Abstract
In this contribution, we discuss state-of-the-art methodology for the collection and analysis of hyperspectral images of tissue that will become useful in complementing classical histopathology. In particular, we discuss sampling strategies, data collection methods, and computational approaches to produce pseudo-color maps of large tissue sections of lymph nodes, up to about 100 mm2 in size. The latter efforts include methods to reduce the presence of dispersion artifacts in IR transflection micro-spectra which can greatly impact the statistical analyzes performed on the data, such as hierarchical cluster analysis and principal components analysis.
Keywords: Cluster analysis, IR spectral imaging, FT-IR micro-spectroscopy, Lymph nodes, Dispersion artifact
1. Introduction
Instrumentation for performing infrared spectral imaging has made many advances over the last few years, enabling rapid analysis of both single cells and tissue samples up to about 100 mm2 in size. The introduction of focal plane array (FPA) detectors into IR spectrometers has rapidly reduced the time required for obtaining IR spectral images and maps, while linear array detectors allow spectral images to be collected whose size is limited only by the memory processing capabilities of the software. However, in contrast to the hybrid HgCdTe/Si technology used for detection/readout hardware in FPA detectors, the photoconductive linear array detectors employ HgCdTe technology only, and therefore, offer higher detector sensitivity.
Up to about 3 years ago, the remaining problems in developing the IR micro-spectral imaging as a medical tool were the expense of the sample substrates, the long data acquisition time, and the long computation times for analysing the data. We have made enormous progress on all fronts, and are now able to collect and analyze data sets from tissue sections as large as entire lymph nodes.
2. Experimental instrumentation
Biological tissue samples were imaged via a Perkin-Elmer Spectrum One/Spotlight 300 Infrared (IR) Spectrometer (Perkin-Elmer Corp., Sheldon, Connecticut). This system is a totally integrated infrared micro-spectrometer, and incorporates a 16 × 1 element (400μm × 25 μm) HgCdTe (MCT) array detector for imaging applications. The instrument provides 1:1 or 6:1 imaging on the MCT detector, resulting in nominal resolution of 25 or 6.25 μm, respectively. Visual image collection via a charge coupled device (CCD) camera is completely integrated with the microscope stage motion and IR spectra data acquisition. The visible images are collected under white light LED illumination, and are “quilted” together to give pictures of arbitrary size and aspect ratio. The desired regions for the IR images are selected from these visual images.
2.1. Sample preparation
Samples were cut from paraffin-embedded tissue blocks and de-paraffinized using standard procedures. Unstained tissue sections (6 μm thick) were mounted on ‘low e slides’ (see below) for analysis. Tissue sections were imaged in reflectance mode at 25 μm spatial resolution, and 4 cm−1 spectral resolution. Thus, the collected IR spectra represent transflection data, where the beam passes the sample, is reflected by the silver layer, and passes the sample again before being analyzed.
An infrared spectral imaging data set for a tissue section measuring 5 mm × 5 mm consists of 40,000 spectra at 25 μm spatial resolution, and requires approximately 20 min data collection time. Following infrared spectral acquisition, the tissue sections were stained (H&E, conventional procedure) and imaged using the visible data collection mode of the micro-spectrometer to allow comparison between pseudo-color maps and histopathology.
With the ultimate aim of IR spectral imaging being the eventual integration of the technique in a clinical setting, such as the diagnosis of cancer, the availability of cheap, consumable IR substrates is a necessity. The introduction of ‘low e’ glass slides (Kevley Technologies, Chesterfield, Ohio) as cheap IR substrates (~US$ 1 per slide compared with >US$ 200 for BaF2 or CaF2) offers a solution to this issue. These slides are made of glass coated with a thin Ag/SnO2 layer. They are chemically inert and nearly transparent to visible light. However, they reflect mid-infrared radiation almost completely and thus are ideal and inexpensive substrates for transflection infrared micro-spectroscopy, as they allow both visual and infrared images to be collected from the same sample.
