Abstract
Laccases belong to the group of multicopper oxidases that exhibit wide substrate specificity for polyphenols and aromatic amines. They are found in plants, fungi, bacteria, and insects. In insects the only known role for laccase is in cuticle sclerotization. However, extracting laccase from the insect’s cuticle requires proteolysis, resulting in an enzyme that is missing its amino-terminus. To circumvent this problem, we expressed and purified full-length and amino-terminally truncated recombinant forms of laccase-2 from the tobacco hornworm, Manduca sexta. We also purified the endogenous enzyme from the pharate pupal cuticle and used peptide mass fingerprinting analysis to confirm that it is laccase-2. All three enzymes had pH optima between 5 and 5.5 when using N-acetyldopamine (NADA) or N-β-alanyldopamine (NBAD) as substrates. The laccases exhibited typical Michaelis-Menten kinetics when NADA was used as a substrate, with Km values of 0.46 mM, 0.43 mM, and 0.63 mM, respectively, for the full-length recombinant, truncated recombinant, and cuticular laccases; the apparent kcat values were 100 min−1, 80 min−1, and 290 min−1. The similarity in activity of the two recombinant laccases suggests that laccase-2 is expressed in an active form rather than as a zymogen, as had been previously proposed. This conclusion is consistent with the detection of activity in untanned pupal wing cuticle using the laccase substrate 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS). Immunoblot analysis of proteins extracted from both tanned and untanned cuticle detected only a single protein of 84 kDa, consistent with the full-length enzyme. With NBAD as substrate, the full-length recombinant and cuticular laccases showed kinetics indicative of substrate inhibition, with Km values of 1.9 mM and 0.47 mM, respectively, and apparent kcat values of 200 min−1 and 180 min−1. These results enhance our understanding of cuticle sclerotization, and may aid in the design of insecticides targeting insect laccases.
Keywords: cuticle, insect, multicopper oxidase, laccase, polyphenol oxidase, sclerotization, substrate inhibition
1. Introduction
Laccase (EC 1.10.3.2) is a multicopper oxidase that can oxidize a wide array of substrates including polyphenols, substituted phenols, and aromatic amines. The enzyme functions by coupling the single-electron oxidation of four substrate molecules with the concomitant reduction of molecular oxygen to water (Bento et al., 2006). It is widely distributed in plants, fungi, bacteria and insects. Roles attributed to this enzyme include lignin synthesis by plants, lignin degradation by fungi, leaf wound healing, pigmentation in bacteria and fungi, and morphogenesis in fungi (Mayer and Staples, 2002; Nakamura and Go, 2005; Baldrian, 2006; Hoegger et al., 2006; Sharma et al., 2007). Because of their wide substrate specificity, special attention has been give to the laccases of wood-rotting basidomycetes for biotechnological applications such as the detoxification of organic pollutants in industrial waste water, delignification of wood in the pulp and paper industries, and decolorization of dyes in the textile industry (Mayer and Staples, 2002; Torres et al., 2003; Chiacchierini et al., 2004; Xu, 2005; Riva, 2006; Gianfreda et al., 2006).
Unfortunately, by comparison, relatively little is known of the functions this enzyme has in insects. The best confirmed role for laccase in insects is in cuticle sclerotization. Yamazaki (1969) was the first to establish its presence in the cuticle and correlate its activity with the process of sclerotization. Similar results have since been obtained from several other insect species (reviewed in Andersen, 2005). During the sclerotization process, diphenols such as N-acetyldopamine (NADA) and N-β-alanyldopamine (NBAD) are oxidized by laccase, and the quinones that are generated react with cuticular proteins to form cross-links between the proteins (Andersen, 2005). Through the use of RNA interference experiments, Arakane et al. (2005) were able to identify the specific gene encoding the cuticular laccase in the red flour beetle, Tribolium castaneum. This gene, Tclac2, was essential for proper formation of the larval, pupal, and adult cuticles. Knockdown of Tclac2 resulted in insects whose cuticle was soft and untanned. In addition, developmental abnormalities were observed and individuals died within several days of molting.
The large size of the tobacco hornworm, Manduca sexta, makes it ideal for biochemical studies, including studies of cuticle sclerotization. Previously, an enzyme exhibiting laccase-like activity was partially purified from the pharate pupal cuticle of M. sexta (Thomas et al., 1989). This enzyme was found to oxidize a set of five laccase substrates, including NBAD, and its activity was inhibited by various laccase inhibitors. We subsequently cloned a cDNA of the M. sexta laccase-2 gene (Mslac2, an ortholog of Tclac2) and showed that it was highly expressed in the integument of pharate pupa just prior to molting, consistent with a role in sclerotization of newly formed cuticle (Dittmer et al., 2004). Two alternatively-spliced cDNAs for laccase-2 have been cloned from both T. castaneum (Arakane et al., 2005) and the malaria mosquito, Anopheles gambiae (Gorman et al., 2008). Examination of the genomes of the fruit fly, Drosophila melanogaster, the silkworm, Bombyx mori, and the yellow fever mosquito, Aedes aegypti, indicate the potential for spliced isoforms in these species as well. Currently, only one laccase-2 cDNA has been identified for M. sexta, but the potential for two isoforms in the closely related B. mori suggests that M. sexta may express two isoforms as well.
One of the challenges encountered in studies of insect cuticular laccase has been its intractable nature; only through the use of limited proteolysis had it been possible to solubilize the enzyme (Yamazaki, 1972; Andersen, 1978, Thomas et al., 1989; He et al., 2007) and it is unknown what affects this may have on its biochemical properties. Ashida and Yamazaki (1990) reported that laccase could be solubilized as an inactive enzyme from the cuticle of B. mori through limited digestion with chymotrypsin, and then activated by treatment with trypsin; recently, it was reported that this enzyme could be solubilized with urea and then activated with trypsin (Yatsu and Asano, 2009). Additionally, the deduced amino acid sequences of insect laccase-2 orthologs are very similar but show high variability at their amino-termini (Dittmer et al., 2004; Arakane et al., 2005). This requirement for trypsin treatment to activate the B. mori enzyme and the high sequence variability at the amino-terminus suggest that cuticular laccases may be expressed as zymogens.
The laccase previously identified from M. sexta pupal cuticle was presumably laccase-2, but this had not been confirmed. Our goal then was to verify that the laccase present in the pharate pupal cuticle (cuticular laccase) is indeed laccase-2 and to determine whether the purified enzyme is of a single isoform or a mix of isoforms. We also wished to address the possibility that laccase-2 is synthesized as a zymogen. We examined this question through enzymatic assays and immunoblot analysis of tanned and untanned cuticle extracts to show that a single protein of the same size is present in both types of cuticle. More importantly, we directly addressed this by expressing and purifying both full-length and amino-terminal truncated recombinant forms of laccase-2, and compared their biochemical properties to that of the endogenous cuticular laccase-2.
2. Materials and Methods
2.1 Recombinant protein expression
Two recombinant proteins were made using the Bac-to-Bac Baculovirus Expression System (Invitrogen); a full length and an N-terminal truncated laccase-2. The recombinant virus encoding the full length protein was generated by cloning the MsLac2 cDNA (Dittmer et al., 2004), which contained 116 bp of 5’-untranslated region (UTR), the ~2.3 kb coding region, and 900 bp of 3’-UTR, into the pFastBac vector and then transforming the E. coli strain DH10Bac per the kit directions. The recombinant virus encoding the N-terminal truncated protein, rΔ106, was cloned in a 4 step process. First, the full-length cDNA was digested with EcoRV and SalI to release a 2.2 kb C-terminal fragment containing 1,265 bp of the coding region (the last 421 codons of the protein) and 0.9 kb of 3’-UTR and cloned into the same sites in the plasmid pBluescriptKS. Second, the region encoding the portion of the protein from H107 to Y318 was amplified by PCR using the forward primer 5’- GAATTCCATCTTGACTTTACTAGC -3’ and the reverse primer 5’- GATATCTCTCAGCACGGTCATCGTG -3’ and cloned into the EcoRI and EcoRV sites of the pBluescriptKS vector already containing the 2.2 kb 3’ fragment to produce the cDNA for rΔ106 (minus a signal peptide). The EcoRI sequence (GAATTC) in the forward primer added a glutamic acid (E) and phenylalanine (F) residues to the amino-terminus preceding H107; the EcoRV sequence (GATATC) in the reverse primer was naturally occurring in the cDNA and therefore, did not add any additional residues to the protein. Third, the putative signal peptide of MsLac2 and the 5’-UTR were amplified by PCR using the forward primer 5’-GGATCCGCTCTTCAGTTAGGATTA -3’ and the reverse primer 5’-GAATTCACCGAGAGCAAGCTCAGT -3’ and cloned into the BamHI and EcoRI restriction sites of pFastBac. Last, the rΔ106 cDNA was excised from pBluescriptKS with EcoRI and SphI and cloned into the same restriction sites of pFastBac containing the signal peptide, creating a cDNA similar to the full-length clone except that it was missing residues V1-K106 (of the mature protein) and contained the additional amino acids of E and F between the signal peptide and H107 because of the EcoRI restriction site used in the cloning process. The pFastBac vector containing the rΔ106 cDNA was used to transform E coli strain DH10Bac to create the rΔ106 virus used for protein expression.
Recombinant proteins were expressed in insect Sf9 cells maintained as suspension cultures in a shaking incubator at 150 rpm and 28°C in Sf-900 II serum free medium (Invitrogen). For protein expression, 400 ml cultures (2 × 106 cells/ml) in 1 L culture flasks supplemented with 0.1 mM CuSO4 were infected with recombinant virus at a multiplicity of infection of 2. Generally, 3–5 flasks were used for each expression experiment. Cultures were collected 2 days after infection and centrifuged at 500 × g to pellet cells. The supernatant was then filtered through a 0.45 µm membrane (Magna nylon; GE Osmonics) to remove any cell debris and used for protein purification (see section 2.3).
