Abstract
Sheep intelectin1 and sheep intelectin3 (sITLN1 and sITLN3) were cloned and sequenced. The amino acid sequences of sITLN1 and sITLN3 shared 86% and 91% homology with the previously cloned sheep intelectin2 (sITLN2), respectively. Expression of sITLN1 and sITLN3 transcript was demonstrated in abomasum, lung, colon and gastric lymph node, terminal rectum, skin, jejunum, mesenteric lymph node, ileal peyer’s patches, brain, kidney, liver, spleen, skin, ear pinna, heart and ovary in normal sheep tissues. sITLN2 transcript expression was restricted to the abomasal mucosa in normal sheep tissues. Using a non selective chicken anti-intelectin antibody, tissue intelectin protein was demonstrated in mucus neck cells in the abomasum, mucus cells in the colon, free mucus in ileum, goblet cells in the lung, small intestinal epithelium and brush border, epidermal layer of the skin and skin sebaceous glands. The expression of the three sITLN transcripts was examined in two nematode infections in sheep known to induce a Th2 response; a Teladorsagia circumcincta challenge infection model and a Dictyocaulus filaria natural infection. The three sITLN were absent in unchallenged naïve lambs and present in the abomasal mucosa of both naïve and immune lambs following T. circumcincta challenge infection. Upregulation of sITLN2 and sITLN3 was shown in sheep lung following D. filaria natural infection. Intelectins may play an important role in the mucosal response to nematode infections in ruminants.
Keywords: sheep intelectin, sequence, tissue distribution, parasite response
1. INTRODUCTION
Nematode infections play an important role in economics of the livestock industry worldwide and in the health of humans in third world countries [6, 10, 16]. Allergic conditions have an increasing morbidity worldwide. The identification of novel molecules upregulated in a Th2 environment may have relevance for new treatment strategies [1]. Intelectins (ITLN) are novel molecules that have been shown to upregulate in a Th2 environment. Two ITLN homologues have been described in man [14, 22, 25], mice [12, 17] and sheep [7, 8]. Upregulation has been demonstrated associated with a Th2 environment in both the gastrointestinal and respiratory tract of all three species. The full sequence of sheep intelectin2 (sITLN2) has been published previously. We now present the full sequence of sheep intelectin1 (sITLN1) and a novel sheep intelectin3 (sITLN3). We demonstrate the differential tissue distribution of sITLN transcripts in normal sheep tissues and the expression of the three sITLN transcripts in response to nematode infections affecting the gastrointestinal tract and respiratory tract of sheep. Finally we have determined the distribution of sITLN protein in normal sheep tissues by means of a specific antibody.
2. MATERIALS AND METHODS
2.1. Animals and parasites
2.1.1. Sheep
Lambs (Scottish Blackface × Leicester, 4–5 months old) for Teladorsagia circumcincta infection experiments and worm free yearling sheep (10–12 months old) for D. filaria controls (Dorset × Suffolk) and for demonstration of tissue distribution of sITLN transcript and protein (Scottish Blackface × Leicester) were born and maintained at the Moredun Research Institute (Midlothian, UK) under conditions designed to exclude accidental infection with nematode parasites. All experiments were approved by the Institute’s ethical review committee and were performed under licence, as required by the United Kingdom’s Animals (Scientific Procedures) Act of 1986. Experiments were undertaken in 2005–2006. D. filaria infected yearling sheep (Scottish Blackface) were from a commercial flock with a naturally acquired infection.
2.1.2. Infective larvae
Larvae from an anthelmintic susceptible isolate of T. circumcincta (Moredun Research Institute) were cultured as previously described [21] and stored at 4 °C for up to one month before use. All challenge doses were from the same batch of larvae.
2.2. T. circumcincta parasite infections
Lambs were divided into three groups: unchallenged naïve (unv), challenged naïve (cnv) and challenged previously-infected (cpi). The previously-infected lambs were given 2 000 L3 three times per week for 8 weeks, a protocol which has previously been shown to confer some immunity [11, 21]. All lambs were treated with anthelminthic prior to challenge to clear any residual worms in the previously-infected group, and as a control treatment in the other groups (Tab. I).
Table I.
Experimental protocol for the T. circumcincta challenge infection experiment.
| Animals | Status | Treatments (relative to day of challenge) |
|||||
|---|---|---|---|---|---|---|---|
| Tricklea infection | Levamisoleb |
Number killed |
|||||
| Unchallenged |
Challengedc |
||||||
| Day | −7 | 0 | 5 | 10 | 22 | ||
| Lambs | Previously infected | + | + | 6d | 5d | 5d | |
| Naïve | − | + | 4d | 6d | 5d | 4d | |
2 000 T. circumcincta L3 3 × per week for 8 weeks.
Levamisole 7.5 mg/kg per os.
1 × 50 000 T. circumcincta L3.
Scottish Blackface × Leicester.
2.3. Post-mortem procedure
All sheep were stunned with a captive bolt and exsanguinated. For T. circumcincta infection experiments the abomasum was removed, opened longitudinally and the contents collected. One half was retained for worm counting along with the contents as described previously [11]. Abomasal folds from the fundic region of the other half were used for tissue sample collection.
For sequencing of sITLN1 and sITLN3 and for demonstration of distribution of sITLN, the following tissues were collected from worm free yearling sheep; abomasum, jejunum, colon, rectum, gastric lymph node, mesenteric lymph node, ileal Peyer’s patches, brain, kidney, liver, lung, pancreas, spleen, skin, ear pinna, ovary, uterus, heart and mesenteric fat. For lungworm infected sheep and worm free controls, lung tissue was collected from the left caudal lobe ventrally. Tissue samples were stored in RNA later (Qiagen, Hilden, Germany) for RNA isolation, or fixed in 4% paraformaldehyde for immunohistochemistry.
