Abstract
Invadopodia are matrix-degrading membrane protrusions in invasive carcinoma cells. The mechanisms regulating invadopodium assembly and maturation are not understood. We have dissected the stages of invadopodium assembly and maturation and show that invadopodia use cortactin phosphorylation as a master switch during these processes. In particular, cortactin phosphorylation was found to regulate cofilin and Arp2/3 complex–dependent actin polymerization. Cortactin directly binds cofilin and inhibits its severing activity. Cortactin phosphorylation is required to release this inhibition so cofilin can sever actin filaments to create barbed ends at invadopodia to support Arp2/3-dependent actin polymerization. After barbed end formation, cortactin is dephosphorylated, which blocks cofilin severing activity thereby stabilizing invadopodia. These findings identify novel mechanisms for actin polymerization in the invadopodia of metastatic carcinoma cells and define four distinct stages of invadopodium assembly and maturation consisting of invadopodium precursor formation, actin polymerization, stabilization, and matrix degradation.
Introduction
It is hypothesized that invasive carcinoma cells use invadopodia, subcellular structures that degrade ECM using matrix metalloproteinase (MMP) activity (Gimona and Buccione, 2006), to migrate through the ECM (Gimona et al., 2008) and intravasate through the endothelium into the blood during metastasis (Yamaguchi and Condeelis, 2007). Studies of the most highly invasive tumor cells support this hypothesis (Philippar et al., 2008). Therefore, in order to prevent invasion and metastasis, it is essential to understand the molecular mechanisms that regulate invadopodium maturation.
During tumor cell invasion, actin polymerization produces force for the plasma membrane to protrude through ECM (Wyckoff et al., 2006). Invadopodia are enriched in many proteins that regulate actin polymerization including cortactin (Artym et al., 2006; Clark et al., 2007; Ayala et al., 2008), N-WASp, Arp2/3, and cofilin (Yamaguchi et al., 2005). These proteins function to generate actin-free barbed ends, which are required for actin polymerization. Mammary carcinoma cells use two dominant mechanisms to generate free barbed ends at lamellipodia: (1) actin filament severing by cofilin, and (2) dendritic nucleation by the Arp2/3 complex (Condeelis, 2001), and these mechanisms synergize to amplify barbed ends both in vitro (Ichetovkin et al., 2002) and in vivo (DesMarais et al., 2004). However, the mechanisms used to generate free barbed ends at invadopodia are not understood.
Lamellipodia and invadopodia are known to use different effectors to stimulate Arp2/3 complex–dependent actin polymerization (Yamaguchi et al., 2005; Sarmiento et al., 2008). Unlike lamellipodia (Sarmiento et al., 2008; Desmarais et al., 2009), N-WASp, its activators Cdc42 and Nck1, and the Arp2/3 complex are all important for invadopodium formation and function in mammary carcinoma cells (Yamaguchi et al., 2005). Studies using an N-WASp biosensor show that N-WASp activity is concentrated at invadopodia (Lorenz et al., 2004b).
The F-actin severing protein cofilin is required for barbed end formation at the plasma membrane during lamellipodium protrusion (Sidani et al., 2007). However, the function of cofilin in invadopodia is not well understood. Although it is not important for invadopodium formation, it is necessary for maturation of degradation activity (Yamaguchi et al., 2005; Desmarais et al., 2009).
Lastly, the Arp2/3 complex regulator and scaffolding protein cortactin is essential for invadopodium formation, MMP recruitment, and ECM degradation in many cancer cell types (Artym et al., 2006; Clark et al., 2007; Ayala et al., 2008), but it is not required for the formation of lamellipodia (Bryce et al., 2005; Kempiak et al., 2005). Cortactin contains an N-terminal acidic (NTA) domain and a C-terminal SH3 domain that binds and activates the Arp2/3 complex (Weed et al., 2000; Uruno et al., 2001; Weaver et al., 2001) and N-WASp (Martinez-Quiles et al., 2004), respectively. The binding of cortactin to Arp2/3 and N-WASp is important for invadopodium formation in melanoma cells (Ayala et al., 2008).
Cortactin is phosphorylated by Src and Abl family kinases at three tyrosine residues: 421, 466, and 482 (Head et al., 2003; Boyle et al., 2007), and cortactin tyrosine phosphorylation is important for the degradation activity of invadopodia in melanoma cells (Ayala et al., 2008). The tyrosine phosphorylation of cortactin is also important for efficient actin polymerization in vitro by facilitating the assembly of an Nck1–N-WASp–Arp2/3 signaling complex (Tehrani et al., 2007). N-WASp activation by Nck1, but not Grb2, is important for invadopodium formation and function (Yamaguchi et al., 2005), suggesting that the phosphorylated cortactin–Nck1–N-WASp–Arp2/3 signaling complex may be important for actin polymerization at invadopodia.
Artym et al. (2006) proposed sequential stages in invadopodium maturation beginning with the formation of a cortactin and actin invadopodium precursor and ending with ECM degradation. Here, we have investigated the molecular mechanisms that regulate the stages of invadopodium maturation and provide evidence that cortactin coordinates the activities of cofilin and N-WASp to spatially and temporally control activation of actin polymerization and invadopodium maturation.
Results
Invadopodium precursors form in response to EGF and mature to degrade ECM
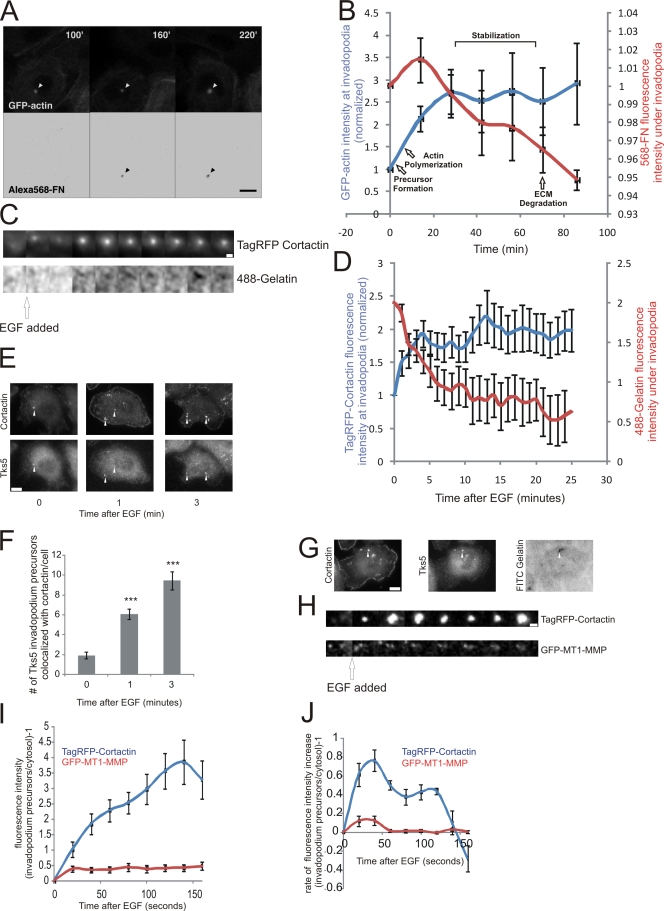
To determine the kinetics of actin recruitment relative to matrix degradation activity at invadopodia in MTLn3 breast cancer cells and to verify that such structures went on to form invadopodia with degradation activity, live cell imaging was performed using MTLn3 cells expressing GFP-actin (Lorenz et al., 2004a; Yamaguchi et al., 2005) plated on 5-μm-thick Alexa 568-fibronectin(FN)/gelatin matrix (Yamaguchi et al., 2005). Our results indicate that although the assembly of the invadopodium precursor takes less than 1 min, ECM degradation occurs 60 min later (Fig. 1, A and B; Video 1). To determine the minimum lag time between assembly and the onset of degradation, cells expressing RFP-cortactin were imaged on a thin 488-gelatin matrix which allows detection of matrix degradation with highest sensitivity (Artym et al., 2006), and cortactin recruitment relative to matrix degradation was quantified. The results show a 2-min lag time from precursor assembly to matrix degradation (Fig. 1, C and D). Clearly, the assembly of a cortactin- and actin-containing protein complex and matrix degradation are uncoupled in time in MTLn3 cells as reported previously in MDA-MB-231 cells (Artym et al., 2006). The GFP-actin accumulation at invadopodia (Fig. 1 B) suggests there are actin polymerization and stabilization events after precursor formation that precede ECM degradation. Here, we will refer to the structures detected during the early stages of maturation that precede matrix degradation as invadopodium precursors and mature structures with degradation activity as invadopodia.
Figure 1.
Invadopodium precursors form in response to EGF and mature to degrade matrix. (A and B) MTLn3 cells expressing GFP-actin were cultured on Alexa 568-FN/gelatin thick matrix and analyzed by time-lapse microscopy. (A) Formation of the precursor before degradation is shown (arrowhead). Bars, 10 μm. (B) Quantification of GFP-actin fluorescence intensity at all stages of invadopodia (blue) vs. that of underlying matrix (red). n = 3 invadopodia; three independent experiments. (C and D) Live cell time-lapse experiments using MTLn3 cells expressing TagRFP-cortactin plated on thin 488-gelatin matrix. (C) Representative montage showing the formation of an invadopodium precursor (top) precedes gelatin matrix degradation (bottom). 3 min/frame. Bar, 1 μm. (D) Quantification of the change in cortactin fluorescence intensity at invadopodia vs. that of the underlying matrix in response to EGF. n = 25 invadopodium precursors, two independent experiments. For cortactin, P < 0.05 for all times compared with 0 min. For 488-gelatin, P < 0.05 for all times compared with 0 min, except 1 min where P > 0.05. (E) Representative images of cortactin and Tks5 antibody staining of MTLn3 cells stimulated with EGF. Arrowheads indicate invadopodium precursors. Bar,10 μm. (F) Quantification of the number of invadopodium precursors per cell that contain Tks5 and cortactin after EGF stimulation. n = 40 (0 min), 37 (1 min), and 17 (3 min) cells; three independent experiments. P values are compared with 0 min. (G) Image showing that Tks5/cortactin punctate structures colocalize with matrix degradation (arrowheads). (H) Representative montage (background subtracted) showing that TagRFP-cortactin (top) and GFP-MT1-MMP (bottom) colocalize at invadopodium precursors formed in response to EGF. 20 seconds/frame. Bar, 1 μm. Quantification of (I) fluorescence intensity and (J) rate of fluorescence intensity increase of TagRFP-cortactin or GFP-MT1-MMP at invadopodium precursors formed in response to EGF. n = 33 invadopodium precursors; three independent experiments. For I, P < 0.05 for all times compared with 0 min.