2.2 Protein preparation from pharate pupal cuticle
M. sexta larvae were reared on an artificial diet at 27°C with a photoperiod of 16 h of light and 8 h of darkness (Bell and Joachim, 1976). The 5th instar larval cuticle was carefully removed from pharate pupae, and the pupae were frozen at −20°C. They were then thawed in ice water, and the cuticle was cut dorsally along the anterior/posterior axis. The pupa was pinned open and internal tissues were removed. This freeze/thaw step greatly facilitated the removal of most adhering tissues (Yamazaki, 1972). Dissected cuticles were stored at −80°C until enough material had been collected for protein purification. Approximately 8 g of dissected cuticles were ground to a powder with a mortar and pestle prechilled with liquid nitrogen. The ground cuticle was then homogenized in a glass tissue grinder with a loose-fitting pestle in 40 ml of buffer consisting of 100 mM Tris (pH 7.8), 500 mM NaCl, 50 mM ascorbic acid, and 10 µM phenylthiourea, at a ratio of 5 ml of buffer per gram of cuticle. The homogenized cuticle was centrifuged at 12,000 × g for 20 min at 4°C. The supernatant was decanted, the pellet was resuspended in buffer, and the homogenization was repeated 5 more times. The pellet was then resuspended in 40 ml of the same buffer containing α-chymotrypsin (0.15 mg/ml, 54 U/mg; Sigma-Aldrich) and stirred at room temperature for 30 min. The sample was centrifuged as described previously, the supernatant was transferred to a new tube and 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF; Sigma-Aldrich) was added to a final concentration of 1 mM to prevent further digestion by the protease. The cuticle pellet was again resuspended in buffer with α-chymotrypsin and this proteolysis treatment was repeated 5 more times. The supernatants were combined and passed successively through 2.5 µm (G8 glass fiber filter; Fisher), 1 µm (GF/B glass microfiber filter; Whatman), and 0.45 µm (Magna nylon; GE Osmonics) filters and used for protein purification (see section 2.3).
2.3 Protein purification
Unless otherwise noted, all purification steps were performed at 4°C. The presence of laccase-2 was followed throughout the purification process by immunoblot analysis using the antiserum described in section 2.7. For full-length recombinant laccase-2, 5 ml of a 50% slurry of ConA Sepharose 4B medium (GE Healthcare) was added to the filtered cell culture medium (see section 2.1) containing secreted recombinant proteins in 1 L Erlenmeyer flasks (500 ml per flask) and shaken overnight at 175 rpm. After the overnight incubation, the beads were allowed to settle and most of the cell culture medium was carefully removed by pipetting. The beads from each flask were then combined together and transferred to a 1.6 cm × 10 cm chromatography column. The column was washed 5 times with 5 bed volumes of 20 mM Tris (pH 7.5), 500 mM NaCl, by gravity flow. Proteins were eluted by resuspending the beads in 5 bed volumes of the same buffer supplemented with 500 mM methyl-α-D-mannopyranoside and incubated for 8 h to overnight with rocking. The supernatant was collected by gravity flow and this process was repeated 2 more times. The elution fractions were pooled and dialyzed against 20 mM Tris (pH 7.5). This sample was then applied to a 2 ml Q-Sepharose fast flow (GE Healthcare) anion exchange column (1 cm × 10 cm) and washed with 20 bed volumes of 20 mM Tris (pH 7.5). Proteins were eluted from the column in 15 bed volumes with 20 mM Tris (pH 7.5), 40 mM NaCl, and a 200-fold dilution of a protease inhibitor cocktail (P-1860; Sigma-Aldrich); the load, wash, and elution steps were performed at a flow rate of 2 ml/min. Ammonium sulfate crystals were added to the combined elution fractions to a final concentration of 20% saturation with stirring for 1 h. The sample was centrifuged at 12,000 × g for 30 min to pellet any precipitated proteins. Ammonium sulfate crystals were then added to the supernatant to a final concentration of 40% saturation and stirred and centrifuged as previously described; a majority of the recombinant protein precipitated in the 20% – 40% saturation fraction. The precipitated protein was resuspended in 100 mM sodium phosphate (pH 6) and dialyzed against the same buffer. An equal volume of glycerol was added to the sample and stored at −20°C.
A similar approach was used to purify rΔ106; the same conditions were used for the affinity chromatography with the ConA Sepharose 4B medium, while the following changes were made to the anion exchange chromatography with the Q-sepharose fast flow medium: the column was washed with 10 bed volumes 20 mM Tris (pH 7.5) followed by 10 bed volumes of 20 mM Tris (pH 7.5), 90 mM NaCl, and eluted with 15 bed volumes of 20 mM Tris (pH 7.5), 130 mM NaCl, and the protease inhibitor cocktail; all flow rates were 2 ml/min. Following the anion exchange chromatography, the sample was dialyzed against 50 mM sodium acetate (pH 5) and applied to a 1 ml prepacked HiTrap SP HP (GE Healthcare) cation exchange column. The column was washed with 10 bed volumes of 50 mM sodium acetate (pH 5), 90 mM NaCl, and eluted in 15 bed volumes of the same buffer with 150 mM NaCl and the protease inhibitor cocktail; all steps were performed with a flow rate of 1 ml/min. The truncated recombinant protein was then precipitated from the SP-column elution fraction by the addition of ammonium sulfate to 50% saturation with stirring and centrifugation as described above.
Purification of the endogenous laccase-2 from the cuticle preparation followed a procedure similar to that for the recombinant proteins. Four ml of a 50% ConA Sepharose slurry was added to the 240 ml of cuticle extract (see section 2.2) in a 500 ml Erlenmeyer flask and shaken overnight at 175 rpm. The beads were allowed to settle, and the supernatant was removed as described above for the recombinant proteins. The beads were transferred to a 1.6 cm × 10 cm column, and the resin washed 5 times with 15 ml of 20 mM Tris (pH 7.5), 500 mM NaCl, by gravity flow. Proteins were eluted from the column and dialyzed as described above. For the anion exchange chromatography a 1 ml prepacked Econo High Q cartridge (Bio-Rad) was used in place of the Q-sepharose fast flow medium. The sample was loaded at 1 ml/min, and the High Q cartridge was washed first with 5 bed volumes of 20 mM Tris (pH 7.5), and then with 5 bed volumes of 20 mM Tris (pH 7.5), 50 mM NaCl, and eluted in 10 bed volumes of 20 mM Tris (pH 7.5), 100 mM NaCl, with the protease inhibitor cocktail. The wash and elution steps were performed with a flow rate of 0.5 ml/min. Purification with a HiTrap SP column was the same as described above for rΔ106 except that the wash and elution buffers contained 120 mM and 210 mM NaCl each, respectively. Following the cation exchange chromatography, in lieu of the ammonium sulfate precipitation, the sample was concentrated by centrifugation in Centriplus YM-10 (Millipore) and Nanosep 10K (Pall Corp.) centrifugal filtration devices.
2.4 Peptide mass fingerprinting analysis of the purified cuticular enzyme
Three hundred ng of the purified cuticular enzyme was run on a 4–12% BisTris polyacrylamide gel (Invitrogen), and the protein was excised and sent to the Proteomics Core Facility at the University of Nevada, Reno for in gel trypsin digestion and peptide mass fingerprinting. Mass spectrometry analysis was performed using an Applied Biosystems 4700 Proteomics Analyzer MALDI TOF/TOF mass spectrometer and ABI’s 4000 Series Explorer software v. 3.6. A peak list of tryptic fragments was created with filter settings included a mass range of 700–4000 Da, a minimum S/N filter equal to 0, and peak density filter of 65 peaks per 200 Da with a maximum number of peaks set to 200. The peak list was checked against predicted tryptic fragments for M. sexta laccase-2 (GenBank accession number AY135186) and B. mori laccase-2 isoforms A and B (GenBank accession numbers EU093074 and BK006378) allowing for 1 missed cleavage and the oxidation of methionine residues and carbamidomethylation of cysteines.
2.5 In vitro enzyme activity assays
Substrate specificity assays contained 0.5 µg of purified cuticular laccase-2 and 1 mM substrate in 100 mM sodium citrate (pH 5) buffer except for assays with syringaldazine, which used 10 µM substrate and 100 mM sodium phosphate (pH 6) buffer. The substrates ABTS, hydroquinone, NADA monohydrate, and syringaldazine were obtained from Sigma-Aldrich (catalog numbers A-1888, H-9003, A-8762, S-7896); L-Tyrosine HCl was obtained from United States Biochemical Corp. (catalog number 22935); NBAD HCl was obtained from the National Institute of Mental Health’s Chemical Synthesis and Drug Supply Program. Each assay was set up in a 96-well plate in a total volume of 200 µl and was performed in triplicate. In assays involving the inhibitors phenylthiourea or diethyldithiocarbamic acid, inhibitor and enzyme were mixed together in 100 µl of buffer and incubated for 10 – 15 min before addition of substrate. Two mM substrate in 100 µl of buffer with inhibitor (to maintain a constant inhibitor concentration) was then added to begin the assay. Absorbance readings were measured once per min for 10 min for all assays except those using NADA and NBAD, in which readings were taken once every 2 min for 20 min. Measurements were taken in a BioTek PowerWave XS Microplate Spectrophotometer and analyzed with Gen5 Software. Enzyme activity was detected as the change in absorbance over time and converted to the concentration of product formed by the Beer-Lambert Law. The wavelengths (nm) and extinction coefficients (ε) used for the assays were as follows: for p-benzoquinone, ε248 = 17,252 (Eggert et al., 1996); ABTS cation, ε414 = 36,000 (Childs and Bardsley, 1975); syringaldazine quinone methide, ε525 = 65,000 (Thomas et al., 1989); NADA and NBAD quinone, ε390 = 1,100 (Thomas et al., 1989). Activity with tyrosine was monitored at 280 nm (hydroxylation of tyrosine to DOPA) and 475 nm (formation of dopachrome from DOPA quinone). A wavelength scan was also performed from 200 nm – 700 nm at 5 min intervals for 30 min to monitor possible changes at other wavelengths; no changes in absorbance at any wavelength were detected with tyrosine as substrate. Statistical significance of the inhibition assays was determined by one-way analysis of variance (ANOVA) and Dunnett’s Multiple Comparison Test using GraphPad Prism version 4.00 for Windows (GraphPad Software).
