhancer | −5 | Transcriptional co-activator involved in regulation of mating-type-specific gene expression | |
| Trehalose biosynthesis | ||||
| UGP1 | suppressor | 4 | UGP2 | UDP-glucose pyrophosphorylase, catalyses the formation of UDP-Glc, a precursor to trehalose |
| TPS3 | suppressor | 3 | Regulatory subunit of trehalose-6-phosphate synthase/phosphatase complex, which synthesizes trehalose | |
| NTH1 | suppressor | 2 | TREH | Neutral trehalase, degrades trehalose; required for thermotolerance and may mediate resistance to other cellular stresses |
| Ubiquitin-related | ||||
| CDC4 | suppressor | 4 | FBXW7 | F-box, associates with Skp1p and Cdc53p to form a complex, SCFCdc4, which acts as ubiquitin-protein ligase |
| UIP5 | suppressor | 4 | Protein of unknown function that interacts with Ulp1p, a Ubl (ubiquitin-like protein)-specific protease | |
| HRD1 | suppressor | 4 | AMFR | Ubiquitin-protein ligase required for endoplasmic reticulum-associated degradation (ERAD) of misfolded proteins |
| SYVNI | ||||
| UBP11 | enhancer | −3 | USP21 | Ubiquitin-specific protease that cleaves ubiquitin from ubiquitinated proteins. |
| UBP7 | enhancer | −4 | USP21 | Ubiquitin-specific protease that cleaves ubiquitin-protein fusions. |
| Vesicular transport, ER-Golgi | ||||
| YPT1 | suppressor | 5 | RAB10 | Ras-like small GTPase, involved in the ER-to-Golgi step of the secretory pathway |
| RAB13 | ||||
| RAB1A | ||||
| RAB1C | ||||
| RAB8A | ||||
| RAB8B | ||||
| YKT6 | suppressor | 4 | YKT6 | v-SNARE involved in trafficking to and within the Golgi, endocytic trafficking to the vacuole, and vacuolar fusion |
| BRE5 | suppressor | 4 | G3BP2 | Ubiquitin protease cofactor, forms deubiquitination complex with Ubp3p to regulate ER-Golgi transport |
| SEC21 | suppressor | 4 | COPG2 | Gamma subunit of coatomer, a heptameric protein complex that together with Arf1p forms the COPI coat |
| COPG | ||||
| UBP3 | suppressor | 3 | USP10 | Ubiquitin-specific protease that interacts with Bre5p to co-regulate anterograde and retrograde transport between ER and Golgi |
| ERV29 | suppressor | 3 | SURF4 | Protein localized to COPII-coated vesicles, involved in vesicle formation and incorporation of specific secretory cargo. |
| SEC28 | suppressor | 3 | COPE | Epsilon-COP subunit of the coatomer; regulates retrograde Golgi-to-ER protein traffic; stabilizes Cop1p |
| SFT1 | suppressor | 2 | mouse BET1 | Intra-Golgi v-SNARE, required for transport of proteins between an early and a later Golgi compartment. |
| GLO3 | enhancer | −1 | ARFGAP3 | ADP-ribosylation factor GTPase activating protein (ARF GAP), involved in ER-Golgi transport |
| ZNF289 | ||||
| TRS120 | enhancer | −2 | NIBP | One of 10 subunits of the transport protein particle (TRAPP) complex of the cis-Golgi which mediates vesicle docking and fusion |
| GYP8 | enhancer | −2 | TBC1D20 | GTPase-activating protein for yeast Rab family members; Ypt1p is the preferred in vitro substrate |
| YIP3 | enhancer | −2 | RABAC1 | Protein localized to COPII vesicles, proposed to be involved in ER to Golgi transport; interacts with Rab GTPases |
| BET4 | enhancer | −3 | RABGGTA | Alpha subunit of Type II geranylgeranyltransferase; provides a membrane attachment moiety to Rab-like proteins Ypt1p and Sec4p |
| SLY41 | enhancer | −5 | SLC35E1 | Protein involved in ER-to-Golgi transport. |
| GOS1 | enhancer | −2 | GOSR1 | v-SNARE protein involved in Golgi transport, homolog of the mammalian protein GOS-28/GS28 |
| SEC31 | enhancer | −2 | SEC31A | Essential phosphoprotein component (p150) of the COPII coat of secretory pathway vesicles, in complex with Sec13p; required for ER-derived transport vesicle formation |
| SEC31B | ||||
| Other cellular processes | ||||
| PFS1 | suppressor | 4 | Sporulation protein required for prospore membrane formation at selected spindle poles | |
| PDE2 | suppressor | 4 | PDE10A | High-affinity cyclic AMP phosphodiesterase, component of the cAMP-dependent protein kinase signaling system |
| PDE11A | ||||
| PDE1A | ||||
| PDE1B | ||||
| PDE1C | ||||
| PDE2A | ||||
| PDE3A | ||||
| PDE3B | ||||
| PDE4A | ||||
| PDE4B | ||||
| PDE4C | ||||
| PDE4D | ||||
| PDE5A | ||||
| PDE6A | ||||
| PDE6B | ||||
| PDE6C | ||||
| PDE7A | ||||
| PDE7B | ||||
| PDE8A | ||||
| PDE8B | ||||
| PDE9A | ||||
| MUM2 | suppressor | 4 | Interacts with Orc2p, which is a component of the origin recognition complex. | |
| OSH3 | suppressor | 3 | OSBPL1A | Member of an oxysterol-binding protein family, functions in sterol metabolism |
| OSBPL2 | ||||
| OSBPL3 | ||||
| OSBPL6 | ||||
| OSBPL7 | ||||
| PHO80 | suppressor | 3 | Cyclin, negatively regulates phosphate metabolism | |
| OSH2 | suppressor | 3 | OSBPL3 | Member of an oxysterol-binding protein family, functions in sterol metabolism |
| OSBP | ||||
| OSBP2 | ||||
| ISN1 | suppressor | 2 | Inosine 5′-monophosphate (IMP)-specific 5′-nucleotidase | |
| EPS1 | enhancer | −1 | Protein disulfide isomerase-related protein involved in endoplasmic reticulum retention of resident ER proteins. | |
| IDS2 | enhancer | −2 | Protein involved in modulation of Ime2p activity during meiosis | |
| QDR3 | suppressor | 4 | Multidrug transporter of the major facilitator superfamily, required for resistance to quinidine, barban, cisplatin, and bleomycin | |
| TPO4 | enhancer | −3 | Polyamine transport protein, recognizes spermine, putrescine, and spermidine; localizes to the plasma membrane; member of the major facilitator superfamily | |
| IZH3 | enhancer | −2 | Membrane protein involved in zinc metabolism, member of the four-protein IZH family, expression induced by zinc deficiency; deletion reduces sensitivity to elevated zinc and shortens lag phase, overexpression reduces Zap1p activity | |
| Unknown Function | ||||
| YKL063C | suppressor | 4 | Uncharacterized, GFP-fusion localizes to the Golgi | |
| YML081W | suppressor | 4 | EGR3 | Uncharacterized, GFP-fusion localizes to the nucleus |
| YNR014W | suppressor | 4 | Uncharacterized, expression is cell-cycle regulated and heat-inducible | |
| YKL088W | suppressor | 4 | PPCDC | Protein required for cell viability. Predicted phosphopantothenoylcysteine decarboxylase |
| YML083C | suppressor | 3 | Uncharacterized, strong increase in transcript abundance during anaerobic growth compared to aerobic growth | |
| YDR374C | suppressor | 3 | YTHDF1 | Uncharacterized |
| YTHDF2 | ||||
| YTHDF3 | ||||
| YOR291W (YPK9) | suppressor | 3 | ATP13A2 | Probable cation-transporting ATPase 2 |
| (PARK9) | ||||
| ATP13A3 | ||||
| ATP13A4 | ||||
| ATP13A5 | ||||
| YDL121C | suppressor | 2 | Uncharacterized, GFP-fusion localizes to the ER | |
| YBR030W | suppressor | 2 | Uncharacterized, predicted to function in phospholipid metabolism | |
| YMR111C | suppressor | 2 | Uncharacterized, GFP-fusion localizes to the nucleus | |
| YOR129C | suppressor | 2 | Putative component of the outer plaque of the spindle pole body; may be involved in cation homeostasis or multidrug resistance. | |
Park9 and the human homologs of seven other genetic modifiers from diverse functional classes (Hrd1, Ubp3, Pde2, Cdc5, Yck3, Sit4 and Pmr1) were found to be efficacious in neuronal models, validating the yeast model as meaningful to α-syn toxicity in neurons (Gitler et al.; manuscript submitted). The genes identified by the screen therefore begin to unravel the surprisingly multifaceted toxicity of α-syn. Importantly, they provide novel causal relations between α-syn expression and toxicities previously associated with PD but not specifically linked to α-syn. A detailed description of the various gene classes and their potential relation to PD appears in the Supplementary Text.