2.4. RNA extraction and reverse transcription for semi-quantitative RT-PCR
Tissues were homogenised using a Stratech Beadbeater-B (Stratech Scientific, Scham, UK) with 1 mm3 Zirconia/silica beads (Thistle Scientific, Glasgow, Scotland, UK) and filtered through a Qiashredder (Qiagen, Crawley, UK). Total RNA was extracted using the RNeasy kit (Qiagen) according to the manufacturer’s instructions. For semi-quantitative RT-PCR RNA was reverse-transcribed using a Promega RT kit (Promega, Southampton, UK).
2.5. Sequencing of sITLN1 and sITLN3
For sITLN1, a 220 bp partial sequence of sITLN1 was amplified from lungs of six normal yearling sheep (control sheep for D. filaria experiment) using the sITLN primers; sITLN ext. GSP1 and 2. Sequences in all sheep were similar and found to share 94% homology with the sITLN2 sequence (GenBank accession number EF521881) obtained previously from T. circumcincta infected sheep abomasum and 98% homology with the sheep respiratory tract ITLN partial sequence (GenBank accession number AM087961).
The 5′ end was amplified from sheep respiratory tissue using the forward primer sITLN F1 with the reverse primer sheep ext. GSP1 which had previously been used in sequencing of sITLN2 [8]. The following conditions were used: 94 °C 2 min then 40 cycles of 72 °C 40 s, 55 °C 40 s, 72 °C 2 min and a final extension at 72 °C for 10 min. Amplification of the 3′ end required preparation of 3′ RACE (rapid amplification of cDNA ends) ready cDNA using BD SMART II™ A oligonucleotide and the BD PowerScript™ Reverse Transcriptase (RT) according to the manufacturer’s recommendations. The following 50 μL PCR reaction was then set up on ice; 5 μL 10 × BD advantage 2 PCR buffer, 1 μL 10 mM dNTP mix, 1 μL 50 × BD advantage 2 polymerase mix and 34.5 μL nuclease free water, 2.5 μL 3′ race ready cDNA, 5 μL universal primer mix, 1 μL sITLN ext. GSP 2 10 μM. The reaction conditions were as follows: 25 cycles of 94 °C for 30 s, 65 °C for 30 s and 72 °C for 3 min. This was followed by a nested PCR for which the following was set up on ice: 5 μL BD advantage polymerase buffer, 1 μL 10 mM dNTP mix, 1 μL BD Advantage™ 2 Polymerase (BD Biosciences, Clontech), 34.5 μL nuclease free water, 2.5 μL PCR product, 5 μL nested universal primer mix, 1 μL sITLN int. GSP 2 10 μM. The reaction conditions were as follows: 20 cycles of 94 °C for 30 s, 65 °C for 30 s and 72 °C for 3 min and a final extension of 72 °C for 7 min. PCR products were cloned into the TA Cloning vector pCR2.1 (Invitrogen, Leek, The Netherlands). Positive clones were identified by restriction enzyme digestion with EcoR1 (Roche, Mannheim, Germany) to excise the insert. All nucleotide sequences were determined using dideoxy chain termination cycle sequencing by the Functional Genomics Unit, Moredun Research Institute. Using the overlapping 5′ and 3′ sequences the full sITLN1 sequence was determined. The full sequence was amplified from respiratory tract tissue of five sheep using the primers sITLN F3 and sITLN R3.
For sITLN3, a novel 3′ end ITLN clone (C0009264A24) was identified from the DEFRA/SHEFC Veterinary Training Research Initiative (VT0102) cDNA library of sheep lymph node tissue (KN511 library data submitted to UniGene). This clone shared 93% homology with sITLN2 and 85% with sITLN1. Using the specific primers sITLN F10 and sITLN R10, this clone was amplified from lymph node tissue of normal sheep and the sequence verified. The reaction conditions were as follows; 94 °C for 2 min, 35 cycles of 94 °C for 40 s, 58 °C for 40 s and 72 °C for 2 min and a final extension of 72 °C for 7 min. The 5′ end of sITLN3 was amplified in lymph node using the forward primer sITLN F1 with the sITLN3 specific reverse primer, sITLN R9-3. Specific primers for sITLN1, 2 and 3 had been designed for a poorly conserved region towards the 3′ end. The full sequence of sITLN3 was obtained from the overlapping sequences of the 5′ and 3′ ends and was amplified in jejunum from three sheep using the full length primers sITLN F7 and sITLN R10. All primers were designed using the primer 3 programme.
For sITLN1 and sITLN3 the PCR products of the full sequence were purified (High Pure PCR Product Purification kit; Roche), the sequences verified, the predicted open reading frame determined, and the predicted ORF, N-glycosylation sites, predicted signal peptide and GPI anchor were identified. PCR products were visualized on ethidium bromide-stained 1.2% agarose gels and images recorded using the Bio-Rad FX imager. All primers used in sequencing and expected product size are shown in Table II.
Table II.
Details of primers used in sequencing sheep genes and for RT-PCR.