2.2. Data analysis
We analyzed spectral imaging data sets, containing between 10,000 and 40,000 individual spectra, using unsupervised hierarchical cluster analysis (HCA). This procedure was discussed previously in detail [1], and will only be briefly reviewed here. All spectra, between 1800 and 800 cm−1, were converted to second derivatives of the absorbance with respect to the wavenumber (15-point Savitzky–Golay function), and vector normalised to remove the effects of sample thickness. Next, the correlation (covariance) matrix C was calculated for all spectra. The two most similar spectra (i.e. the spectra with the largest correlation coefficient) were merged to form a cluster, and a new correlation coefficient was calculated for this new cluster, and all the remaining spectra. This process of merging spectra into clusters and recalculating the C matrix was repeated until all the spectra were clustered into a few groups. Ward’s linkage was utilised to merge spectra and clusters, because this algorithm produces the most homogenous clusters. Pseudo-color maps were produced by assigning a color to each cluster, and plotting this color against the coordinate of the pixel from which the spectrum was collected. Pixels of the same color result from closely related spectra. Mean cluster spectra were extracted in order to interpret chemical or biochemical differences between the clusters. It is important to note that this technique is unsupervised, i.e. no information regarding the nature of the sample, the tissue morphology or the disease state was fed into the cluster analysis. However, this method is not optimal for diagnostic applications, since it is too time consuming, and is not easily transferable from data set to data set. Supervised diagnostic algorithms, based on neural networks and using spectral clusters obtained from HCA and corresponding histopathological input, are presently being developed in our laboratory, and will be reported at a later date.
We carried out all data processing using the CytoSpec, Inc. FT-IR imaging software package [2]. In order to reduce computation times to a reasonable level, very large data sets were reduced from 40,000 to about 20,000 spectra by an averaging/interpolation algorithm [2].
3. Results
The first IR spectral map presented in Fig. 1 illustrates the ability of combining IR spectral imaging with multivariate statistical analysis, in this case HCA, to obtain architectural, morphological and histological information from a biological tissue sample. Panel (a) shows the H&E stained section of a lymph node, typically analyzed by a histopathologist. The primary and secondary follicles containing the B lymphocytes are clearly seen, as are the cortex and paracortical areas, and the medullary cords. The visual image of the lymph node was obtained by quilting together about 70 individual, rectangular images collected via the CCD camera of the Perkin-Elmer micro-spectrometer.
Fig. 1.
(a) Photomicrograph of an H&E stained tissue section of a lymph node (~5 mm × 1.5 mm) used to obtain an infrared image. The section was stained after the IR spectral data were obtained. (b) Five-cluster pseudo-color map generated from an infrared image of the unstained lymph node. The map contains approximately 12,000 pixels, each of which represents an individual infrared spectrum. The spectral region 1800–800 cm−1 was utilised to generate this map. Tissue architecture is reproduced, showing the paracortex containing T-lymphocytes (light blue), medullary cords (orange), follicles containing B-lymphocytes (brown), node capsule (dark blue) and fatty tissue (green). (c) Five-cluster pseudo-color map of the same tissue section using the spectral region 1450–950 cm−1. This pseudo-color map shows further differentiation of the cortex and paracortex and the germinal centers, containing activated B-lymphocytes, of the follicles are observed (brown). (d) Photomicrograph of an H&E stained tissue section of a lymph node (~5 mm × 5 mm) with large colon cancer metastasis. (e) Pseudo-color map in the spectral region 1800–800 cm−1. (f) Pseudo-color map in the spectral region 1580–950 cm−1.
Prior to staining, this section was used to collect the infrared spectral data from which the pseudo-color map, Panel (b) was generated. This pseudo-color map was constructed from about 12,000 pixels, each representing an infrared spectrum. This map, generated by unsupervised hierarchical cluster analysis, is based solely on spectral similarities, and required no reference data set. It accurately reproduces the morphological architecture of the lymph node, seen as the paracortex containing T-lymphocytes (light blue), follicles containing B-lymphocytes (brown), medullary cords (orange), node capsule (dark blue) and fatty tissue (green). The color scheme of the pseudo-color maps is arbitrary and does not permit comparisons of features of the same color in different maps. However, mean cluster spectra extracted from the imaging data set permit a comparison of different tissue types and areas of malignancy between different lymph nodes. Panel (c) shows the pseudo-color map of the same tissue section, however, the wavenumber region has been reduced from 1800–800 to 1450–950 cm−1. In this panel, we can distinguish activated B-lymphocytes in the secondary follicles, from the non-activated B-lymphocytes (blue) and T-lymphocytes (green) in the paracortex. Since the spectral differences between these cell types occur mostly in the low frequency spectral region (900–1300 cm−1), the reduction in spectral range enhances the morphological information seen in the pseudo-color map.