The transcriptional profile occurring in response to α-syn toxicity was determined in a separate study (Supplementary Text; Su et al.; manuscript submitted). Up-regulated genes prominently included genes with oxidoreductase activities (13%, p<10−9). Down-regulated genes included ribosomal genes (28%, p<10−30), as commonly observed under stress 32. More specific to α-syn toxicity, the down-regulated genes were strikingly enriched for genes encoding proteins localized to the mitochondria (60%, p<10−44) and for genes involved in generation of precursor metabolites and energy (18%, p<10−15).
The genetic and transcriptional data obtained in this model system exemplify both the power and the limitations of the current approaches. These technologies reveal the wide range of cellular functions that are altered by α-syn expression. Yet the precise roles of the genetic hits and differentially expressed genes in the cellular response are unclear. For example, we checked whether the ubiquitin-related proteins that emerged from the genetic screen affect α-syn degradation. However, in strains overexpressing these ubiquitin-related genes we did not detect changes by flow cytometry in steady-state α-syn protein levels (Supplementary Figure 2). As with our previous analyses (above), the overlap between the data obtained from the genome-wide genetic screen and mRNA profiling assay was minor and statistically insignificant (four genes, p=0.96).
Applying ResponseNet to these disparate datasets revealed a more coherent view of the cellular response (Supplementary Figure 3). The resulting network provided context to a large portion of the data: 34 (44%) genetic hits and 166 (27%) differentially expressed genes were linked to each other through 106 intermediate connections. These include two thirds of the protein kinase, phosphatase and ubiquitin-related genetic hits, illuminating their intricate role in the response to α-syn. For example, ResponseNet suggests that the genetic suppressor Rck1, a kinase known to respond to oxidative stress, functions through its interactions with the Cad1 transcription factor, and that this sub-network explains the differential transcriptional of seven genes (Supplementary Figure 3J). Similarly, ResponseNet identifies a set of transcriptional changes that it traces back to the genetic hits Bre5 and Ubp3, which form a deubiquitination complex (Supplementary Figure 3C).
The major cellular pathways responding to α-syn toxicity included ubiquitin-dependent protein degradation, cell cycle regulation and vesicle trafficking pathways, all of which have previously been associated with PD (Supplementary text and Supplementary Figure 3). Impairment of the ubiquitin proteasome system33 and mutations in ubiquitin-related genes (parkin and uch-L1) underlie sporadic and familial forms of PD. Interestingly, parkin is associated with the SCF ubiquitin ligase complex 34, components of which were selected by ResponseNet. Inappropriate cell cycle regulation has also been implicated in neuronal cell death in PD 35,36, and ResponseNet predicted several regulators of mitosis and early meiosis. Below we focus on additional ResponseNet predictions that relate to known aspects of PD including nitrosylation, mitochondrial dysfunction and the heat shock response.
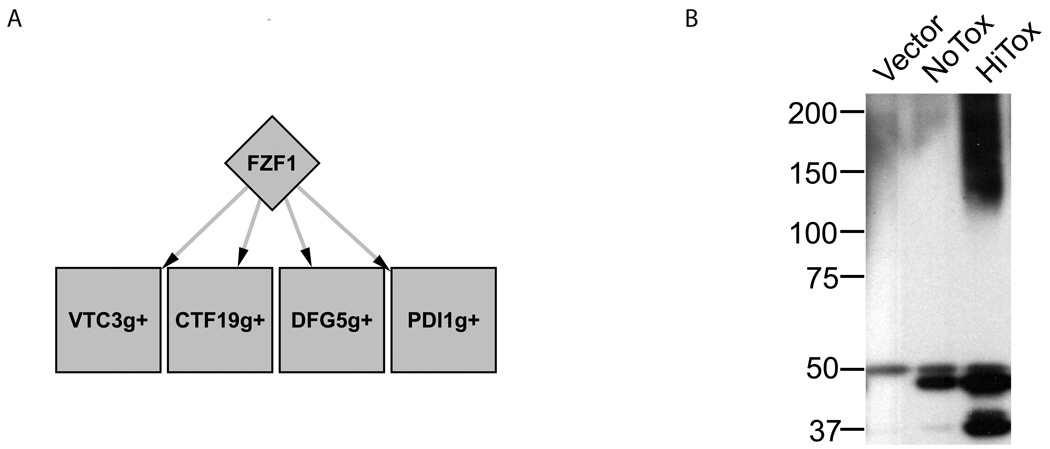
Nitrosative stress
Fzf1 was the only gene identified in the screen related to nitrosative stress 37. However, ResponseNet connected it to four up-regulated transcripts, including Pdi1, a protein disulfide isomerase (PDI) (Figure 3A). Intriguingly, the up-regulation of human PDI protects neuronal cells from neurotoxicity associated with ER stress and protein misfolding (both of which are linked to α-syn expression), and, further, PDI has been found to be S-nitrosylated in PD 38. We found that increased expression of α-syn causes increased S-nitrosylation of proteins (Figure 3B). This result is surprising as nitrosative responses in yeast cells were long thought to represent a defense mechanism against other microbes. Very recently it was shown that yeast synthesize NO in response to exogenous H2O2 39, suggesting that the nitrosylation of specific proteins is a highly conserved response to oxidative stress.
Figure 3. Nitrosative stress response to α-syn expression in yeast.
A. The predicted sub-network containing Fzf1 and its differentially expressed target genes. Graphical representation is similar to Figure 2.
B. Immunoblotting against S-nitrosocysteine performed on a control strain (vector), on a strain expressing one copy of α-syn (NoTox), and on a high-toxicity strain (HiTox) expressing several copies of α-syn, reveals that increasing levels of α-syn increase the amount of S-nitrosylated proteins.
Mitochondrial dysfunction
Mitochondrial dysfunction and oxidative stress have been strongly linked with PD 40, and were recently associated specifically with α-syn (e.g., 41). Although mitochondrial dysfunction was a prominent signature in the microarray data (Su et al.; manuscript submitted), the genetic hits contained only a few genes clearly related to mitochondria. ResponseNet identified two connected components related to mitochondrial dysfunction. One component contained the suppressor Hap4, a transcriptional activator of respiratory genes, directly connected to several of the differentially expressed genes (Supplementary Figure 3B). The other component contained regulators of the retrograde signaling pathway, which senses mitochondrial dysfunction (Mks1, Rtg2 and Grr1 42, Supplementary Figure 3E).