To determine optimum pH, assays were performed as described above in 100 mM sodium citrate-phosphate buffer (pH 4–8). Each assay contained 0.5 µg of purified enzyme and 0.5 mM substrate (NADA or NBAD). Assays to determine kinetic constants were performed in a similar manner using various substrate concentrations in 100 mM sodium citrate buffer (pH 5). One laccase activity unit was defined as a change in the absorbance of 0.001 OD per min at 390 nm. Assays measuring oxygen consumption were performed with 100 mM sodium citrate buffer (pH 5) and 5 µg of the purified cuticular laccase-2 in a total volume of 600 µl in a Micro Oxygen Chamber (Instech Laboratories). The buffer was saturated with air prior to the assay, and oxygen consumption was measured with a Micro Oxygen Probe (Instech Laboratories) and a YSI 5300 Biological Oxygen Monitor (Yellow Springs Instrument Incorporated). The kinetics assays were analyzed by plotting either laccase activity units (spectrophotometric method) or rate of oxygen consumption (biological oxygen monitor) versus substrate concentration, and curves were fitted to the data by non-linear regression using GraphPad Prism. The equation used to fit the data to a Michaelis-Menten curve was:
The equation used to fit the data to a substrate (uncompetitive) inhibition curve was:
2.6 Enzyme activity assays of cuticle extracts
Each assay was performed with cuticle from a pharate pupa (1 insect) or the wing and underlying ventral abdominal cuticle of newly molted pupae (2–3 insects; 0–1 h post molt). Cuticles were dissected as described in section 2.2, frozen in liquid nitrogen and stored at −80°C. Cuticles were weighed and ground to a powder in a mortar and pestle prechilled with liquid nitrogen; the amount of cuticle per experiment varied between 150 ng to 230 ng. The ground cuticle was transferred to a glass tissue grinder and homogenized with ice-cold 100 mM sodium citrate (pH 5) at a ratio of 1 ml buffer per 100 ng of cuticle. The homogenate was transferred to a microcentrifuge tube and centrifuged at 16,000 × g for 5 min. The supernatant was discarded, and the pellet was resuspended in room temperature 100 mM sodium citrate (pH 5) buffer at 1 ml per 100 ng of cuticle. Four hundred µl of cuticle suspension was aliquoted to 3, 2-ml microcentrifuge tubes, and 200 units of catalase (Sigma-Aldrich; C-3155) was added to the third tube and incubated for 30 min at room temperature on a rotary mixer at 20 rpm. Following the 30 minute incubation, ABTS was added to a final concentration of 1 mM to tubes 2 and 3 and all three tubes were incubated at room temperature on a rotary mixer at 20 rpm; tube 1 contained no substrate or catalase and was used to determine the baseline for the assay. Incubation with ABTS was for 30 min for samples containing pharate pupal cuticle, and 90 min for samples containing pupal wing/ventral abdominal cuticle. After incubation with substrate, the samples were centrifuged for 5 min to pellet the cuticle, and 300 µl of supernatant was transferred to a 96-well plate for absorbance readings at 414 nm. The amount of ABTS oxidized was calculated as described in section 2.5.
2.7 Antibody production and purification
A portion of the Mslac2 cDNA encoding amino acids D274 – N586 of the mature protein was amplified by PCR and cloned into the expression vector H6pQE-60 (Lee et al., 1994). The protein was expressed in the E. coli strain XL1-Blue and purified by metal-affinity chromatography on a Ni-NTA agarose column (Qiagen) under denaturing conditions. The protein was then further purified by SDS-PAGE on a 10% acrylamide gel, and the band containing the laccase fragment was excised and sent to Cocalico Biologicals Incorporated (Reamstown, PA) for polyclonal antibody production in New Zealand White rabbits. Antibodies specific to laccase-2 were purified from the antiserum by affinity chromatography. Briefly, a portion of the cDNA encoding amino acids D296 – Q572 was amplified by PCR and cloned into the expression vector pET-32a (Novagen). The protein was expressed in the E. coli strain OrigamiB(DE3) (Novagen) and purified on a Ni-NTA agarose column under denaturing conditions. Following purification, the protein was dialyzed against 3 L of 4 M urea, 100 mM sodium phosphate, 150 mM NaCl, pH 7.2. Six hundred µg of the dialyzed protein was immobilized on 2 ml of AminoLink Plus Coupling Gel (Pierce) following the manufacturer’s instructions. One and a half mlm of the laccase-2 antiserum was applied to the affinity column and binding was allowed to occur for 1 h at room temperature. The column was washed with 12 ml of 100 mM sodium phosphate, 150 mM sodium chloride, pH 7.2. Bound antibodies were eluted with 8 ml of 100 mM glycine (pH 3). Each 1 ml elution fraction was immediately mixed with 50 µl of 1 M Tris (pH 9) to neutralize the pH. Fractions containing antibodies were pooled and dialyzed twice against 2 liters of 100 mM sodium phosphate, 150 mM NaCl, pH 7.2. Glycerol was added to 50%, and the purified antibodies were stored at −20 °C.
2.8 Immunoblot analysis of cuticle extracts
Following the laccase activity assay of cuticle extracts (see section 2.6), all remaining buffer was removed from the control sample (tube 1) and the cuticle was resuspended in 400 µl of an extraction buffer consisting of 5% SDS, 10% glycerol, 4 M urea, 50 mM acetic acid, and 10 mM boric acid (Hopkins et al., 2000). Samples were incubated on a rotary mixer at room temperature for 20–24 h at 20 rpm. Following extraction, the samples were centrifuged for 5 min at 16,000 × g, and the supernatant was stored at −20°C. Abdominal cuticle from 0–1 h and 3 h pupae, as well as wing/ventral abdominal cuticle from 3 h pupae were treated in a similar matter. Protein concentration was determined using the Micro BCA Protein Assay Reagent Kit (Pierce). Fifteen µg of protein were fractionated by SDS-PAGE on duplicate 4–12% BisTris gels (Invitrogen); one gel was stained with Coomassie Brilliant Blue and the other was transferred to a 0.45 µM nitrocellulose membrane for immunoblot analysis. Following blocking for 1 h in 3% dry milk, TBST (25 mM Tris (pH 7.4), 137 mM NaCl, 2.7 mM KCl, 0.05% Tween 20), the membrane was incubated with the purified laccase-2 antibody (see section 2.7) at a 100-fold dilution in blocking buffer for 1 h. The membrane was washed 3 times for 5 min each in TBST and then incubated in blocking buffer with a secondary antibody (goat anti-rabbit AP conjugate; Bio-Rad) at a 3,000-fold dilution for 1 h. The membrane was washed again and developed with the AP Conjugate Substrate Kit (Bio-Rad) for 9 min.
3. Results
3.1 Identification of the cuticular laccase
The endogenous M. sexta cuticular laccase can only be extracted from pharate pupal cuticle in active form via proteolysis (Thomas et al., 1989). Previous research had suggested that a laccase zymogen could be solubilized from the pupal cuticle of the silkworm, B. mori, by limited treatment with chymotrypsin (Ashida and Yamazaki, 1990). Therefore, we tried a similar approach with M. sexta. The molecular mass of the endogenous laccase purified from the cuticle treated with chymotrypsin was 70 kDa (Fig. 1), smaller than the expected size of 81 kDa for the full length laccase-2 based on its amino acid sequence. Varying the amount of chymotrypsin used in the treatment from 5 µg/ml to 500 µg/ml had no effect on the size of the protein extracted from the cuticle (i.e. less protease did not result in a larger protein due to decreased proteolysis); lower enzyme concentrations only reduced the amount of laccase solubilized (data not shown).
Figure 1.
SDS-PAGE analysis of laccase-2. Five hundred nanograms of purified proteins were fractionated by electrophoresis in a 8–16% polyacrylamide gel (Criterion, Bio-Rad) and visualized using a colloidal Coomassie Brilliant Blue G-250 stain (Bio-Safe Coomassie Stain, Bio-Rad). rLac2, purified full-length recombinant laccase-2; rΔ106, purified amino-terminal truncated recombinant laccase-2; cLac2, purified chymotrypsin-solubilized cuticular laccase-2. Mark12 protein standard (Invitrogen) was used for estimation of molecular masses.