To confirm that invadopodium precursors are immature invadopodia without ECM degradation activity and not an unrelated structure, we investigated whether invadopodium precursors contain proteins present in mature invadopodia. MTLn3 cells were stimulated with EGF, shown previously to induce the formation of invadopodia (Yamaguchi et al., 2005), and fixed and stained for cortactin and Tks5, a scaffolding protein that localizes to both immature and mature invadopodia (Seals et al., 2005; Chan et al., 2009; Crowley et al., 2009). Tks5 localized with cortactin to invadopodium precursors at early times (Fig. 1, E and F) that preceded matrix degradation (Fig. 1, B and D), and also localized to invadopodia that were actively degrading matrix (Fig. 1 G). Also, markers of mature invadopodia including cortactin, Arp2, cofilin, and N-WASp all localized with F-actin to invadopodium precursors (Fig. S1 A). Furthermore, these proteins are enriched at invadopodium precursors formed in response to EGF and do not localize with vesicle and other membrane trafficking markers (Desmarais et al., 2009). In addition, we investigated whether invadopodium precursors are enriched with MT1-MMP, an MMP that localizes to invadopodia before ECM degradation (Artym et al., 2006). Live cell imaging revealed that upon EGF stimulation, GFP-MT1-MMP became enriched at invadopodium precursors (Fig. 1, H and I), although at a rate significantly slower than cortactin (Fig. 1 J), both before ECM degradation (Fig. 1, B and D). In summary, invadopodium precursors contain the same markers as mature invadopodia including cortactin, cofilin, N-WASp, the Arp2/3 complex, F-actin, Tks5, and MT1-MMP, but without ECM degradation activity. When followed in time lapse they have the ability to eventually degrade matrix. In this study, we used these colocalization patterns to study the early stages of invadopodium maturation.
The Arp2/3 and N-WASp binding domains of cortactin are important for invadopodium precursor formation
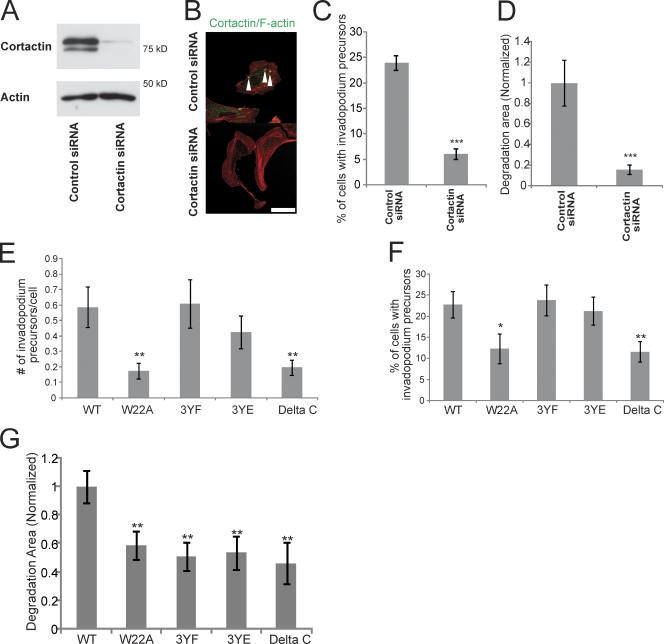
To determine if cortactin is required for invadopodium precursor formation and matrix degradation in MTLn3 cells, cortactin was knocked down using siRNA, which yielded a 95% knockdown (KD) (Fig. 2 A). Invadopodium precursor formation and matrix degradation was dramatically reduced in cortactin KD cells (Fig. 2, B–D; Fig. S1, B and C).
Figure 2.
Multiple domains of cortactin are important for matrix degradation by invadopodia. (A) Western blot of whole-cell lysates from MTLn3 cells transfected with ctrl or cortactin siRNA blotted for cortactin and β-actin. (B) Representative image of cells treated with ctrl or cortactin siRNA and stained for cortactin and F-actin to visualize invadopodium precursors (arrowheads). Bar, 10 μm. (C) Quantification of the percentage of cells that form invadopodium precursors and (D) the degradation area in cells treated with ctrl or cortactin siRNA. (E) Quantification of the number of invadopodium precursors per cell and (F) percentage of cells that form invadopodium precursors in cortactin mutant stable cell lines. P values are compared with WT and graphs show combined data from three independent experiments. (G) Quantification of the Alexa 568-FN degradation area/field normalized to WT cortactin. P values are compared with WT and n = number of fields scored: >80 from more than three independent experiments.
To determine which protein domains of cortactin are necessary for invadopodium precursor formation, cortactin mutants that cannot bind to the Arp2/3 complex (W22A mutant), cannot be tyrosine phosphorylated (3YF mutant), cannot bind to N-WASp (Δ-C terminus mutant), and a 3YE mutant that has three tyrosines (421, 466, and 482) mutated to glutamates to mimic charge were constructed (Fig. S1 D). Silent mutations in the siRNA target sequence were introduced into the cortactin mutants (Fig. S1 E), and stable cell lines were generated. Using this approach, endogenous cortactin could be transiently knocked down using siRNA (Fig. 2 A), and the stable cell lines only express the mutant variant of cortactin (Fig. S1 F).
We analyzed the ability of the cortactin KD cells reconstituted with various cortactin mutants to form invadopodium precursors. The results show that the Arp2/3 and N-WASp binding domains of cortactin are important for the formation of invadopodium precursors (Fig. 2, E and F). In contrast, the tyrosine phosphorylation status of cortactin did not affect invadopodium precursor formation (Fig. 2, E and F).
Multiple domains of cortactin are important for matrix degradation activity at invadopodia
We next investigated which cortactin functions were important for matrix degradation activity at invadopodia. Compared with cells expressing wild-type (WT) cortactin, the cortactin W22A, 3YF, 3YE, and Δ-C terminus mutant cell lines all had partially reduced degradation activity (Fig. 2 G; Fig. S1 G). Interestingly, the tyrosine phosphorylation status of cortactin did not affect the formation of invadopodium precursors (Fig. 2, E and F), despite being necessary for matrix degradation by invadopodia in MTLn3 cells (Fig. 2 G) and melanoma cells (Ayala et al., 2008). This observation suggests that the requirement for cortactin tyrosine phosphorylation in invadopodium maturation occurs after precursor formation, but before maturation to a degradation-competent invadopodium.
Increased actin polymerization occurs as an early event during invadopodium maturation
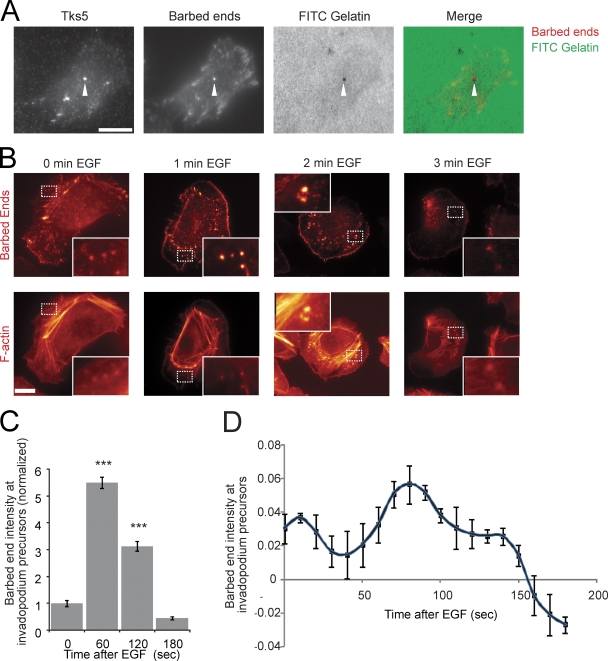
After formation of invadopodium precursors, the proteins localized at the precursor must initiate actin polymerization to push the invadopodium precursor into contact with the ECM. To identify sites of actin polymerization, the presence of actin-free barbed ends in invadopodium precursors was analyzed using a previously described barbed end assay (Chan et al., 1998). The barbed ends in invadopodium precursors localize as punctate structures that contain F-actin and Arp2 (Fig. S2 A). In addition, barbed ends are enriched in mature invadopodia that are actively degrading matrix (Fig. 3 A).
Figure 3.
Barbed ends at invadopodium precursors peak beginning 1 min after EGF stimulation. (A) Image showing that barbed ends localize with Tks5 at areas of matrix degradation (arrowheads). Bar, 10 μm. (B and C) The barbed end assay in response to EGF using MTLn3 cells. (B) Representative images of barbed ends and F-actin at invadopodium precursors in response to EGF. Insets here and throughout the figures show close ups of invadopodium precursors. Bar, 10 μm. (C) Quantification of barbed end intensity at invadopodium precursors normalized to 0 s (sec) EGF. n = number of invadopodium precursors; three independent experiments: 0 s (39), 60 (73), 120 (109), 180 (28). P values are compared with 0 s. (D) Quantification of barbed end intensity at invadopodium precursors in response to EGF measured using a GFP-actin live cell method (Lorenz et al., 2004a). n = 21 invadopodium precursors.
Stimulation of mammary carcinoma cells with EGF results in the amplification of barbed ends at lamellipodia (Mouneimne et al., 2004). To determine when barbed ends are amplified for actin polymerization during invadopodium maturation, cells were stimulated with EGF and barbed ends were measured using two different approaches: the barbed end assay discussed above (Chan et al., 1998) and a live cell approach validated previously using a GFP-actin stable cell line (Lorenz et al., 2004a). Both methods showed a barbed end peak in invadopodium precursors beginning 1 min after EGF stimulation (Fig. 3, B–D; Video 2) before matrix degradation is observed (Fig. 1, B and D) that remained detectable throughout maturation (Fig. 3).
Cortactin tyrosine phosphorylation is important for actin barbed end formation at invadopodium precursors
Cortactin tyrosine phosphorylation is important for the degradation activity of invadopodia, but not precursor formation (Fig. 2, E–G). Because barbed ends are generated early during invadopodium maturation, we hypothesized that cortactin phosphorylation may be important for barbed end formation. To define the time course for cortactin tyrosine phosphorylation at invadopodium precursors in response to EGF, its localization was measured using a cortactin phospho-specific antibody (pY421). The number of invadopodium precursors that contain pY421-cortactin and cortactin significantly increases beginning 1 min after EGF stimulation (Fig. S3, A–C).
To determine whether EGF induces an increase in cortactin tyrosine phosphorylation at invadopodium precursors, FRET acceptor photobleaching experiments were performed between cortactin (donor) and phosphotyrosine (acceptor), a technique which has been proven to work well for the EGF receptor (Wouters and Bastiaens, 1999) (controls are shown in Fig. S4 A and described in detail in Materials and methods). The results show a significant increase in FRET between cortactin and phosphotyrosine at 1 min after EGF stimulation, suggesting that cortactin is tyrosine phosphorylated at invadopodium precursors at this time (Fig. S3, D and E). This result was confirmed by Western blot analysis using triton-insoluble cell lysates (Fig. S3, F and G).