| Gene | Primer sequence | Product size (BP) | |
|---|---|---|---|
| sITLN ext. GSP 2 | Fw | GGT GGC GGC TGG ACC CTG GTG | 309 |
| sITLN ext. GSP 1 | Rv | GCT CCG GAA GAA GCC CGT GTT GG | |
| sITLN int. GSP 2 | Fw | CAC GGT GGG CGA TCG CTG GTC | 176 |
| sITLN int. GSP 1 | Rv | GCA CGT GCC AGA TGC CCA GGT C | |
| sITLN F1 | Fw | GCT CTG AGA CTG CTC CTG GT | 420 |
| sITLN ext. GSP 1 | Rv | GCT CCG GAA GAA GCC CGT GTT GG | |
| sITLN F1 | Fw | GCT CTG AGA CTG CTC CTG GT | 1 182 |
| sITLN R1 | Rv | TTC TCC TTT GCT TGT TGG AGA | |
| sITLN F3 | Fw | GCA TCT GTG AGG AAG GAA GG | 1 112 |
| sITLN R3 | Rv | TTC CAT TTC TGA GGG TCT GG | |
| sITLN F1 | Fw | GCT CTG AGA CTG CTC CTG GT | 1 125 |
| sITLN R9-3 | Rv | TGG GTT CCA CAT CTC ATA GA | |
| sITLN F10 | Fw | GCGAGAATTTGTTGCAGGAT | 417 |
| sITLN R10 | Rv | CCAGGGGTCAGTCAGTTTGT | |
| sITLN F7 | Fw | AGGGGACAGGAGTCAGGTTT | 1 000 |
| sITLN R10 | Rv | CCAGGGGTCAGTCAGTTTGT | |
| sATPase F | Fw | GCT GAC TTG GTC ATC TGC | 167 |
| sATPase R | Rv | CAG GTA GGT TTG AGG GGA TAC | |
| sITLN F10 | Fw | GCGAGAATTTGTTGCAGGAT | 287 |
| sITLN R9-1 | Rv | TGG CTT TCT GTC TCA CAC CA | |
| sITLN F10 | Fw | GCG AGA ATT TGT TGC AGG AT | 286 |
| sITLN R9-2 | Rv | TGG GTT CAC ATC TCA TAG A | |
| sITLN F10 | Fw | GCG AGA ATT TGT TGC AGG AT | 287 |
| sITLN R9-3 | Rv | TGG GTT CCA CAT CTC ATA GA | |
| BD Smart 11 A oligonucleotide | AAGCAGTGGTATCAACGCAGAGTACGCGGG | ||
| 3′-Race CDS primer A | AAGCAGTGGTATCAACGCAGAGTAC(T)30 | ||
| 5′-Race CDS primer | (T)25VN | ||
| 10 × universal primer A long | CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT | ||
| 10 × universal primer A short | CTAATACGACTCACTATAGGGC | ||
Forward (Fw), reverse (Rv), intelectin (ITLN), sheep (s), base pair (BP), external (ext.) and internal (int.).
2.6. Detection of transcripts by RT-PCR
sITLN 1, 2 and 3 specific primers were designed over a poorly conserved area at the 3′ end of the sITLN. The primers were labelled as follows for sITLN1 (sITLN F10, sITLN R9-1), for sITLN2 (sITLN F10, sITLN R9-2) and for sITLN3 (sITLN F10, sITLN R9-3). cDNA (50 ng) was amplified by PCR with the specific primers for sITLN and primers for sheep ATPase using equivalent quantities of non-reversed transcribed RNA as negative controls. For semi-quantitative RT-PCR reaction conditions were optimised to ensure that the number of thermocycles correlated with the amplification stage of the PCR. Reaction conditions were as follows: sITLN 35 cycles of 94 °C for 40 s, 58 °C for 40 s and 72 °C for 2 min; sheep ATPase 31 cycles of 94 °C for 30 s, 60 °C for 30 s and 72 °C for 30 s. The expected product sizes and sequences are shown in Table II. Resultant PCR products were visualized on ethidium bromide-stained 1.6% agarose gels and images recorded using the Bio-Rad FX imager and analysed using the 1D Analysis software.
2.7. Anti-ITLN immunohistochemistry
Tissues fixed in 4% paraformaldehyde were processed into paraffin blocks and sections (4 μm) were placed onto coated slides (Snowcoat X-tra; Surgipath, Winnipeg, Manitoba, Canada). Sections were dewaxed in xylene, rehydrated through graded alcohols and endogenous peroxidase activity was blocked by treatment with methanol containing 3% v/v of 30% hydrogen peroxide (Sigma). After rehydration, sections were placed in 10 mM Tris-HCl pH 9.0 in a pressure cooker, brought to pressure in a microwave and held at pressure for 4 min. Slides were washed, blocked with PBS containing 10% normal rabbit serum, 0.5 M NaCl and 0.5% Tween 80, and incubated with the affinity-purified chicken anti-ITLN peptide (1 μg/mL in blocking buffer) which because of shared homology is thought to be non specific for sheep ITLN or control chicken IgY (1 μg/mL in blocking buffer) for 1 h at 21 °C. After treatment with horseradish peroxidase-conjugated rabbit anti-chicken IgY (Sigma, 1/500), sections were stained with Nova Red (Vector Laboratories) counterstained with hematoxylin, then dehydrated and mounted.
2.8. Statistical analysis of semi-quantitative, quantitative PCR transcripts and ITLN protein
Normality of data distribution was determined using the Kolmogorov Smirnov test. A Kruskal-Wallis was used to compare multiple groups and a Mann–Whitney U test was used as a post hoc test for pair wise comparisons; p value < 0.05 was considered significant.
3. RESULTS
3.1. Cloning and sequencing of sITLN1
The full sequence of sITLN1 was cloned and sequenced. Its presence was confirmed in lung and tracheal mucosal samples from two sheep using the primers sITLN F3 and sITLN R3 (Fig. 1). Using web based programmes the predicted open reading frame was identified, homology ascertained, and predicted signal peptides and N-glycosylation sites determined. The full sequence (1 226 bp) thus obtained had a predicted opening reading frame of 969 bp and a 322 AA deduced amino acid sequence (GenBank accession number AM087961, updated October 2007). The full sequence shares 77% homology with hITLN1 (GenBank accession number NM017625) and 88% homology with sITLN2. The deduced amino acid sequence of sITLN1 shares 80% homology with hITLN1 and 86% homology with sITLN2. A 24 AA predicted signal peptide (probability 1.0) with a predicted cleavage site (probability 0.883) between AA 24 and 25 (GRG-AV) was identified, and a predicted N-glycosylation site was identified at AA 72. There was no evidence of a GPI anchor site. Cysteine residues implicated in oligomer formation are shown in Figure 1.