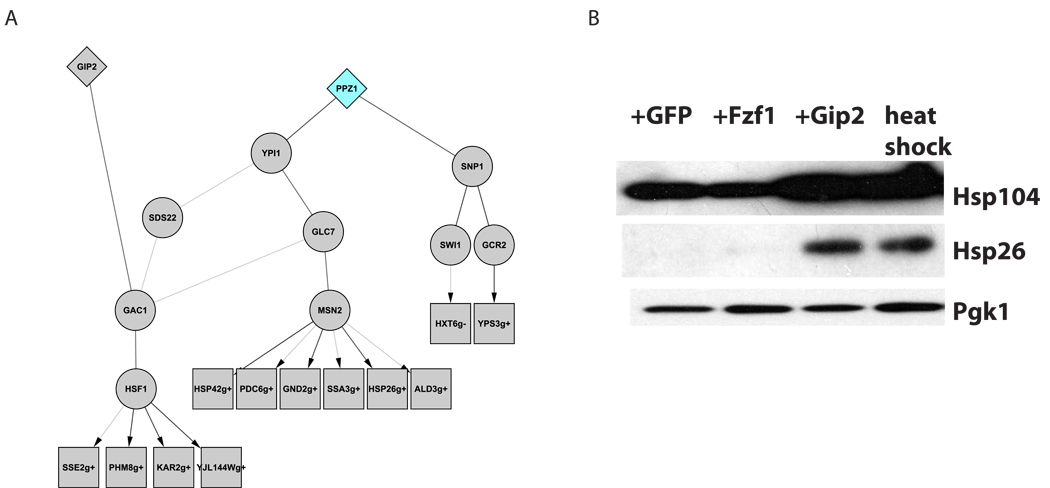
Heat shock
The induction of heat shock response directly or via chemical inhibition of Hsp90 43 suppresses α-syn toxicity in many model systems including yeast, flies, mice and human cells (e.g., 44,45). However, heat shock related genes were conspicuously absent among the list of genetic suppressors. Nonetheless, ResponseNet predicted the involvement of two highly conserved heat shock regulators, the chaperone Hsp90 (isoform Hsp82, Supplementary Figure 3A) and the heat shock transcription factor Hsf1 (Figure 4A). Interestingly, ResponseNet predicted that the toxicity suppressor Gip2, a putative regulatory subunit of the Glc7 phosphatase, interacts with Gac1. Gac1 is a regulatory subunit of the Glc7 complex, which is known to activate Hsf146. This connection suggested that Gip2 overexpression might induce a heat shock response and prompted us to test it. Indeed, we found that strains overexpressing Gip2 show elevated levels of heat shock proteins (Figure 4B). ResponseNet therefore provided a mechanistic explanation for the suppression of α-syn toxicity achieved by Gip2 overexpression and identified a new player in the regulation of the ancient heat shock response.
Figure 4. Overexpression of Gip2 causes induced expression of Hsf1 targets.
A. The predicted sub-network links the toxicity suppressor Gip2 and the toxicity enhancer Ppz1 to Hsf1 and Msn2 via components of type 1 protein phosphatase complex (Gac1, Glc7, Ypi1, Sds22). Graphical representation is similar to Figure 2.
B. Immunoblotting of vector cells overexpressing GFP, Fzf1 or Gip2 with antibodies against Hsp104 and Hsp26. Overexpression of Gip2 is sufficient to activate Hsf1 and induce higher protein levels of both its targets Hsp104 and Hsp26, similar to that of vector cells subjected to heat shock. In contrast, ovexepression of another genetic suppressor, Fzf1, does not activate Hsf1. Immunoblotting against Pgk1 was used as a loading control.
We also identified cellular pathways whose relation to α-syn toxicity was initially obscure, raising the possibility that they may be interesting avenues for future research. Below we focus on two such highly-conserved pathways, the mevalonate/ergosterol pathway that is targeted by the cholesterol lowering statin drugs, and the target of rapamycin (TOR) pathway.
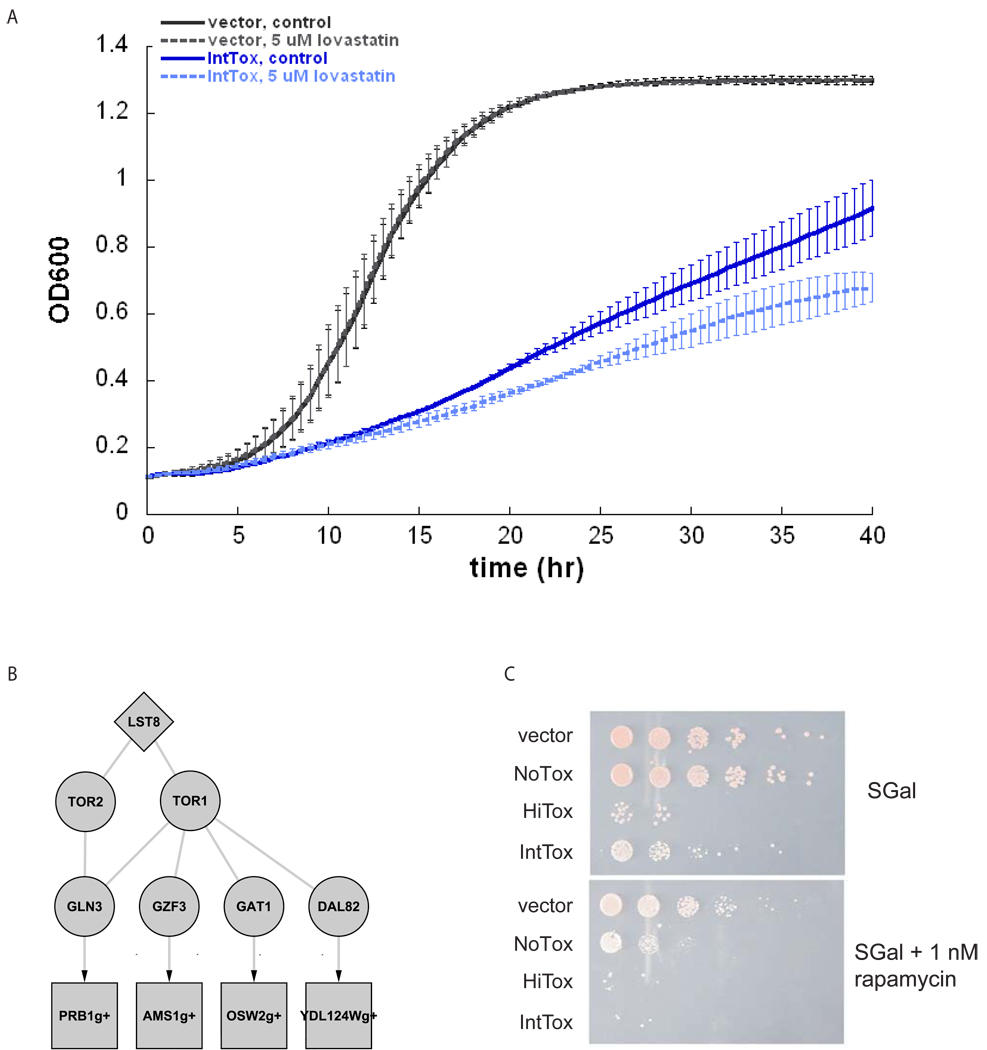
The mevalonate/ergosterol biosynthesis pathway not only synthesizes sterols, but also synthesizes other products with connections to α-syn toxicity such as farnesyl groups required for vesicle trafficking proteins and ubiquinone required for mitochondrial respiration. ResponseNet ranked highly Hrd1, which regulates the protein target of statins, and the predicted intermediary Hap1, a proposed transcriptional regulator of the pathway 47 (Supplementary Figure 3A). In addition, the α-syn mRNA profile was modestly correlated with the profile of yeast treated with lovastatin (r=0.32, p< 10−93, Su et al; manuscript submitted), and several genetic hits could be also associated with products of the pathway (dependent enzymes Bet4 and Cax4, farnesylated proteins Ypt1 and Ykt6 and putative sterol carriers Sut2, Osh2, and Osh3). We therefore tested the effect of lovastatin, which selectively inhibits the highly conserved HMG-CoA reductase of yeast as well as that of mammalian cells, on α-syn toxicity. Addition of 5µM lovastatin to the media caused a further reduction in growth to strains overexpressing α-syn (Figure 5A), but did not reduce growth of either wild-type controls or of cells expressing another toxic protein, a glutamine-expansion variant of huntingtin exon I 48 (Supplementary Figure 4). We further tested ubiquinone, a downstream output of this pathway, reasoning that its down-regulation through the action of α-syn might increase cellular vulnerability. Indeed, the addition of ubiquinone-2 to the media provided a modest suppression against α-syn toxicity. Ubiquinone is an antioxidant, but this was not a non-specific antioxidant response as the antioxidant N-acetylcysteine had no effect (Supplementary Figure 5).