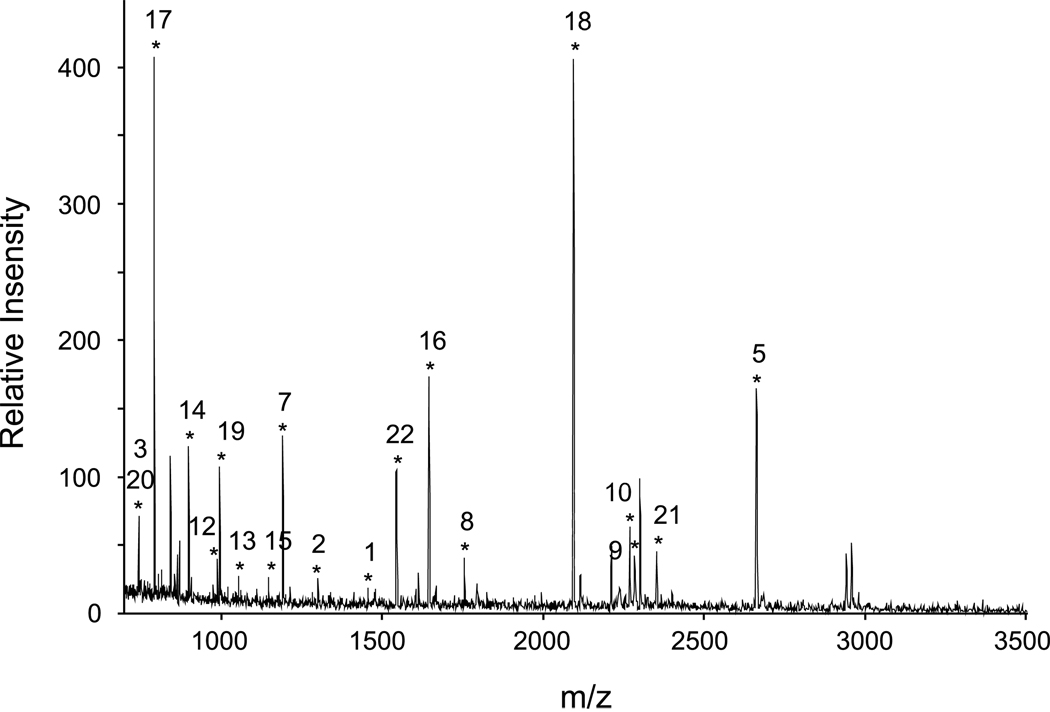
The identity of the cuticular laccase was verified as laccase-2 by two independent methods: amino-terminal sequencing and peptide mass fingerprinting. Amino-terminal sequencing of the endogenous enzyme identified the sequence RRNPALSAPD, which corresponds to residues 119 to 128 of the predicted sequence of mature laccase-2 (Fig. 2). Peptide mass fingerprinting of tryptic fragments was performed by matrix assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) and compared to the predicted tryptic fragments for the deduced amino acid sequence of the Mslac2 cDNA that we cloned and used to make the recombinant proteins. We also compared the MALDI-TOF spectrum to the B. mori laccase-2 isoforms A and B which share 96% and 85% sequence identity with the M. sexta protein. Nineteen peptides (peptides 1–3 and 5–20), representing 34% coverage, matched predicted masses for M. sexta laccase-2 (Table 1 and Fig. 3). An additional three peptides (peptides 4, 21, and 22) matched predicted masses for the B. mori laccase-2A isoform; these peptides differed at only one position from the corresponding sequence in M. sexta and we believe represent allelic variants. Fourteen of the 22 peptides (peptides 1–14) are in the “common” region of laccase-2 and, therefore, cannot be used to distinguish between the two isoforms. Of the remaining 8 peptides, which map to the “variable” region of laccase-2 proteins, 6 match our laccase-2 clone (peptides 15–20) while the remaining 2 match with the B. mori A isoform (peptides 21–22). One peptide mapping to the “variable” region (peptide 17) was common to both B. mori isoforms and therefore, was not isoform-informative. No peptide masses matched predicted masses specific to the B. mori B isoform. Thus, we conclude that most, if not all, of the laccase protein from the cuticle preparation is of a single isoform.
Figure 2.
Alignment of the amino-terminal region of insect laccase-2 orthologs. The amino-terminal regions (with signal peptide removed) from the laccase-2 orthologs of A. gambiae (AgLac2; AY943928), M. sexta (MsLac2; AY135186 ), and T. castaneum (TcLac2; AY884061) were aligned using the program ClustalW (Thompson et al., 1994) available at the European Bioinformatics Institute web site (http://www.ebi.ac.uk/clustalw/). Asterisks below the alignment indicate identical residues, while semicolons and periods represent strong and weak conservative substitutions. The grey box in the MsLac2 sequence shows the residues identified by amino-terminal sequencing of the endogenous cuticular laccase. The arrow indicates where the amino-terminal truncated recombinant enzyme begins.
Table 1.
Mass Spectrometry analysis of the purified cuticular laccase
| Peptide Number |
Sequence a,b | Theoretical Mass (Da) |
Experimental Mass (Da) |
Position c | |
|---|---|---|---|---|---|
| MsLac2 | BmLac2A | ||||
| 1 | RNPALSAPDECAR | 1456.7 | 1456.7 | 120–132 | 123–135 |
| 2 | NPALSAPDECAR | 1300.6 | 1300.61 | 121–132 | 124–135 |
| 3 | GILTANR | 744.44 | 744.44 | 185–191 | 188–194 |
| 4 | GILTANRMLPGPSIQYCENDK | 2256.15 | 2256.1 | 188–208 | |
| 5 | GSQYYDGVPFVTQCPIQQGNTFR | 2662.24 | 2662.23 | 230–252 | 233–255 |
| 6 | YQWQGNAGTHFWHAHTGLQK | 2367.12 | 2367.1 | 253–272 | 256–275 |
| 7 | LDLGLYGSIVVR | 1191.67 | 1191.68 | 273–283 | 276–286 |
| 8 | LAVNTGQDPESVLINGK | 1754.93 | 1754.93 | 322–338 | 325–341 |
| 9 | DPNTGFMTNTPLETFTITAGR | 2284.09 | 2284.15 | 343–363 | |
| 10 | YDFVIEANNIPGAYWIQVR | 2268.15 | 2268.14 | 414–432 | 417–435 |
| 11 | GLGECGIK | 833.42 | 833.44 | 433–440 | 436–443 |
| 12 | GLGECGIKR | 989.52 | 989.52 | 433–441 | 436–444 |
| 13 | RAQQLGILR | 1054.65 | 1054.63 | 441–449 | 444–452 |
| 14 | AQQLGILR | 898.55 | 898.55 | 442–449 | 445–452 |
| 15 | NDAICVSQLK | 1147.58 | 1147.58 | 486–495 | 489–498 |
| 16 | HIDPAILOERPDIK | 1644.91 | 1644.92 | 499–512 | |
| 17 | IFLPFR | 792.48 | 792.48 | 513–518 | 516–521 |
| 18 | FFVYGPETLFQPNTYNR | 2093.01 | 2093.01 | 542–535 | |
| 19 | HALDLDRR | 995.54 | 995.55 | 645–652 | 648–655 |
| 20 | RGLLER | 743.45 | 743.46 | 652–657 | 655–660 |
| 21 | QGDLPPAKDTIAVPNNGYVILR | 2351.27 | 2351.28 | 664–685 | |
| 22 | DTIAVPNNGYVILR | 1544.84 | 1544.86 | 672–685 | |
Bold and underlined residues indicate differences between the Manduca and Bombyx sequences; all other residues in the sequence are identical.
Sequences highlighted in grey are found in the Bombyx A-isoform but not the B-isoform.
Position of peptides within the protein are with respect to the mature protein.
Figure 3.
MALDI-TOF spectrum of trypsinized cuticular laccase-2. A visualization of the mass spectrometry analysis as a plot of signal intensity versus peptide mass. Asterisks indicate peaks whose masses match predicted masses for tryptic fragments of laccase-2 from M. sexta or B. mori. The numbers indicate corresponding peptides listed in Table 1. The peaks for peptides 4, 6, and 11 had low signal intensity and were omitted from the figure for clarity.
3.2 Enzymatic activity of the cuticular laccase
The purified cuticular enzyme was able to oxidize the commonly used laccase substrates syringaldazine and 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS), as well as the o-diphenols NADA and NBAD, and the p-diphenol hydroquinone but not the monophenol tyrosine (Table 2). In addition, the enzyme was insensitive to the chelator diethyldithiocarbamic acid (DETC) but showed mixed sensitivity to phenylthiourea (PTU), exhibiting some inhibition when NADA was the substrate but not with hydroquinone; these results are consistent with previous reports of insect laccase activity and inhibition by Andersen (1978) and Thomas et al. (1989).
Table 2.
Substrate specificity of cuticular laccase
| Substratea | Activityb (nM min−1) |
|---|---|
| ABTS | 4,020 ± 30 |
| Syringaldazine | 410 ± 4 |
| NBAD | 4,180 ± 190 |
| NADA | 5,010 ± 210 |
| 10 µM DETC | 5,070 ± 170 |
| 100 µM DETC | 4,800 ± 330 |
| 1 mM DETC | 4,330 ± 610 |
| 1 µM PTU | 4,890 ± 160 |
| 10 µM PTU | 3,630 ± 190* |
| 100 µM PTU | 160 ± 110* |
| Hydroquinone | 10,060 ± 240 |
| 10 µM DETC | 10,510 ± 470 |
| 100 µM DETC | 10,230 ± 70 |
| 1 µM DETC | 9,380 ± 570 |
| 1 µM PTU | 10,570 ± 400 |
| 10 µM PTU | 10,440 ± 510 |
| 100 µM PTU | 10,750 ± 180 |
| Tryosine | none detected |
All substrate concentrations were 1 mM except for syringaldazine which was 10 µM
Average of 3 replicates ± S.D.
Significantly different from control (p < 0.05)
Insect laccase-2 enzymes are highly variable at their amino-termini. High sequence identity begins at the position equivalent to H107 of the mature protein (Fig. 2). The fact that this residue follows a Lys or Arg suggested that it may be a cleavage site following a prodomain and that the enzyme is produced as a zymogen, as has been reported for B. mori (Ashida and Yamazaki, 1990; Yatsu and Asano, 2009). Because the chymotrypsin-solubilized laccase-2 was missing the first 118 residues, we considered the possibility that the protease treatment was responsible for activating the enzyme. In M. sexta, tanning of the pupal cuticle begins in the pharate stage and is strongly evident in newly molted individuals (Fig. 4A–C). Thus, it is expected that laccase would be present in the pharate pupal cuticle in an already active form. To examine this possibility, we assayed laccase activity in extracts of both pharate pupal abdominal cuticle, in which tanning is already visible, and wing/ventral abdominal cuticle of newly molted pupae in which no tanning is visible. Enzymatic activity was detected in both cuticles using ABTS as the substrate, though the activity in the wing cuticle extract was much lower than the activity in the pharate pupal abdominal cuticle extract (Table 3). No change in activity was observed when the samples were preincubated with catalase, a hydrogen peroxide scavenger, indicating that oxidation of ABTS was not due to peroxidase. These results indicate that active laccase was present in the abdominal cuticle of pharate pupae and the wing cuticle of newly molted pupae, even prior to the initiation of visible tanning.