Figure 1.
Alignment of predicted open reading frame of sITLN 1, 2 and 3. Solid underlines delineate the predicted signal peptides, double underline predicted N-glycosylation sites. The broken underline represents the peptide to which the affinity purified antibody was raised. The hatch indicates the location of the cysteine residues that have been implicated in oligomer formation in hITLN1. The asterices, singles dots and double dots represent homology.
3.2. Cloning and sequencing of sITLN3
The full sequence of sITLN3 was cloned and sequenced. Using web based programmes the predicted open reading frame was identified (Fig. 1), homology ascertained, and predicted signal peptides and N-glycosylation sites determined. The full sequence (1 153 bp) thus obtained has a predicted opening reading frame of 972 bp and a deduced amino acid sequence of 323 AAs (GenBank accession number AM888394). The full sequence shares 73% homology with hITLN1 (GenBank accession number NM017625), 86% homology with sITLN1 and 93% homology with sITLN2. The deduced amino acid sequence of sITLN3 shares 79% homology with hITLN1, 85% homology with sITLN1 and 91% homology with sITLN2. A 26 AA predicted signal peptide was identified (probability 0.992), with a predicted cleavage site (probability 0.509) between AA 26 and 27 (SAA-GT). No predicted N-glycosylation sites were identified using the programme NetNGlyc 1.0. There was no evidence of a GPI anchor site. Cysteine residues implicated in oligomer formation are shown in Figure 1.
3.3. Transcript expression of sITLN1, 2 and 3 in sheep tissues
Expression of these three sITLN transcripts was examined in multiple tissues from three normal yearling sheep (numbered 33, 920 and 1 379) though all tissues were not available from all three.
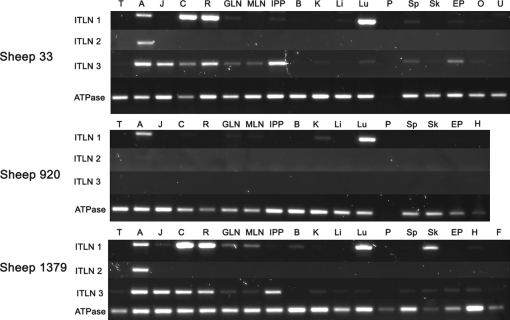
sITLN1 expression was shown in abomasum, lung, colon and gastric lymph node of 3/3 sheep, in terminal rectum of 2/3 sheep and in skin of 1/3 sheep. Weak expression was also shown in jejunum 1/3, mesenteric lymph node 2/3, ileal Peyer’s patches 1/3, brain 1/3, kidney 2/3, liver 1/3, spleen 1/3, skin 1/3, ear pinna 1/3, heart 1/2 and ovary 1/1. There was no evidence of expression in uterus, pancreas or mesenteric fat of the single sheep tested (Fig. 2).
Figure 2.
Tissue distribution of sITLN 1, 2 and 3 transcripts in three sheep (numbered 333, 920 and 1 379). Expression of the house keeping gene ATPase is shown for comparison. Tongue (T), abomasum (A), jejunum (J), colon (C), rectum (R), gastric lymph node (GLN), mesenteric lymph node (MLN), intestinal Peyer’s patches (IPP), brain (B), kidney (K), liver (Li), lung (Lu), pancreas (P), spleen (Sp), skin (Sk), ear pinna (Ep), ovary (O), uterus (U), heart (H), fat (F).
sITLN2 transcript expression was confined to the abomasum. Sheep 920 showed only very weak expression compared to sheep 33 and 1 379 (Fig. 2).
Strong expression of sITLN3 was shown in the abomasum, jejunum, colon, rectum and ileal Peyer’s patches of sheep 33 and 1 379 but not in sheep 920 where no expression was detected in any tissue. Weak expression was also shown in gastric and mesenteric lymph nodes, kidney, lung, spleen, skin and ear pinna of both 33 and 1 379, but the heart, ovary, liver and mesenteric fat was positive in only one animal. There was no evidence of expression in brain, pancreas or uterus.
There was no expression of pancreatic ATPase in sheep 33 and 920 and it was presumed that this tissue had degraded prior to storage (Fig. 2).
3.4. Localisation of ITLN protein expression in sheep tissues
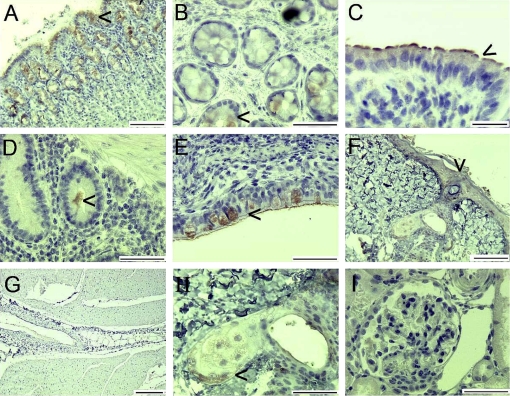
Multiple tissues from sheep 1 379 were immunolabelled using the affinity purified chicken antibody to ITLN peptide 1 as described in Materials and methods. Negative controls were immunolabelled with chicken IgY. There was no staining of negative controls (not shown). There was positive staining of the mucus neck cells in the abomasum (Fig. 3A), the mucus cells in the colon (Fig. 3B), free mucus in ileum (Fig. 3D) and the goblet cells in the lung (Fig. 3E). There was also staining of the small intestinal epithelium and brush border (Fig. 3C). There was weak staining of the epidermal layer of the skin (Fig. 3F) and skin sebaceous glands (Fig. 3H). There was no staining of heart muscle or kidney (Figs. 3G and 3I). There was no staining of lymph nodes, liver, pancreas, spleen or brain (not shown).
