Summary
RVB1/RVB2 are two highly conserved members of the AAA+ family that are present in different protein and nucleoprotein complexes. Recent studies implicate that RVB-containing complexes play a role in variable cellular processes such as transcription, DNA damage response, snoRNP assembly, cellular transformation and cancer metastasis. In this review we discuss recent advances in the understanding of RVB-containing complexes and the functions of RVBs in these pathways.
Introduction
RVB1/RVB2 (also known as Pontin/Reptin, TIP49/TIP48, RuvbL1/RuvbL2, ECP54/ECP51, INO80H/INO80J, TIH1/TIH2, TIP49A/TIP49B) are ATP binding proteins that belong to AAA+ (ATPase Associated with diverse cellular Activities) family of ATPases. RVBs were discovered independently in multiple organisms, first in rat as interactors of TATA-binding protein (named TIP49a and TIP49b) (Kanemaki et al., 1999; Kanemaki et al., 1997), in human cells as interactor of human RPA14 (named as RuvbL1 and RuvbL2)(Qiu et al., 1998), component of a large nuclear protein complex (named ECP-51 and ECP-54) (Salzer et al., 1999) and as essential interactor of β-catenin (named pontin52 and reptin52) (Bauer et al., 2000; Bauer et al., 1998) or c-Myc (Wood et al., 2000). RVB proteins have been implicated in many cellular pathways (Fig. 1). The structure of the RVB1/RVB2 complex that has recently been elucidated suggests that RVBs could act as a scaffolding protein, explaining its appearance in diverse cellular protein complexes (Matias et al., 2006; Puri et al., 2007; Torreira et al., 2008). RVBs are part of various chromatin remodeling complexes (Jha et al., 2008; Jin et al., 2005; Jonsson et al., 2004; Mizuguchi et al., 2004; Shen et al., 2000) and are required for their activities (Jha et al., 2008; Jonsson et al., 2004). As part of chromatin remodeling complexes they regulate the accessibility of DNA to the proteins involved in transcription and DNA damage repair by regulating the position or modification of nucleosomes. The RVB proteins have been implicated in cellular transformation by Myc and β-catenin through its chromatin remodeling function (Feng et al., 2003; Wood et al., 2000). Interestingly, RVBs were also identified as interactor of snoRNA and are essential for assembly and maturation of snoRNPs (Newman et al., 2000; Watkins et al., 2002; Watkins et al., 2004). In this review we discuss the role of RVBs in these complexes and describe important questions to be addressed in the future.
Figure 1.
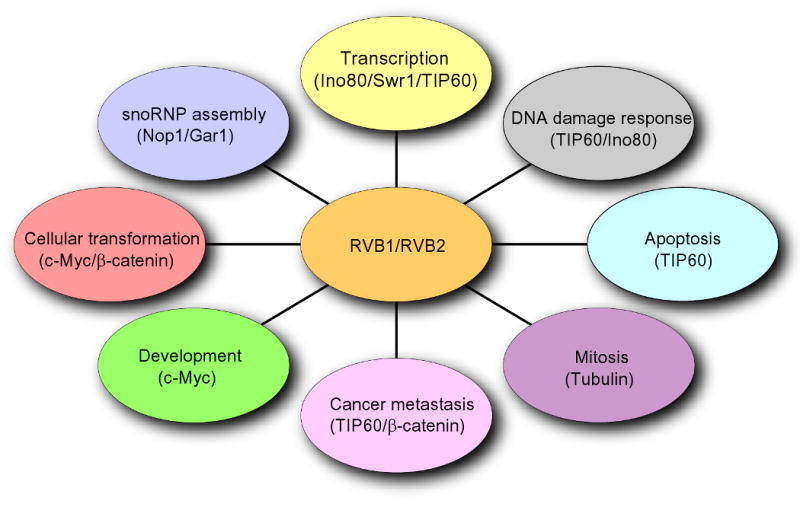
RVB1 and RVB2 are involved in multiple cellular pathways. Schematic showing involvement of RVBs in transcription, DNA damage response, small nucleolar ribonucleotide protein (snoRNPs) assembly, cellular transformation, cancer metastasis, apoptosis, mitosis and development. RVBs act in (i) transcription by regulating function of Ino80, Swr1 and TIP60 complexes, (ii) DNA damage response through TIP60 and Ino80, (iii) snoRNP assembly by affecting maturation of snoRNAs and localization of Nop1 and Gar1, (iv) cellular transformation by c-Myc and β-catenin function, (v) cancer metastasis by regulating expression of KAI1 through TIP60 and β-catenin, (vi) apoptosis through TIP60, (vii) mitosis by regulating assembly of microtubules and (viii) development through c-Myc pathway.
Structural insight on RVB1/RVB2
Sequence analysis of RVB1 and RVB2 shows that both these proteins are very similar to each other and are highly conserved (Fig. 2A). They have conserved Walker A (P-loop, which binds and orients the γ-phosphate for ATP hydrolysis), Walker B box, an arginine-finger (Arg) and sensor domains I and II to sense whether the protein is bound to the di- or tri-nucleotide (Fig. 2A). The crystallographic structure of RVB1 alone and the electron-microscopic structure of RVB1/RVB2 shed new light on these enigmatic proteins (Gribun et al., 2008; Matias et al., 2006; Puri et al., 2007; Torreira et al., 2008). The high resolution crystal structure of human RVB1, solved at 2.2 Å resolution, shows that RVB1 assembles as a hexamer and that each monomer has three distinct domains i.e. domain I (1-120 aa + 296-365 aa), domain II (121-295) and domain III (368-456 aa) (Fig. 2B) (Matias et al., 2006). Domain I of RVB1 forms the core domain similar to the AAA+ module of other family members (Matias et al., 2006; Torreira et al., 2008). A striking difference between RVB1 and other AAA+ family members is that the two halves of domain I are separated by ∼170 amino acids that form domain II which is unique to RVB1. This ATPase-insert domain II is attached to the ATPase domain I by a flexible hairpin-shaped linker composed of two beta strands, and has been proposed to be important for new functions related to DNA/RNA binding (Matias et al., 2006) or oligomerization (Torreira et al., 2008). Domain III is composed of four alpha helices that serve to cap the nucleotide binding pocket in domain I and is similar to the domain II of bacterial RuvB ATPase.
Figure 2.
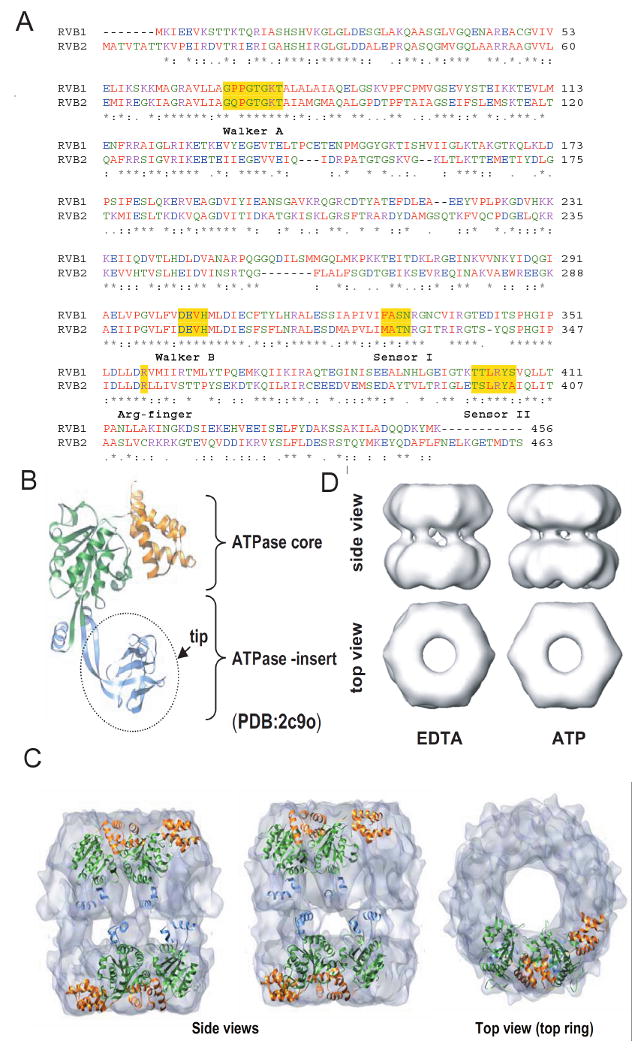
Sequence and structural details of RVBs. (A) Protein sequence alignment of human RVB1 and RVB2 showing regions of similarity where Walker A, Walker B, sensor 1, sensor 2 and arginine finger are highlighted. (B) Structure of monomeric RVB1. ATPase core domain consisting of domain I (1-120 aa + 296-365 aa) and domain III (368-456 aa) and ATPase insert domain II (121-295 aa) are shown. (C) Electron microscopic (EM) image of dodecameric RVB1 and RVB2. RVB1 and RVB2 are arranged as a double hexamer and interact through their ATPase insert domain. (D) Change in conformation after binding of ATP. Different views of the EM images of RVB1 and RVB2 in presence of ATP or EDTA.
The low resolution structure of the RVB1/RVB2 complex has been solved by electron microscopy (EM) (Gribun et al., 2008; Puri et al., 2007; Torreira et al., 2008). In these studies the RVB1/RVB2 complexes were assembled using recombinant proteins from bacteria purified separately (Gribun et al., 2008; Puri et al., 2007; Torreira et al., 2008) or from insect cells expressing both proteins to form a double hexamer (Gribun et al., 2008; Puri et al., 2007; Torreira et al., 2008). Although RVB1 or RVB2 expressed individually are monomeric at low concentration (5 μM) they hexamerize at a higher concentration (40 μM) (Gribun et al., 2008; Puri et al., 2007) and in the presence of ADP or ATP-Mg++ (Gribun et al., 2008; Puri et al., 2007). Mixing RVB1 and RVB2 purified from bacteria promotes oligomerization [(Gribun et al., 2008); S.J. and A.D. unpublished data]. Two groups have reported that human or yeast RVB proteins (1 and 2 together) form a double-hexameric ring, while a third group has suggested that yeast RVB1 & RVB2 form a single hexameric ring. Even when RVB1 and RVB2 form a double-hexamer, the double hexamer can partially dissociate into a single hexamer enriched in RVB1 and antibodies specific for one of the subunits selectively decorate one end of the double-hexamer (Torreira et al., 2008). Taken together these results suggest that the highly similar RVB1 and RVB2 proteins can exist as monomers, homo- or hetero-hexamers and as a double-hexamer (most likely made of two homo-hexamers). RVB1 and RVB2 are individually required for viability in yeast, C. elegans, Drosophila and Xenopus (Etard et al., 2005; Jonsson et al., 2004; Sheaffer et al., 2008), co-immunoprecipitate with each other from cell extracts and decrease of one of the proteins is known to destabilize the other in vivo (S.J and A.D. unpublished data), strongly suggesting that the hetero-oligomers of RVB1 and RVB2 are present in vivo.