The next IR spectral map, shown in Fig. 1d–f, is from a lymph node section which contains a large colon adenocarcinoma metastasis. The H&E stained section of the tissue section is presented in Fig. 1d. The circled areas highlight the region where the colon metastatic cancer cells are located. A pseudo-color map, generated with seven clusters, is shown in Fig. 1e. The differentiation of tissue architecture observed for the tissue section presented in Fig. 1b and c is absent in this pseudo-color map. In fact, the only real distinction of tissue type is that of connective tissue, seen as green in the pseudocolor map and the node capsule, seen as yellow. There is no clear distinction between the lymphocytes in the cortex and the paracortex and there is no differentiation of the colon metastatic cells in the lower right section of the lymph node.
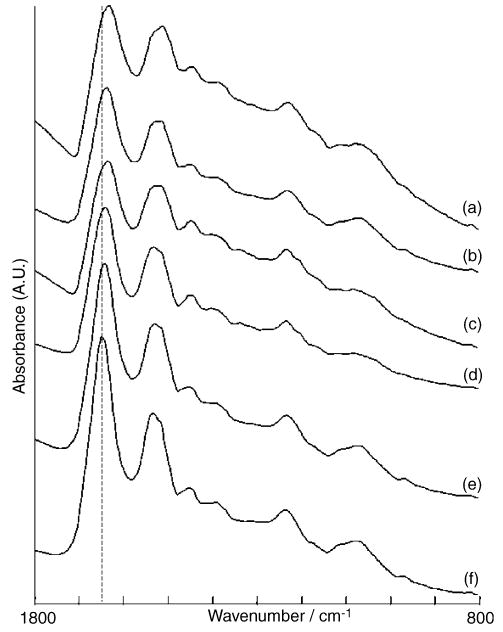
The reason for the failure in distinguishing the tissue types and areas of disease becomes clear upon inspection of the average IR spectra from each cluster, shown in Fig. 2. The main differences between these spectra are the shift in the position of the amide I band, indicated by the dashed line, the ratios between the amide I and II bands, and the sharp dip to the higher wavenumber region of the amide I band. These spectral features, due to reflective components in the spectra [3], and not related to the chemical components of the tissue types, are dominating the cluster analysis, rather than differentiating between the subtle differences between the tissue types. The fact that the node capsule and connective tissue are observable in the pseudo-color map arises from differences in the spectral region from 1300 to 1400 cm−1.
Fig. 2.
Averaged infrared spectra extracted from the pseudo-color map in Fig. 1e. Spectra correspond to the following clusters from this map: (a) brown, (b) red, (c) yellow, (d) green, (e) light blue and (f) dark blue.
When the reflective (dispersive) artifact is removed by one of two methods to be discussed below, detailed spectral maps are obtained that distinguish the different tissue types very well, indeed. An example of a corrected map is shown in Fig. 1f. This map was obtained by simply reducing the spectral range from 1800–800 to 1580–950 cm−1 in an effort to remove the effects of the dispersion artifact on the amide I band. Removal of the affected amide I band significantly improves the distinction between tissue types in the lymph node and further emphasizes the argument that the magnitude of the amide I shift was dominating the cluster analysis. Since second derivative spectra were used throughout this study, the remaining slope in the spectra is of little importance. Inspection of the pseudo-color map, Fig. 1f, reveals quite detailed differentiation of tissue types. Not only are we able to distinguish the follicles (red) from the paracortex (blue) but we are also able to differentiate between the normal and cancerous region of the tissue section (seen as brown). Comparison with the H&E stained section in Fig. 1d shows a high correlation of this cluster with the region of colon adenocarcinoma metastatic cells highlighted in blue. We have shown before that cellular abnormalities affect both the protein vibrations as well as the nucleic acid features observed mostly in the range below 1400 cm−1 [1]; thus, it is not surprising that the reduced range, which eliminates most of the spectral artifact, results in superior images.
We have also applied the dispersion artifact reduction routine, discussed in an accompanying paper, to this data set. This procedure improves the spectral maps substantially, but still suffers from a few minor mathematical problems due to strongly sloping real and imaginary components in a few spectra, which may lead to an overcompensation of the artifact.