Figure 5. Effects of the small molecules lovastatin and rapamycin on α-syn toxicity.
A. Lovastatin inhibits growth of the yeast strain expressing an intermediate level of α-syn. Growth of a control strain (vector) and an intermediate toxicity strain (IntTox) expressing several copies of α-syn was measured in a galactose containing media with and without 5µM lovastatin. Each growth curve reflects the average of 3 individual runs, each of which is indicated by a bar.
B. The predicted sub-network containing TOR pathway components includes the predicted proteins Tor1 and Tor2. Graphical representation is similar to Figure 2.
C. The effect of rapamycin on growth of different yeast strains. The upper panel shows the growth of a control strain (vector), a strain expressing one copy of α- syn (NoTox), a high-toxicity strain (HiTox) and an intermediate toxicity strain (IntTox) both expressing several copies of α-syn, in a galactose containing media (SGal) that is used to induce expression of α-syn. The lower panel shows the same strains grown in media that also contains 1nM rapamycin, showing that rapamycin inhibits growth of all α-syn expressing strains but not the control strain, as observed by the difference in the number of colonies per drop. The different columns correspond to serial dilutions.
The TOR pathway has been related to other neurodegenerative diseases 49,50. ResponseNet identified the TOR pathway proteins Tor1, Tor2 and their target transcription factors as intermediary between the genetic hit Lst8 and several up-regulated genes involved in spore wall formation (a vectorially directed secretory process in yeast) and vacuolar protein degradation (Figure 5B). We found that addition of the TOR-inhibitor rapamycin to the media markedly enhanced the toxicity of α-syn. Indeed, a low dose α-syn, which is otherwise innocuous, became toxic (Figure 5C). Establishing the specificity of this effect to α-syn, rapamycin did not reduce growth of cells expressing glutamine expansion variants of huntingtin exon I (Supplementary Figure 6).
Discussion
We provide a novel framework in which genetic, physical and transcriptional data naturally complement each other in the context of cellular response to biological perturbations. Although the complementary nature of these data has been noted 2,5–9,51–55, a systematic analysis of the relationship between stimulus-specific genetic modifiers and transcriptional responses has been lacking. Here we find that in response to over 150 distinct stimuli differentially expressed genes and genetic hits are consistently disparate (Table 1).
Our analysis suggests some interesting possible explanations for the discrepancy between these two types of experiments. We have observed that differentially transcribed genes are disproportionately involved in metabolic processes. These genes are less likely to appear as genetic hits because metabolic processes tend to be robust against single mutations 56. We have also found that genetic hits are biased towards regulatory proteins. Since deletion or over-expression of a regulatory protein is likely to dramatically alter cellular signaling, this bias is, in hindsight, quite reasonable. However, why are regulatory proteins rarely found to be differentially transcribed?
The lack of an observed transcriptional response by regulatory proteins could arise from either technical or biological reasons. From a technical perspective, regulatory proteins are commonly controlled post-transcriptionally, and therefore show no change in mRNA levels. For example, purely post-transcriptional changes in HSF1 activity can produce enormous changes in the expression of protein chaperones. Further, since many of these proteins have very low transcript concentrations57, alterations in their mRNA levels may be below the detection limits of microarray platforms.
Several possible biological explanations for the dearth of regulatory proteins among the differentially transcribed genes would add to this effect. Proteins that initiate a signaling response such as the components of the “sliding clamp checkpoint,” which detects DNA damage, must be present at all times in order to detect environmental changes. In addition, proteins that transmit signaling information but that do not initiate a response (such as kinases) may also need to be kept at relatively constant levels, as changes in the levels of these proteins can dramatically alter the systems-level properties of a signaling pathway, resulting in surprising biological effects58. In such cases, it is precisely because changes in the expression level of a genetic hit produces such a readily detectable phenotype that the expression of the gene is maintained at a constant level.
The discordance between genetic hits and differentially expressed genes has implications for the search for therapeutic strategies. In yeast, inactivating a differentially expressed gene is no more likely to affect cell viability than targeting a randomly chosen gene. Yet bridging the gap between these data can potentially reveal additional intervention points that may be targeted by drugs.
Deciphering the role of genetic hits identified under conditions such as stress or disease is a complex task. Our analysis indicates that this task can be facilitated by incorporating molecular interactions data. Previous interactions-based interpretations often focused on graph-related properties of the data, such as neighboring genes representing functionally related proteins59,60, connected components representing functional modules 61 and network hubs representing key proteins 62. However, these approaches have limited power to reveal mechanistic insights especially when the underlying networks become dense.
We provide a novel scheme to interpret the functional role of genetic hits. By focusing on their regulatory relationship with differentially expressed genes and using the interactome as the underlying architecture, our approach represents an important step toward fully mechanistic models for the regulation of cellular responses. As described for the deletion of Ste5 and the response to DNA damage, ResponseNet can provide a richer framework for previously explored pathways (Figure 2). More importantly, ResponseNet has the power to uncover connections between high-throughput data sets that reveal underlying biological processes in cases for which little is known, as was true for α-syn. The resulting networks provide extended views of cellular responses as they incorporate relevant proteins not discovered in the high-throughput assays themselves (Figure 2).
Our computational approach is based on a flow algorithm to connect the genetic hits and differentially expressed genes. Unlike intriguing studies that link a target gene with its causal transcriptional change13,15,16,63–66, a flow-based approach allows for a global, efficient and simultaneous solution for multiple target genes that puts no a priori bounds on the structure of the output. In fact, the predicted output networks have rich structures with half of all paths of length of 3 or more. The ability of ResponseNet to analyze interactome data containing tens of thousands of nodes and edges make it well-suited to analyzing the accumulating data from other species or other techniques67.
We applied our approach to a yeast model for α-syn pathobiology implicated in PD. The complexity of PD and the multifaceted nature of the toxicities associated with just one protein, α-syn, mandate their investigation via systems biology approaches. We identified 77 genes whose overexpression altered α-syn toxicity (Table 3). In addition to genes involved in vesicle trafficking (as previously reported), these included genes involved in protein degradation, cell cycle regulation, nitrosative stress, osmolyte biosynthesis and manganese transport. This screen established an interface between α-syn and a large number of cellular and environmental factors previously linked to neuropathology and, in some cases, specifically to Parkinsonism, but not specifically linked to α-syn. Many of the genes we identified are highly conserved in humans, where they may exert similar affects. Indeed, eight out of nine toxicity modifiers we have now tested had similar effects on α-syn toxicity in yeast and in neuronal systems (Gitler et al; manuscript submitted).
Application of ResponseNet to the disparate high-throughput genetic and transcriptional data of the α-syn model succeeded in providing a functional context to many of the genetic hits identified in our yeast screen (Supplementary Figure 3). It uncovered the involvement of the heat shock response, the TOR pathway and the mevalonate/ergosterol pathway in the response to α-syn expression, which we experimentally validated (Figure 3–Figure 5). Of these, the mevalonate/ergosterol pathway is of special interest as its perturbation could potentially alter a variety of downstream pathways, including protein farnesylation and ubiquinone biosynthesis that are closely related to the vesicle trafficking defects and mitochondrial dysfunction observed in the yeast model. The fact that several of the yeast modifiers of α-syn toxicity act similarly in neuronal systems suggests that the pathways we identified in yeast are relevant in neuronal systems. Indeed, a link between sterol biosynthesis and the etiology of PD surfaced recently in man. PD patients have significantly lower levels of low-density lipoprotein (LDL) cholesterol than their spouses 68, and low levels of LDL preceded the appearance of PD in a group of men of Japanese ancestry 69.