When RVB1 and RVB2 are observed in a double-hexamer conformation, domains I and III appear to form hexameric rings, similar to that observed for other AAA+ ATPases. The two rings are asymmetric, as the top ring is slightly wider than the bottom and the two RVB rings interact in a back to back fashion through their domain II (also known as ATPase insert domain) (Fig. 2C) (Torreira et al., 2008). The interaction domains are located closer to one of the two rings formed by the AAA+ core domains, contributing to the asymmetry between the rings.
An isolated Domain II has a structure similar to that of the single-stranded DNA binding domains of RPA and can bind single-stranded DNA, double-stranded DNA and RNA (Matias et al., 2006). While domain II is available for binding nucleic acid when the RVB1 protein forms a single hexamer, it is unclear whether the inter-ring interactions in the double-hexamer permit domain II to interact with nucleic acid. Thus oligomerization of RVB may regulate its ability to bind nucleic acids, and conversely, nucleic acid binding might affect the ability of RVB to form a double-hexamer. Comparison of the hexameric RVB1 with that of the double-hexameric RVB1 & RVB2 also suggests that the central channel widens from 15 Å for the hexamer (large enough to accommodate single-stranded, but not double-stranded DNA) to 25 Å for the double hexamer (large enough of accommodate double-strand DNA) (Matias et al., 2006; Puri et al., 2007; Torreira et al., 2008).
As RVBs have conserved ATP binding and hydrolysis motifs and mutations within these motifs affect the viability of the organism (Jonsson et al., 2001), there have been a number of experiments performed to understand the importance and significance of nucleotide binding and hydrolysis. From the crystal structure of RVB1-ADP complex it appears that ADP binds tightly between domains I of the adjacent subunits, with the Arg-finger of one subunit positioned to interact with the ATP bound to the adjoining subunit (Matias et al., 2006). Consistent with the importance of this inter-subunit interaction, the RVB1 & RVB2 double-hexamer or the RVB1 single-hexmer have stronger ATPase activities than monomers of RVB proteins. The tight fit between adjoining subunits and the presence of the capping domain III make access to the ATPase site difficult and thus affect the exchange of ADP for ATP. This may be responsible for the weak ATPase activity of RVB complexes and raises the possibility that co-factors may stimulate the ATPase activity of RVBs. Such stimulation has been reported by double-stranded DNA with a 5′ overhang (Gribun et al., 2008). Since, however, RVB proteins are known to co-purify with a bacterial ATPase that binds to single-stranded DNA (Puri et al., 2007), recombinant RVB with mutations in the Walker B motif should be tested in parallel experiments before concluding that RVB is the nucleic-acid-stimulated ATPase (Gribun et al., 2008). Interestingly, changes in conformation of the RVB1 & RVB2 complex in EM images were observed after exposure to nucleotide (Fig. 2D) (Gribun et al., 2008; Puri et al., 2007; Torreira et al., 2008). These changes are consistent with the idea that RVB proteins function as molecular motors that utilize the energy of ATP hydrolysis for their function.
In summary, although RVB1 and RVB2 are very similar to each other and to other members of AAA+ family at the sequence level, recent attempts at understanding the architecture of these proteins have identified several unique and distinctive feature of RVB1/RVB2.
RVBs in transcription activation
Transcription of genes involves the coordinated action of many different factors, which require transcription regulatory proteins to facilitate access to DNA (Ruthenburg et al., 2007). These transcription regulatory proteins can be distinguished into two classes (Narlikar et al., 2002). The first class comprises the ATP-dependent chromatin remodeling enzymes that utilize the energy of ATP hydrolysis both to mobilize nucleosomes and to exchange histones from the DNA. The second class consists of enzymes that add or remove covalent modifications on histones like acetylation, methylation or ubiqutiylation resulting in a change in chromatin state by recruiting various protein complexes that recognize these modification on histones [reviewed in (Ruthenburg et al., 2007)]. RVB1/RVB2 have been identified in both classes of chromatin remodeling complexes and several studies have implicated them in regulating transcription. In the following section we will discuss various RVB-containing chromatin remodeling complexes involved in transcription.
Ino80 complex
One of the most studied and well characterized RVB1/RVB2 containing chromatin remodeling complex contains Ino80. The multi-subunit Ino80 complex was identified in yeast (Fig. 3). The Ino80 protein, a member of the Swi2/Snf2 superfamily, is the catalytically active subunit as mutation of lysine 737 to alanine abolishes the chromatin remodeling activity of the complex (Jin et al., 2005; Jonsson et al., 2004; Shen et al., 2000). The energy of ATP hydrolysis is utilized to slide nucleosomes on DNA, increasing the accessibility of the DNA to various proteins. The Ino80 complex has been identified in different organisms and is conserved within eukaryotes [(Jin et al., 2005; Jonsson et al., 2004; Shen et al., 2000) and reviewed in (Bao and Shen, 2007)]. The Ino80 complex has both RVB1 and RVB2 in a 6:1 stoichiometry relative to the other subunits. Thus stoichiometry is consistent with the double hexameric structure of RVBs (Jonsson et al., 2004). Although the Ino80 complex has been identified, purified and characterized in both yeast and human and has similar nucleosome-stimulated ATPase activity and ATP dependent chromatin remodeling activity in vitro, there are some differences in the composition of the complex (Jin et al., 2005; Jonsson et al., 2004; Shen et al., 2000) (Fig. 3). The human Ino80 complex contain orthologs of yeast Ino80, RVB1, RVB2, Arp4, Arp5, Arp8, Ies2 and Ies6 (Jin et al., 2005). In addition, the b-ZIP domain-containing Amida protein, the forkhead-associated domain-containing MCRS1 protein, the NFRKB protein, and proteins encoded by the FLJ90652 and FLJ20309 ORFs are found only in the human Ino80 complex (Fig. 3). The functional significance of the human-specific subunits is yet to be determined (Jin et al., 2005).
Figure 3.
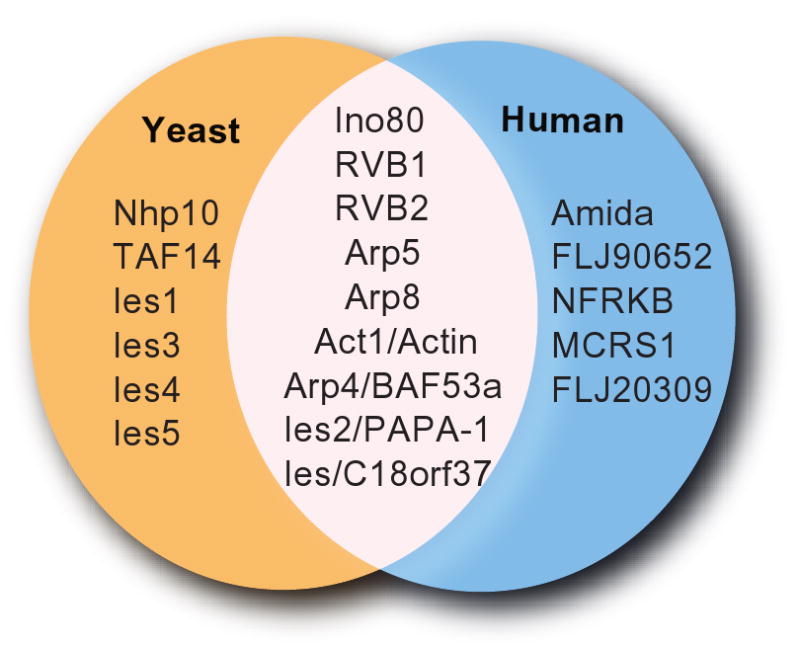
Composition of human and yeast Ino80 complexes. Subunits that are common and different between human and yeast Ino80 complex are shown.
In yeast, both Ino80 and RVBs regulate transcription of a similar set of genes, and in vitro assays show that RVBs are essential for Ino80-dependent chromatin remodeling activity but not required for binding of Ino80 complex to the promoters (Jonsson et al., 2004). However, binding of RVBs to the promoter could not be detected by chromatin immunoprecipitation assay (Jonsson et al., 2004). Ino80 complex purified from cells lacking RVB2 also lacked RVB1 and addition of recombinant RVB1/RVB2 to the Ino80 complex in vitro did not restore chromatin remodeling activity. The Ino80 complex purified in the absence of RVBs was also depleted of an actin-like Arp5 subunit that is essential for the chromatin remodeling activity (Jonsson et al., 2004). In vitro association studies indicate that ATP-bound RVBs promotes the assembly of Arp5 into the Ino80 complex. Collectively, these results suggest that the primary function of RVBs is to nucleate the association of Arp5 with the Ino80 complex in an ATP dependent manner. Thus, RVBs are required for the proper assembly and function of the Ino80 complex, but we do not know yet whether this function is conserved in humans.
SWR1/SRCAP complex
Swr1 (SRCAP, mammalian homolog of Swr1) is also a Swi2/Snf2-related ATPase and the Swr1 complex exhibits nucleosome stimulated ATPase activity (Mizuguchi et al., 2004). Mutation at the catalytic site of Swr1 (lysine 727 to glycine) impairs the function of the Swr1 complex, confirming that the Swr1 polypeptide is the catalytic subunit. The Swr1 complex is specifically involved in the loading of histone H2B-H2AZ dimers on to chromatin (Kobor et al., 2004; Krogan et al., 2003; Mizuguchi et al., 2004) to generate a structurally and functionally distinct chromatin region. Studies in various systems have implicated H2AZ in transcriptional activation, antagonizing gene-silencing and chromosome stability [reviewed in (Raisner and Madhani, 2006)]. Genome-wide studies demonstrate that H2AZ is globally localized to gene promoters near transcription initiation sites and Swr1 is required for the H2AZ deposition (Raisner et al., 2005; Zhang et al., 2005).