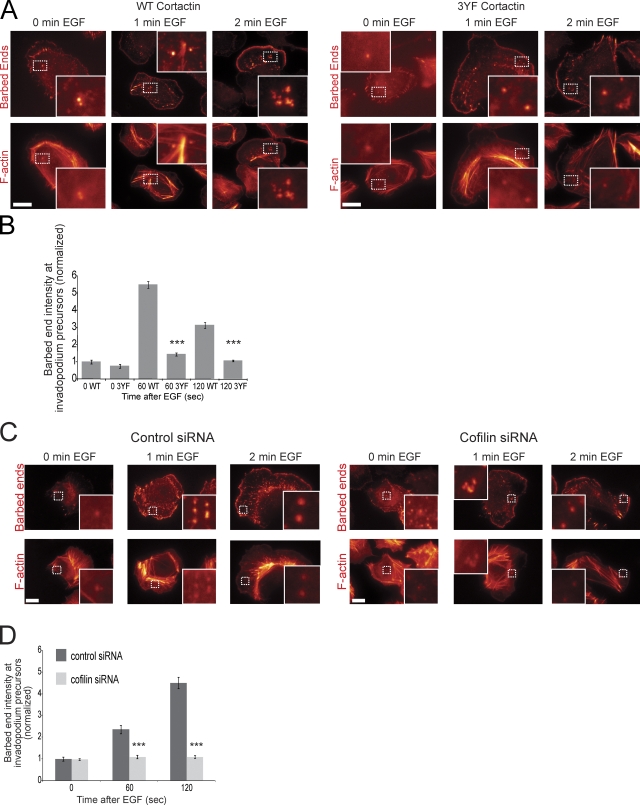
To determine whether cortactin tyrosine phosphorylation is important for barbed end formation at invadopodium precursors, the barbed end assay was performed using cortactin WT or 3YF cell lines. Interestingly, cells expressing 3YF cortactin had reduced barbed end formation at invadopodium precursors after EGF stimulation (Fig. 4, A and B) and during steady-state serum stimulation (Fig. S2, B and C). This result was confirmed using the GFP-actin live cell method for measuring barbed ends (Fig. S2, D–F). These results indicate that EGF-induced signaling leading to cortactin tyrosine phosphorylation is required for the efficient formation of free barbed ends at invadopodium precursors.
Figure 4.
Cortactin tyrosine phosphorylation and cofilin are important for barbed end formation at invadopodium precursors. (A and B) Barbed end assay in response to EGF with cortactin WT or 3YF cells, treated with cortactin siRNA. (A) Representative images and (B) quantification of barbed end intensity in response to EGF at invadopodium precursors normalized to WT at 0 s. Bar, 10 μm. P values are compared with WT at the same time point. Throughout the figure, n = number of invadopodium precursors; three independent experiments: WT 0 (39), 3YF 0 (24), WT 60 (255), 3YF 60 (103), WT 120 (192), 3YF 120 (171). (C) Representative images and (D) quantification of barbed ends in response to EGF in ctrl vs. cofilin siRNA treated cells normalized to 0 s. Bar, 10 μm. P values are compared with ctrl siRNA at the same time point. n: ctrl siRNA 0 (33), 60 (42), 120 (60) and cofilin siRNA 0 (57), 60 (49), 120 (110).
The tyrosine phosphorylation of cortactin controls barbed end formation at invadopodium precursors through a direct interaction with cofilin
The F-actin severing protein cofilin is important for the initial generation of free barbed ends formed in response to EGF at the leading edge of lamellipodia (Mouneimne et al., 2004). To investigate whether cofilin is important for EGF-induced barbed end formation at invadopodium precursors, the barbed end assay was performed using MTLn3 cells treated with control (ctrl) or cofilin siRNA that specifically targets cofilin-1 (Fig. S5 A), the major cofilin isoform in these cells (Sidani et al., 2007). The results show that cofilin is required for the formation of barbed ends at invadopodium precursors (Fig. 4, C and D).
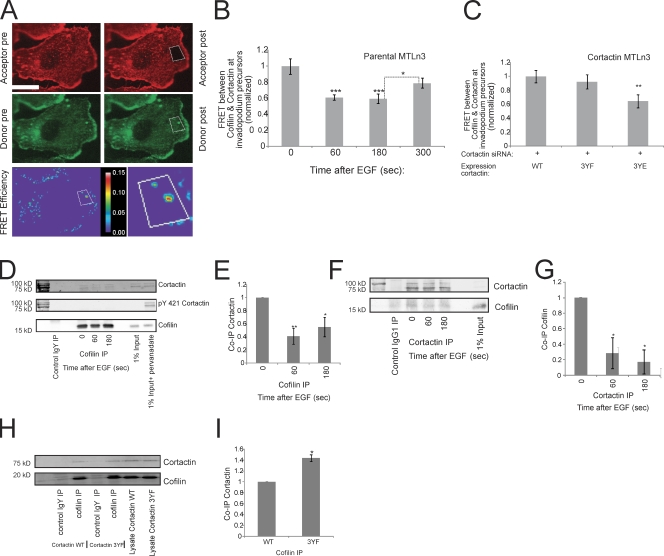
Because cortactin tyrosine phosphorylation and cofilin are both important for barbed end generation at invadopodium precursors, we investigated whether cortactin phosphorylation regulates cofilin's severing activity. To determine whether cofilin and cortactin interact at invadopodium precursors and whether the interaction is EGF dependent, FRET acceptor photobleaching experiments were performed. Cells were stimulated with EGF for various times, fixed, and stained with antibodies against cofilin (donor) and cortactin (acceptor) using secondary antibodies that have potential to yield FRET (controls are shown in Fig. S4 B and described in detail in Materials and methods). The FRET results indicate that cofilin and cortactin interact at invadopodium precursors in starved cells, the interaction decreases after EGF stimulation when the barbed end peak occurs, and increases again after 5 min of EGF stimulation (Fig. 5, A and B). To confirm that the FRET interaction between cofilin and cortactin was not an artifact of fixation, both acceptor photobleaching and sensitized emission FRET experiments were performed between cofilin and cortactin in live cells expressing GFP-cofilin and TagRFP-cortactin. In both live cell FRET methods, FRET between cofilin and cortactin was observed in invadopodia (Fig. S4, C and D).
Figure 5.
Cortactin interacts with cofilin at invadopodium precursors through a phosphorylation-dependent mechanism. (A) Representative cofilin-cortactin FRET efficiency image of a resting cell (0 s EGF). Red = cortactin, green = cofilin. Box indicates bleached area. Bottom right inset shows close up of FRET efficiency between cofilin and cortactin at invadopodium precursors. Bar, 10 μm. (B) Quantification of FRET between cofilin and cortactin at invadopodium precursors in response to EGF normalized to 0 s (FRET efficiency at 0 s = 11.8% ± 1.1). P values are compared with 0 s unless indicated. n = number of invadopodium precursors: 0 (25), 60 (24), 180 (22), 300 (17); three independent experiments. (C) Quantification of cofilin-cortactin FRET at invadopodium precursors in starved MTLn3 cells lines expressing WT, 3YF, and 3YE cortactin with endogenous cortactin knocked down normalized to WT (FRET efficiency of WT cortactin = 4.2% ± 0.36). P values are compared with WT. n = number of invadopodium precursors: WT (22), 3YF (22) 3YE (17); three independent experiments. (D–G) IP experiments between cofilin and cortactin in response to EGF. (D) Representative Western blot showing IP of cofilin and coIP of cortactin. (E) Quantification of the protein level of Co-IP cortactin/IP cofilin normalized to 0 s. n = number of independent experiments: 6. P values are compared with 0 s (F) Representative Western blot showing IP of cortactin and coIP of cofilin. (G) Quantification of the protein level of the Co-IP cofilin/IP cortactin normalized to 0 s. P values are compared with 0 s. n = number of independent experiments: 4. (H) Representative Western blot showing IP of cofilin and coIP of cortactin in cortactin WT vs. 3YF cell lines treated with cortactin siRNA. (I) Quantification of the protein level of Co-IP cortactin/IP cofilin normalized to WT. n = number of independent experiments: 3.
To test whether the cofilin–cortactin interaction is altered by cortactin tyrosine phosphorylation, acceptor photobleaching FRET experiments between cofilin and cortactin were performed using cell lines expressing WT, 3YF, or 3YE cortactin. Interestingly, FRET between cofilin and cortactin in cells expressing 3YE cortactin was significantly lower than the FRET in either the WT or 3YF cell lines (Fig. 5 C). Together, the decrease in cofilin–cortactin FRET in the 3YE cortactin cell line (Fig. 5 C) and after 1 min of EGF stimulation (Fig. 5 B)—the time when cortactin is tyrosine phosphorylated (Fig. S3)—strongly suggests that the in vivo interaction between cofilin and cortactin at invadopodium precursors is weakened when cortactin is phosphorylated.
To further investigate the interaction between cofilin and cortactin, immunoprecipitation (IP) experiments were performed using MTLn3 cells stimulated with EGF. In line with the FRET data, the coIP of endogenous cortactin with cofilin (Fig. 5, D and E) and the reverse coIP of endogenous cofilin with cortactin (Fig. 5, F and G) was highest in starved cells and significantly decreases during the barbed end peak at invadopodium precursors. Furthermore, there was no detectable phospho-cortactin in the cofilin IP, suggesting that cofilin is interacting with a pool of dephosphorylated cortactin (Fig. 5 D). To confirm this, cofilin IP experiments were performed using cell lines expressing WT or 3YF cortactin. Significantly more cortactin coimmunoprecipitated with cofilin in cells expressing cortactin 3YF compared with WT (Fig. 5, H and I), showing that cofilin preferentially interacts with dephosphorylated cortactin in vivo. This result was confirmed with the reverse coIP of cofilin with mRFP-cortactin WT or 3YF (Fig. S5, B and C).
Cofilin and cortactin directly bind in vitro
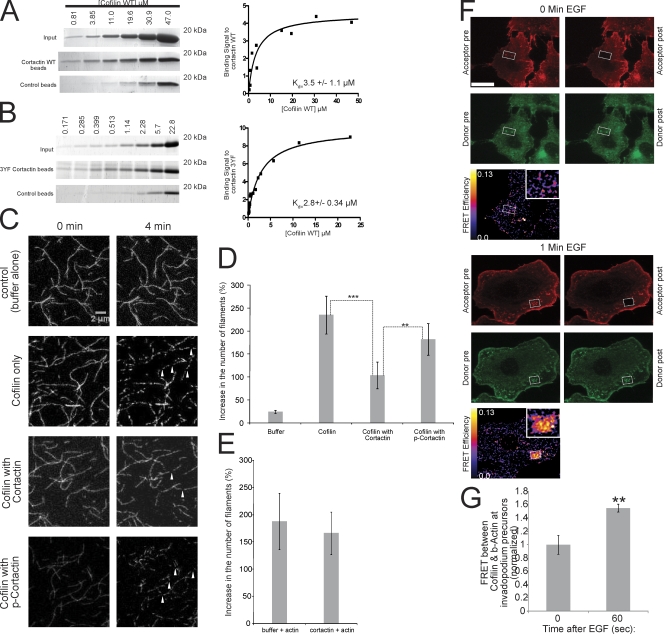
To investigate if the in vivo FRET results are detecting a direct binding interaction between cofilin and cortactin, we tested whether they bind directly in vitro. To determine this, in vitro pull-down assays were performed using purified recombinant cortactin WT or 3YF, and cofilin. Cortactin WT and cofilin directly bind in vitro with a Kd of 3.5 ± 1.1 μM (Fig. 6 A). Furthermore, cortactin 3YF and cofilin bind in vitro with a Kd of 2.8 ± 0.34 μM, confirming that cofilin directly binds to this cortactin mutant in vitro (Fig. 6 B). Given that cofilin and cortactin directly bind in vitro and that FRET is only observed when two molecules are <8 nm apart, the cofilin–cortactin FRET at invadopodium precursors likely measures a direct interaction between these proteins in vivo.
Figure 6.