The global picture we obtained by integrating high-throughput genetic, transcriptional and physical yeast data demonstrates the power of integrative approaches to illuminate under-explored cellular processes. As high-throughput assays are becoming routine in the study of complex disease and developmental processes, approaches for deciphering these data based on their underlying characteristics are vital.
Materials and Methods
Genetic and transcriptional datasets
Datasets for chemical perturbations were downloaded from original papers. Genetic hits for gene inactivation were defined as the set of proteins found to genetically interact with the inactivated gene. These data were downloaded from SGD 19 and included all types of genetic interactions. Transcriptional data consisting of differentially expressed genes showing at least a two-fold change in expression with a p-value ≤ 0.05 were extracted from 20, or else were defined according to original papers. Chemical perturbation assays were paired if the concentrations of the chemical were comparable in both assays.
Interactome data description
The interactome is represented as a graph G = (V,E) that consists of nodes (vertices) V representing genes and proteins, and a set of bidirectional and directed edges E representing their interactions. Different nodes in the network represent a gene and its corresponding protein.
Bidirectional edges between protein nodes in the interactome consisted of:
Physical protein-protein interactions, which were downloaded from 70 and from BioGRID release 2.0.30.
Interactions between two proteins if they both appeared in the same literature-curated protein complex, downloaded from MIPS 71.
Metabolic interactions between two enzymes, if the substrate of one was the product of the other, based on the metabolic map of S. cerevisiae 17.
Directed edges in the interactome consisted of:
Edges from a protein node to a gene node if there was evidence from either literature or ChIP-chip assays 72–74 that the protein was a probable transcriptional regulator of the gene.
Edges from one protein node to another if both proteins acted as transcriptional regulators and the first regulated the second.
Supplementary Table 4 lists the number of interacting pairs per interaction type in the interactome.
Weighting scheme for interactome edges
Each edge (i,j) ∈ E between node i and node j of the interactome is characterized by a weight wij calculated as follows:
Interactions between protein nodes
We developed a Bayesian weighting scheme that favors interactions between proteins functioning within a common response pathway (RP). Each interacting protein pair pi,pj was associated with an interaction vector Ipi,pj, where vector entry Ik pi,pj serves as an indicator function for interaction evidence of type k. For example, I ”two-hybrid HTP” pi,pj was set to 1 if pi interacted with pj in a high-throughput two-hybrid experiment. Each interacting protein pair pi,pj was assigned a weight wij reflecting the probability that pi,pj function in a randomly selected response pathway (denoted RPpi,pj=1) based on their interaction evidence vector Ipi,pj. By Bayes’ rule,
We assumed that different types of evidence are conditionally independent, so that . To estimate the prior probability P(RP) and the conditional probability table associated with each evidence type P(Ik| RP) we compiled the following:
A set of response pathways containing 54 response-specific processes according to GO process annotations (e.g., response to osmotic stress GO:0006970).
A set of positive examples containing all interacting protein pairs functioning in a common response pathway (see 1 above) based on reliable GO process annotations. To exclude less reliable sources of annotation we used only GO evidence relying on direct assay or expert knowledge (GO evidence codes IC, IDA and TAS).
A set of negative examples composed of interacting protein pairs known not be in a common response pathway similar to 75.
Supplementary Table 6 lists the resulting weights associated with individual evidence types.
Some edge weights wij were close to 1, which was unrealistic biologically and could instead indicate unusually well-studied proteins 76 or imperfectness of the assumption of conditional independence. To prevent such edges from dominating the predicting response networks, and to place all edges with high enough weights on equal footing, the weights wij were capped to a maximum value of 0.7. Notably, small changes in this value (0.7±0.1) gave similar results in the subsequent analyses.
Interactions between protein and gene nodes
These weights were designed to reflect the interaction’s reliability based on experimental evidence and conservation. “ChIP-chip interactions” refer to interactions discovered by the ChIP-chip method. “ChIP-chip motif interactions” refer to those ChIP-chip interactions for which the gene’s upstream sequence contained the binding motif of the specific transcription factor. “Reliable interactions” included those ChIP-chip motif interactions for which the motif occurrence in the gene’s upstream sequence was conserved in at least two other Saccharomyces sensu stricto species, as well as literature-curated interactions. The weight of reliable interactions was set to 0.7. The weight of remaining “ChIP-chip interactions” was set to the fraction of “ChIP-chip interactions” that were also reliable (0.51), and similarly the weight of remaining “ChIP-chip motif interactions” was set to the fraction of “ChIP-chip motif interactions” that were also reliable (0.59).
Linear programming formulation
The inputs to ResponseNet consist of the weighted interactome G = (V,E), the genetic hits data set Gen ⊂ V and the transcriptional data set Tra ⊂ V identified following a specific perturbation. Each edge (i,j) ∈ E is characterized by a weight wij representing its probability (as described above), and by a capacity cij = 1.
For each perturbation the graph G is updated as follows:
V'=V∪{S,T}, where S and T are auxiliary nodes representing the source and sink, respectively.
E'= E∪(S,i)∀i∈Gen ∪ (i, T)∀i∈Tra, thus connecting S to the genetic hits and T to the differentially expressed genes data using directed edges.
. The strength of each genetic hit is measured by the variation it confers on the number of colonies per drop, if available, otherwise strengths are taken to be uniform. Thus the capacities associated with the edges between the source S and the genetic hits are proportional to the strength of each genetic hit, and can be viewed as a prior for including the gene in the output.
. The strength is measured by either the fold-change in its transcript level or the p-value associated with it, depending on their availability. Thus the capacities associated to the edges between each transcriptional hit and the sink T are proportional to the logarithm of the gene strength, and can be viewed as a prior for including the gene in the output.
wSi = cSi ∀i ∈ Gen and wiT = ciT ∀i ∈ Tra.
Let fij denote the flow from node i to node j, and let F = {fij} denote the solution to the following optimization problem: min
| (1) |
subject to:
| (2) |
| (3) |
| (4) |
The objective function
The expression being minimized in (1) reflects the objectives of increasing the total flow in the network, given by ∑ieGenfSi, while at the same favoring high-probability interactions via the term ∑ij−log(wij*fij.
To better explain this second term, suppose that the integer variable xij indicates the presence of an interaction between node i and node j in the solution (xij = 1 if the interaction between node i and node j is in the solution and xij = 0 otherwise). Note that minimizing ∑ij−log(wij*xij is equivalent to maximizingΠij wijXij, which is the overall probability of all the interactions in the solution for which xij=1. In the optimization problem given by (1)–(4) above, we allow the xij variable to take continuous values fij rather than binary values xij. This relaxes an integer programming problem, which can be computationally intractable, to a linear programming problem, for which efficient algorithms are available.
The objective function in (1) contains one tunable parameter, γ, which controls the size of the input being connected by ResponseNet, with effective values of 7 to 20 (γ <7 typically results in an empty solution, γ >20 typically saturates the maximum flow attainable). Smaller values of γ identify the subset of input connected by the highest-probability paths. As γ increases additional input components that are connected by lower-probability paths are added. Notably, a change in the tunable parameter values (γ±1) does not typically affect the highest ranking proteins, which provide a skeleton around which the network is built; it does affect the coverage of the input data and consequently determines the inclusion or exclusion of more lowly ranked intermediary proteins (Supplementary Text and Supplementary Figure 8). A protocol for setting γ value appears in the Supplementary text. In the assessment of ResponseNet γ =10 was used because it gave intermediate sized networks.
The constraints
Constraint (2) requires the conservation of flow for each node in the interactome; constraint (3) requires that all flow out of the source must arrive at the sink; constraint (4) enforces that the flow for each interaction is non-negative and does not exceed the capacity of the interaction.