The Swr1 complex purified from yeast is comprised of 16 subunits: Swr1, Swc2, Swc3, Swc4, Swc5, Swc6 Swc7, Yaf9, Bdf1, Act1, Arp4, Arp6, H2AZ, H2B, RVB1 and RVB2 (Mizuguchi et al., 2004) (Table 1). The N-terminal region of Swr1 is responsible for binding of Arp4, Act1, Swc4, Swc5 and Yaf9, whereas the ATPase domain is crucial for the association of Swc2, Swc3, RVB1, RVB2, Arp6 and Swc6 (Wu et al., 2005). Out of the six subunits of the Swr1 complex studied, only Swc5 is dispensable for complex assembly while Swc2, Arp6, Swc6, Yaf9 and Swr1 are required for the association of the other subunits (Wu et al., 2005). Swc2 directly binds to H2AZ through its N-terminal region (1-281) thus facilitating the association of the Swr1 complex with H2AZ. Arp6 and Swc6 are required for nucleosomal binding and exchange whereas Yaf9 is required for H2AZ exchange but not for nucleosomal binding (Wu et al., 2005). Note that in these experiments the function of subunits essential for survival (RVB1, RVB2, Swc4, Arp4 and Act1) were not tested. As RVBs interacts through the ATPase domain of Swr1 that is shared as an interaction domain with other subunits (Swc2, Swc3, Arp6 and Swc6) it would be interesting to know whether RVBs regulates Swr1-dependent ATPase activity or nucleosomal exchange.
Table 1.
Composition of yeast Swr1, yeast NuA4, human SRCAP and human TIP60.
| Species | Yeast | Human | ||
|---|---|---|---|---|
| Complex | Swr1 | NuA4 | SRCAP | TIP60 |
| Catalytic subunit | Swr1 | Esa1 | Srcap | TIP60 |
| Homologous subunits | RVB1 | TIP49 | ||
| RVB2 | TIP48 | |||
| Swc2 | YL-1 | |||
| Arp4 | Baf53a | |||
| Yaf9 | GAS41 | |||
| Act1 | Act1 | |||
| Swc4 | DAMP1 | |||
| Arp6 | Arp6 | |||
| H2AZ | H2AZ | |||
| H2B | H2B | |||
| Swc6 | ZnF-HIT1 | |||
| Tra1 | TRRAP | |||
| Eaf3 | MRG15 | |||
| Eaf6 | Eaf6 | |||
| Eaf7 | MRGBP | |||
| Epl1 | EPC1 | |||
| Yng2 | ING3 | |||
| Bdf1 | Brd8 | |||
| Unique subunits | Swc3 | |||
| Swc5 | ||||
| Swc7 | ||||
| Eaf1 | ||||
| Eaf5 | ||||
| p400 | ||||
TIP60/NuA4 complex
TIP60/NuA4 (Nucleosomal Acetyltransferase of H4) complex is a member of a large group of evolutionary related histone acetyltransferase enzymes (HAT) and belong to the MYST (Moz, Ybf2/Sas3, Sas2 and TIP60) family. This complex was initially purified from yeast and had nucleosomal H4/H2A HAT activity. TIP60 also acetylates several non-histone proteins. RVBs are component of the TIP60 complex and are required for its acetyltransferase activity (Jha et al., 2008) thus influencing wide range of cellular pathways.
A high degree of conservation is seen between the subunits of yeast NuA4 and human TIP60 complex (Table 1) (Doyon et al., 2004; Ikura et al., 2000). Esa1 in yeast and TIP60 in humans are the catalytic subunit of the NuA4 complex (Ikura et al., 2000; Smith et al., 1998). An interesting feature shared by both Esa1 and TIP60 is the presence of a chromodomain at the N-terminal of the protein. Chromodomains have been suggested to be involved in binding to methylated histones and RNA-binding modules (Brehm et al., 2004). However, the chromodomain's role in Esa1/TIP60 function is still unclear. Although a recent study suggests that TIP60 binds to methylated p53 in a Set7 dependent manner (Kurash et al., 2008), whether the chromodomain has a role in this interaction has yet to be ascertained.
Although many subunits are conserved between yeast NuA4 and human TIP60 complexes, Brd8, YL-l, RVB1 and RVB2 of the human TIP60 complex are absent in yeast NuA4 complex (Table 1). The presence and requirement of RVBs in the human TIP60 complex is at odds with their absence in the yeast NuA4 complex. However, the presence of RVBs in the yeast Swr1 complex raises the possibility that some of the subunits of yeast Swr1 and NuA4 complexes have been interchanged to constitute the human TIP60 complex (Table 1). Yeast Bdf1 (a bromodomain containing subunit of Swr1 complex) shows synthetic lethality with yeast Esa1 (Matangkasombut and Buratowski, 2003) suggesting the two complexes may carry out redundant functions. In addition, human p400/Domino is important for H2AZ exchange at mammalian promoters (Gevry et al., 2007), a function that in yeast is attributed solely to the Swr1 complex and not to the NuA4 complex that actually contains yeast p400/Domino. These observations suggest that at least some of the subunits and functions of two separate yeast complexes, Swr1 and NuA4, have been integrated into a single TIP60 complex in humans (Table 1). Further studies are necessary to test this hypothesis.
TIP60 and other conserved subunits of the human TIP60 complex have a well-demonstrated role in transcription. Following serum stimulation, the human TIP60 complex is recruited by c-Myc to its target promoters (Frank et al., 2003). c-Myc and TIP60 (along with other subunits TRRAP, p400, RVB1, RVB2, BAF53 and actin) appear rapidly on the nucleolin (NUC) intron 1 and this recruitment is c-Myc dependent. Myc-/-cells failed to induce nucleolin expression or recruit TIP60 to the nucleolin gene in these cells (Frank et al., 2003). The E2F family of transcription factors also utilize the TIP60 complex. In higher eukaryotes “activating” and “repressive” E2F species have been identified: E2F1, -2, -3 are transactivators whereas E2F4, -5 repress the expression of the target genes (reviewed in (van den Heuvel and Dyson, 2008)). These activating and repressive functions of E2Fs are primarily achieved by recruiting either activating complexes like HATs or repressive complexes such as histone deacetylase (HDACs) or histone methyltransferase (HMTs). Activating E2F interacts with PCAF/GCN5 and p300/CBP. In addition, the human TIP60 complex is recruited to the chromatin in late G1 after mitogenic stimulation in an E2F-dependent manner that results in acetylation of histone H3 and H4 (Taubert et al., 2004). Furthermore, by performing chromatin immunoprecipitation assays (ChIP) binding of TRRAP, p400, RVB1 and RVB2 was also observed. In summary RVB-containing human TIP60 complex acts as a co-factor that regulates transcription through the acetylation of histones.
TIP60 not only acetylates histones but also acetylates other cellular proteins involved in transcription. It plays an important role in modulating Androgen Receptor (AR) function. Intriguingly TIP60 interacts with AR (Gaughan et al., 2001), directly acetylates AR (Gaughan et al., 2002) and is required for AR-mediated regulation of prostate cellular growth by regulating the AR-mediated gene expression program. Another important TIP60 target protein is p53. Recent studies identified TIP60-dependent acetylation of p53 on K120, with this modification helping to decide between the cell-cycle arrest and apoptotic functions of p53 (Sykes et al., 2006; Tang et al., 2006). Acetylation on K120 of p53 is required for binding of p53 on promoters of pro-apoptotic genes. Additionally, mutation of K120R has been identified in human cancers suggesting that this mutation may affect p53 mediated apoptotic function. Given the fact that RVBs associate stably with the human TIP60 complex, their role in regulating transcription through the modulation of histone acetylation or the acetylation of AR or p53 is virtually unexplored.
RVBs in transcription repression
RVBs are also found in protein complexes involved in repressor function. c-Myc acts as a repressor by binding to Miz-1 transcription factor (Wanzel et al., 2003). Among the well characterized targets of c-Myc/Miz-1 repression is p21, a cyclin dependent kinase inhibitor protein. Recent studies in Drosophila (Bellosta et al., 2005) and Xenopus (Etard et al., 2005) implicate RVBs in regulating Myc function. Overexpression of RVB1 and RVB2 in fertilized Xenopus eggs increases cell proliferation while depletion of RVB1 and RVB2 reduces proliferation (Etard et al., 2005). Interestingly, expression of c-Myc, but not c-Myc mutant (MycV394D) defective in Miz-1 binding, rescues the cell proliferation defect seen after depletion of RVB. Additionally, overexpression of a dominant negative form of Miz-1 (Miz1ΔZn lacking the DNA binding domain) mimicked the RVB-depletion phenotype of reduced growth. Furthermore, c-Myc interactes with full length RVB, but not with N-terminally-deleted RVBs that were unable to form homo- or heterodimers of RVB1 and RVB2. Together, these results indicate that in Xenopus, RVB oligomers associate and cooperate with c-Myc/Miz-1 to promote cell proliferation, possibly by repressing p21 gene expression. These experiments, however, do not give any mechanistic insight on how RVB regulate cMyc/Miz-1. One interesting possibility is that just as RVB function as a scaffolding protein in yeast Ino80 complex or the human TIP60 complex, the RVB oligomer promotes the assembly of a repressor complex nucleated by c-Myc/Miz-1. RVB are also present in other repressor complexes like Polycomb, β-catenin and nuclear factor (NF)-κB, and so could have a similar function in these complexes (Bauer et al., 2000; Diop et al., 2008; Kim et al., 2005). Interestingly, RVB1 and RVB2 function antagonistically to each other in these complexes and function independent of each other in different complexes. The reason for this will be discussed later.
RVBs in DNA damage response
For the purpose of discussing the role of RVB-containing complexes in DNA damage response (DDR) we have divided DDR into four steps: 1) sensing and activation of DDR by sensors, 2) amplification of DDR signal, 3) recruitment of repair proteins at the site of damage and 4) signaling to activate cell cycle checkpoints (Fig. 4A).
Figure 4.
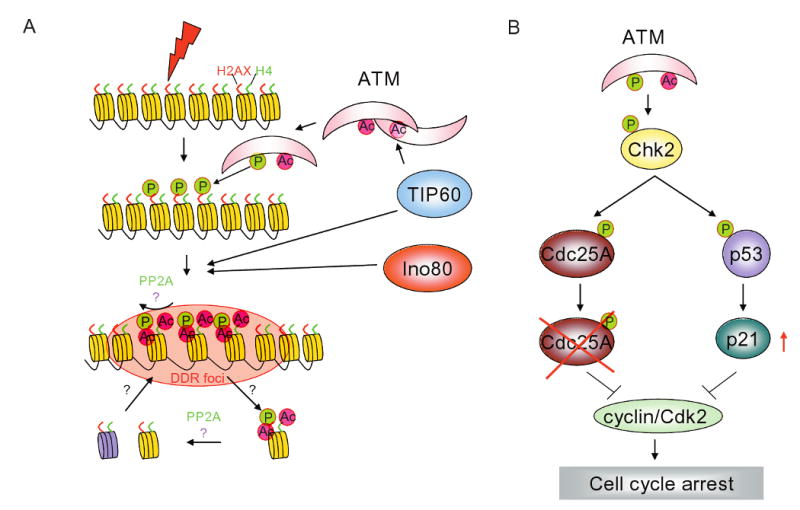
RVBs are involved at multiple steps in the DNA damage response pathways. (A) Schematic showing various steps of DNA damage response (DDR). Step 1) sensing and activation of DDR by sensors involves acetylation of ATM by TIP60 complex, 2) amplification of DDR signal by phosphorylation of H2AX by ATM and acetylation of histone H2AX and H4 by TIP60 complex, 3) recruitment of repair proteins at the site of damage is facilitated by chromatin de-condensation by action of TIP60 and Ino80 complexes. Relaxation of chromatin at the site of DNA damage enables binding of DNA damage repair protein and 4) signaling to activate cell cycle checkpoints. Activation of ATM results in phosphorylation of Chk2 that activates a fast and slow checkpoint signaling by either degrading Cdc25A or increasing levels of p21, respectively.