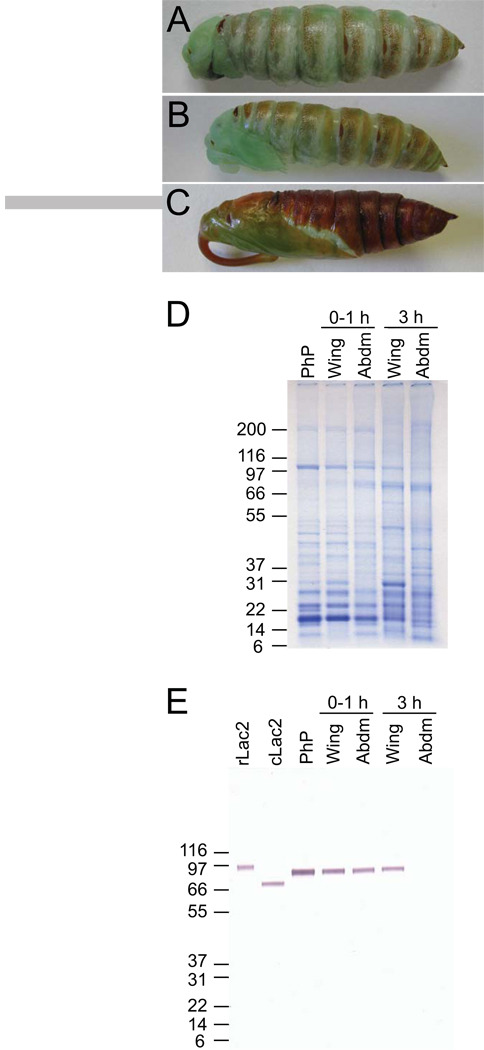
Figure 4.
Correlation between tanning and presence of laccase-2 in early pupal development. Photograph of (A) pharate pupa (with larval cuticle removed), (B) pupa 0 h post molt, and (C) pupa 3 h post molt. Proteins were extracted from the abdominal (abdm) cuticle or wing/ventral abdominal (wing) cuticle of pharate pupa (PhP) or pupa at 0–1 h and 3 h post molt. Fifteen µg of protein extract was subjected to SDS-PAGE in duplicate gels and were either (D) stained with Coomassie brilliant blue or (E) transferred to a nitrocellulose membrane for immunoblot analysis with the affinity-purified MsLac2 antibody (1:100). rLac2, 50 ng of purified full-length recombinant laccase-2; cLac2, 50 ng purified chymotrypsin-solubilized cuticular laccase-2. Mark12 protein standard (Invitrogen) was used for estimation of molecular masses.
Table 3.
In vivo assay for laccase activity
| Tissue | Sample | Actvitya (nM) |
|---|---|---|
| Pharate Pupa | 1 mM ABTS | 30,200 ± 9,800 |
| 1 mM ABTS | 31,700 ± 8,400 | |
| w/catalase | ||
| Pupal Wing | 1 mM ABTS | 8,300 ± 1,040 |
| 1 mM ABTS | 9,400 ± 2,080 | |
| w/catalase |
Average of 3 replicates ± SD. The incubation time for pharate pupal cuticle with substrate was 30 min; the incubation time for pupal wing cuticle with substrate was 90 min.
n.d. not determined
3.3 Immunoblot analysis of cuticle extracts
To determine the size of the laccase-2 present in the cuticle we extracted total protein from the abdominal cuticle of pharate pupae, and separately from the wings and abdominal cuticle of newly molted pupae at 0–1 h and 3 h post eclosion, using an SDS/urea buffer and subjected the samples to immunoblot analysis using an affinity-purified antibody to recombinant laccase-2 (Fig. 4E). A single band of ~84 kDa was detected in all samples except for the 3 h abdominal cuticle (in which no band was detected). This mass is larger than the chymotrypsin-solubilized purified cuticular enzyme and similar to the predicted mass of 81 kDa for the mature protein based on the amino acid sequence. These results suggest that the form of laccase-2 present in the cuticle during the tanning process is the full-length form, and that chymotrypsin cleavage after Leu118 released the protein from the cuticle.
The immunoblot also indicates that the amount of laccase-2 in the cuticle is greater in the abdomen than in the wing/ventral abdomen (Fig 4E; compare lane PhP with 0–1 h wing). However, quantitation by immunoblot is problematic as protein extraction becomes more difficult as sclerotization proceeds. This can be seen by the reduced detection in 0–1 h pupal abdomen, and no detection in the heavily sclerotized 3 h pupal abdomen, when compared to the less sclerotized pharate pupal cuticle. Thus, the amount of laccase-2 in sclerotizing tissue may be underestimated by this method.
3.4 Kinetic properties of recombinant and cuticular laccase-2
To directly test whether laccase-2 is expressed as a zymogen and activated by cleavage, we expressed and purified two recombinant forms of laccase-2: a full-length and an amino-terminally truncated form, rΔ106. Both the full-length and truncated recombinant proteins were produced using a baculovirus expression system. The full-length recombinant laccase-2 had an apparent molecular mass of approximately 90 kDa as determined by SDS-PAGE under reducing conditions (Fig. 1). This was greater than the predicted mass of 81 kDa, based on the deduced amino acid sequence of the cDNA, suggesting that the protein was glycosylated. Glycosylation was also indicated by the binding to a lectin (Concanavalin A) affinity column. The truncated recombinant laccase-2, had a molecular mass of approximately 70 kDa (Fig. 1), closely matching the predicted mass of 71 kDa, and also the apparent mass of the purified cuticular laccase-2, which is just 12 residues smaller (Fig. 2). It also bound to concanavalin A, indicating that it was glycosylated even though its apparent mass was not greater than it’s predicted mass.
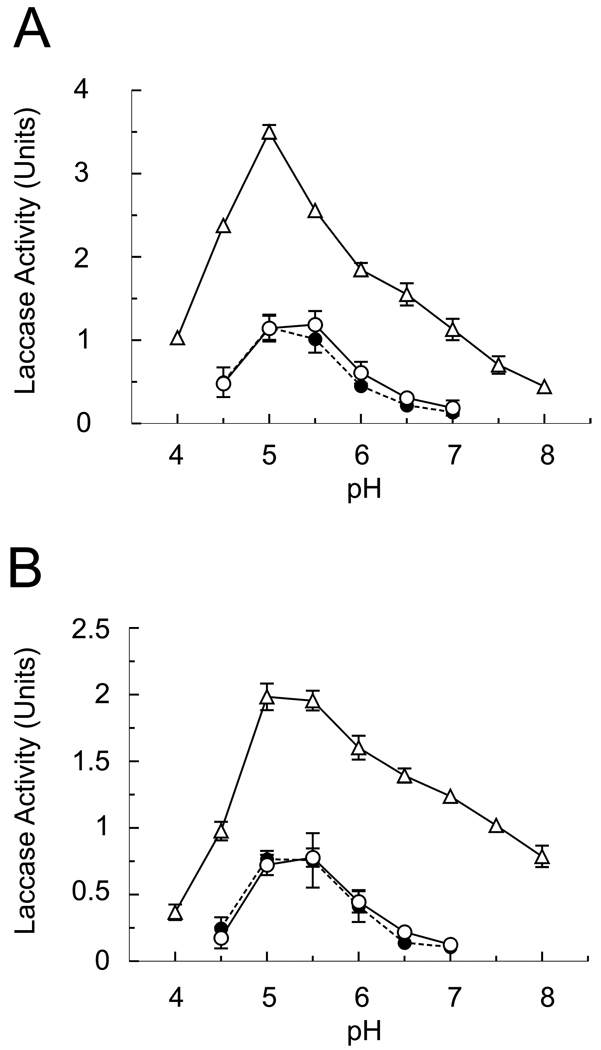
All three forms of laccase-2 showed similar pH profiles when tested in a citrate-phosphate buffer with the natural substrates NADA and NBAD, with peak activity between pH 5 to 5.5 (Fig. 5). However, the cuticular enzyme was three to four times more active than either of the recombinant proteins. All three forms of laccase-2 had slightly higher activity with NADA as the substrate than with NBAD.
Figure 5.
pH profile of laccase-2 activity. Assays were performed with 0.5 µg of purified laccase-2 and 0.5 mM NADA (A) or NBAD (B) in 100 mM sodium citrate-phosphate buffer of various pH. Activity was determined by monitoring the formation of the corresponding quinone at 390 nm. Each data point indicates the mean of 3 replicates ± standard deviation. Filled circle (●), full-length recombinant laccase-2; open circle (○), amino-terminal truncated recombinant laccase-2; open triangle (△), endogenous cuticular laccase-2.