The optimization problem was solved using LOQO 77. F = {fij>0} was interpreted as a set of weighted interactions that connects the genetic and the transcriptional data sets, defining the predicted response network. This network does not contain proteins and genes from the input sets, except for genetic hits that receive flow from nodes other than the source. Network proteins were ranked in decreasing order according to the total amount of their incoming flow (calculated for each protein as the sum of the flow value for each incoming edge). Notably, although the solution to the optimization problem is a directed network, the directionality of the interactions in the network, except for the interactions between transcription factors and their targets, was ignored. This directionality only reflects the way in which the algorithm directed flow from the genetic hits to the differentially expressed genes and is not intended to represent the causal order of events (Figure S1 and Supplementary Text).
ResponseNet reports the optimal solution to the flow problem. To map the space of sub-optimal solutions and the stability of the optimal solution we perturbed the edge weights in network and compared the solutions of the perturbed and unperturbed networks. We found that the solution is quite stable even when edge weights are perturbed by a random scaling factor with a mean of one and a standard deviation of 25% (Supplementary Text).
Statistical analysis
Probabilities of overlap between genetic and transcriptional data were calculated using Fisher’s Exact Test, based on a total of 6000 yeast genes.
Enrichment analysis was performed using the Gene Ontology Term Finder from SGD. Assessment of ResponseNet on genetic perturbations was based on the subset of 101 genetic perturbations for which the inactivated gene had a reliable GO process annotation (based on direct assays, manual curation or explicit citation, denoted as GO evidence codes IC, IDA or TAS). A specific ResponseNet solution was considered successful when: (i) the predicted network contained the inactivated gene, or (ii) the predicted network was significantly enriched for a biological process to which the inactivated gene was reliably attributed.
Significance was computed against two null hypotheses: (1) relative to the number of genes with the same annotation that could be found by random selection from the genome (p-value ≤ 10−2 using the hyper-geometric approach and correcting for multiple hypothesis testing), and (2) relative to the enrichment that could be found in 100 perturbation-specific solutions based on randomized inputs (empirical p-value ≤ 0.05). In both cases enrichments were calculated based on genes with reliable process annotation (evidence codes IC, IDA or TAS).
The randomizations were conducted separately for each perturbation as follows: we created 100 pairs of inputs sets of the same sizes as the original genetic and transcriptional data, containing either proteins or genes randomly chosen from the interactome. The interactome data remained fixed so that all predicted networks relied on real interaction data. Each random input set was solved using ResponseNet and the significant GO process annotation enrichments were recorded (p-value ≤ 10−2 by Fisher’s Exact Test; 0.05 FDR). Process annotations enriched in the original solution were considered significant if at least 95 random input solutions had lower significance scores. Interactome data was as described above. Physical interactions relied upon BioGRID release 2.0 70. Additional information appears in Supplementary Text.
Yeast Strains and Media
The α-syn overexpressing yeast strain we used in the modifier screen was W303 with α-syn integrated into HIS3 and TRP1 loci (IntTox): MATa can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 pRS303Gal-α–synWTYFPpRS304Gal-αSynWT-YFP. The α-syn overexpressing yeast strain we used for drug assays and microarray experiments was W303 with α-syn integrated into TRP1 and URA3 loci (HiTox): MATa can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 pRS304Gal-α-synWT-GFP pRS306Gal-α-synWT-GFP. Controls in the drug assays and microarray experiments were W303 with two copies of empty vector integrated into TRP1 and URA3 loci (2x vector): MATa can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 pRS304Gal pRS306Gal, and one copy of a-syn integrated into TRP1 locus (1x α-syn): MATa can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 pRS304Gal-α–synWTGFP. The Gal promoter reporter strain used to determine the effect of modifier genes on expression from galactose regulated promoter was W303 with YFP integrated into HIS3 locus: MATa can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1pRS303Gal-YFP. Strains were manipulated and media prepared using standard techniques.
α–Syn toxicity modifier screen
We performed the high-throughput yeast transformation protocol as described previously for a smaller library of genes 12. 5,000 full-length yeast ORFs were amplified by polymerase chain reaction and captured by recombination cloning into a Gateway™ pDONR221 vector (Invitrogen). The clones were sequenced from N-terminus to C-terminus and verified to be wild type. For the expression screen, the clones were transfered into a galactose-inducible expression plasmid (pBY011; CEN, URA3, AmpR) using the Gateway™ technology (Invitrogen). Additional information about the Yeast FLEXGene collection is available at http://www.hip.harvard.edu/research/yeast_flexgene/. Plasmid DNAs from the expression clones were isolated using the REALTM miniprep kit (Qiagen). DNA was dried in individual wells of 96-well microtiter plates and transformed into a strain expressing α–syn integrated at the HIS3 and TRP1 loci. A standard lithium acetate transformation protocol was modified for automation and used by employing a BIOROBOT Rapidplate 96-well pipettor (Qiagen). The transformants were grown in synthetic deficient media lacking uracil (SD-Ura) with glucose overnight. The overnight cultures were inoculated into fresh SD-Ura media with raffinose and allowed to reach stationary phase. The cells were spotted on to SD-Ura + glucose and SD-Ura + galactose agar plates. Suppressors of α–syn induced toxicity were identified on galactose plates after 2–3 days of growth at 30°C. We repeated the screen 3 independent times and candidate modifier genes were retested at least twice to confirm their authenticity. To exclude the possibility of false positive toxicity suppressor genes caused simply by a reduction in α–syn expression, the amount of α–syn protein was monitored by flow cytometry. To exclude false positive enhancer genes caused by a general inhibition of growth unrelated to α–syn expression, these genes were transformed into wild type yeast cells and their effect on growth determined.
Immunoblotting
Yeast lysates were subjected to SDS/PAGE (4–12% gradient, Invitrogen) and transferred to a PVDF membrane (Invitrogen). Membranes were blocked with 5% nonfat dry milk in PBS for 1 hr at room temperature. Primary antibody incubations were performed overnight at 4°C or at room temperature for 1–2 hours. After washing with PBS, membranes were incubated with a horseradish peroxidase-conjugated secondary antibody for 1 hour at room temperature, followed by washing in PBS+0.1% Tween 20 (PBST). Proteins were detected with SuperSignal West Dura (Pierce). Phosphoglycerase kinase 1(Pgk 1) mouse monoclonal antibody was used at 1:5000. Hsp26 rabbit polyclonal antibody(gift from Dr. Johannes Buchner) was used at 1:5000. Hsp104 mouse monoclonal antibody (4B; 78) was used at 1:5000.
α-Syn ResponseNet analysis
The α-syn transcriptional data consisted of genes showing at least a two-fold change in expression with a p-value ≤ 0.05 (Su et al.; manuscript submitted, Supplementary Table 7). ResponseNet was run with γ=12. Capacities of edges connecting the source node to the genetic hits were relative to the absolute strength of the genetic hits (Table 3). Capacities of edges connecting the differentially expressed genes to the sink node were relative to the absolute value of the fold change. In an effort to exclude non-specific stress response from our predictions, we ran ResponseNet with the complete genetic hits data, but using only a subset of the transcriptional data from which 111 environmental stress response genes 32 were excluded. This resulted in an almost identical network (Supplementary text).
Western blot with S-nitrosocysteine antibody
Yeast cells were harvested, spun down and snap froze prior to cell lysis via bead beating in a buffer containing 50 mM HEPES, pH 7.4, 150 mM NaCl, 1% Triton X-100, 5% glycerol, 1 mM PMSF, and EDTA-free complete protease inhibitor cocktail tablet. Protein concentration was determined via bicinchonic acid assay prior to resolution of products on SDS-PAGE, followed by transfer onto nitrocellulose membrane and probe with S-nitrosocysteine antibody (1:10,000, Sigma).