Sensors
Two groups of proteins have been identified as DNA damage sensors; the members of the phosphatidylinositol 3-kinase-related protein kinase (PIKK) family and RFC/PCNA (clamp loader/polymerase clamp) related Rad17-RFC/9-1-1 complex (Sancar et al., 2004). Among the PIKK family member ataxia-telangiectasia mutated (ATM), ataxia telangiectasia and Rad3 related (ATR) and DNA-dependent protein kinase catalytic subunit (DNA-PKcs) have been implicated in sensing and activating the DNA damage signal (Sancar et al., 2004).
Ataxia-telangiectasia mutant (ATM) protein kinase plays a central role in sensing and activation of DNA damage signaling. Under normal conditions ATM is present as a soluble inactive dimer. Upon DNA damage, autophosphorylation at Ser1981 results in dissociation of the dimer into active monomers (Bakkenist and Kastan, 2003). Even a few double strand breaks (DSBs) resulting from stalled replication forks or from local change in chromatin topology is sufficient to activate nuclear ATM (Bakkenist and Kastan, 2003). Once activated, ATM binds to the chromatin aided by the MRN complex and phosphorylates its substrates. Interestingly, ATM is acetylated in response to DNA damage and the kinetics of acetylation correlates with autophosphorylation on Ser 1981 indicating that acetylation may regulate ATM activation (Sun et al., 2005; Sun et al., 2007). Consistent with this, overexpression of wild-type TIP60 increases acetylation and autophosphorylation of ATM while overexpression of mutant TIP60 (defective in HAT activity) reduces acetylation and autophosphorylation of ATM and reduces phosphorylation of ATM substrates such as Chk2 and p53 (Sun et al., 2005). These data suggest that the acetyltransferase activity of TIP60 is required for efficient activation of the DNA damage signal through ATM. Since, RVBs are required for HAT activity of the TIP60 complex on histone H4 (Jha et al., 2008), it is conceivable that RVBs are also required for the sensing of the DNA damage response. However, downregulation of RVB1 does not decrease the phosphorylation of ATM substrates (Jha et al., 2008), raising the possibility that the requirement of RVB for the acetyltransferase activity of the TIP60 complex is substrate-specific.
Amplification
The amplification step is primarily achieved by ATM/ATR mediated phosphorylation of histone variant H2AX on Ser 139. Phosphorylation of H2AX spreads over megabases of DNA around the damage sites and serves as a marker for damaged DNA that recruits other proteins like MDC1, MRN complex, 53BP1 and BRCA1 to either amplify the damage signal or for DNA damage repair (Bonner et al., 2008). Interestingly, the RVB-containing NuA4/TIP60 complex of Drosophila, yeast or human is recruited to DNA damage sites (Bird et al., 2002; Ikura et al., 2007; Jha et al., 2008; Kusch et al., 2004), and in the higher eukaryotes is required for the dephosphorylation of H2AX that terminates the DDR signal. Drosophila NuA4 complex co-purifies with phosphoH2AV (the fly equivalent of phosphoH2AX) and acetylates nucleosomal phosphoH2AV at lysine 5, a modification that is essential for exchange with the unmodified H2AV (Kusch et al., 2004). Human TIP60 complex is also required to remodel phosphoH2AX containing nucleosomes at sites of DNA damage in vivo but acetylation of histone H4 is necessary for this remodeling prior to dephosphorylation of phosphoH2AX (Jha et al., 2008). Additionally, RVB1 is critical for the dephosphorylation of phosphoH2AX due to its role in maintaining the HAT activity of TIP60 (Jha et al., 2008). RVB1 is required for maintaining the HAT activity of the TIP60 complex, but not of the isolated TIP60 polypeptide (Jha et al., 2008). Addition of recombinant RVB1 to the inactive TIP60 complex from RVB1 depleted cells restores HAT activity, suggesting that RVB1 is required for the proper assembly of the TIP60 complex. Another group has also reported that TIP60 associates with H2AX and is required for the mobilization of H2AX in the first five minutes after induction of double-strand breaks with ionizing radiation (Ikura et al., 2007). However, a different mechanism was suggested where TIP60 acetylates H2AX on K5 and promotes UBC13 mediated polyubiquitination of H2AX on K119 prior to release of phosphoH2AX from chromatin. Thus both H4 and H2AX are important substrates of the NuA4/Tip60 complex for the down-modulation of the phosphoH2AX response, with H2AX acetylation and ubiquitinylation being more important early in the DNA damage response (within minutes) while H4 acetylation is more important on a longer time scale. In summary, once ATM/ATR kinases are activated the DNA damage signal is amplified by phosphorylating H2AX. This amplification signal is down-modulated through the dephosphorylation of phosphoH2AX by human PP2A (Chowdhury et al., 2005; Jha et al., 2008) or yeast PPH3 (Keogh et al., 2006) but for this to occur in higher eukaryotes nucleosomal remodeling activity of RVB1-containing TIP60 complex is required. Although the yeast NuA4 complex is recruited to the sites of DNA damage, it is not known whether it is required for the dephosphorylation of phosphoH2A.
Repair
Phosphorylation of H2AX recruits not only DNA damage repair proteins but also chromatin modifiers and remodelers that alter the chromatin structure and facilitate repair. Yeast NuA4 complex interacts with phosphorylated H2A and is recruited to the sites of double strand breaks (Bird et al., 2002). The N-terminal tail of histone H4 is acetylated by the NuA4 complex. Mutations in these lysines, deletion of the N-terminal tail or mutations in NuA4 subunits (Esa1 or Yng2) result in hypersensitivity of the yeast to DNA damaging agents without a significant change in expression of any of the repair proteins. The requirement of histone H4 acetylation for recovery from DNA damage, the recruitment of NuA4 to sites of DNA damage and the higher activity of NuA4 on linear nucleosomal arrays (mimicking a break site) relative to circular arrays suggest that chromatin around the site of damage needs to be opened by the NuA4/TIP60 complex before repair can take place. Of course, this conclusion may need to be modified if yeast NuA4 (like the TIP60 complex in higher eukaryotes) is required for down-modulating phosphoH2A rather than for facilitating repair.
Two RVB-containing ATPase type chromatin remodeling complexes in yeast, Ino80 and Swr1, have also been implicated in chromatin remodeling around the site of DNA damage (Downs et al., 2004; Morrison et al., 2004; van Attikum et al., 2007; van Attikum et al., 2004). Yeast cells lacking either functional Ino80 or Swr1 complex are hypersensitive to DNA damaging agents (Shen et al., 2000; van Attikum et al., 2007). ChIP assays show that Ino80 and Swr1 complexes are recruited to the site of damage and this recruitment is dependent on activities of Tel1 and Mec1 (yeast ATM and ATR homolog) (van Attikum et al., 2007). Recruitment of Ino80 and Swr1 complex to the site of damage is impaired in a strain with a non-phosphorylatable H2A. The association of yeast Ino80 complex with phosphoH2A is dependent on the Nhp10 and Arp4 subunits of the Ino80 complex (Downs et al., 2004; Morrison et al., 2004; van Attikum et al., 2007). Although both Ino80 and Swr1 complex are recruited at the sites of damage, there are differences in their function. Eviction of histones, processing of DNA ends and checkpoint activation depend on functional Ino80 but is independent of Swr1 (van Attikum et al., 2007). Additionally, Ino80 seems to be involved in homologous repair whereas Swr1 is involved in non-homologous end repair. Following recruitment, the RVB-containing chromatin remodelers reposition nucleosomes at the site of DNA damage. Histones are evicted from these sites followed by binding of repair proteins. The absence of functional Ino80 complex not only delays histone eviction but also the recruitment of the DNA damage repair protein, Rad51 (Tsukuda et al., 2005). Despite the delay, however, chromatin remodeling at the sites of damage is not completely abrogated suggesting redundancy in the chromatin remodeling complexes involved in these process. Although not formally tested the role of RVBs in the biochemical activity of the yeast Ino80 complex (Jonsson et al., 2004), suggests that lack of RVBs should impair the repair of DNA damage in yeast.
Human Ino80 and RVB were also found in the YY1 complex (Wu et al., 2005). Depletion of YY1 or Ino80 decreases cell survival after DNA damage due to an impairment of homologous recombination. Depletion of RVB2 has a similar effect, although it is currently not known whether RVB2 is required for the activity of mammalian Ino80 or YY1 complexes. In addition, non-functional human TIP60 complex impaires the recruitment of the DNA damage repair protein Rad51 to the sites of DNA damage (Murr et al., 2006). Interestingly, relaxing the chromatin by addition of chloroquine, sodium butyrate or hypotonic conditions restores the recruitment of Rad51. Thus, besides its role in down-modulating phosphoH2AX, the human TIP60 complex (whose HAT activity is dependent on RVB1) is involved in remodeling chromatin around DNA damage sites to facilitate access of the repair machinery (Murr et al., 2006). All these lines of evidence prompt the suggestion that mammalian RVBs, like yeast RVBs, is important for the repair of DNA damage at least partly due to its requirement for chromatin remodeling at the sites of DNA damage.
Checkpoint pathways
DNA damage activates checkpoint pathways that block cell-cycle progression to ensure that the damage is repaired before the progression of the cell cycle to the next stage (Sancar et al., 2004). There are two ATM/ATR dependent pathways involved in checkpoint activation. The ATM-Chk2-Cdc25a pathway is a fast response that inhibits DNA synthesis in response to DNA damage (Fig. 4B). This inhibition occurs due to an accelerated proteolysis of Cdc25A leading to the inhibition of the cyclin/Cdk2 kinase complex through the persistence of inhibitory phosphorylation on T14, Y15 of Cdk2. On the other hand the ATM-Chk2-p53 signaling pathway is a slower response to DNA damage that phosphorylates p53, attenuates its interaction with Mdm2 and thus stabilizes p53. Accumulation of p53 induces the cyclin dependent kinase inhibitor p21 which in turn inhibits Cdk activity. Human TIP60 complex is involved in activation of ATM and thus sits at the apex of both these checkpoint pathways (Sun et al., 2005).