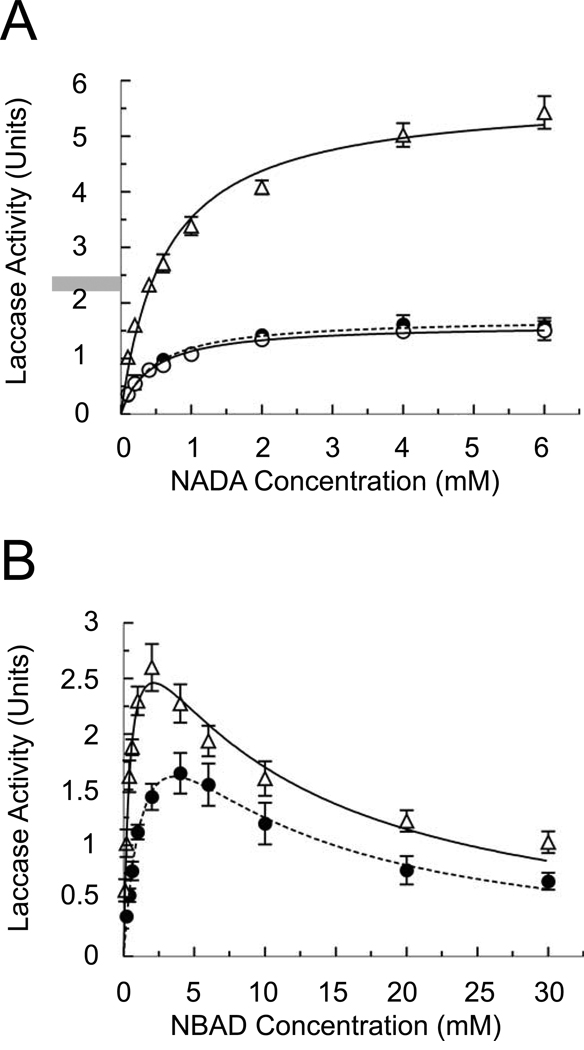
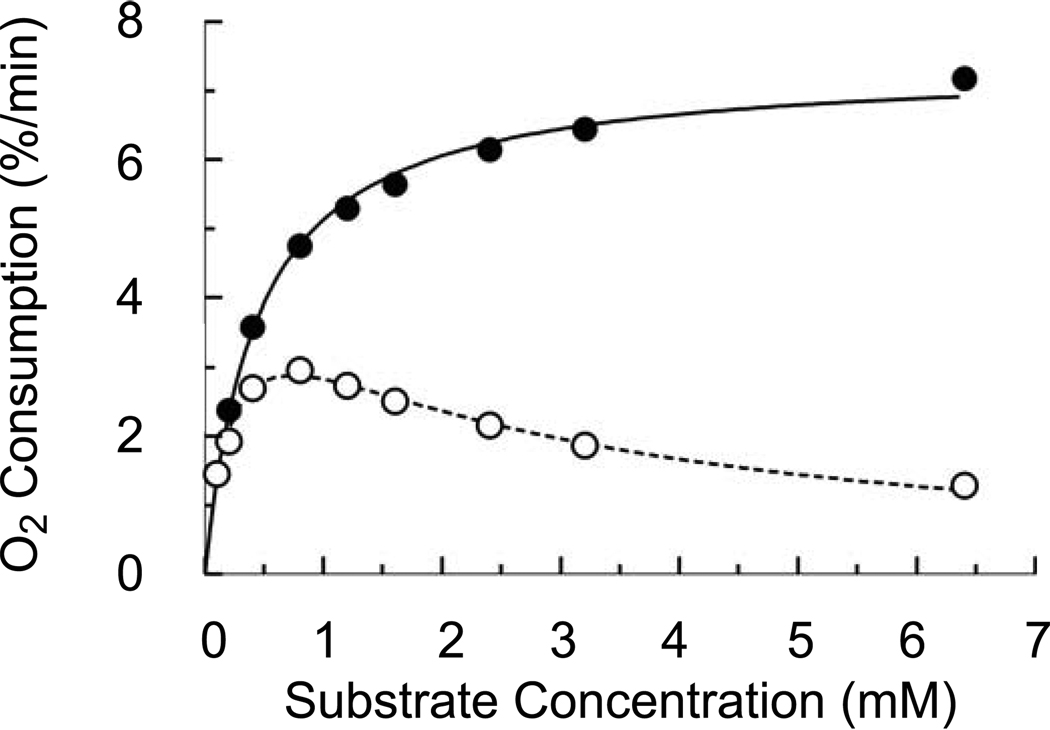
When NADA was used as the substrate, both the recombinant and cuticular enzymes exhibited classic Michaelis-Menten kinetics (Fig. 6A), with similar Km values for NADA of approximately 0.5 mM (Table 4). However, much different results were observed when NBAD was used as the substrate. Peak activity occurred at 2 mM substrate with the cuticular laccase-2, and at 4 mM with the full-length recombinant enzyme, but the activity then declined as substrate concentration increased, indicative of substrate inhibition (Fig. 6B). The apparent Km of the full-length recombinant laccase-2 with NBAD was 4-fold higher than that of the endogenous protein (Table 4). To verify that this apparent decrease in activity at higher substrate concentrations was not due to further reaction of the NBAD quinone product resulting in lower absorbance at 390 nm, we also examined activity of the cuticular laccase-2 by measuring consumption of oxygen, a second substrate used by laccase in the redox reaction. This experiment confirmed the spectrophotometric assays, demonstrating that the oxidation of NADA conformed to typical Michaelis-Menten kinetics, whereas the oxidation of NBAD is consistent with substrate inhibition. (Fig. 7).
Figure 6.
Laccase-2 kinetics. Assays were performed with 0.5 µg of purified laccase-2 and various concentrations of NADA (A) or NBAD (B) in 100 mM sodium citrate-phosphate buffer (pH 5). Activity was determined by monitoring the formation of the corresponding quinone at 390 nm. Each data point indicates the mean of 3 replicates ± standard deviation. Curves were fitted to the data by non-linear regression using the GraphPad Prism software with the equations described in Materials and Methods. Designations are the same as listed in Figure 5.
Table 4.
Kinetic parameters for laccase-2.
| Enzyme | Substrate |
Vmax (nM min−1) |
Km (mM) |
kcat (min−1) |
kcat/Km (min−1 mM−1) |
Ksi (mM) |
|---|---|---|---|---|---|---|
| rLac2 | NADA | 3,100 ± 100 | 0.46 ± 0.05 | 100 ± 3 | 220 | n.a. |
| rLac2 | NBAD | 6,100 ± 760 | 1.90 ± 0.39 | 200 ± 25 | 103 | 6.7 ± 1.4 |
| rΔ106 | NADA | 2,900 ± 76 | 0.43 ± 0.04 | 80 ± 2 | 195 | n.a. |
| cLac2 | NADA | 10,500 ± 250 | 0.63 ± 0.05 | 290 ± 7 | 460 | n.a. |
| cLac2 | NBAD | 6,400 ± 350 | 0.47 ± 0.06 | 180 ± 10 | 380 | 9.7 ± 1.3 |
rLac2: full-length recombinant laccase-2
rΔ106: N-terminal truncated recombinant laccase-2
cLac2: endogenous cuticular laccase-2
n.a. not applicable
Figure 7.
Oxygen consumption by cuticular laccase-2. Assays were performed with 5 µg of purified cuticular laccase-2 and various concentrations of NADA (●) or NBAD (○) in 100 mM sodium citrate buffer (pH 5). Relative oxygen levels were measured using a Micro oxygen probe. Curves were fitted to the data by non-linear regression using the GraphPad Prism software with the equations described in Experimental Procedures. The graph is derived from a single assay at each data point and was used to confirm the kinetics observed spectrophotometrically.
The catalytic constant (kcat) of cuticular laccase-2 was approximately three times greater than that of the recombinant proteins (~300 min−1 vs. 100 min−1) when NADA was used as the substrate (Table 4). The cuticular enzyme was also more efficient as indicated by the specificity constant (kcat/Km). When NBAD was tested as a substrate, both the full-length recombinant and endogenous forms of laccase-2 had similar apparent kcat values of ~200 min−1 (Table 4). When ABTS was used as substrate, the cuticular laccase-2 behaved in typical Michaelis-Menten fashion with kinetic parameters of 1.5 mM (Km), 460 min−1 (kcat), and 307 min−1 mM−1 (kcat/Km) (data not shown).
4. Discussion
Laccase has long been postulated to play an important role in the sclerotization of the insect cuticle. RNAi experiments in the red flour beetle, T. castaneum, have shown that a specific gene, Tclac2, is required for cuticle sclerotization (Arakane et al., 2005). Our goal was to express a recombinant form of M. sexta laccase-2 and compare its biochemical properties to the laccase isolated from the pupal cuticle, which previously had been only partially purified and characterized (Thomas et al., 1989), and determine if the cuticular enzyme is indeed laccase-2. In addition, because laccase can only be solubilized from the cuticle by limited proteolysis, we made an amino-terminally truncated recombinant laccase that would be similar to the cuticular enzyme in sequence. Therefore, any differences observed in activity can be attributed to how the proteins were produced (i.e. recombinant versus endogenous) and not due to the protease treatment needed to release laccase from the cuticle. It has been reported that laccase can be purified from the cuticle of the silk moth B. mori as an inactive zymogen (Ashida and Yamazaki, 1990; Yatsu and Asano, 2009), and we sought to address this question as well by comparing the activities of the full-length and truncated recombinant proteins.
The recombinant laccases were produced via a baculovirus expression system in insect Sf9 cells. The full-length recombinant protein was ~10 kDa larger than predicted from its amino acid sequence, as determined by SDS-PAGE, indicating glycosylation. This is in contrast to the SDS/urea-solubilized cuticular laccase and the truncated recombinant laccase, which had molecular weights similar to those of their predicted masses. These results indicate that the full-length recombinant laccase-2 may be more highly glycosylated than the amino-terminally truncated forms. Therefore, much of the glycosylation of the full-length laccase-2 must occur in the initial 106 residues. This region is enriched in Ser and Thr (24%), signifying possible O-linked glycosylation.