Figure 3.
Immunoperoxidase labelling of sections with affinity purified chicken anti-ITLN peptide 1. Abomasum (A), colon (B), small intestine (C), Peyer’s patches and ileum (D), large bronchus (E), dermis/epidermis (F), heart muscle and fat (G), skin sebaceous gland (H), kidney (I). Arrows indicate positive immunolabelling. Control IgY labelling was negative (not shown). Bars represent 50 μm (B, D, E, H, I), 100 μm (A, C, F), 200 μm (G).
3.5. Expression of sITLN1, 2 and 3 in lamb abomasum after infection with T. circumcincta
Worm counts are not included in this paper. In brief, significantly fewer worms were found in the cpi lambs compared with the cnv lambs (p < 0.01) as has been described previously [21]. In addition, the previously-infected lambs contained a significantly higher proportion of arrested early L4s than the naïve sheep after the challenge infection.
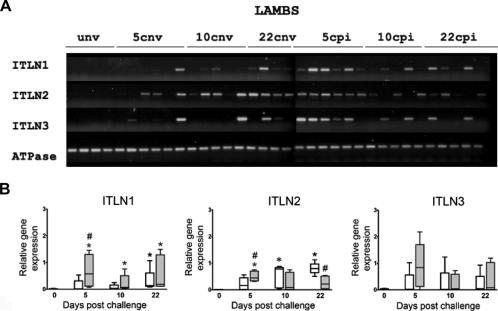
Abomasal mucosal tissue was collected from the following groups of 4–5 month old lambs; unv (n = 4), cnv; days 5 (n = 6), 10 (n = 5), 22 (n = 4) and cpi; days 5 (n = 6), 10 (n = 5), 22 (n = 5) in total n = 35. The relative expression of transcripts of sITLN1, sITLN2 and sITLN3 was determined. Sheep ATPase was used as the housekeeping gene. Challenged groups (previously infected and naïve) were compared to the unv group. Full details of molecular biology techniques used have been described previously. Data were not normally distributed and a Kruskal–Wallis test was used to determine significant difference between groups. For sITLN1 and sITLN2, groups were significantly different and a Mann–Whitney U test was applied as a post hoc test. There was no expression of sITLN1, sITLN2 or sITLN3 in unv lambs. There was significantly higher expression of sITLN1 transcript in cpi lambs compared to unv at all time points (days 5, 10 and 22) whilst significantly higher expression was only seen at day 22 when cnv were compared to unv. For sITLN2 significantly higher transcript expression was seen at day 5 when cpi were compared to cnv and at days 10 and 22 when cnv were compared to unv. There was significantly higher expression of transcript in the cpi compared to the cnv sheep at day 5 for sITLN 1 and 2 and also at day 22 for sITLN2. For sITLN3 no significant difference in expression was shown between groups and thus no post hoc tests were applied (Fig. 4).
Figure 4.
(A) Ethidium bromide gel (1.4%) of sITLN 1, 2 and 3 transcripts in lambs; unv, cnv (days 5, 10, 22) and cpi (days 5, 10 and 22). Expression of the housekeeping gene sheep ATPase is shown for comparison. Group numbers were as follows: unv, 22 cnv (n = 4), groups 10 cnv, 10 cpi, 22 cpi (n = 5) and groups 5 cnv, 5 cpi (n = 6). (B) Box and whisker plots showing relative transcript expression by RT-PCR in abomasal mucosa of unv, T. circumcincta cnv and T. circumcincta cpi lambs. No unchallenged previously infected group was present in this experiment. The house keeping gene was sheep ATPase. Semi-quantitative RT-PCR was carried out as described in Materials and methods. The naïve groups are shown as clear black boxes, whilst the previously infected groups as hatched grey boxes. Boxes represent the 25th and 75th quartiles, median is shown within the box, whiskers represent the range of the data. Asterix represents significant difference between cnv (*) or cpi (*) and unv group, whilst hatch (#) represents significant difference between cnv and cpi groups at the same time points post challenge. A Kruskal-Wallis test was used to compare all groups and a Mann–Whitney U as a post hoc test. p < 0.05 was considered significant.
3.6. Expression of sITLN1, 2 and 3 in sheep lungs after infection with D. filaria
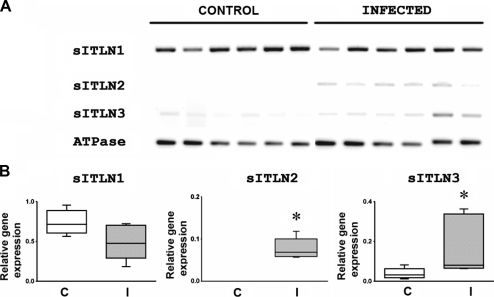
The relative expression of transcripts for the three sITLN was determined by RT-PCR in 6 D. filaria infected sheep and 6 control yearling sheep as described previously. Expression was determined relative to the house keeping gene sheep ATPase. sITLN1, sITLN2 and sITLN3 transcripts were expressed in lung samples from D. filaria infected sheep whilst only expression of sITLN1 and sITLN3 was shown in lungs from control sheep. There was no significant difference in expression of sITLN1 between infected and control sheep (p = 0.0649) whilst there was a significant difference in expression of sITLN2 (p = 0.0022) and sITLN3 (p = 0.0152) between infected and control sheep with induction of sITLN2 and upregulation of sITLN3 in the infected sheep (Fig. 5).
Figure 5.
Ethidium bromide gel showing relative expression of sITLN 1, 2 and 3 transcripts in lung from normal and Dictyocaulus filaria infected sheep (A) and corresponding graphs showing relative transcript expression in control (C) and infected (I) lungs (B). Transcript expression is relative to the house keeping gene sheep ATPase. Boxes represent 25th and 75th quartile range, lines represents median and whiskers range of data. * Represents significant difference compared to controls. Groups were compared using a Mann–Whitney U test. p < 0.05 was considered significant.
