Abstract
Microtubule (MT)-destabilizing kinesin 13s perform fundamental roles throughout the cell cycle. In this study, we show that the Drosophila melanogaster kinesin 13, KLP10A, is phosphorylated in vivo at a conserved serine (S573) positioned within the α-helix 5 of the motor domain. In vitro, a phosphomimic KLP10A S573E mutant displays a reduced capacity to depolymerize MTs but normal affinity for the MT lattice. In cells, replacement of endogenous KLP10A with KLP10A S573E dampens MT plus end dynamics throughout the cell cycle, whereas a nonphosphorylatable S573A mutant apparently enhances activity during mitosis. Electron microscopy suggests that KLP10A S573 phosphorylation alters its association with the MT lattice, whereas molecular dynamics simulations reveal how KLP10A phosphorylation can alter the kinesin–MT interface without changing important structural features within the motor’s core. Finally, we identify casein kinase 1α as a possible candidate for KLP10A phosphorylation. We propose a model in which phosphorylation of the KLP10A motor domain provides a regulatory switch controlling the time and place of MT depolymerization.
Introduction
Kinesins are defined by their “motor” domains, which are catalytically active domains of ∼350 amino acids that display ATPase- and microtubule (MT)-binding activities (Vale and Fletterick, 1997). The motor domains of most kinesins convert the energy of ATP hydrolysis into movement along the MT lattice. However, some kinesins, particularly the kinesin 13s, do not actively translocate along MTs but instead bind to and destabilize their ends (Desai et al., 1999).
Kinesin 13–catalyzed MT depolymerization contributes to cellular processes ranging from mitosis to axon branching (Wordeman, 2005). Individual kinesin 13s are also reutilized for different tasks throughout the cell cycle, a characteristic exemplified by the Drosophila melanogaster kinesin 13, KLP10A, which regulates spindle assembly and poleward flux during mitosis and interactions between MTs and the cell cortex during interphase (Rogers et al., 2004; Mennella et al., 2005). It is now clear that the localization and catalytic activities of at least some kinesin 13s are subject to tight spatial and temporal control via phosphorylation (Andrews et al., 2004; Lan et al., 2004). Somewhat surprisingly, phosphorylation within the motor domain of kinesin 13s, or any other kinesin, has not been previously reported.
This study focuses on the phospho-regulation of KLP10A, which is a topic of interest given that kinesin 13 associates with polymerizing MT plus ends but stimulates their catastrophe only near the cortex (Mennella et al., 2005; Moore et al., 2005). This suggests that KLP10A associates with MTs in an inactive state and becomes activated only at specific cellular sites. The vertebrate kinesin 13, mitotic centromere-associated kinesin, behaves similarly (Moore et al., 2005). In this study, we describe the identification and characterization of a novel phosphorylation site within the conserved α-helix 5 of the KLP10A motor domain. In addition, we identify casein kinase 1α (CK1α) as a potential kinase for KLP10A serine 573 (S573) phosphorylation and link this kinase to KLP10A’s activity in vivo.
Results and discussion
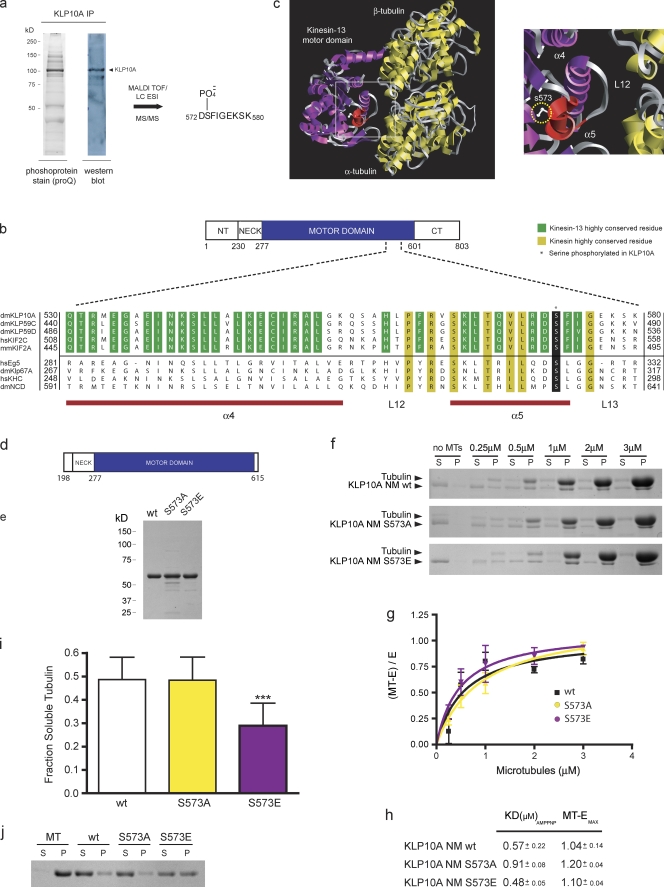