All three forms of laccase-2 had very similar pH profiles with peak activity between pH 5 – 5.5. While this value is similar to many fungal laccases, it was somewhat surprising as previous results reported a broad optimum between pH 5.5 – 7 for the laccase extracted from M. sexta cuticle (Thomas et al., 1989). That study used trypsin instead of chymotrypsin to extract laccase, and the enzyme was enriched but not completely purified; therefore, differences in cleavage sites or the presence of contaminating proteins may account for the difference in outcomes of the pH assays. Of significance here is the fact that in this study the recombinant and cuticular forms of laccase-2 had very similar pH profiles. These results are consistent with previous studies on insect laccases that have shown pH optima tend to be between 4.5 – 5.5 (see Table 4 in Thomas et al. (1989) and references within; Sugumaran et al., 1992; Hattori et al., 2005).
All three forms of laccase-2 exhibited standard Michaelis-Menten kinetics when NADA was the substrate, with Km values between 0.43 mM – 0.63 mM. These results are similar to the Km observed for the laccase-like “enzyme B” from the blowfly, Calliphora vicina (0.53 mM; Barrett and Andersen, 1981), but it is lower than a Km reported for laccase from the locust, Schistocecra gregaria (1.3 mM; Andersen, 1978). Significantly, both recombinant forms of laccase-2 had essentially identical activity with NADA as determined by the kinetic parameters shown in Table 4. In addition, a cuticle suspension from untanned pupal wing/ventral abdominal cuticle oxidized the laccase substrate ABTS, suggesting that laccase is present in an active form in these samples. Similar results using syringaldazine were observed previously by Thomas et al. (1989). Tanning of the pupal cuticle in M. sexta begins in the pharate stage and is quite extensive by 3 h post molt. Therefore, there is a correlation in timing between expression of the laccase-2 gene in the pharate pupa (Dittmer et al., 2004) and detection of enzyme activity. Furthermore, the detection of only a single protein with an apparent mass of 84 kDa by immunoblot analysis in both tanned and untanned cuticle is consistent with the hypothesis that the full-length enzyme is active; no smaller cleaved form was detected. Thus, it does not appear that M. sexta laccase-2 is produced as a zymogen requiring proteolytic activation as was suggested for B. mori (Ashida and Yamazaki, 1990; Yatsu and Asano, 2009).
While this paper was undergoing revisions, the work of Yatsu and Asano (2009) on B. mori laccase-2 was published. Similar to M. sexta, the B. mori laccase-2 gene was expressed in the pharate pupal stage. However, unlike M. sexta, no visible tanning or enzyme activity could be detected until after the larval-to-pupal molt. These authors reported that B. mori laccase could be solubilized from the cuticle using a Tris-buffered urea solution and then activated through proteolysis of the crude extract with trypsin. Cuticle extract treated in this way oxidized both dopamine and methyl hydroquinone, as determined by a spectrophotometric assay (dopamine) or native PAGE coupled with in-gel activity staining (methyl hydroquinone), whereas cuticle extract that was not trypsinized showed no activity. In addition, direct staining of pupal cuticle with dopamine showed no visible enzymatic activity until approximately 2 h post molt. From these experiments they concluded that laccase-2 is most likely synthesized as an inactive proenzyme and activated at a later time through proteolytic cleavage. These results are seemingly contradictory to ours. Our work with recombinant proteins clearly demonstrates that the full-length M. sexta laccase-2 is active. Given the high sequence identity between the M. sexta and B. mori laccase-2 proteins it is surprising that two such different conclusions were reached. However, as was noted by Yatsu and Asano (2009), an alternative hypothesis for the inactivity of laccase-2 in the pharate pupal cuticle of B. mori may rely on the presence of inhibitor(s). Therefore, the action of trypsin may have been to disrupt an interaction between inhibitor(s) and enzyme.
The difference in laccase activity between the lightly tanned pharate pupal cuticle and the untanned pupal wing/ventral abdominal cuticle may be explained by the amount of laccase present. The immunoblot analysis indicated higher amount of laccase in the abdomen than in the wing and underlying abdominal cuticle. This difference may be even greater than indicated as it is difficult to extract protein from sclerotized cuticle (Hopkins et al., 2000). A higher amount of laccase-2 in the abdominal cuticle would correlate with the higher enzymatic activity observed.
The different rates of tanning seen in vivo between the wing and abdominal cuticle is likely also affected by the amount of substrate available. Hopkins and coworkers (1984) determined catecholamine levels in the cuticle of M. sexta throughout development and found that the combined levels of NADA and NBAD in the pupa were twice as high in the abdominal cuticle as compared to the wing cuticle in the first 6 h after molting, but were similar by 24 h post molt. Levels in the abdominal cuticle were ~150 nmol/g of cuticle at the time of molting (0 h), increased to ~1,200 nmol/g at 6 h post molt, and remained at 1,000 nmol/g by 24 h post molt. In contrast, catecholamine levels in the wing were at only 60 nmol/g at 3 h post molt, 600 nmol/g at 6 h post molt, and ~1,000 nmol/g at 24 h post molt. These data, taken together with the laccase activity and immunoblot results, suggest that laccase and substrate are present at higher concentrations in the abdominal cuticle, where tanning is first visible, than in the wing and underlying abdominal cuticle where tanning occurs later.
The cuticular enzyme exhibited standard Michaelis-Menten kinetics with ABTS, with kcat and kcat/Km values less than 2-fold different from those obtained with NADA, although greater affinity was observed for the natural substrate. In general, these parameters differ greatly from those reported for fungal laccases that frequently have 10-fold lower Km values and activity 3–4 orders of magnitude higher when oxidizing ABTS (Baldrian, 2006). However, some fungal laccases, as well as one from the Japanese lacquer tree Rhus vernicifera, have exhibited low activity toward ABTS (Xu et al., 1996), a result that is similar to that observed with M. sexta laccase-2.
A novel finding in this study was the apparent substrate inhibition observed with NBAD. This inhibition became noticeable when the NBAD concentration was in the low millimolar range (1–2 mM for the cuticular enzyme), but the physiological significance of this is unclear. NBAD levels in the pupal cuticle at the time of molting are only in the range of 60 – 150 nmol/g of cuticle (Hopkins et al., 1984), while the water content has been estimated at 80% of the total weight (see Note 10 in Schaefer et al., 1987). This would give NBAD concentrations of less than 0.2 mM, which is 5 – 10 fold lower than necessary for inhibition. Nevertheless, NBAD levels quickly rise to approximately 1,000 nmol/g by 24 hours after ecdysis and 2,000 nmol/g by 3 days post-ecdysis, with the water content concurrently dropping to 50%. Under these conditions NBAD concentrations would be in the range of 2 – 4 mM (assuming an even distribution of water and NBAD in the cuticle). We can envision then that the low levels of NBAD in the pupal cuticle at the time of ecdysis allows the insect to first fully expand its cuticle and then to rapidly sclerotize as the NBAD level rises. As the water content of the cuticle decreases, the concentration of NBAD increases into the millimolar range, and the corresponding inhibitory effect on the enzyme would allow sclerotization to continue but at a slower, and perhaps more controlled, rate.
The cuticular laccase-2 consistently had 2–4 times greater activity than either of the recombinant proteins. This could be due to the heterologous system in which the recombinant proteins were produced resulting in some differences in post-translational processing as compared to the cuticular laccase. Lower activity for recombinant laccases has been reported previously. For example, Sigoillot and coworkers (2004) noticed a 2-fold decrease in activity of a Pyncoporrus cinnabarinus laccase produced in Aspergillus oryzae or A. niger when compared to that of the natural enzyme. Butler et al. (2003) found a 10 – 25 fold difference in activity (depending on substrate) between a Myceliophthora thermophila laccase produced in A. oryzae versus Saccharomyces cerevisiae. de Wilde and coworkers (2007) noticed an ~10-fold lower activity for a Melanocarpus albomyces laccase, and an ~30-fold lower activity for a P. cinnabarinus laccase produced in rice when compared to the natural fungal enzyme.
It is also possible that the difference in activity between the recombinant and cuticular laccase-2 forms could be due to the extent in which they were loaded with copper. For example, a recombinant form of the bacterial laccase CotA when produced in E. coli had on average only 0.5 moles of copper per mole of laccase protein when expressed under aerobic conditions (Durão et al., 2008). Under microaerobic conditions, which allowed for greater intracellular levels of copper, the copper content of recombinant CotA increased to 3.7, close to the expected number of 4 copper atoms per laccase molecule. The resulting differences in kcat values ranged from 4–200 fold, depending on the substrate, while the Km values remained similar.
In addition, a mixture of laccase-2 isoforms present in the cuticle preparation could also explain the differences seen in activity. However, analysis by MALDI-TOF mass spectrometry could confirm the presence of only one isoform, the same as that encoded by the cDNA used to produce the recombinant enzymes. These results are also consistent with RNAi experiments performed in T. castaneum. Arakane and coworkers (2005) showed that while both the A and B isoforms of laccase-2 are necessary for cuticle tanning, knock-down of the Tclac2A transcript resulted in a much more severe phenotype with individuals dying sooner. The Mslac2 cDNA we have cloned is more similar in sequence to the T. castaneum laccase-2 A isoform than to the B isoform (Gorman et al., 2008), and therefore, we would predict this to be the predominant isoform in the cuticle.
Our results demonstrate the suitability of using recombinant insect laccases in place of the endogenous enzymes for biochemical assays, though further work is necessary to optimize this production. This will allow for a more extensive analysis of the enzymatic properties of M. sexta laccase-2 and other insect laccases. The knowledge gained will significantly add to our understanding of cuticle sclerotization, and may help in the design of insecticides specific to insect laccases.