The Ino80 and Swr1 complexes have also been implicated in cell cycle checkpoint activation. Yeast Ino80 and Swr1 were known to be involved in DDR but this was primarily thought to be through their role in transcription. Recent studies have, however, shown that both these chromatin remodeling complexes are recruited to DNA damage sites and are required for cellular response to damage. Ies4, one of the subunit of Ino80 complex is phosphorylated by Mec1/Tel1 (yeast homolog of ATM/ATR) kinases after DNA damage and mutation of Ies4 on the phosphoacceptor site affects the DNA damage checkpoint response without affecting the DNA repair pathway (Morrison et al., 2007). Additionally, human RVB2 was identified as an ATM/ATR target but the significance of this modification is yet to discovered (Matsuoka et al., 2007).
In summary, RVB-containing complexes have been implicated in different steps of the DNA damage response and further studies are expected to decipher the role of RVBs in these processes.
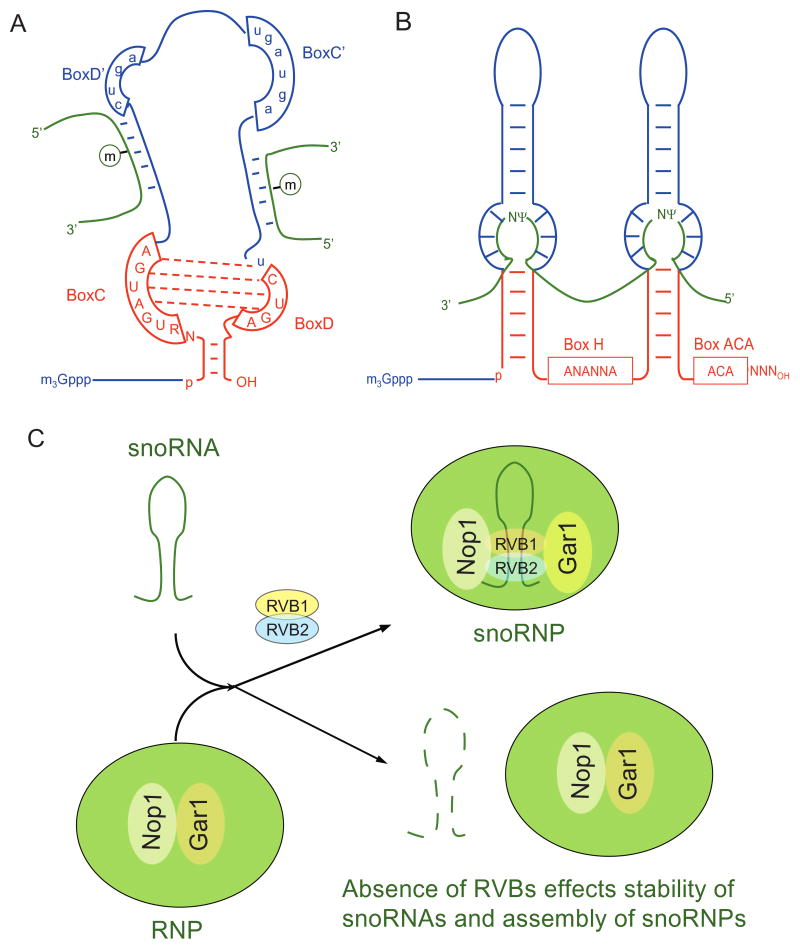
RVBs in snoRNP assembly
Small nucleolar RNPs (snoRNPs) are ribonucleolar protein (RNP) complexes made up by either box C/D or box H/ACA (Fig. 5A and 5B) containing small nucleolar RNAs (snoRNAs) complexed with proteins. snoRNPs are involved in cleavage and modification of small nuclear RNA (snRNA), ribosomal RNA (rRNA) and tRNAs (Kiss, 2002). RVB proteins associate with snoRNPs. Cellular proteins binding in vitro to immobilized U14 and U15 RNAs (examples of box C/D snoRNAs) include the mouse homologs of RVB1 and RVB2 (Newman et al., 2000). The biological significance of these interactions were discovered from two different studies. In the first study, depletion of yeast RVB2 by use of a galactose regulated allele of RVB2 suppresses the levels of snoRNAs (King et al., 2001). RVB2-GFP localizes in the nucleoplasm suggesting that RVB2 might be involved in snoRNP assembly or trafficking. Nop1 and Gar1 are two core snoRNP proteins that associate with snoRNAs of box C/D family and H/ACA family, respectively and are localized to the nucleolus, the site of rRNA gene transcription and processing. However, after RVB2 depletion both Nop1 and Gar1 were dispersed from the nucleolus to the nucleoplasm, suggesting that RVB2 is required for localization and perhaps the assembly of the snoRNP proteins (King et al., 2001) (Fig. 5C). In a different approach (Watkins et al., 2002), wild type and various mutants of U14 snoRNA were used to characterize the binding of the RVB proteins along with other snoRNP proteins like 15.5 K, NOP56, NOP58 and fibrillarin [reviewed in (Terns and Terns, 2002)]. 15.5 K binds to stem I and II in a structure-dependent but sequence-independent manner but is required for the subsequent assembly of the RVB proteins. Mutations in stem II dramatically affect binding of RVB1 and RVB2 and of other snoRNP proteins (NOP56, NOP58 and fibrillarin). Wild type U14 snoRNA localized to the nucleoli within 30 minutes after microinjection into cells, but mutations in the U14 snoRNA that disrupt binding of RVBs and other proteins resulted in mis-localization of the snoRNPs (Watkins et al., 2002). In human cell lines depletion of endogenous RVB proteins by siRNAs decreased the levels of snoRNAs (Watkins et al., 2004). These experimental results suggest that RVB proteins play an important role in the assembly of snoRNPs and stability of snoRNAs (Fig. 5C).
Figure 5.
RVBs are required for small nucleoprotein (snoRNP) assembly. (A) and (B) Schematics of box C/D and H/ACA small nucleolar ribonuleotide (snoRNA) family. (C) RVB bind to the stem of snoRNA and are required maturation of snoRNAs. Loss of RVB affects proper localization of two core snoRNP proteins, Nop1 and Gar1 that associate with snoRNAs of box C/D family and H/ACA family, respectively.
Recently RVB proteins have been implicated in the assembly of yet another RNP, the telomerase complex (Venteicher et al., 2008). Telomerase adds DNA repeats to the end of the chromosomes to stabilize the chromosomal ends. Telomerase is a ribonucleoprotein complex that consists of the TElomerase Reverse Transcriptase (TERT) and dyskerin protein subunits and the TElomerase RNA Component (TERC) [reviewed in (Kiss, 2002)]. To identify other proteins associated with telomerase, epitope tagged telomerase was expressed and affinity purified for mass spectrometry. RVB1 and RVB2 were identified in the complex and these interactions were confirmed by co-immunoprecipitation experiments. Furthermore, RVB proteins are recruited into TERT complex through its interaction with RVB2 but not with RVB1. Loss of RVB proteins decreased telomerase activity by 80-90% and this was due to a decrease in TERC RNA. In dyskeratosis congenita patients, mutations in dyskerin also decreased TERC levels because dyskerin binds TERC at its 3′ H/ACA motif to stabilize TERC (Mitchell et al., 1999). As depletion of RVB proteins also decreased TERC levels the functional relationship between dyskerin and RVBs were investigated. Dyskerin and RVB proteins interacted with each other independent of TERC. Interestingly, depletion of RVBs also reduced dyskerin levels thus explaining why the RVB proteins are required to maintain the levels of TERC. The association of RVBs with the TERT complex increased during the S phase of the cell cycle suggesting their role in maintaining telomerase activity in the phase of cell cycle where the function of telomerase is most critical. RVB proteins also interacted directly with TERT and surprisingly, in contrast to TERT/TERC/dyskerin, TERT/RVB1/RVB2 complex showed decreased telomerase activity. These results indicate that RVB proteins are present in a pre-telomerase complex that promotes the assembly of an active telomerase complex from which RVB are lost. Such transient association of RVB with TERT during the assembly of the telomerase complex is consistent with the hypothesis that they are scaffolding proteins important for assembly of molecular machines. The involvement of RVB in the assembly of snoRNPs and telomerase RNP, adds a new direction of research for these enigmatic proteins and raises the potential usefulness of anti-RVB agents for chemotherapy of cancers.
RVB proteins in cell transformation and metastasis
RVBs have been implicated in cellular transformation through their role in regulating the functions of c-Myc and β-catenin oncogenes (Feng et al., 2003; Wood et al., 2000). c-Myc is one of the best-characterized oncogenic transcription factor (Meyer and Penn, 2008). Mutations in the DNA binding domain of c-Myc abolish oncogenic activity (Brough et al., 1995). The N-terminal half of Myc family proteins contain two highly conserved domains: Myc homology box I (MbI) and Myc homology box II (MbII). The MbII domain of c-Myc is essential for oncogenic transformation and induction of apoptosis. Nuclear proteins affinity purified on this N-terminal half of c-Myc reveal that RVB1 and RVB2 interact with c-Myc, The interaction is through the MbII domain of c-Myc as deletion (Δ118–152 and Δ129–145) or point mutation (W136E) within the MbII domain abolish interaction with RVBs (Wood et al., 2000). The significance of this interaction was tested in a c-Myc mediated cellular transformation assay. Wild type RVB1 or Walker B mutant RVB1 (D302N) was cotransfected into rat embryo fibroblasts along with c-Myc and H-rasG12V oncogene (Fig. 6A). Interestingly, over-expression of the Walker B mutant form of RVB1 inhibited c-Myc mediated transformation, whereas wild type RVB1 had no effect. This inhibition was specific for c-Myc mediated transformation pathways as expression of mutant RVB1 did not inhibit general growth of the cells. It is not clear, however, whether the suppression of transformation is mediated through the direct inhibition of the biochemical activity of the Myc complex or through the interference of function of the many chromatin remodeling factors, RNPs and other molecular machines that depend on RVB proteins for normal assembly.
Figure 6.