To identify sites of phosphorylation within KLP10A, endogenous KLP10A was immunoprecipitated from Drosophila S2 cell lysates and stained with the fluorescent dye ProQ Diamond to selectively recognize phosphoproteins. ProQ stained a prominent band of ∼100 kD, which was confirmed to be KLP10A by Western blotting and matrix-assisted laser desorption/ionization (MALDI) time of flight (TOF) mass spectrometry (MS; Fig. 1 a). The gel band corresponding to phosphorylated KLP10A was trypsin digested and analyzed by electrospray ionization MS. This identified a phosphopeptide corresponding to KLP10A sequence 572–579 (DS-PO4−-FIGEKSK), which was confirmed by MS/MS sequencing (Fig. 1 a and Fig. S1 a). The S573 phosphorylated in KLP10A is located within the conserved α-helix 5 of the core kinesin motor domain (Fig. 1 b) and near the kinesin–MT interface according to cryo-EM–based models (Tan et al., 2008). Mutagenesis experiments have indicated that α-helix 5 is important for MT binding in conventional kinesin (Fig. 1 c; Woehlke et al., 1997). Interestingly, an alignment of the corresponding region of other kinesins revealed a serine at the analogous position in all three Drosophila kinesin 13s as well the vertebrate kinesin 13s, Kif2C and Kif2A, and some members of other kinesin subfamilies (Fig. 1 b).
Figure 1.
Identification of a novel phosphorylation site within the KLP10A motor domain. (a) Anti-KLP10A immunoprecipitate from S2 cell lysate separated by SDS-PAGE and stained with ProQ or transferred to nitrocellulose for Western blotting with anti-KLP10A antibodies. (b) Partial sequence alignment of the identified region from the motor domains of different kinesin family members. (c) The cryo-EM map of the Kif1A head–MT complex structure (Protein Data Bank accession no. 2hxh; Kikkawa and Hirokawa, 2006) was used as a model for kinesin motor domain–α-β–tubulin interaction. The motor domain of Kif1A was replaced with the crystal structure of the kinesin 13 Kif2C (Protein Data Bank accession no. 1v8j; Ogawa et al., 2004) using homology-modeling features of Chimera software (University of California, San Francisco, San Francisco, CA). A magnified view of the boxed region is show on the right. (d) Linear map of KLP10A neck/motor construct used for biochemical studies. (e) Coomassie-stained SDS-PAGE gel of purified, bacterially expressed KLP10A proteins used in our assays. (f) Representative KLP10A/MT cosedimentation assay. MT depolymerization assay. n = 18 (wt), 17 (S573A), and 19 (S573E). S, supernatant; P, pellet. MT concentrations are indicated in micromolars. (g) Mean measurements from three independent experiments were fit to rectangular hyperbolas. MT-E, MT-bound KLP10A; E, soluble KLP10A. (h) Apparent dissociation constants of KLP10A constructs for MTs (mean ± SEM). (i and j) Measurement of depolymerase activities and representative MT sedimentation assay, respectively. ***, P < 0.001. Error bars indicate SD.
The discovery of a phosphorylated serine in the minimal motor domain of a kinesin 13 is significant. Although phosphorylation of numerous kinesins from a variety of families has been reported, all previously identified phosphorylation sites lie outside of the core kinesin motor domain. Zen-4, a kinesin 6 in Caenorhabditis elegans, is phosphorylated within a class-specific N-terminal extension to the motor domain, and this regulates its affinity for MTs (Mishima et al., 2004).
Phosphomimic KLP10A mutants display reduced MT depolymerase activity
The position of S573 suggests a mechanism of regulation based on a change of KLP10A’s interaction with MTs. To evaluate this possibility, the enzymatic properties of purified neck/motor wild-type (wt), phosphomimic S573E, or nonphosphomimic S573A KLP10A constructs were studied (Fig. 1, d and e). Somewhat surprisingly, MT cosedimentation assays indicated that all three constructs bound to MTs with a similar affinity (Fig. 1, f and g). However, KLP10A S573E displayed a significant reduction (∼40%) in its ability to destabilize MTs compared with KLP10A wt and the S573A mutant (Fig. 1, h–j). In a similar vein, myosin I is also phosphorylated in its motor domain on a residue positioned near the actin-binding site, and this phosphorylation decreases motor activity without influencing binding to the actin filament (Ostap et al., 2002).