4. Discussion
In this paper, we demonstrate that the methodology used in our laboratory is capable of detecting various tissue types, and stages of disease, in large sections of tissue sections obtained from biopsy. The use of “low e” slides as sample substrates greatly reduces the cost of these experiments, while providing nearly the same flexibility and ruggedness of glass slides normally used in histopathology.
The main advantage of modern IR micro-spectrometers, incorporating array detectors, is the speed and sensitivity of data acquisition. As pointed out, maps consisting of about 40,000 spectra can be collected in about 20 min. The main problem, however, is the increased size of the resulting IR data sets, which may exceed ~250–300 MB of memory. The size of the correlation matrix required for multivariate statistical analysis, such as hierarchical cluster analysis or principal component analysis, exceeds 4 GB, which is more than the direct address space of current Pentium processors. Thus, computations are best carried out using 64-bit processors with large memory access. Some of the maps represented here were obtained using a 64 bit Athlon-equipped machine incorporating 8 GB RAM.
One disadvantage of the “low e” sample substrate used in this study is the appearance of a dispersion or reflectance artifact in the IR spectra. This dispersion artifact has been observed around the edges of cells and tissues and in tissues where the sample deposit is sparse. We find that the artifact occurs predominantly in the abnormal regions of tissue. This is because the particular colon adenocarcinoma studied produces glandular metastases with many voids, whereas the lymph node itself consists of dense tissue with relatively uniform coverage. It is at the edges of the glandular tissue that most of the artifact is observed.
The presence of the artifact causes an unusual ratio of the amide I/II bands, a distorted, sloping baseline and a significant shift (to lower wavenumbers) of the bands. In cases where the artifact is not as strong, the manifested effects are reduced, but still noticeable, particularly in the position of the amide I band. The peak shifts and amide ratio will have a significant influence on the statistical analysis of IR spectra. In many cases, these effects totally dominate the statistical analysis, for example HCA, as the magnitude of the changes caused by the artifact are more significant than the subtle spectral changes that we are trying to detect. The dispersion artifact has also been noted for IR spectral images of biological tissue samples performed in transmittance mode on BaF2 substrates. Again, the presence of this artifact is thought to arise from edges of the tissue, and regions of the tissue where the tissue deposit is sparse. For a discussion of the mathematical theory behind the presence of the dispersive line shape in IR micro-spectroscopy the reader is referred to our paper in this special issue [4].
Since a spectral data set contains over 10,000 spectra, a visual elimination of contaminated spectra is impossible. However, inspection of the mean cluster spectra demonstrates clearly that the dispersion artifact plays a major role in the spectra of the abnormal regions, and needs to be eliminated. This aspect is important in that at times where we rely more and more on the automatic analysis of spectra, the eye of the spectroscopist to detect and interpret spurious effects is invaluable.
5. Conclusions
This paper discusses the recent advances in infrared spectral imaging, allowing us to rapidly analyze large samples previously thought impossible. This paper also demonstrates the importance of recognising artifacts in infrared spectra. It is dangerous to claim that spectral shifts are real and sample dependent if underlying artefacts, such as dispersion, are not investigated and corrected for.
Acknowledgments
Partial support of this research from grants GM 60654 and CA 090346 from the National Institutes of Health is gratefully acknowledged. A “Research Centers in Minority Institutions” award RR-03037 from the NCRR of the NIH, which supports the infrastructure of the Chemistry Department at Hunter, is also acknowledged. We also acknowledge the expertise of Dr. Raj Emmadi, for her histopathological diagnosis of the tissue samples, which were also supplied by her.
References
- 1.Wood BR, Chiriboga L, Yee H, Quinn MA, McNaughton D, Diem M. Gynecol Oncol. 2004;93:59–68. doi: 10.1016/j.ygyno.2003.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.P. Lasch, A Matlab based application for infrared imaging, see http://www.cytospec.com for details.
- 3.White Jenkins. Fundamentals of Optics. 3. McGraw-Hill Book Company, Inc; NY: 1957. [Google Scholar]
- 4.Romeo MJ, Diem M. Vibrat Spectrosc. 2005;38:129–132. doi: 10.1016/j.vibspec.2005.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]