Cortactin directly binds to cofilin and inhibits cofilin's severing activity, and this inhibition is relieved when cortactin is tyrosine phosphorylated. (A and B) (left) Coomassie-stained gels and (right) quantification of the binding signal of cofilin to (A) WT cortactin (Kd = 3.5 ± 1.1 μM) or (B) 3YF cortactin (Kd = 2.8 ± 0.34 μM) from in vitro pull-down assays at increasing concentrations of cofilin. Number of data points for Kd calculation: WT = 12, 3YF = 16. (C) Representative images showing cofilin's severing activity with cofilin alone, in the presence of cortactin, or phospho-cortactin. Arrowheads show actin filaments severed by cofilin. Bar, 2 μm. (D) Quantification of the percent increase in the number of actin filaments after incubation with cofilin alone, cofilin with cortactin, or cofilin with phospho-cortactin. P > 0.05 for cofilin alone vs. cofilin with p-cortactin. (E) Quantification of the percent increase in the number of actin filaments after preincubation of actin filaments with cortactin or buffer alone as described in Materials and methods. (F) Representative cofilin/β-actin FRET efficiency images of cells stimulated with EGF for 0 (top) and 1 min (bottom). Red = β-actin, green = cofilin. Bar, 10 μm. (G) Quantification of FRET between cofilin and β-actin at invadopodium precursors in response to EGF normalized to time 0 (FRET efficiency at 0 s = 4.2% ± 0.6). n = number of invadopodium precursors: 0 (34), 60 (46); three independent experiments.
Cortactin, but not tyrosine phosphorylated cortactin, inhibits cofilin's severing activity
We hypothesized that cortactin binds to cofilin and inhibits cofilin's actin binding activity, and when cortactin is tyrosine phosphorylated, this inhibition is relieved, and cofilin can bind F-actin to create free barbed ends by severing. To test this directly, a cofilin severing assay was performed using cofilin alone, cofilin incubated with nonphosphorylated cortactin, or cofilin incubated with tyrosine phosphorylated cortactin. Interestingly, when cofilin is preincubated with nonphosphorylated cortactin, its severing activity is significantly reduced (Fig. 6, C and D). This was not due to cortactin binding to actin filaments and blocking cofilin from severing because preincubation of cortactin with actin filaments failed to inhibit severing by cofilin (Fig. 6 E). In contrast, when cofilin is preincubated with phosphorylated cortactin (Fig. S5 D), this inhibition is not observed and cofilin's severing activity is similar to that observed in the absence of cortactin (Fig. 6, C and D). This shows that the binding of cortactin to cofilin inhibits cofilin's severing activity and that cortactin phosphorylation functions as a switch to turn off this inhibitory effect.
We then tested whether cortactin phosphorylation facilitates the binding of cofilin to F-actin at invadopodium precursors using acceptor photobleaching FRET. The results show a significant increase in FRET between cofilin and β-actin at invadopodium precursors 1 min after EGF stimulation (Fig. 6, F and G)—the same time when cortactin is tyrosine phosphorylated and the barbed end peak begins.
The tyrosine phosphorylation of cortactin recruits Nck1 and N-WASp activity to invadopodium precursors leading to Arp2/3 complex–dependent actin polymerization
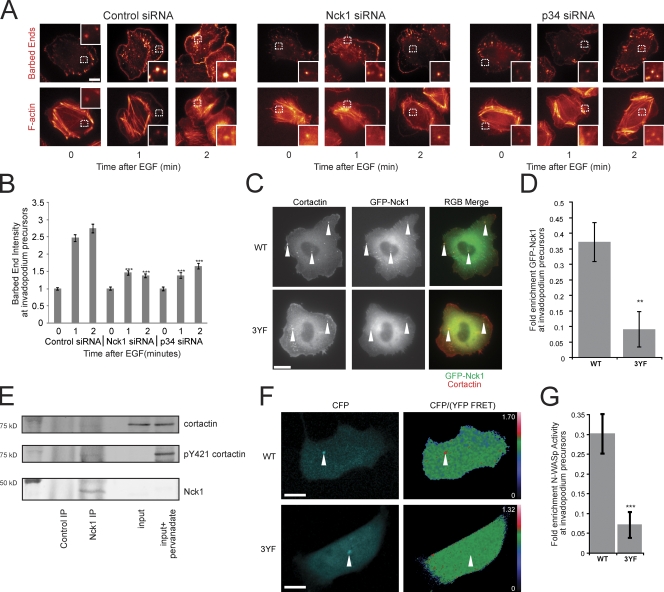
To determine whether the Arp2/3 complex contributes to the cofilin-generated barbed ends at invadopodium precursors (Fig. 4), cells were treated with p34 siRNA (Fig. S5 F), an Arp2/3 complex subunit, and the EGF-induced formation of barbed ends was analyzed. The results indicate that, in addition to cofilin, the Arp2/3 complex contributes to barbed end formation at invadopodium precursors (Fig. 7, A and B).
Figure 7.
The tyrosine phosphorylation of cortactin is important for Nck1 localization and N-WASp activity at invadopodium precursors for Arp2/3-dependent barbed end formation. (A) Representative images of EGF-induced barbed ends at invadopodium precursors in ctrl vs. Nck1 vs. p34 KD cells. Bar, 10 μm. (B) Quantification (normalized to 0 min) of barbed ends formed in response to EGF at invadopodium precursors in ctrl vs. Nck1 vs. p34 KD cells. P values are compared with ctrl siRNA at the same time point. n = number of invadopodium precursors from two independent experiments: ctrl 0 (264), 1 (287), 2 (275), Nck1 0 (223), 1 (356), 2 (375), p34 0 (130), 1 (201), 2 (259). (C) Representative images of GFP-Nck1 and cortactin in WT and 3YF cortactin cell lines (arrowheads show invadopodium precursors). Bar, 10 μm. (D) Quantification of fold enrichment of GFP-Nck1 at invadopodium precursors in WT and 3YF cell lines. n = number of invadopodium precursors: WT (29), 3YF (34). (E) Representative Western blot showing the coIP of endogenous cortactin and pY421-cortactin with endogenous Nck1. n = 3 independent experiments. (F) Representative images of N-WASp activity measured using the CFP/YFP FRET ratio in live cells expressing cortactin WT or 3YF (arrowheads show invadopodium precursors). Bar, 10 μm. (G) Quantification of the fold enrichment of N-WASp activity at invadopodium precursors in WT and 3YF cell lines. n = number of invadopodium precursors: WT (20), 3YF (25).
Because Nck1 and N-WASp are both important for invadopodium function in MTLn3 cells (Yamaguchi et al., 2005), we investigated whether invadopodium precursors use cortactin phosphorylation to activate a Nck1–N-WASp–Arp2/3 signaling complex in vivo (Tehrani et al., 2007). Similar to the cortactin 3YF phenotype, Nck1 KD cells (Fig. S5 E) had reduced barbed end formation at invadopodium precursors (Fig. 7, A and B). We then analyzed Nck1 localization in cells expressing cortactin WT or 3YF. Interestingly, cortactin 3YF cells had reduced enrichment of GFP-Nck1 at invadopodium precursors (Fig. 7, C and D), suggesting that cortactin phosphorylation is important for recruitment of Nck1 to invadopodium precursors. To further investigate the cortactin–Nck1 interaction, IP experiments were performed. Our results indicate that cortactin coIPs with Nck1 and the cortactin in the coIP is tyrosine phosphorylated (Fig. 7 E), suggesting that Nck1 interacts with a pool of tyrosine phosphorylated cortactin. In contrast, cortactin tyrosine phosphorylation was not detectable in the cortactin–cofilin IP complex (Fig. 5 D).
Activation of a Nck1–N-WASp–Arp2/3 signaling complex would require N-WASp activation at invadopodium precursors. Therefore, N-WASp activity was measured in cells expressing cortactin WT or 3YF using a CFP-YFP N-WASp intramolecular FRET biosensor (Lorenz et al., 2004b). In cells expressing WT cortactin, there was a significant increase in N-WASp activity at invadopodium precursors compared with the cytosol (Fig. 7, F and G). In contrast, cortactin 3YF cells had greatly reduced enrichment of N-WASp activity at invadopodium precursors (Fig. 7, F and G) and this was not due to decreased N-WASp localization (Fig. 8 B). Consistent with previously described in vitro findings (Tehrani et al., 2007), our results suggest that a phospho-cortactin–Nck1–N-WASp–Arp2/3 signaling complex contributes to the generation of barbed ends at invadopodia. These results support a synergy between the cofilin and Arp2/3-generated barbed ends at invadopodia as observed previously in lamellipodia (DesMarais et al., 2004).
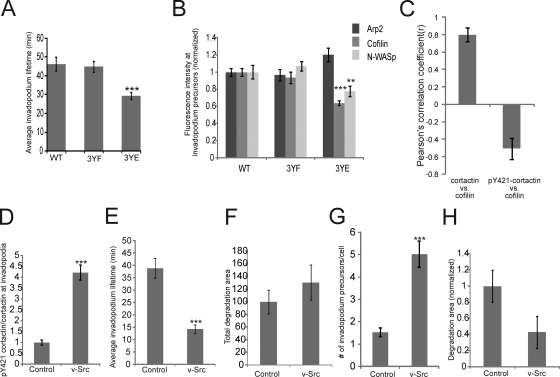
Figure 8.
The tyrosine dephosphorylation of cortactin is required for stable long-lived invadopodia. (A) Quantification of the average lifetimes of invadopodium precursors in cortactin WT, 3YF, and 3YE GFP-actin cell lines. n = number of invadopodium precursors: WT (133), 3YF (183), 3YE (152) from more than three independent experiments. (B) The fluorescence intensity of Arp2, cofilin, and N-WASp at invadopodium precursors in WT, 3YF, and 3YE cell lines. P values for A and B are compared with WT. n = number of invadopodium precursors: >30, three independent experiments. (C) Pearson's correlation analysis between cortactin and cofilin (r = 0.80) vs. pY421-cortactin/total cortactin and cofilin (r = −0.51) in invadopodium precursors. Error bars represent ± 95% confidence interval. (D) Quantification of the fluorescence intensity of pY421-cortactin/total cortactin at invadopodium precursors in ctrl vs. v-Src expressing cells. (E) Quantification of average invadopodium precursors lifetimes in GFP-actin cells expressing v-Src. (F) Quantification of the average FN degradation area/field, (G) number of invadopodium precursors/cell, and (H) FN degradation area normalized to the number of invadopodium precursors/cell in ctrl vs. v-Src expressing cells.
The tyrosine dephosphorylation of cortactin is important for stable long-lived invadopodia
Invadopodium stability is important for an invadopodium precursor to mature into an invadopodium capable of degrading ECM (Fig. 1). To determine whether cortactin phosphorylation regulates invadopodium precursor stability, time-lapse microscopy was performed to analyze invadopodium lifetimes using cortactin WT, 3YF, or 3YE cells expressing GFP-actin. Interestingly, cortactin 3YE cells formed invadopodia with shorter lifetimes than both WT and 3YF cells (Fig. 8 A; Videos 3, 4, and 5).