KLP10A S573E/A mutants alter MT dynamics in cells
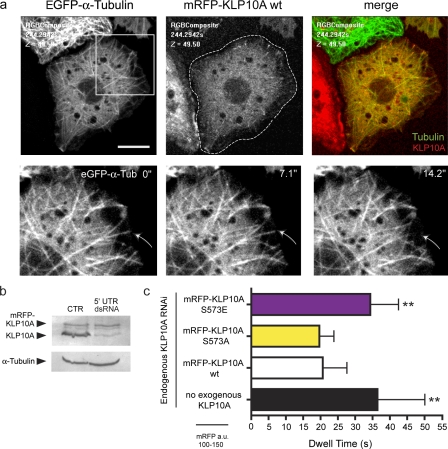
To determine whether and how S573 phosphorylation alters KLP10A’s activity in cells, we analyzed MT dynamics in cell lines in which the endogenous protein was replaced by monomeric RFP (mRFP)–KLP10A wt, S573E, or S573A mutants. We generated three cell lines stably transfected with (a) a constitutively expressed EGFP–α-tubulin construct, allowing visualization of MTs, and (b) a construct encoding mRFP-KLP10A wt, S573E, or S573A protein whose expression was driven from an inducible promoter. Endogenous KLP10A was first depleted by RNAi using a double-stranded RNA (dsRNA) oligomer against the 5′ untranslated region (UTR) of the KLP10A mRNA (Goshima and Vale, 2005). As the transgenic mRNA does not contain UTR sequences, mRFP-KLP10A levels remained unaltered after RNAi treatment (Fig. 2 b). To ensure the analysis of cells expressing similar titers of mRFP-KLP10A (wt or S573A/E mutants), the level of mRFP fluorescence was measured for each cell under identical imaging conditions (Fig. S1 b).
Figure 2.
Expression of the phosphomimetic KLP10A S573E mutant dampens MT dynamics during interphase S2 cells. (a, top) Interphase cell expressing EGFP–α-tubulin and mRFP-KLP10A wt. (bottom) Magnified images (of the boxed region) from a time-lapse video were used to analyze MT dwell times. Numbers are shown in elapsed time (seconds). Arrows indicate a catastrophe. (b) Western blot of S2 cell lysates treated with control (CTR) dsRNA or KLP10A 5′ UTR dsRNA. (c) Mean dwell times of MT ends in the vicinity of the cell cortex. Cells expressing comparable levels of mRFP-KLP10A were selected for analysis. S573E, n = 200 (10); S573A, n = 200 (11); wt, n = 201 (10); no KLP10A, n = 199 (10); with numbers in parentheses indicating the number of cells analyzed. **, P < 0.01. Error bars indicate SD. Bar, 10 µm.
As in our earlier work, depletion of KLP10A significantly increased the dwell time of MT plus ends positioned at the cortex of interphase cells. Replacement with mRFP-KLP10A wt or S573A restored this aspect of MT dynamics to normal, suggesting that either protein can functionally mimic endogenous KLP10A (Fig. 2 c). Both proteins also tip track on polymerizing plus ends (Fig. 2 a, Fig. S2 a, and Video 1).
In contrast, cells expressing S573E displayed dampened MT plus end dynamics with a 60% increase in cortical dwell time relative to the wt or S573A proteins (Fig. 2 c). This effect was both qualitatively and quantitatively indistinguishable from the knockdown condition (with no exogenous KLP10A). S573E also failed to tip track on plus ends and instead labeled longer stretches of the MT (Fig. S2 b). Altered tip-tracking behavior has also been reported for a phosphomimetic mitotic centromere-associated kinesin mutant displaying decreased depolymerase activity in vitro (Moore et al., 2005). These data suggest that phosphorylation of KLP10A at S573 can largely inactivate its ability to promote MT plus end depolymerization in cells perhaps by altering its association with MT tips. However, additional inhibitory mechanisms are likely to exist, at least during interphase.
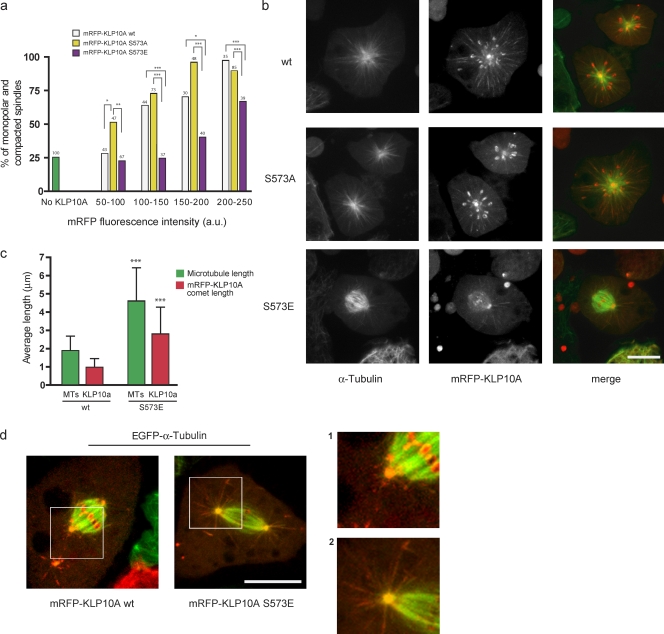
We also compared the impacts of KLP10A wt and S573E/A expression on mitosis. KLP10A is believed to depolymerize pole-focused MT minus ends to constrain mitotic spindle length and promote poleward tubulin flux (Rogers et al., 2004). KLP10A also tracks the polymerizing plus ends of astral MTs and may regulate their dynamic behaviors (Goshima and Vale, 2005). The KLP10A wt protein localized normally, concentrating on centrosomes, spindle poles, centromeres, and the tips of astral MTs (Fig. 3, Fig. S2, and Video 2), and had little impact on spindle morphology when present at low concentrations. However, increasing expression levels induced a corresponding elevation in the frequency of monopolar or collapsed spindles, which is an expected result given KLP10A’s proposed role in the regulation of spindle length. By comparison, defects in spindle morphology were significantly reduced in cells expressing S573E and exacerbated in cells with S573A (Fig. 3, a and b). With regard to the latter, even very low expression levels strongly altered spindle morphology, suggesting that nonphosphorylatable S573A is not subject to normal regulatory control in mitotic cells.