4. DISCUSSION
ITLN homologues are known in sea squirts, fishes [4, 9], frogs [3] and mammals [2, 12, 14, 15, 17, 22, 25]. Within mammalian species two homologues have been described in mice [12, 17], man [14], sheep [7, 8] and recently in the bovine [2]. In the present work the full sequence of sITLN1 has been determined and furthermore a novel third sITLN has been cloned and sequenced. The three sITLN homologues share 85–91% identity with each other and 53–81% homology with human and murine sequences. For the three sITLN the predicted AA sequences are poorly conserved at the N-terminus and better conserved towards the middle and C-terminus end. sITLN1 and sITLN3 have predicted signal peptides, as found with sITLN2 suggestive that all three sheep ITLN can be secreted. Interestingly sITLN2 has a predicted GPI anchor site at the C-terminus however this is not present in sITLN1 or sITLN3 which might suggest a difference in function for sITLN2.
Recent studies have shown that recombinant hITLN1 is a 120 kDa trimer, however despite 84% homology with mITLN1, recombinant and intestinal mITLN1 exist as unglycosylated 30 kDa monomers [26]. In the present study, sITLN1 and sITLN3 possess Cys-39/40 and Cys-57/58 respectively, which align with Cys-31 and Cys-48 in hITLN1, that have been implicated in oligomer formation. However sITLN2 differs, and only possesses Cys-58. Again this may suggest a difference in structure and possibly function for sITLN2 compared to sITLN1 and sITLN3. No work has been undertaken to examine the carbohydrate specificity of the sITLN. However it cannot be presumed that they are identical to other species, as recent work has shown that hITLN1 and mITLN1 have different saccharide binding specificities from each other [26].
The three sheep ITLN differed in number of predicted N-glycosylation sites with one present in sITLN1, two in sITLN2 and none in sITLN3. This is another factor that might suggest different functional roles for the three sheep ITLN.
The three sITLN differed in transcript tissue distribution pattern, as has been reported previously for the two murine [12, 17, 23, 24] and two human analogues [14]. Transcripts for sITLN1 and sITLN3 were present in several tissues whilst the sITLN2 transcript was restricted to the abomasum in the normal animal. It is of interest that transcripts for hITLN2 [14] are also restricted to the gastrointestinal tract whilst for mITLN1b expression is restricted to the respiratory tract in the normal animal [27].
The variation in expression of the three sITLN transcripts between sheep was of particular interest. It is known that some in-bred murine strains show genomic variation, with the relatively poor T. spiralis expelling strain, C57BL10, lacking mITLN1b [17]. It is unknown if similar variation occurs in sheep or people and the significance of this finding regarding expression patterns of disease.
In the present study sITLN was immunolocalised to epithelial cells and mucus producing cells with expression shown in the gastrointestinal tract, respiratory tract and skin of normal sheep. Whilst strong immunolabelling was present in mucus producing cells of the abomasum and colon, and goblet cells in the respiratory tract, protein expression was also present on the small intestinal brush border as has been reported for mice and pigs [23, 30]. There was also faint immunolabelling of the enterocytes as reported in pigs [30]. These findings are suggestive that ITLN is not only produced by mucus producing cells. In mice, ITLN has been immunolocalised to a greater range of tissues with expression reported in gastrointestinal, respiratory, reproductive, urinary tract, lymphatic, nervous and endocrine tissue (pancreas) [23] whilst in man protein expression has been shown in endothelial cells, respiratory epithelial cells, mesothelial cells, omental fat and serum [13, 28, 32].
The presence of ITLN transcript and protein in sheep skin tissue is very interesting as there are no reports of expression of ITLN in skin of other mammalian species, however recently expression has been shown in rainbow trout skin [19]. In the present work ITLN appeared to be immunolocalised to skin sebaceous gland and to the dermis. ITLN has been recognised as a lactoferrin receptor in several species [15, 22, 23]. Expression of lactoferrin has been shown in the skin in man [5, 29] and furthermore using competitive binding assays has been shown to bind to keratinocyte epithelial cells. This finding supports the presence of lactoferrin receptors in keratinocytes [29]. ITLN may play an independent role in skin defence or have a role as a lactoferrin receptor in this tissue.
ITLN1 and 3 transcripts were shown in lymphoid tissue in the sheep however there was no convincing evidence of protein expression on immunohistochemistry. Studies in mice have shown immunolabelling of small lymphocytes in the lymph node and spleen [23]. In man, whilst hITLN1 transcript was shown in lymph node, spleen and thymus, protein expression was immunolocalised to endothelial cells [14]. Interestingly in carp, whilst there was immunolabelling of endothelial cells in the spleen, small lymphocytes were also immunolabelled as well as clusters of immunoreactive cells (macrophages/lymphocytes) in the kidney [4]. Thus, there is evidence for a role for ITLN in lymphoid tissue, further work may clarify this role.
Omentin, shown to be identical to hITLN1, is thought to have a link to obesity and type II diabetes mellitus and expression has been shown in mesenteric fat cells [20, 31, 32]. In the present experiment there was evidence of sITLN3 transcript expression in mesenteric fat tissue and faint immunolabelling of mesenteric fat. Further work is required to verify these findings. Immunolabelling of neurological tissue has been shown in mice and in carp [4, 23] however whilst sITLN1 transcript was shown in brain from sheep 1 379 in the present experiment, there was no evidence of immunolabelling of cells. This disparity may reflect a species difference or difference in immunolabelling techniques or part of the brain examined.