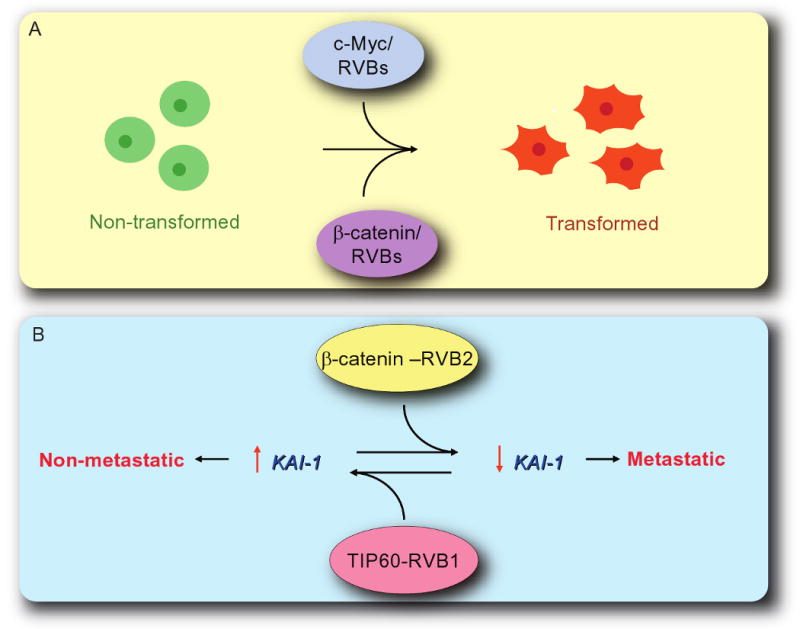
RVBs are involved in cellular transformation and cancer metastasis. (A) Cellular transformation by c-Myc and β-catenin requires functional RVBs. Expression of Walker B mutant of RVB1 acts as a dominant negative form of RVB1 and inhibits cellular transformation. (B) RVB1-TIP60 and RVB2-β-catenin complexes function antagonistically. RVB1-TIP60 complex act as an activator of KAI1 expression whereas RVB2-β-catenin complex act as a repressor. Inhibition of expression of KAI1 transforms cells from non-metaststic to metastatic state.
RVB proteins are also implicated in the Wnt-β-catenin signaling pathway that regulates various cellular processes such as the polarity of cell division, proliferation of cells and cell-fate determination (Cadigan and Nusse, 1997). Many different cancers show deregulation of Wnt-β-catenin signaling pathway primarily through changes in subcellular localization of β-catenin and its levels (Polakis, 2000). In the absence of Wnt stimulus, glycogen synthase kinase 3β (GSK3β) phosphorylates β-catenin in an N-terminal sequence that serves as a phosphodegron that is recognized by the β-TrCP1 ubiquitin ligase (Aberle et al., 1997; Su et al., 2003). Polyubiquitination of β-catenin leads to degradation of cytosolic β-catenin by the 26S proteosome. Activation of Wnt signaling pathway inhibits GSK3β and stabilizes unphosphorylated β-catenin that accumulates in the nucleus, binds to T-cell factor (TCF) family of transcription factors and promotes target gene expression. Mutations in the Wnt signaling pathway stabilize β-catenin in many cancers (Polakis, 2000). To identify proteins that are involved in β-catenin mediated transcriptional control, GST pull-down experiments were performed (Bauer et al., 1998). The GST-β-catenin fusion protein (1-284 of β-catenin) purified proteins of 102, 52 and 44 kD from metabolically labeled lysates of SW480 colon carcinoma cell. These proteins bind to the N-terminus of β-catenin and include RVB1. β-catenin interactes with RVB1 both in vivo and in vitro and amino acids from 187-284 of β-catenin are required for this interaction. To establish the role of RVB1 in β-catenin function, overexpression of wild type or Walker B mutant of RVB1 and depletion of endogenous RVB1 were performed (Feng et al., 2003). RK3E cell line, an adenovirus E1A-immortalized rat epithelial line were stably transfected with wild type or Walker B mutant of RVB1. Introduction of S33Y oncogenic mutant form of β-catenin in the control and wild type RVB1 expressing cells showed similar number of transformed foci, but cells expressing Walker B mutant form of RVB1 had 70-80 % fewer foci (Feng et al., 2003). The target genes downstream from β-catenin such as ITF-2 and Axil were repressed by the Walker B mutant form of RVB1. Cells depleted of RVB1 by siRNA also failed to activate ITF-2 and Axil expression. Furthermore, ChIP assays suggest that the level of acetylation of histone H4 at the proximal promoter of the target genes is important for the function of RVB1 in β-catenin driven promoters. Thus RVB1 regulates the output of the Wnt-β-catenin signaling pathway by modulating the chromatin structure of the genes targeted by β-catenin (Fig. 6A).
Identification of Hint1/PKCI (histidine triad nucleotide-binding protein 1/ protein kinase C inhibitor 1) as an interactor and a regulator of RVB suggests that RVB1 may regulate β-catenin target genes (Weiske and Huber, 2005). Hint1 is a member of histidine triad (HIT) protein family, which is characterized by a common His-X-His-X-His-XX motif (where X is a hydrophobic amino acid). Hint1 was identified as an interactor of RVB1 in a yeast two hybrid screen. GST pull down experiments demonstrated that Hint1 bound directly to both RVB1 and RVB2 (Weiske and Huber, 2005). Hint1 binding site was mapped to central region on RVBs (214-297 in RVB1 and 218-289 in RVB2). Interestingly, in an in vitro binding study Hint1 disrupted interaction between RVB1 and RVB2 suggesting that it shares the RVB1-RVB2 interaction domain. Consistent with this overexpression of Hint1 modulates RVB1-β-catenin regulated genes. These data identifies Hint1 as a regulator of RVB and Wnt-β-catenin signaling pathway.
Metastasis of a cancer seriously impairs successful therapy of cancers by chemotherapy, radiation or surgery. KAI1 is one among a handful of metastasis suppressor genes that have been identified [reviewed in (Shevde and Welch, 2003; Steeg, 2003)]. KAI1 is a member of tetraspanin family, which inhibits metastasis by promoting cell adhesion and interacting with many plasma membrane receptors. Expression of KAI1 is repressed in prostate cancer cell lines that are highly metastatic in nature compared to normal epithelial or non-metastatic cancer cells (Dong et al., 1996). An intricate interplay of RVB associated transcription factors governs the regulation of KAI1.
TIP60 along with RVB1 (but not RVB2) binds to the KAI1 promoter and acts as a co-activator of KAI1 expression through the acetylation of histones at the promoter (Baek et al., 2002). In metastatic prostate cancer cells the TIP60 co-activator complex is not bound to the KAI1 promoter, at least partly because of a decrease in TIP60 RNA and protein in the cells. Overexpressing TIP60 in these metastatic cells restores both binding of TIP60 to the KAI1 promoter and levels of KAI1 (Baek et al., 2002).
In contrast, in metastatic cells RVB2 along with β-catenin occupied the KAI1 promoter and repressed gene expression (Kim et al., 2005). The preference for β-catenin was again partly dictated by the higher levels of β-catenin in the metastatic cells, because overexpression of β-catenin in the non-metastatic cells will reverse the dominance of the naturally occurring TIP60 at the KAI1 promoter. The antagonistic activities of TIP60-RVB1 (activator) and β-catenin-RVB2 (repressor) (Fig. 6B) is consistent with the fact that the two complexes occupy the KAI1 promoter in a mutually exclusive manner even when the levels of TIP60 or β-catenin are manipulated to correct for the original predominance of TIP60 in the non-metastatic cells (Kim et al., 2005).
The antagonist effect of the two RVB proteins on KAI1 expression and on metastatic potential of the cancer cells on the surface appears to be in conflict with the double hexameric complex containing both RVB1 and RVB2. Similar antagonism has been reported in other contexts (Bauer et al., 2000; Diop et al., 2008; Rottbauer et al., 2002), and suggests that the proteins could work in certain contexts independent of each other.
Purification of the repressive RVB2 complex from cells and mass spectrometry of the interactors revealed yet another surprise. Besides the known interactors like RVB1 and β-catenin the experiment identified novel interactors like HDAC-1 and two small ubiquitin-like modifier (SUMO) processing enzymes, SUMO-sentrin-specific protease 1 (SENP1) and SUMO1-specific proteases 1 (SUSP1) (Kim et al., 2007). SUMOylation is a multistep process that involves the E1-activating molecules SAE1/SAE2, the E2-conjugating enzyme Ubc9 and E3 ligases that results in addition of SUMO, an 11-kDa protein to the target protein. SUMOylation of a protein plays an important regulatory role in many cellular processes such as signal transduction, transcriptional regulation, chromatin structure and nuclear/cytoplasmic shuttling. Consistent with the presence of SUMO processing enzymes in the RVB2 complex, both in vitro and in vivo data suggest that RVB2 is SUMOylated on lysine 456. Mutations in the SUMO acceptor site and fusion of a SUMO to the mutant reveal that SUMOylation of RVB2 is essential before it can translocate to the nucleus and repress a promoter. Consistent with this, increase in SUMOylation (by overexpressing the SUMO conjugating enzyme, Ubc9) enhances that transcriptional repression by RVB2. Decrease in SUMOylation by over-expressing catalytically active SENP1 reverses the RVB2 mediated transcriptional repression. KAI1 expression is affected by these manipulations in a manner consistent with this model. Surprisingly, when SUMO-acceptor mutant of RVB2 is forced into nucleus by fusing a strong constitutive nuclear localization sequence (NLS) TIP60 localizes to the KAI1 promoter and activates its expression. Thus, SUMOylation of RVB2 is critical for promoter repression independent of its role in nuclear transport, most likely because the SUMO modification is required to recruit HDAC1 at the KAI1 promoter. Taken together these data suggest that SUMOylation of RVB2 is required both for nuclear localization and for recruiting a histone deacetylase to the β-catenin repressor complex.
RVBs in DNA replication, mitosis and URI/prifoldin complexes
The chromatin is remodeled during DNA replication. As the replication fork moves along the chromatin the nucleosomes are disrupted and re-assembled on both the parental and sister strand by various chromatin assembly factors. The first clue of RVB being involved in the DNA associated processes was when RVB1 was identified as an interactor of RPA14, one of the subunit of RPA trimeric complex, a single strand binding protein (Qiu et al., 1998). Recent studies have implicated RVB containing chromatin remodeling complex, Ino80 in recovery from DNA replication stress (Papamichos-Chronakis and Peterson, 2008; Shimada et al., 2008; Vincent et al., 2008). Addition of hydroxyurea (HU), an inhibitor of ribonucleotide reductase, decreases the free nucleotide pool, stalling replication forks and activating the checkpoint. In yeast, deletion of Ino80 delays S-phase progression (Papamichos-Chronakis and Peterson, 2008; Shimada et al., 2008; Vincent et al., 2008). Interestingly, in the absence of Ino80 cells are sensitized to HU (Shen et al., 2000) due to the collapse of replication fork (Papamichos-Chronakis and Peterson, 2008; Shimada et al., 2008; Vincent et al., 2008). These data suggest that Ino80, along with RVB) are required for the maintenance and restart of replication forks stalled after replication stress.