α-Syn growth in presence of small molecules
α-syn strains as well as control strains were grown overnight to saturation in media containing raffinose. Yeast cultures were normalized for their OD and serially diluted by five-fold prior to spotting onto yeast media plates containing galactose, and where necessary, rapamycin. Growth curves were monitored using Bioscreen (www.bioscreen.fi). Yeast strains were pre-grown in 2% raffinose medium and induced in 2% galactose medium with starting OD600 of 0.1. 300 µl of induced cells were dispensed to individual wells, in presence of either the compound or vehicle control (1% DMSO final). Each growth condition was analyzed in triplicate wells per run, and at least 3 independent runs were conducted for each growth condition. Cells were grown at 30°C, with plates shaken every 30 seconds to ensure proper aeration and OD600 measurements taken every half hour over a two-day period. The resulting data (OD600 versus time) were plotted using Kaleidagraph.
Supplementary Material
Acknowledgements
E.Y-L has been supported by an EMBO long term post-doctoral fellowship and by a research grant from the National Parkinson Foundation. L.R. has been supported by Roberto Rocca doctoral fellowship and the CSBi Merck-MIT postdoctoral fellowship. L.J.S. was supported by an American Cancer Society postdoctoral fellowship. A.D.G. was a Lilly Fellow of the Life Sciences Research Foundation. M.L.G is supported by a research grant from the National Parkinson Foundation. S.L. is a founder of and has received consulting fees from FoldRx Pharmaceuticals, a company that investigates drugs to treat protein folding diseases. A.D.G. and S.L. are inventors on patents and patent applications that have been licensed to FoldRx. E.F. is the recipient of the Eugene Bell Career Development Chair. This work was supported in part by MGH/MIT Morris Udall Center of Excellence in PD Research NS38372. We thank Mikko Taipale, Sebastian Treusch and Gabriela Caraveo Piso for helpful discussions and comments, and Tom DiCesare for help with figures. L.R. thanks Giorgio Casari and Sergio Cerutti for support and helpful discussions.
Footnotes
Summary: A novel approach that integrates genetic hits, differentially expressed genes and known molecular interactions reveals a dramatically enhanced view of cellular responses and was used to create the first cellular map of alpha-synuclein toxicity.
References
- 1.Calvano SE, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 2.Haugen AC, et al. Integrating phenotypic and expression profiles to map arsenic-response networks. Genome Biol. 2004;5:R95. doi: 10.1186/gb-2004-5-12-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parsons AB, et al. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat Biotechnol. 2004;22:62–69. doi: 10.1038/nbt919. [DOI] [PubMed] [Google Scholar]
- 4.Begley TJ, Rosenbach AS, Ideker T, Samson LD. Hot spots for modulating toxicity identified by genomic phenotyping and localization mapping. Mol Cell. 2004;16:117–125. doi: 10.1016/j.molcel.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Deutschbauer AM, Williams RM, Chu AM, Davis RW. Parallel phenotypic analysis of sporulation and postgermination growth in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2002;99:15530–15535. doi: 10.1073/pnas.202604399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fry RC, Begley TJ, Samson LD. Genome-wide responses to DNA-damaging agents. Annu Rev Microbiol. 2005;59:357–377. doi: 10.1146/annurev.micro.59.031805.133658. [DOI] [PubMed] [Google Scholar]
- 7.Birrell GW, et al. Transcriptional response of Saccharomyces cerevisiae to DNA-damaging agents does not identify the genes that protect against these agents. Proc Natl Acad Sci U S A. 2002;99:8778–8783. doi: 10.1073/pnas.132275199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 9.Smith JJ, et al. Expression and functional profiling reveal distinct gene classes involved in fatty acid metabolism. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100051. 2006 0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schiesling C, Kieper N, Seidel K, Kruger R. Review: Familial Parkinson's disease--genetics, clinical phenotype and neuropathology in relation to the common sporadic form of the disease. Neuropathol Appl Neurobiol. 2008;34:255–271. doi: 10.1111/j.1365-2990.2008.00952.x. [DOI] [PubMed] [Google Scholar]
- 11.Outeiro TF, Lindquist S. Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science. 2003;302:1772–1775. doi: 10.1126/science.1090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper AA, et al. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson's models. Science. 2006;313:324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Workman CT, et al. A systems approach to mapping DNA damage response pathways. Science. 2006;312:1054–1059. doi: 10.1126/science.1122088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beyer A, Bandyopadhyay S, Ideker T. Integrating physical and genetic maps: from genomes to interaction networks. Nat Rev Genet. 2007;8:699–710. doi: 10.1038/nrg2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeang CH, Ideker T, Jaakkola T. Physical network models. J Comput Biol. 2004;11:243–262. doi: 10.1089/1066527041410382. [DOI] [PubMed] [Google Scholar]
- 16.Ourfali O, Shlomi T, Ideker T, Ruppin E, Sharan R. SPINE: a framework for signaling-regulatory pathway inference from cause-effect experiments. Bioinformatics. 2007;23:i359–i366. doi: 10.1093/bioinformatics/btm170. [DOI] [PubMed] [Google Scholar]
- 17.Dasika MS, Burgard A, Maranas CD. A computational framework for the topological analysis and targeted disruption of signal transduction networks. Biophys J. 2006;91:382–398. doi: 10.1529/biophysj.105.069724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cormen TH, Leiserson CE, Rivest RL, Stein C. Introduction to Algorithms. 2001 [Google Scholar]
- 19.SGD project. “Saccharomyces Genome Database” [Google Scholar]
- 20.Hughes TR, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 21.Chang M, Bellaoui M, Boone C, Brown GW. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc Natl Acad Sci U S A. 2002;99:16934–16939. doi: 10.1073/pnas.262669299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gasch AP, et al. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol Biol Cell. 2001;12:2987–3003. doi: 10.1091/mbc.12.10.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tofaris GK, Spillantini MG. Physiological and pathological properties of alpha-synuclein. Cell Mol Life Sci. 2007;64:2194–2201. doi: 10.1007/s00018-007-7217-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spillantini MG, et al. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 25.Polymeropoulos MH, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 26.Singleton AB, et al. alpha-Synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 27.Lee VM, Trojanowski JQ. Mechanisms of Parkinson's disease linked to pathological alpha-synuclein: new targets for drug discovery. Neuron. 2006;52:33–38. doi: 10.1016/j.neuron.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 28.Gitler AD, et al. The Parkinson's disease protein alpha-synuclein disrupts cellular Rab homeostasis. Proc Natl Acad Sci U S A. 2008;105:145–150. doi: 10.1073/pnas.0710685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schapira AH. Mitochondria in the aetiology and pathogenesis of Parkinson's disease. Lancet Neurol. 2008;7:97–109. doi: 10.1016/S1474-4422(07)70327-7. [DOI] [PubMed] [Google Scholar]
- 30.Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 31.Olanow CW. Manganese-induced parkinsonism and Parkinson's disease. Ann N Y Acad Sci. 2004;1012:209–223. doi: 10.1196/annals.1306.018. [DOI] [PubMed] [Google Scholar]
- 32.Gasch AP, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dawson TM. Parkin and defective ubiquitination in Parkinson's disease. J Neural Transm Suppl. 2006:209–213. doi: 10.1007/978-3-211-45295-0_32. [DOI] [PubMed] [Google Scholar]
- 34.Staropoli JF, et al. Parkin is a component of an SCF-like ubiquitin ligase complex and protects postmitotic neurons from kainate excitotoxicity. Neuron. 2003;37:735–749. doi: 10.1016/s0896-6273(03)00084-9. [DOI] [PubMed] [Google Scholar]