Previous studies have shown that cofilin is important for invadopodium stability (Yamaguchi et al., 2005). We hypothesized that invadopodia are short lived in the cortactin 3YE mutant due to a defect in cofilin recruitment. To test whether cortactin phosphorylation influences the enrichment of cofilin, N-WASp, and the Arp2/3 complex at invadopodium precursors, immunofluorescence experiments were performed. Significantly less cofilin and N-WASp localized to invadopodium precursors in cells expressing cortactin 3YE, but not 3YF (Fig. 8 B). To confirm this, the intensity of cofilin in invadopodium precursors was analyzed in cells co-stained for pY421-cortactin and cortactin. A Pearson's correlation analysis revealed a negative correlation between pY421-cortactin and cofilin (r = −0.51) compared with a positive correlation between total cortactin and cofilin (r = 0.80) (Fig. 8 C; Fig. S5 G). These results suggest that the tyrosine dephosphorylation of cortactin may be important for the recruitment of cofilin to invadopodium precursors for stabilization.
v-Src overexpression induces formation of short-lived invadopodium precursors each with less matrix degradation activity
To further investigate whether cortactin tyrosine dephosphorylation is important for invadopodium stability, we studied Src—a nonreceptor tyrosine kinase that phosphorylates cortactin. Overexpression of v-Src increased cortactin tyrosine phosphorylation at invadopodium precursors by fourfold (Fig. 8 D), and the invadopodium precursors formed had dramatically reduced lifetimes (Fig. 8 E; Video 6). Overall, v-Src overexpression increased matrix degradation by invadopodia (Fig. 8 F), as has been reported previously (Artym et al., 2006). However, when matrix degradation was normalized to the number of invadopodium precursors formed in ctrl vs. v-Src overexpressing cells (Fig. 8 G), on an individual basis, invadopodium precursors formed in v-Src–overexpressing cells exhibited less ECM degradation (Fig. 8 H). In summary, the cortactin 3YE and v-Src invadopodium lifetime data suggest that constitutive cortactin phosphorylation decreases invadopodium stability.
Discussion
As shown here and in previous work (Artym et al., 2006), invadopodia exhibit several discrete stages of maturation from assembly of a precursor to development into a degradation-competent invadopodium. Here, we have investigated the molecular mechanisms that regulate the stages of invadopodium maturation and have made several novel findings. We find that invadopodium precursors generate barbed ends for actin polymerization at a temporal stage preceding matrix degradation activity. Although cortactin tyrosine phosphorylation is not required for invadopodium precursor formation in mammary carcinoma cells, it is critical for the formation of barbed ends. A novel finding that explains these results is the observation that cofilin and cortactin bind to each other in vitro and interact at invadopodia in vivo. The interaction between cofilin and cortactin in invadopodia is disrupted when cortactin is tyrosine phosphorylated. In addition, using an in vitro severing assay, we found that cortactin can directly inhibit cofilin's severing activity, and when cortactin is phosphorylated, cofilin's severing activity is not inhibited. Finally, we found that the tyrosine dephosphorylation of cortactin is important for invadopodium stability and involves the rebinding of cofilin to cortactin. This allows the invadopodium lifetime to increase, thereby facilitating ECM degradation.
Cortactin functions as an essential scaffold to regulate the activities of cofilin and N-WASp
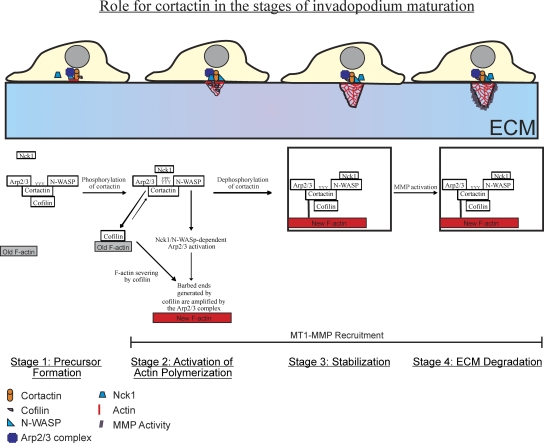
Overall, our study indicates that there are distinct stages of invadopodium maturation (Fig. 9, model). Initially, formation of an invadopodium precursor, consisting of cortactin, N-WASp, cofilin, and the Arp2/3 complex, requires the Arp2/3 and N-WASp binding domains of cortactin (stage 1: formation). This suggests that cortactin acts as a scaffold to bring together N-WASp and Arp2/3 to form a nucleus that becomes the invadopodium precursor. Numerous biochemical studies have demonstrated that cortactin binds to Arp2/3 directly via its NTA domain (Weed et al., 2000; Uruno et al., 2001) and N-WASp directly via its SH3 domain (Martinez-Quiles et al., 2004), and that both cortactin and N-WASp can simultaneously interact with the Arp2/3 complex (Weaver et al., 2002). Our in vivo results support these in vitro findings.
Figure 9.
Cortactin regulates the activities of cofilin and N-WASp to control the stages of invadopodium assembly and maturation (model). During precursor formation (stage 1), cortactin, N-WASp, cofilin, and Arp2/3 form a complex involving cortactin's Arp2/3 and N-WASp binding domains. Cortactin is then tyrosine phosphorylated, which activates cofilin's severing activity to generate free barbed ends and the Arp2/3 complex can use these cofilin-generated barbed ends for efficient actin polymerization (stage 2). Cortactin is then dephosphorylated, which stabilizes the invadopodium precursor for maturation (stage 3). Box at stage 3 and 4 indicates stabilization. MT1-MMP can be recruited at stages 2–4. Stages 1–3 are required for a precursor to become a mature invadopodium that efficienctly degrades ECM (stage 4).
Whether cortactin is the initial scaffold that forms the precursor or whether it is recruited by another protein remains an open question. In this study, we show that Tks5 also localizes with cortactin to invadopodium precursors during precursor formation, suggesting that Tks5 may be the scaffold that recruits cortactin. Tks5 is thought to be the master scaffold for podosome formation (Courtneidge et al., 2005; Oikawa et al., 2008) and our data suggest that invadopodia may use a similar mechanism for precursor formation.
The interaction between cofilin and cortactin regulates the maturation of invadopodia
After formation of invadopodium precursors, cortactin binds to cofilin, sequestering it and inhibiting its severing activity. However, when cortactin becomes tyrosine phosphorylated, release of cofilin from binding to cortactin occurs and cofilin severing activity generates amplification of free barbed ends for actin polymerization at invadopodia (stage 2: activation). Previous studies have shown that cofilin is dispensible for invadopodium precursor formation, but is required for degradation activity (Yamaguchi et al., 2005; Desmarais et al., 2009). Here, we show that cofilin is specifically required for actin polymerization in invadopodia—the stage after precursor formation during invadopodium maturation.
Cortactin function has been implicated in many other subcellular structures including lamellipodia (Bryce et al., 2005), endosomes (Zhu et al., 2005), cell–cell junctions (Helwani et al., 2004), synapses (Hering and Sheng, 2003), and podosomes (Zhou et al., 2006). One mechanism that could account for cortactin function in these subcellular structures is cortactin's direct activation of the Arp2/3 complex and stabilization of newly formed actin filaments (Weaver et al., 2001). Our results elucidate a previously unknown function for cortactin: potent amplification of barbed ends through its regulation of cofilin. Future work will be required to determine whether this function of cortactin makes invadopodia unique from these other subcellular structures.
Cortactin tyrosine phosphorylation also functions to localize Nck1 and N-WASp activity selectively at invadopodium precursors for Arp2/3-dependent barbed end formation. In vitro studies showed that cortactin tyrosine phosphorylation is required for maximal activation of barbed ends via a Nck1–N-WASp–Arp2/3 signaling complex, independent of cortactin's ability to bind directly to the Arp2/3 complex and N-WASp (Tehrani et al., 2007), and we show here that invadopodia use this signaling complex in vivo. We hypothesize that dendritic nucleation by the Arp2/3 complex amplifies the barbed ends generated by cofilin, and that cofilin and the Arp2/3 complex function synergistically to polymerize actin at invadopodia. The synergy between cofilin and the Arp2/3 complex has been previously shown in vitro (Ichetovkin et al., 2002) and in vivo (DesMarais et al., 2004). Future studies will help to better understand whether there are distinct pools of cortactin that selectively regulate cofilin vs. Arp2/3-dependent actin polymerization.
Here and previously (Yamaguchi et al., 2005), we showed that Nck1, but not Grb2, localizes to invadopodia and is important for invadopodium function. In contrast, a recent report (Oikawa et al., 2008) shows that Grb2, but not Nck1, localizes and is important for podosome function in Src-transformed fibroblasts, supporting the speculation that invadopodia and podosomes are structurally and functionally distinct (Linder, 2007). It would be interesting to determine whether the localization of Nck1 vs. Grb2 could be used as specific markers to distinguish invadopodia from podosomes in other cell types.
According to our model, after the barbed end peak and actin polymerization is generated by cofilin and the Arp2/3 complex, cortactin is dephosphorylated, which is required for the stabilization of invadopodium precursors (stage 3: stabilization). In turn, stabilization is required for an invadopodium precursor to mature into a matrix-degrading invadopodium (stage 4: ECM degradation). We hypothesize that cortactin dephosphorylation sequesters cofilin and prevents cofilin from excessively severing and depolymerizing actin, and this stabilizes invadopodium precursors. Which phosphatases are involved in the dephosphorylation of cortactin at this step is unknown. One candidate is the phosphatase PTP1B, which can bind directly to cortactin (Stuible et al., 2008) and is important for breast cancer cell invasion and invadopodium precursor formation (Cortesio et al., 2008). Future studies will be required to identify the mechanisms that coordinate the phosphorylation and dephosphorylation of cortactin at the tyrosine residues implicated in our study.
Materials and methods
Cell culture
For all experiments, MTLn3 cells, derived from the 13762NF rat mammary adenocarcinoma, were cultured in α-MEM supplemented with 5% FBS and antibiotics and starved and stimulated with EGF as described previously (Mouneimne et al., 2004). The MTLn3 cell line stably expressing GFP-actin at physiological levels was described previously (Lorenz et al., 2004a).
Invadopodium degradation assay and immunofluorescence
All experiments in this study were performed using MTLn3 cells cultured on FN/gelatin matrix unless indicated. FN/gelatin matrix was prepared as described previously (Chen, 1989). In brief, MatTek dishes were treated with 2.5% gelatin/2.5% sucrose, cross-linked with 0.5% glutaraldehyde, treated with 10 μg/ml of fluorescently labeled fibronectin (FN) (Alexa 568 [Invitrogen]) or unlabeled FN (Sigma-Aldrich), and then with 1 mg/ml NaBH4 in PBS. 100,000 MTLn3 cells were plated on FN/gelatin matrix 16 h before fixation. The cells were fixed and immunofluorescence was performed as described previously (Eddy et al., 2000). FN degradation was analyzed by quantifying the average area of degraded FN pixels per field. For live cell thin-matrix experiments, Oregon Green 488-gelatin (Invitrogen) was used and the thin-matrix coverslips were prepared as described previously (Artym et al., 2006). MTLn3 cells were stimulated with EGF and images were acquired every 1 min. Gelatin degradation was analyzed by measuring the change in 488-gelatin fluorescence over time in the cortactin-containing invadopodia region corrected for background.