Figure 3.
KLP10A S573 phosphomimetic and nonphosphorylatable mutants alter MT dynamics in mitosis. (a) Percentage of KLP10A wt–, S573A–, or S573E–expressing mitotic cells with collapsed or monopolar spindles. Control cells were not transfected with any KLP10A construct (green bar). Cells expressing similar levels of the different transgenic KLP10A constructs are grouped together for comparative analysis. Numbers at tops of bars indicate n. Fig. S1 b shows the distributions of mRFP expression levels in the analyzed cells. (b) Representative confocal images of mitotic figures observed in the presence of similar amounts of mRFP-KLP10A wt, S573A, and S573E. (c) Mean astral MT lengths and mRFP-KLP10A comet lengths in mitotic cells expressing mRFP-KLP10A wt (n = 9 for 40.17 MTs) or S573E (n = 9 for 88.71 comets). (d) Images showing astral MTs of spindles from cells expressing either the wt or S573E protein. Magnified views of the boxed regions are shown on the right. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars indicate SD. Bars, 5 µm.
We examined the impacts of KLP10A wt and S573E on finer aspects of spindle dynamics and most notably astral MTs (analyses could not be extended to S573A because of its particularly strong impact on spindle architecture). As with the wt protein, S573E associated with centrosomes, spindle poles, and centromeres (Fig. S2 d). However, S573E was quite different in its effects on and localization to astral MTs. Expression of S573E significantly increased the number and length of astral MTs (Fig. 3 c), and these lengthy astral MTs were extensively labeled by S573E in a fashion quite similar to interphase (Fig. 3, c and d; Fig. S2; and Video 2). This was distinguishable from the short comets of mRFP-KLP10A wt typically observed at MT plus ends (Fig. 3, c and d; Mennella et al., 2005; Moore et al., 2005).
Finally, we found that mRFP-KLP10A wt and S573E had distinct impacts on poleward tubulin flux. For a variety of reasons, these experiments were difficult to perform using our loss of function/rescue approach, so, in this case, they were performed in cells also containing endogenous KLP10A. Flux velocities were positively correlated with the level of expression of KLP10A wt protein, whereas the S573E protein significantly reduced flux rates when present at higher concentrations (Fig. S3, a and b). Interpreting these data are problematic because analyzed cells also contain endogenous KLP10A but are generally consistent with the notion that phosphorylation of KLP10A at S573 impacts the cellular functions of the protein and could regulate KLP10A’s activity at MT minus and plus ends.
KLP10A S573E fails to form MT-associated rings
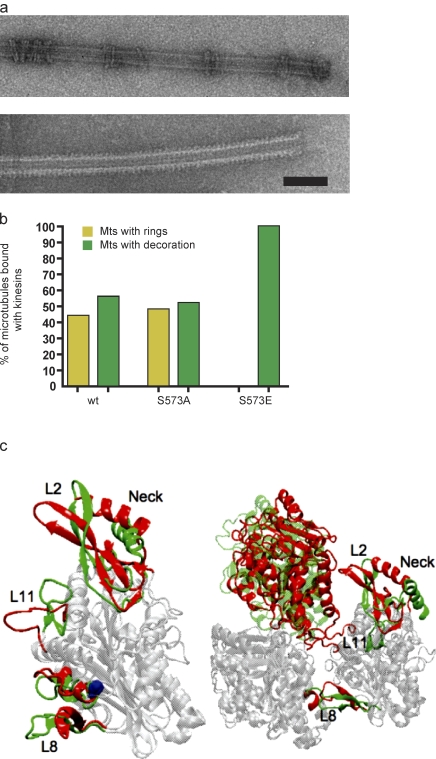
To determine whether phosphorylation alters the structural relationship between KLP10A and the MT, we used EM to visualize MTs incubated with KLP10A wt or S573E/A neck/motor constructs. To enhance binding, experiments were performed in the presence of the nonhydrolyzable ATP analogue AMPPNP. Under these conditions, KLP10A, along with other kinesin 13s, has been found to assemble into rings or spirals around MTs; no other kinesins have been observed to form similar structures (Moores et al., 2006; Tan et al., 2006). Although the relevance of these rings remains unclear, they may represent a class-specific intermediate of KLP10A’s enzymatic activity.