Apart from their presence in various chromatin remodeling complexes RVBs have been also identified in complexes involved in mitosis. RVB1 was found to interact with tubulin and to localize at the mitotic spindles and spindle poles (Gartner et al., 2003). Interestingly, RVB2 was found to localize to the midzone during telophase and to the midbody during cytokinesis in addition to the mitotic spindles and spindle poles (Sigala et al., 2005). A proteome-based approach and an siRNA based screen to identify the mitotic regulators also implicated RVB in mitotic function (Ducat et al., 2008). RVB1 and RVB2 were found in the complex involved in mitosis. Depletion of RVBs either by siRNA in cultured cells or by immunodepletion in Xenopus extracts revealed that RVBs are required in regulating microtubule assembly and organization (Ducat et al., 2008).
Another protein complex that contains RVBs is URI complex (Unconventional prefoldin RPB5 Interactor) (Gstaiger et al., 2003). URI is a multi-subunit complex that includes F-box protein SKP2, prefoldins (PFDs), RPB5 (the core subunit of RNA polymerase II) and RVB (RVB1 and RVB2) (Gstaiger et al., 2003). URI complex is involved in the regulation of nutrient-sensitivity through the TOR-(Target Of Rapamycin) dependent transcription pathway [reviewed in (Raught et al., 2001)]. Although RVB were identified in URI complex, the role of RVB in this complex is yet to be determined.
Biochemical activities associated with RVBs and future perspectives
One of the biochemical activities of RVB is their ability to bind and hydrolyze ATP as each subunit has a conserved Walker A and Walker B motif. Indeed, mutations in Walker A or B motif of either RVB protein renders it inactive (Jonsson et al., 2001; Qiu et al., 1998). However, studies to demonstrate their ability to hydrolyze ATP are controversial. Experiments showing ATP hydrolysis by recombinant RVBs lack an important negative control i.e. Walker A or Walker B mutant of these proteins (Gribun et al., 2008; Puri et al., 2007; Torreira et al., 2008). To rule out the possibility that any ATPase activity is not from an associated bacterial ATPase this negative control is very important. On the other hand the failure to detect ATPase activity of a recombinant RVBs could result from inactivation of the protein during purification or requirement of additional partners.
Another activity associated with RVB is its helicase activity. As RVB are structural homologs of the bacterial RuvB proteins, which posseses DNA helicase activity, their ability to separate DNA strands was tested. Although it was demonstrated that RVB are active as a helicase with opposite polarity (Kanemaki et al., 1999; Makino et al., 1999), this result has been controversial. Multiple groups were unable to detect the helicase activity attributed to RVB (Ikura et al., 2000; Matias et al., 2006; Qiu et al., 1998).
RVB play important roles in cellular growth and depletion of these proteins sensitizes cells to DNA damage [(Wu et al., 2007), S.J. and A.D. unpublished data]. Also they are required for cellular transformation and assembly of functional telomerase. Thus, these proteins may be good therapeutic targets particularly if a robust and RVB specific ATPase activity is established. One caveat, of course, lies in the numerous complexes associated with RVB, so that a future RVB inhibitor may have unacceptable toxicity. Over the next few years we expect to see more instances where the depletion of one of the RVB proteins leads to a specific molecular lesion in a biochemical complex. A close examination of these defects should yield in vitro systems where the mechanism by which RVB promotes formation of a protein complexes involved in transcription or DNA damage response can be dissected. Only then will it become clear how these single and dual protein rings carry out so many disparate functions in such a variety of molecular pathways.
Acknowledgments
We apologize to all authors whose important contributions could not be acknowledged due to space limitations. This work was partly supported by RO1 GM84465 to A.D.
References
- Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. beta-catenin is a target for the ubiquitin-proteasome pathway. Embo J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and beta-amyloid precursor protein. Cell. 2002;110:55–67. doi: 10.1016/s0092-8674(02)00809-7. [DOI] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Bao Y, Shen X. INO80 subfamily of chromatin remodeling complexes. Mutat Res. 2007;618:18–29. doi: 10.1016/j.mrfmmm.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer A, Chauvet S, Huber O, Usseglio F, Rothbacher U, Aragnol D, Kemler R, Pradel J. Pontin52 and reptin52 function as antagonistic regulators of beta-catenin signalling activity. Embo J. 2000;19:6121–6130. doi: 10.1093/emboj/19.22.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer A, Huber O, Kemler R. Pontin52, an interaction partner of beta-catenin, binds to the TATA box binding protein. Proc Natl Acad Sci U S A. 1998;95:14787–14792. doi: 10.1073/pnas.95.25.14787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellosta P, Hulf T, Balla Diop S, Usseglio F, Pradel J, Aragnol D, Gallant P. Myc interacts genetically with Tip48/Reptin and Tip49/Pontin to control growth and proliferation during Drosophila development. Proc Natl Acad Sci U S A. 2005;102:11799–11804. doi: 10.1073/pnas.0408945102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–415. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. GammaH2AX and cancer. Nat Rev Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm A, Tufteland KR, Aasland R, Becker PB. The many colours of chromodomains. Bioessays. 2004;26:133–140. doi: 10.1002/bies.10392. [DOI] [PubMed] [Google Scholar]
- Brough DE, Hofmann TJ, Ellwood KB, Townley RA, Cole MD. An essential domain of the c-myc protein interacts with a nuclear factor that is also required for E1A-mediated transformation. Mol Cell Biol. 1995;15:1536–1544. doi: 10.1128/mcb.15.3.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- Chowdhury D, Keogh MC, Ishii H, Peterson CL, Buratowski S, Lieberman J. gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol Cell. 2005;20:801–809. doi: 10.1016/j.molcel.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Diop SB, Bertaux K, Vasanthi D, Sarkeshik A, Goirand B, Aragnol D, Tolwinski NS, Cole MD, Pradel J, Yates JR, 3rd, et al. Reptin and Pontin function antagonistically with PcG and TrxG complexes to mediate Hox gene control. EMBO Rep. 2008;9:260–266. doi: 10.1038/embor.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong JT, Suzuki H, Pin SS, Bova GS, Schalken JA, Isaacs WB, Barrett JC, Isaacs JT. Down-regulation of the KAI1 metastasis suppressor gene during the progression of human prostatic cancer infrequently involves gene mutation or allelic loss. Cancer Res. 1996;56:4387–4390. [PubMed] [Google Scholar]
- Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, Kron SJ, Jackson SP, Cote J. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Doyon Y, Selleck W, Lane WS, Tan S, Cote J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol. 2004;24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducat D, Kawaguchi S, Liu H, Yates JR, 3rd, Zheng Y. Regulation of microtubule assembly and organization in mitosis by the AAA+ ATPase Pontin. Mol Biol Cell. 2008;19:3097–3110. doi: 10.1091/mbc.E07-11-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etard C, Gradl D, Kunz M, Eilers M, Wedlich D. Pontin and Reptin regulate cell proliferation in early Xenopus embryos in collaboration with c-Myc and Miz-1. Mech Dev. 2005;122:545–556. doi: 10.1016/j.mod.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Feng Y, Lee N, Fearon ER. TIP49 regulates beta-catenin-mediated neoplastic transformation and T-cell factor target gene induction via effects on chromatin remodeling. Cancer Res. 2003;63:8726–8734. [PubMed] [Google Scholar]
- Frank SR, Parisi T, Taubert S, Fernandez P, Fuchs M, Chan HM, Livingston DM, Amati B. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 2003;4:575–580. doi: 10.1038/sj.embor.embor861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner W, Rossbacher J, Zierhut B, Daneva T, Base W, Weissel M, Waldhausl W, Pasternack MS, Wagner L. The ATP-dependent helicase RUVBL1/TIP49a associates with tubulin during mitosis. Cell Motil Cytoskeleton. 2003;56:79–93. doi: 10.1002/cm.10136. [DOI] [PubMed] [Google Scholar]
- Gaughan L, Brady ME, Cook S, Neal DE, Robson CN. Tip60 is a co-activator specific for class I nuclear hormone receptors. J Biol Chem. 2001;276:46841–46848. doi: 10.1074/jbc.M103710200. [DOI] [PubMed] [Google Scholar]
- Gaughan L, Logan IR, Cook S, Neal DE, Robson CN. Tip60 and histone deacetylase 1 regulate androgen receptor activity through changes to the acetylation status of the receptor. J Biol Chem. 2002;277:25904–25913. doi: 10.1074/jbc.M203423200. [DOI] [PubMed] [Google Scholar]
- Gevry N, Chan HM, Laflamme L, Livingston DM, Gaudreau L. p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev. 2007;21:1869–1881. doi: 10.1101/gad.1545707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribun A, Cheung KL, Huen J, Ortega J, Houry WA. Yeast Rvb1 and Rvb2 are ATP-dependent DNA helicases that form a heterohexameric complex. J Mol Biol. 2008;376:1320–1333. doi: 10.1016/j.jmb.2007.12.049. [DOI] [PubMed] [Google Scholar]
- Gstaiger M, Luke B, Hess D, Oakeley EJ, Wirbelauer C, Blondel M, Vigneron M, Peter M, Krek W. Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science. 2003;302:1208–1212. doi: 10.1126/science.1088401. [DOI] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, Yoder K, Izumi S, Kuraoka I, Tanaka K, et al. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol Cell Biol. 2007;27:7028–7040. doi: 10.1128/MCB.00579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S, Shibata E, Dutta A. Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol Cell Biol. 2008;28:2690–2700. doi: 10.1128/MCB.01983-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Cai Y, Yao T, Gottschalk AJ, Florens L, Swanson SK, Gutierrez JL, Coleman MK, Workman JL, Mushegian A, et al. A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J Biol Chem. 2005;280:41207–41212. doi: 10.1074/jbc.M509128200. [DOI] [PubMed] [Google Scholar]
- Jonsson ZO, Dhar SK, Narlikar GJ, Auty R, Wagle N, Pellman D, Pratt RE, Kingston R, Dutta A. Rvb1p and Rvb2p are essential components of a chromatin remodeling complex that regulates transcription of over 5% of yeast genes. J Biol Chem. 2001;276:16279–16288. doi: 10.1074/jbc.M011523200. [DOI] [PubMed] [Google Scholar]