Barbed end assay
The barbed end assay was performed using biotin-actin as described previously (Chan et al., 1998). In brief, cells were starved, stimulated with EGF, permeabilized with a permeabilization buffer (PB) (20 mM Hepes, pH 7.5, 138 mM KCl, 4 mM MgCl2, 3 mM EGTA, 0.2 mg/ml of saponin, 1 mM ATP, and 1% BSA) containing 0.4 μM biotin-actin (Cytoskeleton, Inc.) for 1 min at 37°C. The cells were fixed in 3.7% formaldehyde for 5 min, blocked in 1%FBS/1%BSA/PBS containing 3 μM phalloidin, stained with FITC anti-biotin to visualize barbed ends, and rhodamine-phalloidin and Arp2 to identify invadopodium precursors. Alternatively, cells plated on fluorescently labeled matrix were co-stained with Tks5 to identify actively degrading invadopodia. The barbed end intensity at invadopodium precursors was quantified by measuring the mean gray value (mgv) at invadopodium precursors − mgv of the background. The data were normalized to the control condition for each experiment.
FRET acceptor photobleaching experiments
The cells were starved and stimulated with EGF as described previously (Mouneimne et al., 2004) and immunofluorescence was performed using: (1) Cofilin 774, Alexa 488-goat anti–chicken (donor) and cortactin ab-33333, Alexa 555-goat anti–mouse (acceptor) for the cofilin-cortactin experiments; (2) Cofilin 774, Alexa 488-goat anti–chicken (donor) and β-actin AC15, Alexa 555-goat anti–mouse (acceptor) for the cofilin-actin experiments. Based on the fixation method (Eddy et al., 2000), F-actin, but not G-actin is fixed. For the cortactin-phosphotyrosine FRET experiments, MTLn3 cells transfected with TagRFP-cortactin WT or 3YF (donor), were fixed and stained with 4G10, cy5-goat anti–mouse (acceptor). For all fixed cell acceptor photobleaching FRET and immunofluorescence experiments, all samples were stored in PBS and imaged the following day after fixation. Cortactin, β-actin, or phosphotyrosine was bleached from invadopodium precursors and images before and after photobleaching were acquired. The images were analyzed and the FRET efficiency was calculated as E = 1 − (Donor pre/Donor post). All images were corrected for laser fluctuations, overall sample bleaching, and background. Controls are shown in Fig. S4. Included are controls for photoconversion, laser fluctuation, and nonspecific molecular interactions. To exclude fixation artifacts, the data were confirmed in live cells using sensitized emission FRET as described previously (van Rheenen et al., 2004). TagRFP and Cy5 FRET had components of photoactivation and photoconversion that were corrected for by subtracting the FRET observed in the donor and acceptor only control samples from each experimental sample. For microscopy setup including laser lines and filter settings, see the Microscopy section in Materials and methods.
RNAi
Control nonsilencing siRNA, cortactin siRNA (5′-CAAGCTTCGAGAGAATGTCTT-3′), p34 siRNA (5′-AAGGAACTTCAGGCACACGGA-3′), and Nck1 siRNA (5′-GATGATAGCTTTGTTGATCCA-3′) sequences were from QIAGEN and described previously (Kempiak et al., 2005). Control scrambled siRNA and cofilin siRNA (5′-AAGGTGTTCAATGACATGAAA-3′) sequences were described previously (Sidani et al., 2007). MTLn3 cells were transfected with 100 nM siRNA using Oligofectamine (Invitrogen) as described previously (Kempiak et al., 2005). For all experiments, cells were transfected with siRNA 48 h before each experiment.
Transfection
2 μg DNA was mixed with Lipofectamine and added to MTLn3 cells. Transfections were performed 24 h before each experiment.
Light microscopy severing assay
The ability of cofilin to sever F-actin was observed directly using a light microscopy severing assay as described previously (Ichetovkin et al., 2002). In brief, 500 nM cofilin and 500 nM cortactin or p-cortactin were mixed in perfusion buffer (PB) (50 mM Hepes, pH 6.8, 2 mM MgCl2, 5 mM EGTA, 50 mM KCl, 1 mM ATP, and 1 mM DTT) on ice for 1 h. Chambers with prebound actin filaments (unlabeled, Alexa 488, and biotin-labeled actin filaments: ratio of 68:22:10) were perfused with PB alone as a control, cofilin alone, or cofilin preincubated with cortactin or p-cortactin. The filaments were observed before (0 min) and after (4 min) addition of proteins and images were acquired. Cofilin's severing activity was scored as the percent increase in the number of actin filaments after the 4-min incubation ([number of filaments at 4 min/number of filaments at 0 min − 1] × 100) (Ichetovkin et al., 2002). In Fig. 6 E, actin filaments were incubated with the PB alone or the PB with 500 nM cortactin for 4 min. The filaments were then washed once with PB and cofilin was added. Images were taken before (0 min) and after addition of cofilin (1 min) and quantified as described above.
Immunoprecipitation and Western blotting
For all IP experiments, 106 MTLn3 cells were plated on plastic tissue culture dishes 16 h before the experiment. After EGF stimulation, the cells were washed with ice-cold PBS and lysed in lysis buffer containing 10 mM Hepes, 50 mM NaCl, 5 mM EGTA, 0.5 mM MgCl2, 1% TX-100, and phosphatase (NaF and NaVO4) and protease inhibitors. The lysates were centrifuged at 14,000 rpm for 10 min at 4°C and precleaned with chicken-IgY beads (Precipiphen) or protein A/G Agarose beads for 30 min. The IP was rotated overnight with 2 μg cofilin antibody or 2 μg control IgY antibody conjugated to chicken IgY beads for cofilin IPs, or 2 μg cortactin 4F11 antibody or 2 μg control mouse IgG1 antibody conjugated to protein A/G Agarose beads for cortactin IPs. For RFP IPs, 106 MTLn3 cells were transfected with mRFP cortactin WT or 3YF and 5 μg of RFP antibody or 5 μg control IgG1 antibody conjugated to protein A/G Agarose beads. For Nck1 IPs, 5 μg Nck1 antibody or 5 μg control IgG1 conjugated to protein A/G Agarose beads was used. The IPs were washed 3× in lysis buffer and analyzed by Western blot. For quantification, control antibody IP mgv or coIP mgv was subtracted from the IP mgv or coIP mgv. All values were corrected for area. The coIPs were normalized to the IP at each time point and then normalized to the 0-s EGF or WT sample.
For Western blot analysis, whole-cell lysates were prepared by washing 2× with cold PBS before direct extraction in SDS-PAGE sample buffer. Triton-insoluble cell lysates were prepared by resuspending the triton-insoluble pellet by sonication using the IP lysis buffer. Western blots were performed as follows: the samples were resolved by SDS-PAGE, transferred to nitrocellulose, blocked in odyssey blocking solution (LiCor), incubated with 1° antibodies, 2° antibodies (Mouse 680 and Rabbit 800 [Licor] and chicken 800 [Rockland]), and analyzed using the Odyssey (LiCor). For quantification, the mgv of each sample background corrected for area was calculated.
Protein purification
Recombinant cortactin protein was covalently coupled at a concentration of 1 mg/ml to Affigel 15 beads as per manufacturer's protocol. Cofilin protein was purified as described previously (Ghosh et al., 2004). Murine cortactin WT or 3YF was expressed and purified as described previously (Boyle et al., 2007). Recombinant phosphorylated cortactin was generated by coexpressing 6x His-tagged cortactin with untagged Arg kinase using baculoviral vectors in insect cells. Phosphorylated cortactin was eluted off a nickel column using 100 mM imidazole and its phosphorylation status verified via Western blotting with a cocktail of anti-phosphotyrosine antibodies (4G10, Y20, Y99).
Measurements of cortactin–cofilin binding
Cofilin protein was buffer exchanged into cofilin binding buffer (50 mM Hepes, pH 6.8, 20 mM NaCl, 0.01% NP-40, and 5% glycerol) using a G25 Sephadex column by gravity flow. 500 μl of a serial dilution of cofilin was incubated with 50 μl of cortactin beads or 50 μl of blank beads blocked with an excess of ethanolamine. The reaction was incubated at 4°C while rotating for 30 min. The supernatant was removed, beads washed, and bound material was recovered by boiling in Laemmli sample buffer and analyzed on 12% SDS-PAGE gels. Analysis and determination of Kd was described previously (Sirotkin et al., 2005). In brief, using Quantity One software (Bio-Rad Laboratories) and rectangular hyperbola binding curves were generated using GraphPad Prism software using the following equation: Y = (Bmax * X/X + Kd)+NS*X. In this equation, Y equals specific binding signal, X equals concentration of cofilin added to the cortactin beads, and NS equals the slope of the least-squares linear regression fit of the nonspecific cofilin binding as measured with the ethanolamine beads. The Kd is then solved as the value of X when Y equals 50% of Bmax. Cofilin concentrations of at least five times the Kd were used to ensure saturation of the curve.
Invadopodium precursor formation analysis
MTLn3 cell lines stably expressing cortactin mutants were treated with cortactin siRNA to deplete endogenous cortactin. The cells were fixed, immunofluorescence was performed, and invadopodium precursors were identified as colocalized Arp2 and F-actin (in cortactin cell line experiments) or cortactin and F-actin (in v-Src experiments) punctate structures. Invadopodium precursor formation was scored as the number of invadopodium precursors per cell and the percentage of cells that form invadopodium precursors.
Invadopodium lifetime analysis
Cortactin WT, 3YF, or 3YE MTLn3 cell lines stably expressing GFP-actin were treated with cortactin siRNA to knock down endogenous cortactin. The cells were placed in a 37°C heated chamber and images were collected using an autofocus system operated by IP-laboratory Spectrum software (VayTek). Eight random fields of GFP-actin cells were chosen and phase and fluorescence images were taken every 2 min for 16 h. During the 16 h, all MTLn3 cells formed invadopodia and 25% of cells formed invadopodia at a given time. Invadopodium lifetimes were analyzed by the appearance and disappearance of GFP-actin at invadopodia. Individual lifetimes were combined to calculate average lifetimes.
GFP-Nck1 and GFP-MT1-MMP localization analysis
For GFP-Nck1 experiments, MTLn3 cells expressing cortactin WT or 3YF, with endogenous cortactin knocked down, were transiently transfected with GFP-Nck1. The cells were fixed and stained with a cortactin antibody to visualize invadopodium precursors. Enrichment of GFP-Nck1 at invadopodium precursors in both WT and 3YF cortactin MTLn3 cell lines was calculated by measuring: (GFP-Nck1 mgv at invadopodium precursors-background/GFP-Nck1 mgv in the cytosol-background) − 1. For GFP-MT1-MMP live cell experiments, MTLn3 cells were transiently transfected with GFP-MT1-MMP and TagRFP-cortactin. Images were acquired every 20 s before and after EGF stimulation. Enrichment of GFP-MT1-MMP and TagRFP-cortactin at invadopodium precursors was calculated by measuring: (GFP-MT1-MMP or TagRFP-cortactin mgv at invadopodium precursors-background/mgv in the cytosol-background) − 1. The rate of fluorescence intensity increase was calculated by measuring the slope of: (GFP-MT1-MMP or TagRFP-cortactin mgv at invadopodium precursors-background/mgv in the cytosol-background) vs. time.