Using non specific primers we have previously demonstrated the upregulation of sITLN transcript in the ovine respiratory tract [7] in response to culture of tracheal explants with the Th2 cytokine IL-4 and the upregulation of sITLN2 transcript in the sheep abomasum in response to challenge infection with T. circumcincta [8]. More recently sITLN2 has also been shown to be highly upregulated in the abomasum of sheep following challenge with Haemonchus contortus [18]. We now demonstrate that sITLN2 is induced and sITLN3 upregulated in response to D. filaria infection in sheep. Furthermore, although no sITLN were detected in the abomasal mucosa of worm free lambs, all three were upregulated in this tissue following infection with T. circumcincta. Interestingly, significant upregulation of sITLN1 and sITLN2 transcripts occurred by day 5 in the immune (cpi) lambs. This was fast enough for ITLN to be implicated in the mechanisms of worm loss and arrested development as reported before [11].
ITLN may play a role in altering the characteristics of mucus leading to worm entrapment, however the recognition of bacterial specific carbohydrate residues [25] supports an anti-bacterial role. Of particular interest is recent work in rainbow trout where ITLN was found to be one of the acute phase proteins upregulated in the liver 24 h after intraperitoneal injection with Listorella anguillarum. It was postulated that ITLN may have been involved in the sequestration of iron in its role as a lactoferrin receptor.
Further studies are required to elucidate the exact role of these recently described molecules and to determine whether they play a protective role in nematode infections.
Acknowledgments
We would like to acknowledge the following people: Kevin McLean and colleagues at Functional Genomics Unit, Moredun Research Institute; Aileen Halliday and Steve Smith at Moredun Research Institute for the post-mortem material; Elisabeth Thornton and Steven Wright for technical assistance at Easter Bush Veterinary Centre and Neil McIntrye and everyone in the Department of Pathology, Easter Bush Veterinary Centre. This work was supported by grants from the Department for Environment, Food and Rural Affairs/Scottish Higher Education Funding Council (DEFRA/SHEFC) Veterinary Training and Research Initiative (VTRI; VT0102), the Wellcome Trust (Award No. 060312), the Norman Salvesen Emphysema Research Trust and the Scottish Executive Environment and Rural Affairs Department.
References
- 1.Artis D.. New weapons in the war on worms: identification of putative mechanisms of immune-mediated expulsion of gastrointestinal nematodes. Int. J. Parasitol. 2006;36:723–733. doi: 10.1016/j.ijpara.2006.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blease S.C., French A.T., Knight P.A., Gally D.L., Pemberton A.D.. Bovine intelectins: cDNA sequencing and expression in the bovine intestine. Res. Vet. Sci. 2008;86:254–256. doi: 10.1016/j.rvsc.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Chang B.Y., Peavy T.R., Wardrip N.J., Hedrick J.L.. The Xenopus laevis cortical granule lectin: cDNA cloning, developmental expression, and identification of the eglectin family of lectins. Comp. Biochem. Physiol. 2004;137:115–129. doi: 10.1016/s1095-6433(03)00269-1. [DOI] [PubMed] [Google Scholar]
- 4.Chang M.X., Nie P.. Intelectin gene from the grass carp Ctenopharyngodon idella: cDNA cloning, tissue expression, and immunohistochemical localization. Fish Shellfish Immunol. 2007;23:128–140. doi: 10.1016/j.fsi.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 5.Cumberbatch M., Dearman R.J., Uribe-Luna S., Headon D.R., Ward P.P., Conneely O.M., Kimber I.. Regulation of epidermal Langerhans cell migration by lactoferrin. Immunology. 2000;100:21–28. doi: 10.1046/j.1365-2567.2000.00014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Veer M.J., Kemp J.M., Meeusen E.N.. The innate host defence against nematode parasites. Parasite Immunol. 2007;29:1–9. doi: 10.1111/j.1365-3024.2006.00910.x. [DOI] [PubMed] [Google Scholar]
- 7.French A.T., Bethune J.A., Knight P.A., McNeilly T.N., Wattegedera S., Rhind S.. et al. The expression of intelectin in sheep goblet cells and upregulation by interleukin-4. Vet. Immunol. Immunopathol. 2007;120:41–46. doi: 10.1016/j.vetimm.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 8.French A.T., Knight P.A., Smith W.D., Brown J.K., Craig N.M., Pate J.A.. et al. Up-regulation of intelectin in sheep after infection with Teladorsagia circumcincta. Int. J. Parasitol. 2007;38:467–475. doi: 10.1016/j.ijpara.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 9.Gerwick L., Corley-Smith G., Bayne C.J.. Gene transcript changes in individual rainbow trout livers following an inflammatory stimulus. Fish Shellfish Immunol. 2007;22:157–171. doi: 10.1016/j.fsi.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Guyatt H.. Do intestinal nematodes affect productivity in adulthood? Parasitol. Today. 2000;16:153–158. doi: 10.1016/s0169-4758(99)01634-8. [DOI] [PubMed] [Google Scholar]
- 11.Halliday A.M., Routledge C.M., Smith S.K., Matthews J.B., Smith W.D.. Parasite loss and inhibited development of Teladorsagia circumcincta in relation to the kinetics of the local IgA response in sheep. Parasite Immunol. 2007;29:425–434. doi: 10.1111/j.1365-3024.2007.00959.x. [DOI] [PubMed] [Google Scholar]
- 12.Komiya T., Tanigawa Y., Hirohashi S.. Cloning of the novel gene intelectin, which is expressed in intestinal paneth cells in mice. Biochem. Biophys. Res. Commun. 1998;251:759–762. doi: 10.1006/bbrc.1998.9513. [DOI] [PubMed] [Google Scholar]