- Jonsson ZO, Jha S, Wohlschlegel JA, Dutta A. Rvb1p/Rvb2p recruit Arp5p and assemble a functional Ino80 chromatin remodeling complex. Mol Cell. 2004;16:465–477. doi: 10.1016/j.molcel.2004.09.033. [DOI] [PubMed] [Google Scholar]
- Kanemaki M, Kurokawa Y, Matsu-ura T, Makino Y, Masani A, Okazaki K, Morishita T, Tamura TA. TIP49b, a new RuvB-like DNA helicase, is included in a complex together with another RuvB-like DNA helicase, TIP49a. J Biol Chem. 1999;274:22437–22444. doi: 10.1074/jbc.274.32.22437. [DOI] [PubMed] [Google Scholar]
- Kanemaki M, Makino Y, Yoshida T, Kishimoto T, Koga A, Yamamoto K, Yamamoto M, Moncollin V, Egly JM, Muramatsu M, Tamura T. Molecular cloning of a rat 49-kDa TBP-interacting protein (TIP49) that is highly homologous to the bacterial RuvB. Biochem Biophys Res Commun. 1997;235:64–68. doi: 10.1006/bbrc.1997.6729. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kim JA, Downey M, Fillingham J, Chowdhury D, Harrison JC, Onishi M, Datta N, Galicia S, Emili A, et al. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature. 2006;439:497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- Kim JH, Kim B, Cai L, Choi HJ, Ohgi KA, Tran C, Chen C, Chung CH, Huber O, Rose DW, et al. Transcriptional regulation of a metastasis suppressor gene by Tip60 and beta-catenin complexes. Nature. 2005;434:921–926. doi: 10.1038/nature03452. [DOI] [PubMed] [Google Scholar]
- Kim JH, Lee JM, Nam HJ, Choi HJ, Yang JW, Lee JS, Kim MH, Kim SI, Chung CH, Kim KI, Baek SH. SUMOylation of pontin chromatin-remodeling complex reveals a signal integration code in prostate cancer cells. Proc Natl Acad Sci U S A. 2007;104:20793–20798. doi: 10.1073/pnas.0710343105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King TH, Decatur WA, Bertrand E, Maxwell ES, Fournier MJ. A well-connected and conserved nucleoplasmic helicase is required for production of box C/D and H/ACA snoRNAs and localization of snoRNP proteins. Mol Cell Biol. 2001;21:7731–7746. doi: 10.1128/MCB.21.22.7731-7746.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- Kurash JK, Lei H, Shen Q, Marston WL, Granda BW, Fan H, Wall D, Li E, Gaudet F. Methylation of p53 by Set7/9 mediates p53 acetylation and activity in vivo. Mol Cell. 2008;29:392–400. doi: 10.1016/j.molcel.2007.12.025. [DOI] [PubMed] [Google Scholar]
- Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, 3rd, Abmayr SM, Washburn MP, Workman JL. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- Makino Y, Kanemaki M, Kurokawa Y, Koji T, Tamura T. A rat RuvB-like protein, TIP49a, is a germ cell-enriched novel DNA helicase. J Biol Chem. 1999;274:15329–15335. doi: 10.1074/jbc.274.22.15329. [DOI] [PubMed] [Google Scholar]
- Matangkasombut O, Buratowski S. Different sensitivities of bromodomain factors 1 and 2 to histone H4 acetylation. Mol Cell. 2003;11:353–363. doi: 10.1016/s1097-2765(03)00033-9. [DOI] [PubMed] [Google Scholar]
- Matias PM, Gorynia S, Donner P, Carrondo MA. Crystal structure of the human AAA+ protein RuvBL1. J Biol Chem. 2006;281:38918–38929. doi: 10.1074/jbc.M605625200. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, Shen X. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell. 2004;119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- Morrison AJ, Kim JA, Person MD, Highland J, Xiao J, Wehr TS, Hensley S, Bao Y, Shen J, Collins SR, et al. Mec1/Tel1 phosphorylation of the INO80 chromatin remodeling complex influences DNA damage checkpoint responses. Cell. 2007;130:499–511. doi: 10.1016/j.cell.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- Newman DR, Kuhn JF, Shanab GM, Maxwell ES. Box C/D snoRNA-associated proteins: two pairs of evolutionarily ancient proteins and possible links to replication and transcription. Rna. 2000;6:861–879. doi: 10.1017/s1355838200992446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Peterson CL. The Ino80 chromatin-remodeling enzyme regulates replisome function and stability. Nat Struct Mol Biol. 2008;15:338–345. doi: 10.1038/nsmb.1413. [DOI] [PubMed] [Google Scholar]
- Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- Puri T, Wendler P, Sigala B, Saibil H, Tsaneva IR. Dodecameric structure and ATPase activity of the human TIP48/TIP49 complex. J Mol Biol. 2007;366:179–192. doi: 10.1016/j.jmb.2006.11.030. [DOI] [PubMed] [Google Scholar]
- Qiu XB, Lin YL, Thome KC, Pian P, Schlegel BP, Weremowicz S, Parvin JD, Dutta A. An eukaryotic RuvB-like protein (RUVBL1) essential for growth. J Biol Chem. 1998;273:27786–27793. doi: 10.1074/jbc.273.43.27786. [DOI] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Madhani HD. Patterning chromatin: form and function for H2A.Z variant nucleosomes. Curr Opin Genet Dev. 2006;16:119–124. doi: 10.1016/j.gde.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Raught B, Gingras AC, Sonenberg N. The target of rapamycin (TOR) proteins. Proc Natl Acad Sci U S A. 2001;98:7037–7044. doi: 10.1073/pnas.121145898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottbauer W, Saurin AJ, Lickert H, Shen X, Burns CG, Wo ZG, Kemler R, Kingston R, Wu C, Fishman M. Reptin and pontin antagonistically regulate heart growth in zebrafish embryos. Cell. 2002;111:661–672. doi: 10.1016/s0092-8674(02)01112-1. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzer U, Kubicek M, Prohaska R. Isolation, molecular characterization, and tissue-specific expression of ECP-51 and ECP-54 (TIP49), two homologous, interacting erythroid cytosolic proteins. Biochim Biophys Acta. 1999;1446:365–370. doi: 10.1016/s0167-4781(99)00104-9. [DOI] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Sheaffer KL, Updike DL, Mango SE. The Target of Rapamycin pathway antagonizes pha-4/FoxA to control development and aging. Curr Biol. 2008;18:1355–1364. doi: 10.1016/j.cub.2008.07.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- Shevde LA, Welch DR. Metastasis suppressor pathways--an evolving paradigm. Cancer Lett. 2003;198:1–20. doi: 10.1016/s0304-3835(03)00304-5. [DOI] [PubMed] [Google Scholar]
- Shimada K, Oma Y, Schleker T, Kugou K, Ohta K, Harata M, Gasser SM. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr Biol. 2008;18:566–575. doi: 10.1016/j.cub.2008.03.049. [DOI] [PubMed] [Google Scholar]
- Sigala B, Edwards M, Puri T, Tsaneva IR. Relocalization of human chromatin remodeling cofactor TIP48 in mitosis. Exp Cell Res. 2005;310:357–369. doi: 10.1016/j.yexcr.2005.07.030. [DOI] [PubMed] [Google Scholar]
- Smith ER, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, Cook RG, Lucchesi JC, Allis CD. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc Natl Acad Sci U S A. 1998;95:3561–3565. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeg PS. Metastasis suppressors alter the signal transduction of cancer cells. Nat Rev Cancer. 2003;3:55–63. doi: 10.1038/nrc967. [DOI] [PubMed] [Google Scholar]
- Su Y, Ishikawa S, Kojima M, Liu B. Eradication of pathogenic beta-catenin by Skp1/Cullin/F box ubiquitination machinery. Proc Natl Acad Sci U S A. 2003;100:12729–12734. doi: 10.1073/pnas.2133261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Xu Y, Roy K, Price BD. DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol Cell Biol. 2007;27:8502–8509. doi: 10.1128/MCB.01382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- Taubert S, Gorrini C, Frank SR, Parisi T, Fuchs M, Chan HM, Livingston DM, Amati B. E2F-dependent histone acetylation and recruitment of the Tip60 acetyltransferase complex to chromatin in late G1. Mol Cell Biol. 2004;24:4546–4556. doi: 10.1128/MCB.24.10.4546-4556.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns MP, Terns RM. Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 2002;10:17–39. [PMC free article] [PubMed] [Google Scholar]
- Torreira E, Jha S, Lopez-Blanco JR, Arias-Palomo E, Chacon P, Canas C, Ayora S, Dutta A, Llorca O. Architecture of the pontin/reptin complex, essential in the assembly of several macromolecular complexes. Structure. 2008;16:1511–1520. doi: 10.1016/j.str.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H, Fritsch O, Gasser SM. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. Embo J. 2007;26:4113–4125. doi: 10.1038/sj.emboj.7601835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H, Fritsch O, Hohn B, Gasser SM. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004;119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- van den Heuvel S, Dyson NJ. Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol. 2008;9:713–724. doi: 10.1038/nrm2469. [DOI] [PubMed] [Google Scholar]
- Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132:945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JA, Kwong TJ, Tsukiyama T. ATP-dependent chromatin remodeling shapes the DNA replication landscape. Nat Struct Mol Biol. 2008;15:477–484. doi: 10.1038/nsmb.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanzel M, Herold S, Eilers M. Transcriptional repression by Myc. Trends Cell Biol. 2003;13:146–150. doi: 10.1016/s0962-8924(03)00003-5. [DOI] [PubMed] [Google Scholar]
- Watkins NJ, Dickmanns A, Luhrmann R. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol Cell Biol. 2002;22:8342–8352. doi: 10.1128/MCB.22.23.8342-8352.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Lemm I, Ingelfinger D, Schneider C, Hossbach M, Urlaub H, Luhrmann R. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol Cell. 2004;16:789–798. doi: 10.1016/j.molcel.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Weiske J, Huber O. The histidine triad protein Hint1 interacts with Pontin and Reptin and inhibits TCF-beta-catenin-mediated transcription. J Cell Sci. 2005;118:3117–3129. doi: 10.1242/jcs.02437. [DOI] [PubMed] [Google Scholar]
- Wood MA, McMahon SB, Cole MD. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol Cell. 2000;5:321–330. doi: 10.1016/s1097-2765(00)80427-x. [DOI] [PubMed] [Google Scholar]
- Wu S, Shi Y, Mulligan P, Gay F, Landry J, Liu H, Lu J, Qi HH, Wang W, Nickoloff JA, Wu C. A YY1-INO80 complex regulates genomic stability through homologous recombination-based repair. Nat Struct Mol Biol. 2007;14:1165–1172. doi: 10.1038/nsmb1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WH, Alami S, Luk E, Wu CH, Sen S, Mizuguchi G, Wei D, Wu C. Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nat Struct Mol Biol. 2005;12:1064–1071. doi: 10.1038/nsmb1023. [DOI] [PubMed] [Google Scholar]
- Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]