- 35.West AB, Dawson VL, Dawson TM. To die or grow: Parkinson's disease and cancer. Trends Neurosci. 2005;28:348–352. doi: 10.1016/j.tins.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 36.Hoglinger GU, et al. The pRb/E2F cell-cycle pathway mediates cell death in Parkinson's disease. Proc Natl Acad Sci U S A. 2007;104:3585–3590. doi: 10.1073/pnas.0611671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarver A, DeRisi J. Fzf1p regulates an inducible response to nitrosative stress in Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:4781–4791. doi: 10.1091/mbc.E05-05-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uehara T, et al. S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature. 2006;441:513–517. doi: 10.1038/nature04782. [DOI] [PubMed] [Google Scholar]
- 39.Almeida B, et al. NO-mediated apoptosis in yeast. J Cell Sci. 2007;120:3279–3288. doi: 10.1242/jcs.010926. [DOI] [PubMed] [Google Scholar]
- 40.Mandemakers W, Morais VA, De Strooper B. A cell biological perspective on mitochondrial dysfunction in Parkinson disease and other neurodegenerative diseases. J Cell Sci. 2007;120:1707–1716. doi: 10.1242/jcs.03443. [DOI] [PubMed] [Google Scholar]
- 41.Martin LJ, et al. Parkinson's disease alpha-synuclein transgenic mice develop neuronal mitochondrial degeneration and cell death. J Neurosci. 2006;26:41–50. doi: 10.1523/JNEUROSCI.4308-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Z, Spirek M, Thornton J, Butow RA. A novel degron-mediated degradation of the RTG pathway regulator, Mks1p, by SCFGrr1. Mol Biol Cell. 2005;16:4893–4904. doi: 10.1091/mbc.E05-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94:471–480. doi: 10.1016/s0092-8674(00)81588-3. [DOI] [PubMed] [Google Scholar]
- 44.Flower TR, Chesnokova LS, Froelich CA, Dixon C, Witt SN. Heat shock prevents alpha-synuclein-induced apoptosis in a yeast model of Parkinson's disease. J Mol Biol. 2005;351:1081–1100. doi: 10.1016/j.jmb.2005.06.060. [DOI] [PubMed] [Google Scholar]
- 45.Auluck PK, Meulener MC, Bonini NM. Mechanisms of Suppression of {alpha}-Synuclein Neurotoxicity by Geldanamycin in Drosophila. J Biol Chem. 2005;280:2873–2878. doi: 10.1074/jbc.M412106200. [DOI] [PubMed] [Google Scholar]
- 46.Lin JT, Lis JT. Glycogen synthase phosphatase interacts with heat shock factor to activate CUP1 gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:3237–3245. doi: 10.1128/mcb.19.5.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hickman MJ, Winston F. Heme levels switch the function of Hap1 of Saccharomyces cerevisiae between transcriptional activator and transcriptional repressor. Mol Cell Biol. 2007;27:7414–7424. doi: 10.1128/MCB.00887-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duennwald ML, Jagadish S, Muchowski PJ, Lindquist S. Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc Natl Acad Sci U S A. 2006;103:11045–11050. doi: 10.1073/pnas.0604547103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ravikumar B, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 50.Khurana V, et al. TOR-mediated cell-cycle activation causes neurodegeneration in a Drosophila tauopathy model. Curr Biol. 2006;16:230–241. doi: 10.1016/j.cub.2005.12.042. [DOI] [PubMed] [Google Scholar]
- 51.Kelley R, Ideker T. Systematic interpretation of genetic interactions using protein networks. Nat Biotechnol. 2005;23:561–566. doi: 10.1038/nbt1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schuldiner M, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 53.Yeger-Lotem E, et al. Network motifs in integrated cellular networks of transcription-regulation and protein-protein interaction. Proc Natl Acad Sci U S A. 2004;101:5934–5939. doi: 10.1073/pnas.0306752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao R, et al. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 55.Ulitsky I, Shlomi T, Kupiec M, Shamir R. From E-MAPs to module maps: dissecting quantitative genetic interactions using physical interactions. Mol Syst Biol. 2008;4:209. doi: 10.1038/msb.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deutscher D, Meilijson I, Kupiec M, Ruppin E. Multiple knockout analysis of genetic robustness in the yeast metabolic network. Nat Genet. 2006;38:993–998. doi: 10.1038/ng1856. [DOI] [PubMed] [Google Scholar]
- 57.Holstege FC, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 58.Santos SD, Verveer PJ, Bastiaens PI. Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nat Cell Biol. 2007;9:324–330. doi: 10.1038/ncb1543. [DOI] [PubMed] [Google Scholar]
- 59.Goehler H, et al. A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington's disease. Mol Cell. 2004;15:853–865. doi: 10.1016/j.molcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 60.Pujana MA, et al. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat Genet. 2007;39:1338–1349. doi: 10.1038/ng.2007.2. [DOI] [PubMed] [Google Scholar]
- 61.Lim J, et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 62.Said MR, Begley TJ, Oppenheim AV, Lauffenburger DA, Samson LD. Global network analysis of phenotypic effects: protein networks and toxicity modulation in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2004;101:18006–18011. doi: 10.1073/pnas.0405996101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shachar R, Ungar L, Kupiec M, Ruppin E, Sharan R. A systems-level approach to mapping the telomere length maintenance gene circuitry. Mol Syst Biol. 2008;4:172. doi: 10.1038/msb.2008.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bromberg KD, Ma'ayan A, Neves SR, Iyengar R. Design logic of a cannabinoid receptor signaling network that triggers neurite outgrowth. Science. 2008;320:903–909. doi: 10.1126/science.1152662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tu Z, Wang L, Arbeitman MN, Chen T, Sun F. An integrative approach for causal gene identification and gene regulatory pathway inference. Bioinformatics. 2006;22:e489–e496. doi: 10.1093/bioinformatics/btl234. [DOI] [PubMed] [Google Scholar]
- 66.Suthram S, Beyer A, Karp RM, Eldar Y, Ideker T. eQED: an efficient method for interpreting eQTL associations using protein networks. Mol Syst Biol. 2008;4:162. doi: 10.1038/msb.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Silva JM, et al. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science. 2008;319:617–620. doi: 10.1126/science.1149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang X, et al. Lower low-density lipoprotein cholesterol levels are associated with Parkinson's disease. Mov Disord. 2007;22:377–381. doi: 10.1002/mds.21290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang X, Abbott RD, Petrovitch H, Mailman RB, Ross GW. Low LDL cholesterol and increased risk of Parkinson's disease: prospective results from Honolulu-Asia Aging Study. Mov Disord. 2008;23:1013–1018. doi: 10.1002/mds.22013. [DOI] [PubMed] [Google Scholar]
- 70.Reguly T, et al. Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J Biol. 2006;5:11. doi: 10.1186/jbiol36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mewes HW, et al. MIPS: a database for genomes and protein sequences. Nucleic Acids Res. 2002;30:31–34. doi: 10.1093/nar/30.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.MacIsaac KD, et al. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Milo R, et al. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 74.Harbison CT, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Myers CL, et al. Discovery of biological networks from diverse functional genomic data. Genome Biol. 2005;6:R114. doi: 10.1186/gb-2005-6-13-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hoffmann R, Valencia A. Life cycles of successful genes. Trends Genet. 2003;19:79–81. doi: 10.1016/s0168-9525(02)00014-8. [DOI] [PubMed] [Google Scholar]
- 77.Vanderbei RJ. LOQO User's Manual--Version 3.10. Optimization Methods and Software. 1999;12:485–514. [Google Scholar]
- 78.Cashikar AG, et al. Defining a pathway of communication from the C-terminal peptide binding domain to the N-terminal ATPase domain in a AAA protein. Mol Cell. 2002;9:751–760. doi: 10.1016/s1097-2765(02)00499-9. [DOI] [PubMed] [Google Scholar]
- 79.Dudley AM, Janse DM, Tanay A, Shamir R, Church GM. A global view of pleiotropy and phenotypically derived gene function in yeast. Mol Syst Biol. 2005;1 doi: 10.1038/msb4100004. 2005 0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Koerkamp MG, et al. Dissection of transient oxidative stress response in Saccharomyces cerevisiae by using DNA microarrays. Mol Biol Cell. 2002;13:2783–2794. doi: 10.1091/mbc.E02-02-0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tong AH, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 82.Tong AH, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.