Although both KLP10A wt and S573A neck/motor constructs could form rings or uniformly decorate MTs and did so with roughly equal probability (Fig. 4, a and b), rings were never observed on MTs incubated with S573E, which always uniformly decorated the MT lattice (Fig. 4 b). Thus, although KLP10A S573 phosphorylation does not appear to significantly alter the motor’s affinity for the MT lattice, it does alter some aspect of the kinesin–MT interaction specific to this kinesin subfamily.
Figure 4.
Phosphorylation of KLP10A at S573 alters its interactions with the MT lattice. (a) Electron micrographs of MTs incubated with either KLP10A S573A (top) or S573E (bottom) neck/motor constructs. (b) Three independent protein preparations for each construct were analyzed, and only MTs with KLP10A bound were included in the measurements. n = 352 (wt), 359 (S573A), and 393 (S573E). (c, left) Overlay of the structures of an unphospho- and phospho-KLP10A motor domain obtained from molecular dynamics simulations by minimizing the root mean square difference between the two structures. Regions of the molecule that remain unchanged are shown in light gray, whereas those that undergo significant conformational changes are shown in color (unphospho, red; phospho, green). The phosphorylation site is shown in blue. (right) Overlay of the structures of a tubulin heterodimer in complex with the unphospho- or phospho-KLP10A motor domain. We note that the core regions of the two KLP10A do not overlay very well because of the difference in their relative positions to β-tubulin. Bar, 125 nm.
S573 phosphorylation may alter the KLP10A–MT interface
To identify potential molecular mechanisms underlying the changes in KLP10A activity brought on by S573 phosphorylation, we used molecular dynamics simulations to compare the structures of a phospho- versus unphospho-KLP10A motor domain. Our findings suggest that phosphorylation induces significant conformational changes in the three portions of the molecule that directly interface with tubulin dimers (L2, L11, and L8), although leaving the structural core relatively unchanged (Fig. 4 C and Video 3). The most prominent phosphorylation-induced effects are described in the following paragraphs.
L2. Upon S573 phosphorylation, loop L2 and the two β sheets separated by it, β1b and β1c, bend away from the tubulin dimer. Because L2 is the so-called KVD (lysine-valine-aspartic acid) loop, a class-specific domain required for normal MT depolymerization (Ovechkina et al., 2002), its reduced interaction with tubulin could also help explain the observed decrease in the depolymerization activity of the S573E protein. Meanwhile, the neck helix undergoes a significant rotation, causing the neck to fold into the back of the molecule, closer to the kinesin 13 class-specific tubulin-binding site involved in the formation of MT-associated rings (Tan et al., 2008). This repositioning of the neck may underlie the observed inability of KLP10A S573E to form MT-associated rings. Consistent with this possibility, we found that removal of the neck restored robust ring assembly in S573E constructs (Fig. S3 c).
L11. Although L11 is a coiled loop protruding toward the tubulin dimer in unphospho-KLP10A, it forms two short β sheets and bends away from tubulin in the phosphorylated protein. We noted that L11 was missing in the original crystal structure as a result of disorder; thus, its coordinates were constructed from ideal internal coordinates for the residues comprising it.
L8. The two β sheets connected by loops L8, β5a, and β5b undergo partial unfolding upon S573 phosphorylation, causing L8 to become more extended and stick out further toward the tubulin dimer. The partial unfolding of the short helix (α5) containing S573 and its movements toward L8 might be responsible for the observed conformational changes in L8.
KLP10A is regulated by CK1α
Finally, to identify the kinase-phosphorylating KLP10A S573 in vivo, we used a bioinformatic approach to search for consensus sequences. The primary sequence surrounding S573 in KLP10A did not resemble any known kinase consensus sequence. However, Netphos (Blom et al., 2004), a program that recognizes nonconventional consensus sequences by an artificial neural network, predicted KLP10A S573 as a substrate for CK1.