Ratio FRET N-WASp biosensor analysis
MTLn3 cells, expressing cortactin WT or 3YF, with endogenous cortactin knocked down, were transiently transfected with the CFP-YFP N-WASp biosensor (Lorenz et al., 2004b). The cells were imaged live at 37°C and CFP and YFP emission images were collected after excitation of CFP. N-WASp activity was calculated using the CFP/(YFP FRET) ratio. The fold enrichment of N-WASp activity at invadopodium precursors was calculated as: (N-WASp activity at invadopodium precursors/N-WASp activity in the cytosol) − 1.
Immunofluorescence quantitative analysis
The images from the fixed cell experiments were acquired using the same exposure time. Invadopodium precursors were identified as punctate structures containing F-actin and one invadopodium marker (either cortactin, cofilin, N-WASp, or Arp2). The average mgv background was calculated for each protein in the invadopodium precursor. Values were normalized to the average mgv of cortactin at invadopodium precursors for each cortactin mutant cell line and then normalized to the WT cortactin cell line. For the cortactin, pY421-cortactin, and cofilin Pearson's correlation analysis, the intensity (mgv-bk) of cortactin, pY421-cortactin, and cofilin in invadopodia was quantified. The pY421-cortactin/cortactin ratio was calculated to ensure that changes in pY421-cortactin were due to phosphorylation and not to changes in total cortactin. The Pearson's correlation analysis was performed as described previously (van Rheenen et al., 2007). The variation of the Pearson's correlation coefficient (r) was estimated by bootstrapping 2,000 samples from the datasets to obtain the mean correlation coefficient and the 95% confidence interval according to:
where and are mean values of x and y; Sx and Sy are the standard deviations in x and y.
Barbed end analysis using GFP-actin MTLn3 cells
Barbed ends were analyzed using a live cell approach with GFP-actin MTLn3 cells as described previously (Lorenz et al., 2004a). In brief, GFP-actin MTLn3 cells were starved and stimulated with EGF, and imaged live for 10 min acquiring phase and fluorescence images every 10 s. The fluorescence intensity of GFP-actin at invadopodium precursors was quantified by measuring the mgv of GFP-actin at invadopodium precursors − background for each frame of the time lapse. The first derivative of the GFP-actin fluorescence intensity was calculated and used as a measure of barbed ends in live cells (Lorenz et al., 2004a).
GFP-cofilin and TagRFP-cortactin sensitized emission FRET
MTLn3 cells transfected with GFP-cofilin (donor) and/or TagRFP-cortactin (acceptor) were imaged live at 37°C using the Deltavision Core Microscope (Applied Precision, LLC). Sensitized emission was calculated from images using a highly corrected algorithm as recently described (van Rheenen et al., 2004), in a custom-made Visual Basic program. In short, three images were collected; a donor (donor excitation, collection of donor fluorescence), FRET (donor excitation, collection of acceptor fluorescence), and an acceptor image (acceptor excitation, collection of acceptor fluorescence). Upon background subtraction, sensitized emission (sens) was calculated as follows: sens = (FRET – βDonor − (γ − αβ)Acceptor)/(1 − βδ), where α is the correction factor for acceptor fluorescence excitation at donor excitation wavelength and detected in the donor channel, β is the leak-through of donor in the acceptor channel, δ that for leak-through of acceptor back into the donor channel, and γ relates the FRET-independent acceptor fluorescence detected in the FRET channel to the acceptor fluorescence detected in the acceptor channel. The α, β, γ, and δ correction factors were determined on-line from cells expressing either donor or acceptor fluorescence proteins alone, present on the same coverslip. For each image, the correction factors were redetermined to correct for laser fluctuations.
Antibodies, constructs, and cell lines
Anti-cofilin (AE774) and Anti-N-WASp (AE922) were custom made and described previously (Yamaguchi et al., 2005). Anti-cortactin 4F11 for Western blots and IP experiments and anti-p34-arc/arpC2 (07–227) were from Millipore. Anti-cortactin (ab-33333) for immunofluorescence and anti-RFP (ab-62341) and anti-Nck1 (ab-14588) for IP's were from Abcam. The Arp2 (H-84), GAPDH (sc-47724), and Tks5 (sc-30122) antibodies for immunofluorescence were from Santa Cruz Biotechnology, Inc. Anti-biotin FITC for the barbed end assay was from Jackson ImmunoResearch Laboratories. Fibronectin, anti-β-actin antibody (AC15), and phospho-421 cortactin antibody were from Sigma-Aldrich. 4G10, phosphotyrosine antibody was from Millipore. Anti-Nck1 was from Cell Signaling Technology (15B9). The Alexa 2° antibodies were from Invitrogen and the cy5 2° antibody was from Millipore. The CFP-N-WASp-YFP FRET biosensor was described previously (Lorenz et al., 2004b). GFP-Nck1 is a gift from M. Way (Cancer Research UK, London, UK). GFP-MT1-MMP was described previously (Bravo-Cordero et al., 2007). v-Src expression construct is a gift from Dr. Tadashi Yamamoto (University of Tokyo, Tokyo, Japan). For the mRFP-cortactin constructs, cDNAs encoding the rat cortactin-A were amplified by RT-PCR from total RNA extracted from MTLn3 cells and then subcloned into mRFP vector. QuikChange Multi Site-directed mutagenesis kit (Stratagene) was used to generate cortactin mutants with point mutations. Standard PCR method was used to generate deletion mutants of cortactin. For generation of stable cell lines, cortactin cDNAs were inserted into pBabe puro vector and recombinant retroviruses were produced with Phoenix packaging cell line. The supernatant was used to infect MTLn3 cells or GFP-actin MTLn3 cells and the cell lines were selected with puromycin and maintained under constant selection. For TagRFP-cortactin constructs, murine cortactin WT or 3YF TagRFP fusion protein constructs were subcloned into the N1 mammalian expression vector (Clontech Laboratories, Inc.).
Microscopy
The fixed cell experiments were performed on a wide-field electronically motorized microscope (model IX81; Olympus) and images were acquired with a cooled CCD camera (model QE; Sensicam) using IP Laboratory 4.0. The GFP-actin barbed-end live cell, the GFP-actin overnight time lapses, and the cofilin severing experiments were performed on an inverted microscope (model IX70; Olympus) with a cooled CCD camera (model QE; Sensicam) using IP Laboratory 4.0. TagRFP-cortactin thin-matrix live cell experiments, GFP-MT1-MMP localization experiments, and GFP-cofilin/TagRFP-cortactin sensitized emission FRET experiments were performed using the Deltavision Core microscope (Applied Precision) with the GFP and mCherry live cell filter sets with a CCD coolsnap camera using softWoRx software. All Deltavision images were deconvulved using softWoRx software. The cofilin/cortactin, cofilin/β-actin, and GFP-cofilin/mRFP-cortactin acceptor photobleaching FRET experiments were performed on a laser scanning microscope (LSM 5 LIVE DuoScan; Carl Zeiss, Inc.) using a 561-nm laser to bleach and excite the acceptor and a 488-nm laser to excite the donor. The acceptor emission was collected using a 550–615-nm band pass filter and the donor emission was collected using a 495–525-nm band pass filter with a CCD camera using LSM 5 Live Duoscan software. The N-WASp biosensor sensitized emission FRET experiments were performed on a confocal microscope (model SP2; Leica). The cortactin-phosphotyrosine acceptor photobleaching FRET experiments were performed on a microscope (model SP5; Leica) using a 633-nm laser to bleach and excite the acceptor and a 543-nm laser to excite the donor. The emission detection window was set between 640–750 nm for the acceptor and 550–600 nm for the donor and images were acquired using LAS AF software (Leica). All imaging was done using 60X or 63X NA 1.4 oil objectives. All live cell experiments were imaged at 37°C in L15 media. All fixed cell experiments were imaged at 25°C in PBS.
Statistical analysis
Statistical significance was calculated using unpaired, two-tailed t test. Values were considered statistically significant if the P value was <0.05. For all figures, * indicates P value <0.05; ** indicates P value <0.01; and *** indicates P value <0.001. Error bars represent SEM.
Online supplemental material
Fig. S1 shows markers to identify invadopodium precursors and cortactin constructs used for this study. Fig. S2 shows that barbed ends at invadopodium precursors are identified by costaining with Arp2 and F-actin, and cortactin tyrosine phosphorylation is important for EGF-induced barbed end formation at invadopodium precursors. Fig. S3 shows that cortactin is tyrosine phosphorylated at invadopodium precursors beginning 1 min after EGF stimulation. Fig. S4 shows the controls for acceptor photobleaching FRET experiments and sensitized emission FRET between GFP-cofilin and TagRFP-cortactin. Fig. S5 shows cofilin, Nck1, and p34 siRNA Western blots, cofilin-cortactin IP data, and pY421-cortactin/cofilin Pearson's correlation analysis. Video 1 shows that invadopodium precursor formation is temporally distinct from matrix degradation. Video 2 shows that increased actin polymerization occurs as an early event during invadopodium maturation. Video 3 shows invadopodium lifetimes in MTLn3 cells expressing WT cortactin. Video 4 shows invadopodium lifetimes in MTLn3 cells expressing 3YF cortactin. Video 5 shows invadopodium lifetimes in MTLn3 cells expressing 3YE cortactin. Video 6 shows that dephosphorylation is required for invadopodium precursor stability. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200812176/DC1.
Acknowledgments
We thank Condeelis, Cox, and Segall laboratories for helpful discussions; the Analytical Imaging Facility of the Gruss Lipper Biophotonics Center for technical help; and Dr. Ved Sharma for help with FRET experiments.
This work was funded by NIHGM38511 (J. Condeelis), NIHNS39475, an award from the Elsa U. Pardee Foundation (A.J. Koleske), and NIHGM064346 (J. van Rheenen).