- 13.Kuperman D.A., Lewis C.C., Woodruff P.G., Rodriguez M.W., Yang Y.H., Dolganov G.M.. et al. Dissecting asthma using focused transgenic modeling and functional genomics. J. Allergy Clin. Immunol. 2005;116:305–311. doi: 10.1016/j.jaci.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 14.Lee J.K., Schnee J., Pang M., Wolfert M., Baum L.G., Moremen K.W., Pierce K.M.. Human homologs of the Xenopus oocyte cortical granule lectin XL35. Glycobiology. 2001;11:65–73. doi: 10.1093/glycob/11.1.65. [DOI] [PubMed] [Google Scholar]
- 15.Liao Y., Lopez V., Shafizadeh T.B., Halsted C.H., Lonnerdal B.. Cloning of a pig homologue of the human lactoferrin receptor: expression and localization during intestinal maturation in piglets. Comp. Biochem. Physiol. 2007;148:584–590. doi: 10.1016/j.cbpa.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonald V.. Parasites in the gastrointestinal tract. Parasite Immunol. 2003;25:231–234. doi: 10.1046/j.1365-3024.2003.00629.x. [DOI] [PubMed] [Google Scholar]
- 17.Pemberton A.D., Knight P.A., Gamble J., Colledge W.H., Lee J.K., Pierce M., Miller H.R.. Innate BALB/c enteric epithelial responses to Trichinella spiralis: inducible expression of a novel goblet cell lectin, intelectin-2, and its natural deletion in C57BL/10 mice. J. Immunol. 2004;173:1894–1901. doi: 10.4049/jimmunol.173.3.1894. [DOI] [PubMed] [Google Scholar]
- 18.Rowe A., Gondro C., Emery D., Sangster N.. Sequential microarray to identify timing of molecular responses to Haemonchus contortus infection in sheep. Vet. Parasitol. 2009;161:76–87. doi: 10.1016/j.vetpar.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 19.Russell S., Young K.M., Smith M., Hayes M.A., Lumsden J.S.. Identification, cloning and tissue localization of a rainbow trout (Oncorhynchus mykiss) intelectin-like protein that binds bacteria and chitin. Fish Shellfish Immunol. 2008;25:91–105. doi: 10.1016/j.fsi.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 20.Schaffler A., Neumeier M., Herfarth H., Furst A., Scholmerich J., Buchler C.. Genomic structure of human omentin, a new adipocytokine expressed in omental adipose tissue. Biochim. Biophys. Acta. 2005;1732:96–102. doi: 10.1016/j.bbaexp.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 21.Smith W.D., Jackson F., Jackson E., Williams J.. Local immunity and Ostertagia circumcincta: changes in the gastric lymph of immune sheep after a challenge infection. J. Comp. Pathol. 1983;93:479–488. doi: 10.1016/0021-9975(83)90035-x. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki Y.A., Shin K., Lonnerdal B.. Molecular cloning and functional expression of a human intestinal lactoferrin receptor. Biochemistry. 2001;40:15771–15779. doi: 10.1021/bi0155899. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki Y.A., Lonnerdal B.. Baculovirus expression of mouse lactoferrin receptor and tissue distribution in the mouse. Biometals. 2004;17:301–309. doi: 10.1023/b:biom.0000027709.42733.e4. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki Y.A., Lopez V., Lonnerdal B.. Mammalian lactoferrin receptors: structure and function. Cell. Mol. Life Sci. 2005;62:2560–2575. doi: 10.1007/s00018-005-5371-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsuji S., Uehori J., Matsumoto M., Suzuki Y., Matsuhisa A., Toyoshima K., Seya T.. Human intelectin is a novel soluble lectin that recognizes galactofuranose in carbohydrate chains of bacterial cell wall. J. Biol. Chem. 2001;276:23456–23463. doi: 10.1074/jbc.M103162200. [DOI] [PubMed] [Google Scholar]
- 26.Tsuji S., Yamashita M., Nishiyama A., Shinohara T., Li Z., Myrvik Q.N.. et al. Differential structure and activity between human and mouse intelectin-1: human intelectin-1 is a disulfide-linked trimer, whereas mouse homologue is a monomer. Glycobiology. 2007;17:1045–1051. doi: 10.1093/glycob/cwm075. [DOI] [PubMed] [Google Scholar]
- 27.Voehringer D., Stanley S.A., Cox J.S., Completo G.C., Lowary T.L., Locksley R.M.. Nippostrongylus brasiliensis: identification of intelectin-1 and -2 as Stat6-dependent genes expressed in lung and intestine during infection. Exp. Parasitol. 2007;116:458–466. doi: 10.1016/j.exppara.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wali A., Morin P.J., Hough C.D., Lonardo F., Seya T., Carbone M., Pass H.I.. Identification of intelectin overexpression in malignant pleural mesothelioma by serial analysis of gene expression (SAGE) Lung Cancer. 2005;48:19–29. doi: 10.1016/j.lungcan.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 29.Ward P.P., Uribe-Luna S., Conneely O.M.. Lactoferrin and host defense. Biochem. Cell Biol. 2002;80:95–102. doi: 10.1139/o01-214. [DOI] [PubMed] [Google Scholar]
- 30.Wrackmeyer U., Hansen G.H., Seya T., Danielsen E.M.. Intelectin: a novel lipid raft-associated protein in the enterocyte brush border. Biochemistry. 2006;45:9188–9197. doi: 10.1021/bi060570x. [DOI] [PubMed] [Google Scholar]
- 31.Wurm S., Neumeier M., Weigert J., Schaffler A., Buechler C.. Plasma levels of leptin, omentin, collagenous repeat-containing sequence of 26-kDa protein (CORS-26) and adiponectin before and after oral glucose uptake in slim adults. Cardiovasc. Diabetol. 2007;6:7. doi: 10.1186/1475-2840-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang R.Z., Lee M.J., Hu H., Pray J., Wu H.B., Hansen B.C.. et al. Identification of omentin as a novel depot-specific adipokine in human adipose tissue: possible role in modulating insulin action. Am. J. Physiol. 2006;290:E1253–E1261. doi: 10.1152/ajpendo.00572.2004. [DOI] [PubMed] [Google Scholar]