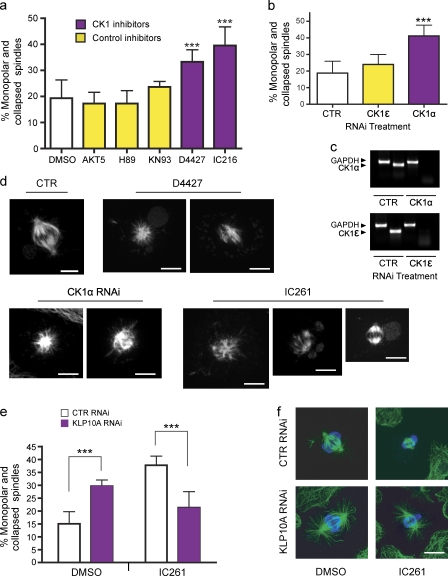
If CK1 phosphorylates S573 and thereby inhibits KLP10A in vivo, inactivation of CK1 should phenocopy the effects of KLP10A overexpression. To test this prediction, we added a series of cell-permeable kinase inhibitors to S2 cells and subsequently observed the impacts on spindle morphology (Fig. 5). After treatment with the CK1 inhibitors IC261 and D4427, the percentage of monopolar and collapsed spindles significantly increased similarly to KLP10A overexpression (Fig. 5 a, P < 0.001; and Video 4). To ensure that this effect was specific to CK1 inhibition, we repeated this assay using inhibitors that target kinases that may be affected by high concentrations of IC261 and D4427 (Table S1).
Figure 5.
Regulation of KLP10A by CK1α. (a) The concentrations of kinase inhibitors applied to medium were chosen according to their reported Ki values (Table S1). DMSO, n = 6 (600); AKT5, n = 4 (400); H89, n = 4 (400); KN93, n = 3 (300); D4427, n = 4 (400); and IC216, n = 4 (400); with numbers in parentheses indicating the total number of cells analyzed. (b) RNAi treatment was performed to deplete the CK1α and -ϵ isoforms in S2 cells. Only CK1α RNAi significantly increased the percentage of monopolar spindles. Control (CTR), n = 4 (400); CK1ϵ, n = 6 (600); and CK1α, n = 8 (800). (c) Semiquantitative PCR showing the efficacy of RNAi. The housekeeping gene, GAPDH, was used as a control. (d) Gallery of mitotic phenotypes obtained after inhibition of CK1. (e) Depletion of KLP10A by RNAi rescues the increase in monopolar/collapsed spindles caused by CK1 inhibition. After RNAi, S2 cells were incubated in the presence of 8 µM IC261 for 3 h and processed for immunostaining with antitubulin and antiphospho-histone antibodies. DMSO control RNAi, n = 2 (400); DMSO 10A RNAi, n = 3 (600); IC261 control RNAi, n = 2 (400); and IC261 10A RNAi, n = 3 (600). (f) Representative images of mitotic spindles obtained from the rescue experiment shown in e. Green, antitubulin; blue, antiphospho-histone. ***, P < 0.001. Error bars indicate SD. Bars, 5 µm.
IC261 and D4427 are specific for only two of the eight CK1 isoforms in Drosophila (Table S1). To further distinguish the specific CK1 isoform responsible for the increase in monopolar spindles, we performed RNAi against each of the two candidates, ε and α. When S2 cells were depleted of CK1α, but not CK1ϵ, we also observed a dramatic increase in monopolar and compacted spindles (Fig. 5 b, P < 0.001; see Materials and methods).
To establish whether this spindle collapse phenotype was mediated by an elevated activation of KLP10A, CK1 was also inhibited in S2 cells depleted of KLP10A. Spindles of control RNAi–treated cells were frequently collapsed in the presence of the CK1 inhibitor IC261, whereas in S2 cells depleted of KLP10A, normal spindle bipolar architecture was maintained in the presence of IC261 (Fig. 5, e and f). Thus, KLP10A depletion rescues the spindle collapse caused by CK1 inhibition. Spindle collapse induced by the MT-destabilizing drug colchicine was not rescued by KLP10A RNAi (unpublished data).
The CK1 inhibitor IC261 also had a dramatic effect on interphase MTs (Fig. S3 d), causing the plus ends of interphase MTs to rapidly shrink toward the cell interior, eventually resulting in the almost complete disappearance of MTs (Video 4). This effect was dependent on KLP10A activity, but not stathmin, another MT-destabilizing protein (Fig. S3 d). We note that CK1α is predicted to phosphorylate two additional sites within KLP10A (S699 and S697), which may explain why CK1 inhibitors produce a much stronger impact on interphase MT arrays than the expression of KLP10A S573A (which should presumably induce the same effect). These data are consistent with the hypothesis that CK1 acts upstream of KLP10A, inhibiting its activity by phosphorylation.
In summary, our findings provide new insights into the manner in which phosphorylation regulates the structure and function of kinesin 13s. The rapid and transient activation of the kinesin 13 motor domain provides an appealing mechanism for controlling the timing and place of efficient kinesin 13–driven MT depolymerization. We propose that the majority of cytoplasmic KLP10A is phosphorylated at S573 by CK1α (and perhaps at other sites by additional kinases) and thus held in reserve in a low activity state. Local dephosphorylation of the protein (e.g., at the cell cortex) activates a small pool of the protein to induce the depolymerization of a subset of MT ends. KLP10A, like other kinesin 13s, is likely phosphorylated at multiple sites, and its specific mechanism of regulation involves the integration of these events.