Acknowledgements
We wish to thank Rebekah Woolsey and the Nevada Proteomics Center for the peptide mass fingerprinting, and Dr. Emily Ragan for assistance with the analysis. We also wish to thank Dr. Karl Kramer for critical review of the manuscript. This research was supported by NSF grant I050726425 and NIH grant AI070864. The Nevada Proteomics Center is supported by NIH Grant Number P20 RR-016464 from the INBRE Program of the National Center for Research Resources. This is contribution 08-290-J of the Kansas Agricultural Experiment Station.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen SO. Characterization of a trypsin-solubilized phenoloxidase from locust cuticle. Insect Biochem. 1978;8:143–148. [Google Scholar]
- Andersen SO. Cuticular sclerotization and tanning. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive Molecular Insect Science. Vol. 4. Oxford: Elsevier; 2005. pp. 145–170. [Google Scholar]
- Arakane Y, Muthukrishnan S, Beeman RW, Kanost MR, Kramer KJ. Laccase 2 is the phenoloxidase gene required for beetle cuticle tanning. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11337–11342. doi: 10.1073/pnas.0504982102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida M, Yamazaki HI. Biochemistry of the phenoloxidase system in insects: with special reference to its activation. In: Ohnishi E, Ishizaki H, editors. Molting and Metamorphosis. Tokyo/Berlin: Japan Scientific Societies Press/Springer-Verlag; 1990. pp. 239–265. [Google Scholar]
- Baldrian P. Fungal laccases-occurrence and properties. FEMS Microbiol. Rev. 2006;30:215–242. doi: 10.1111/j.1574-4976.2005.00010.x. [DOI] [PubMed] [Google Scholar]
- Barrett FM, Andersen SO. Phenoloxidases in larval cuticle of the blowfly, Calliphora vicina. Insect Biochem. 1981;11:17–23. [Google Scholar]
- Bell RA, Joachim FG. Techniques for rearing laboratory colonies of tobacco hornworms and pink bollworms. Ann. Ent. Soc. Am. 1976;69:365–373. [Google Scholar]
- Bento I, Carrondo MA, Lindley PF. Reduction of dioxygen by enzymes containing copper. J. Biol. Inorg. Chem. 2006;11:539–547. doi: 10.1007/s00775-006-0114-9. [DOI] [PubMed] [Google Scholar]
- Butler T, Alcalde M, Sieber V, Meinhold P, Schlachtbauer C, Arnold FH. Functional expression of a fungal laccase in Saccharomyces cerevisiae by directed evolution. Appl. Environ. Microbiol. 2003;69:987–995. doi: 10.1128/AEM.69.2.987-995.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiacchierini E, Restuccia E, Vinci G. Bioremediation of food industry effluents: recent applications of free and immobilised polyphenoloxidases. Food Sci. Tech. Int. 2004;10:373–382. [Google Scholar]
- Childs RE, Bardsley WG. The steady-state kinetics of peroxidase with 2,2’-azino-di-(3-ethyl-bendthiazoline-6-sulphonic acid) as chromogen. Biochem. J. 1975;145:93–103. doi: 10.1042/bj1450093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde C, Uzan E, Zhou Z, Kruus K, Andberg M, Buchert J, Record E, Asther M, Lomascolo A. Transgenic rice as a novel production system for Melanocarpus and Pycnoporus laccases. Transgenic Res. 2007 doi: 10.1007/s11248-007-9124-9. [DOI] [PubMed] [Google Scholar]
- Dittmer NT, Suderman RJ, Jiang H, Zhu Y-C, Gorman MJ, Kramer KJ, Kanost MR. Characterization of cDNAs encoding putative laccase-like multicopper oxidases and developmental expression in the tobacco hornworm, Manduca sexta, and the malaria mosquito, Anopheles gambiae. Insect Biochem. and Mol. Biol. 2004;34:29–41. doi: 10.1016/j.ibmb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Durão P, Chen Z, Fernandes AT, Hildebrandt P, Murgida DH, Todorovic S, Pererira MM, Melo EP, Martins LO. Copper incorporation into recombinant CotA laccase from Bacillus subtilis: characterization of fully copper loaded enzymes. J. Biol. Inorg. Chem. 2008;13:183–193. doi: 10.1007/s00775-007-0312-0. [DOI] [PubMed] [Google Scholar]
- Eggert C, Temp U, Eriksson K-EL. The ligninolytic system of the white rot fungus Pycnoporus cinnabarinus: purification and characterization of the laccase. Appl. Environ. Microbiol. 1996;62:1151–1158. doi: 10.1128/aem.62.4.1151-1158.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianfreda L, Iamarino G, Scelza R, Rao MA. Oxidative catalysts for the transformation of phenolic pollutants: a brief review. Biocatal. Biotransform. 2006;24:177–187. [Google Scholar]
- Gorman MJ, Dittmer NT, Marshall JL, Kanost MR. Characterization of the multicopper oxidase gene family in Anopheles gambiae. Insect Biochem. Molec. Biol. 2008;38:817–824. doi: 10.1016/j.ibmb.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M, Konishi H, Tamura Y, Konno K, Sogawa K. Laccase-type phenoloxidase in salivary glands and watery saliva of the green rice leafhopper, Nephotettix cincticeps. J. Insect Physiol. 2005;51:1359–1365. doi: 10.1016/j.jinsphys.2005.08.010. [DOI] [PubMed] [Google Scholar]
- He N, Botelho JMC, McNall RJ, Belozerov V, Dunn WA, Mize T, Orlando R, Willis JH. Proteomic analysis of cast cuticles from Anopheles gambiae by tandem mass spectrometry. Insect Biochem. Mol. Biol. 2007;37:135–146. doi: 10.1016/j.ibmb.2006.10.011. [DOI] [PubMed] [Google Scholar]
- Hoegger PJ, Kilaru S, James TY, Thacker JR, Kües U. Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. FEBS J. 2006;273:2308–2326. doi: 10.1111/j.1742-4658.2006.05247.x. [DOI] [PubMed] [Google Scholar]
- Hopkins TL, Morgan TD, Kramer KJ. Catecholamines in haemolymph and cuticle during larval, pupal and adult development of Manduca sexta (L.) Insect Biochem. 1984;14:533–540. [Google Scholar]
- Hopkins TL, Krchma LJ, Ahmad SA, Kramer KJ. Pupal cuticle proteins of Manduca sexta: characterization and profiles during sclerotization. Insect Biochem. Molec. Biol. 2000;30:19–27. doi: 10.1016/s0965-1748(99)00091-0. [DOI] [PubMed] [Google Scholar]
- Lee E, Linder ME, Gilman AG. Expression of G-protein α subunits in Escherichia coli. Methods Enzymol. 1994;237:146–164. doi: 10.1016/s0076-6879(94)37059-1. [DOI] [PubMed] [Google Scholar]
- Mayer AM, Staples RC. Laccase: new functions for an old enzyme. Phytochemistry. 2002;60:551–565. doi: 10.1016/s0031-9422(02)00171-1. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Go N. Function and molecular evolution of multicopper blue proteins. Cell. Mol. Life Sci. 2005;62:2050–2066. doi: 10.1007/s00018-004-5076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva S. Laccases: blue enzymes for green chemistry. Trends in Biotechnol. 2006;24:219–226. doi: 10.1016/j.tibtech.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Schaefer J, Kramer KJ, Garbow JR, Jacob GS, Stejskal EO, Hopkins TL, Speirs RD. Aromatic cross-links in insect cuticle: detection by solid-state 13C and 15N NMR. Science. 1987;235:1200–1204. doi: 10.1126/science.3823880. [DOI] [PubMed] [Google Scholar]
- Sharma P, Goel R, Capalash N. Bacterial laccases. Wrold J. Microbiol. Biotechnol. 2007;23:823–832. [Google Scholar]
- Sigoillot C, Record E, Belle V, Robert JL, Levasseur A, Punt PJ, van den Hondel CAMJJ, Fournel A, Sigoillot JC, Asther M. Natural and recombinant fungal laccases for paper pulp bleaching. Appl. Microbiol. Biotechnol. 2004;64:346–352. doi: 10.1007/s00253-003-1468-3. [DOI] [PubMed] [Google Scholar]
- Sugumaran M, Giglio L, Kundzicz H, Saul S, Semensi V. Studies on the enzymes involved in puparial cuticle sclerotization in Drosophila melanogaster. Arch. Insect Biochem. Physiol. 1992;19:271–283. doi: 10.1002/arch.940190406. [DOI] [PubMed] [Google Scholar]
- Thomas BR, Yonekura M, Morgan TD, Czapla HT, Hopkins TL, Kramer KH. A trypsin-solubilized laccase from pharate pupal integument of the tobacco hornworm, Manduca sexta. Insect Biochem. 1989;19:611–622. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres E, Bustos-Jaimes I, La Borgne S. Potential use of oxidative enzymes for the detoxification of organic pollutants. Appl. Catal. B. 2003;46:1–15. [Google Scholar]
- Xu F. Applications of oxidoreductases: recent progress. Ind. Biotechnol. 2005;1:38–50. [Google Scholar]
- Xu F, Shin W, Brown SH, Wahleithner JA, Sundaram UM, Solomon EI. A study of a series of recombinant fungal laccases and bilirubin oxidase that exhibit significant differences in redox potential, substrate specificity, and stability. Biochim. Biophys. Acta. 1996;1292:303–311. doi: 10.1016/0167-4838(95)00210-3. [DOI] [PubMed] [Google Scholar]
- Yamazaki HI. The cuticular phenoloxidase in Drosophila virilis. J. Insect Physiol. 1969;15:2203–2211. [Google Scholar]
- Yamazaki HI. Cuticular phenoloxidase from the silkworm Bombyx mori: properties, solubilization, and purification. Insect Biochem. 1972;2:431–444. [Google Scholar]
- Yatsu J, Asano T. Cuticle laccase of the silkworm, Bombyx mori: purification, gene identification and presence of its inactive precursor in the cuticle. Insect Biochem. Molec. Biol. 2009 doi: 10.1016/j.ibmb.2008.12.005. [DOI] [PubMed] [Google Scholar]