Footnotes
Abbreviations used in this paper:
- FN
- fibronectin
- IP
- immunoprecipitation
- KD
- knockdown
- mgv
- mean gray value
- MMP
- matrix metalloproteinase
- NTA domain
- N-terminal acidic domain
- WT
- wild type
References
- Artym V.V., Zhang Y., Seillier-Moiseiwitsch F., Yamada K.M., Mueller S.C. 2006. Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function.Cancer Res. 66:3034–3043 [DOI] [PubMed] [Google Scholar]
- Ayala I., Baldassarre M., Giacchetti G., Caldieri G., Tetè S., Luini A., Buccione R. 2008. Multiple regulatory inputs converge on cortactin to control invadopodia biogenesis and extracellular matrix degradation.J. Cell Sci. 121:369–378 [DOI] [PubMed] [Google Scholar]
- Boyle S.N., Michaud G.A., Schweitzer B., Predki P.F., Koleske A.J. 2007. A critical role for cortactin phosphorylation by Abl-family kinases in PDGF-induced dorsal-wave formation.Curr. Biol. 17:445–451 [DOI] [PubMed] [Google Scholar]
- Bravo-Cordero J.J., Marrero-Diaz R., Megías D., Genís L., García-Grande A., García M.A., Arroyo A.G., Montoya M.C. 2007. MT1-MMP proinvasive activity is regulated by a novel Rab8-dependent exocytic pathway.EMBO J. 26:1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryce N.S., Clark E.S., Leysath J.L., Currie J.D., Webb D.J., Weaver A.M. 2005. Cortactin promotes cell motility by enhancing lamellipodial persistence.Curr. Biol. 15:1276–1285 [DOI] [PubMed] [Google Scholar]
- Chan A.Y., Raft S., Bailly M., Wyckoff J.B., Segall J.E., Condeelis J.S. 1998. EGF stimulates an increase in actin nucleation and filament number at the leading edge of the lamellipod in mammary adenocarcinoma cells.J. Cell Sci. 111:199–211 [DOI] [PubMed] [Google Scholar]
- Chan K.T., Cortesio C.L., Huttenlocher A. 2009. FAK alters invadopodia and focal adhesion composition and dynamics to regulate breast cancer invasion.J. Cell Biol. 185:357–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.T. 1989. Proteolytic activity of specialized surface protrusions formed at rosette contact sites of transformed cells.J. Exp. Zool. 251:167–185 [DOI] [PubMed] [Google Scholar]
- Clark E.S., Whigham A.S., Yarbrough W.G., Weaver A.M. 2007. Cortactin is an essential regulator of matrix metalloproteinase secretion and extracellular matrix degradation in invadopodia.Cancer Res. 67:4227–4235 [DOI] [PubMed] [Google Scholar]
- Condeelis J. 2001. How is actin polymerization nucleated in vivo? Trends Cell Biol. 11:288–293 [DOI] [PubMed] [Google Scholar]
- Cortesio C.L., Chan K.T., Perrin B.J., Burton N.O., Zhang S., Zhang Z.Y., Huttenlocher A. 2008. Calpain 2 and PTP1B function in a novel pathway with Src to regulate invadopodia dynamics and breast cancer cell invasion.J. Cell Biol. 180:957–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtneidge S.A., Azucena E.F., Pass I., Seals D.F., Tesfay L. 2005. The SRC substrate Tks5, podosomes (invadopodia), and cancer cell invasion.Cold Spring Harb. Symp. Quant. Biol. 70:167–171 [DOI] [PubMed] [Google Scholar]
- Crowley J.L., Smith T.C., Fang Z., Takizawa N., Luna E.J. 2009. Supervillin reorganizes the actin cytoskeleton and increases invadopodial efficiency.Mol. Biol. Cell. 20:948–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesMarais V., Macaluso F., Condeelis J., Bailly M. 2004. Synergistic interaction between the Arp2/3 complex and cofilin drives stimulated lamellipod extension.J. Cell Sci. 117:3499–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmarais V., Yamaguchi H., Oser M., Soon L., Mouneimne G., Sarmiento C., Eddy R., Condeelis J. 2009. N-WASP and cortactin are involved in invadopodium-dependent chemotaxis to EGF in breast tumor cells.Cell Motil. Cytoskeleton. 66:303–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy R.J., Pierini L.M., Matsumura F., Maxfield F.R. 2000. Ca2+-dependent myosin II activation is required for uropod retraction during neutrophil migration.J. Cell Sci. 113:1287–1298 [DOI] [PubMed] [Google Scholar]
- Ghosh M., Song X., Mouneimne G., Sidani M., Lawrence D.S., Condeelis J.S. 2004. Cofilin promotes actin polymerization and defines the direction of cell motility.Science. 304:743–746 [DOI] [PubMed] [Google Scholar]
- Gimona M., Buccione R. 2006. Adhesions that mediate invasion.Int. J. Biochem. Cell Biol. 38:1875–1892 [DOI] [PubMed] [Google Scholar]
- Gimona M., Buccione R., Courtneidge S.A., Linder S. 2008. Assembly and biological role of podosomes and invadopodia.Curr. Opin. Cell Biol. 20:235–241 [DOI] [PubMed] [Google Scholar]
- Head J.A., Jiang D., Li M., Zorn L.J., Schaefer E.M., Parsons J.T., Weed S.A. 2003. Cortactin tyrosine phosphorylation requires Rac1 activity and association with the cortical actin cytoskeleton.Mol. Biol. Cell. 14:3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helwani F.M., Kovacs E.M., Paterson A.D., Verma S., Ali R.G., Fanning A.S., Weed S.A., Yap A.S. 2004. Cortactin is necessary for E-cadherin-mediated contact formation and actin reorganization.J. Cell Biol. 164:899–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hering H., Sheng M. 2003. Activity-dependent redistribution and essential role of cortactin in dendritic spine morphogenesis.J. Neurosci. 23:11759–11769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichetovkin I., Grant W., Condeelis J. 2002. Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex.Curr. Biol. 12:79–84 [DOI] [PubMed] [Google Scholar]
- Kempiak S.J., Yamaguchi H., Sarmiento C., Sidani M., Ghosh M., Eddy R.J., Desmarais V., Way M., Condeelis J., Segall J.E. 2005. A neural Wiskott-Aldrich Syndrome protein-mediated pathway for localized activation of actin polymerization that is regulated by cortactin.J. Biol. Chem. 280:5836–5842 [DOI] [PubMed] [Google Scholar]
- Linder S. 2007. The matrix corroded: podosomes and invadopodia in extracellular matrix degradation.Trends Cell Biol. 17:107–117 [DOI] [PubMed] [Google Scholar]
- Lorenz M., DesMarais V., Macaluso F., Singer R.H., Condeelis J. 2004a. Measurement of barbed ends, actin polymerization, and motility in live carcinoma cells after growth factor stimulation.Cell Motil. Cytoskeleton. 57:207–217 [DOI] [PubMed] [Google Scholar]
- Lorenz M., Yamaguchi H., Wang Y., Singer R.H., Condeelis J. 2004b. Imaging sites of N-wasp activity in lamellipodia and invadopodia of carcinoma cells.Curr. Biol. 14:697–703 [DOI] [PubMed] [Google Scholar]
- Martinez-Quiles N., Ho H.Y., Kirschner M.W., Ramesh N., Geha R.S. 2004. Erk/Src phosphorylation of cortactin acts as a switch on-switch off mechanism that controls its ability to activate N-WASP.Mol. Cell. Biol. 24:5269–5280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouneimne G., Soon L., DesMarais V., Sidani M., Song X., Yip S.C., Ghosh M., Eddy R., Backer J.M., Condeelis J. 2004. Phospholipase C and cofilin are required for carcinoma cell directionality in response to EGF stimulation.J. Cell Biol. 166:697–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa T., Itoh T., Takenawa T. 2008. Sequential signals toward podosome formation in NIH-src cells.J. Cell Biol. 182:157–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippar U., Roussos E.T., Oser M., Yamaguchi H., Kim H.D., Giampieri S., Wang Y., Goswami S., Wyckoff J.B., Lauffenburger D.A., et al. 2008. A Mena invasion isoform potentiates EGF-induced carcinoma cell invasion and metastasis.Dev. Cell. 15:813–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmiento C., Wang W., Dovas A., Yamaguchi H., Sidani M., El-Sibai M., Desmarais V., Holman H.A., Kitchen S., Backer J.M., et al. 2008. WASP family members and formin proteins coordinate regulation of cell protrusions in carcinoma cells.J. Cell Biol. 180:1245–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seals D.F., Azucena E.F., Jr., Pass I., Tesfay L., Gordon R., Woodrow M., Resau J.H., Courtneidge S.A. 2005. The adaptor protein Tks5/Fish is required for podosome formation and function, and for the protease-driven invasion of cancer cells.Cancer Cell. 7:155–165 [DOI] [PubMed] [Google Scholar]
- Sidani M., Wessels D., Mouneimne G., Ghosh M., Goswami S., Sarmiento C., Wang W., Kuhl S., El-Sibai M., Backer J.M., et al. 2007. Cofilin determines the migration behavior and turning frequency of metastatic cancer cells.J. Cell Biol. 179:777–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirotkin V., Beltzner C.C., Marchand J.B., Pollard T.D. 2005. Interactions of WASp, myosin-I, and verprolin with Arp2/3 complex during actin patch assembly in fission yeast.J. Cell Biol. 170:637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuible M., Dubé N., Tremblay M.L. 2008. PTP1B regulates cortactin tyrosine phosphorylation by targeting Tyr446.J. Biol. Chem. 283:15740–15746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehrani S., Tomasevic N., Weed S., Sakowicz R., Cooper J.A. 2007. Src phosphorylation of cortactin enhances actin assembly.Proc. Natl. Acad. Sci. USA. 104:11933–11938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uruno T., Liu J., Zhang P., Fan Yx, Egile C., Li R., Mueller S.C., Zhan X. 2001. Activation of Arp2/3 complex-mediated actin polymerization by cortactin.Nat. Cell Biol. 3:259–266 [DOI] [PubMed] [Google Scholar]
- van Rheenen J., Langeslag M., Jalink K. 2004. Correcting confocal acquisition to optimize imaging of fluorescence resonance energy transfer by sensitized emission.Biophys. J. 86:2517–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rheenen J., Song X., van Roosmalen W., Cammer M., Chen X., Desmarais V., Yip S.C., Backer J.M., Eddy R.J., Condeelis J.S. 2007. EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells.J. Cell Biol. 179:1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver A.M., Karginov A.V., Kinley A.W., Weed S.A., Li Y., Parsons J.T., Cooper J.A. 2001. Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation.Curr. Biol. 11:370–374 [DOI] [PubMed] [Google Scholar]
- Weaver A.M., Heuser J.E., Karginov A.V., Lee W.L., Parsons J.T., Cooper J.A. 2002. Interaction of cortactin and N-WASp with Arp2/3 complex.Curr. Biol. 12:1270–1278 [DOI] [PubMed] [Google Scholar]
- Weed S.A., Karginov A.V., Schafer D.A., Weaver A.M., Kinley A.W., Cooper J.A., Parsons J.T. 2000. Cortactin localization to sites of actin assembly in lamellipodia requires interactions with F-actin and the Arp2/3 complex.J. Cell Biol. 151:29–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wouters F.S., Bastiaens P.I. 1999. Fluorescence lifetime imaging of receptor tyrosine kinase activity in cells.Curr. Biol. 9:1127–1130 [DOI] [PubMed] [Google Scholar]
- Wyckoff J.B., Pinner S.E., Gschmeissner S., Condeelis J.S., Sahai E. 2006. ROCK- and myosin-dependent matrix deformation enables protease-independent tumor-cell invasion in vivo.Curr. Biol. 16:1515–1523 [DOI] [PubMed] [Google Scholar]
- Yamaguchi H., Condeelis J. 2007. Regulation of the actin cytoskeleton in cancer cell migration and invasion.Biochim. Biophys. Acta. 1773:642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H., Lorenz M., Kempiak S., Sarmiento C., Coniglio S., Symons M., Segall J., Eddy R., Miki H., Takenawa T., Condeelis J. 2005. Molecular mechanisms of invadopodium formation: the role of the N-WASP-Arp2/3 complex pathway and cofilin.J. Cell Biol. 168:441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Webb B.A., Eves R., Mak A.S. 2006. Effects of tyrosine phosphorylation of cortactin on podosome formation in A7r5 vascular smooth muscle cells.Am. J. Physiol. Cell Physiol. 290:C463–C471 [DOI] [PubMed] [Google Scholar]
- Zhu J., Zhou K., Hao J.J., Liu J., Smith N., Zhan X. 2005. Regulation of cortactin/dynamin interaction by actin polymerization during the fission of clathrin-coated pits.J. Cell Sci. 118:807–817 [DOI] [PubMed] [Google Scholar]