Materials and methods
Cell culture and RNAi
Schneider S2 cells were cultured in Schneider’s Drosophila medium supplemented with 10% heat-inactivated FCS (Invitrogen) and penicillin/streptomycin. Stably transfected cells were depleted of endogenous KLP10A by RNAi using a dsRNA against the 5′ UTR of endogenous KLP10A mRNA for 5 d (40 µg dsRNA applied on days 0, 2, and 4; Goshima and Vale, 2005; Mennella et al., 2005) and prepared for imaging (see Cell lines, imaging, and measurements). CK1 inhibition by RNAi was performed similarly except that 40 µg dsRNA was applied on days 0, 2, 4, and 6 of a 7-d treatment regimen. For in vitro transcription of the dsRNA oligonucleotides targeting the CK1α (CG2028) and -ε (CG2048) genes, DNA templates were amplified from S2 cell cDNA using gene-specific primers. The following primer sequences were used in this study: CK1α (forward), 5′-ATGTAAATGACGTTGATGTGGG-3′; CK1α (reverse), 5′-TTATTGAGTTTCGCTCG-3′; CK1ε (forward), 5′-ATTTTGTTGCGTAAACAAAGGG-3′; and CK1ε (reverse), 5′-ATTTTGTTGCGTAAACAAAGGG-3′. DsRNA was generated with commercial transcription kits (Applied Biosystems; Promega) following the manufacturers’ instructions.
Immunoprecipitation and phosphoprotein staining
Affinity-purified KLP10A antibodies (Rogers et al., 2004) were preincubated with protein G beads (GE Healthcare) for 1 h at 4°C and subsequently mixed with S2 cell lysate (5 mg/ml in 50 mM Tris HCl, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, and 1 mM DTT) in the presence of protease inhibitor cocktail (Roche) and phosphatase inhibitors mix I and II (Sigma-Aldrich) for 4 h at 4°C. After extensive washing, bead-bound proteins were resolved on a 7.5% SDS-PAGE gel. Subsequently, gels were stained with ProQ Diamond (Invitrogen) to detect phosphorylated proteins or transferred to nitrocellulose for Western blotting with 2 mg/ml anti-KLP10A antibody (1:1,000). The band corresponding to KLP10A was excised for MS.
MS
MS was performed at the MS Facility at the Rockefeller University (New York, NY) as follows: the gel bands corresponding to the phospho and unphospho proteins were excised from the gel, reduced with 10 mM DTT, alkylated with 55 mM iodoacetamide, and digested with sequence grade modified trypsin (Promega) in ammonium bicarbonate buffer at 37°C overnight. The digestion products were analyzed by MALDI-TOF to identify peptides that were modified with a MALDI-TOF mass spectrometer (DE-STR; Applied Biosystems). Half of the digestion products for each sample were also analyzed by liquid chromatography (LC)–MS/MS. For LC-MS/MS analysis, each digestion product was separated by gradient elution with a capillary/nano-HPLC system (Dionex) and analyzed by a mass spectrometer (QSTAR XL; Applied Biosystems) using information-dependent, automated acquisition.
Cell lines, imaging, and measurements
Analysis of MT dynamics in transfected S2 cells.
Three different stably transfected S2 cell lines were generated. Each S2 cell line was transfected with a plasmid encoding EGFP–α-tubulin in the pAc5.1/V5-HisB vector (Invitrogen) together with one other plasmid encoding wt, S573A, or S573E mRFP-KLP10A in the pMT/V5-HisC (Invitrogen). Stably transfected cells were depleted of endogenous KLP10A by RNAi using a dsRNA against the 5′ UTR of endogenous KLP10A mRNA for 5 d (40 µg dsRNA applied on days 0, 2, and 4). Before microscopy, cells were plated on concanavalin A–coated (to promote cell spreading) glass-bottom culture dishes and treated with 300 µM CuS04 to induce the expression of exogenous mRFP-KLP10A. Images and time-lapse videos were acquired at 20°C with a spinning-disk system (UltraView RS3; PerkinElmer) mounted on an inverted microscope (TE-200S; Nikon) using a Plan Apo 100× 1.4 NA objective (imaging was performed in the presence of Schneider’s medium). Images were captured with a cooled charge-coupled device camera (OrcaER; Hamamatsu Photonics) controlled by the UltraView acquisition software. Exposure time, image gain, and laser intensity were kept constant to allow the mRFP-KLP10A expression levels to be measured and compared between experiments and cell lines. Four 0.5-µm z sections were obtained for 4D data collection (x, y, z, and time). Images were acquired at 2.68-s intervals for ∼300 s. MT ends were tracked using ImageJ software (National Institutes of Health) and analyzed for the time spent pausing while close to the cell cortex (dwell time). Images of mitotic spindles were acquired by spinning-disk confocal microscopy (UltraView) in the presence of endogenous KLP10A.
Poleward flux measurements.
Double transgenic S2 cells constitutively expressing EGFP–α-tubulin were induced to express exogenous mRFP-KLP10A with the addition of 300 µM CuS04 to the S2 cell medium (Invitrogen) and simultaneously plated on concanavalin A–coated dishes. Narrow rectangular bars were photobleached across the fluorescent mitotic spindles of preanaphase cells using a confocal system (TCS SP2; Leica) on an inverted microscope with a Plan Apo 63× 1.4 NA objective (DMIRE2; Leica). Time-lapse videos of the photobleached spindles were captured at 1.8–3.6-s frame intervals. The movement of the bleach mark through the spindle was measured with the MetaMorph calipers tool (MDS Analytical Technologies), and flux rate was calculated from the change of distance between bleach mark and spindle pole as a function of time. Before photobleaching, single images of each cell were captured (with constant exposure time, gain, laser intensity, etc.) to allow measurement of the fluorescence intensities of the expressed exogenous mRFP-KLP10A. Cells were categorized by their mean expressed mRFP-KLP10A intensity values, and cells with similar levels of mRFP-KLP10A could then be compared between experiments and between different cell lines.
MT cosedimentation
Assays were performed as described previously (Hertzer et al., 2006) using 1 µM protein of purified, recombinant KLP10A neck/motor (T198-I615) of wt, S573A, or S573E. The relative quantities of KLP10A in the supernatant and pellet fractions were determined by densitometry using ImageJ. To determine the apparent Kd of each construct for MTs, the data from at least three independent experiments were fit to a hyperbola equation MT-E = (Bmax × MT)/(Kd + MT) with Prism software (GraphPad Software, Inc.), where MT-E is the fraction of KLP10A bound to MTs, and MT is the total concentration of MTs.
MT depolymerization
1.0 µM GMPCPP MTs was incubated with a fixed concentration (50 nM) of a KLP10A neck/motor recombinant protein in BRB80 buffer containing 2 mM MgATP (Sigma-Aldrich) and 2 mM MgCl2 for 20 min at 25°C and centrifuged at 239,000 g for 5 min at 25°C. Each pellet was resuspended in a volume of BRB80 equal to the supernatant. Samples were resolved by SDS-PAGE and stained with Coomassie blue. The percentage of unpolymerized tubulin was analyzed by densitometry using ImageJ. The quantity of soluble tubulin in the sample supernatants without KLP10A was subtracted from the total amount of unpolymerized tubulin in the experimental samples to obtain a measure of the MT depolymerization resulting only from KLP10A activity. Each KLP10A construct was analyzed using two independent preparations of the protein. The means of several independent reactions from separate experiments were analyzed statistically.
EM
Purified recombinant proteins (KLP10A neck/motor of wt, S573A, or S573E mutants) were incubated in BRB80 with MTs in the presence of 1 mM AMPPNP (Sigma-Aldrich) at room temperature for 20 min. MTs and attached motor proteins were centrifuged at 239,000 g for 15 min at 25°C, and the pellet was used to generate negatively stained grids. Randomly selected pictures of EM micrographs were examined and scored for the presence of KLP10A rings on MTs and for regular lattice decoration. For quantitation, we considered only MTs that showed kinesin binding from three independent protein preparations for each construct.
Molecular dynamics simulations
Protein molecules (both the kinesin 13 monomers and the kinesin–tubulin complexes) are simulated in aqueous solution with the CHARMM polar hydrogen (c19) topology and parameter sets (Brooks et al., 1983) together with the ACE2 implicit solvent model (Schaefer et al., 1998, 2001) to reduce the size of the system so that simulations can be carried out for longer time and gain better convergence. The starting structure for the KLP10A monomer is the crystal structure of a human KIF2C (Protein Data Bank accession no. 2HEH) molecule because no crystal structure for KLP10A is available. As a result of disorder, coordinates of several residues, 360–370, 457–465, 498–502 (loop L11), and 541–555, were missing in the original crystal structure. Therefore, these coordinates were constructed from idealized internal coordinate values in simulations. The initial structure for the mutant KLP10A monomer was obtained by substituting S551, which structurally corresponds to S573 in Drosophila KLP10A, with its phosphorylated version in the crystal structure. The initial structures were subjected first to energy minimizations and then to molecular dynamics simulations using the Langevin dynamics algorithm until the total potential energy of the system become equilibrated, which took ∼5 ns in the wt system (because of L11) and 13 ns in the phospho mutant. Then, both systems were simulated for an additional few nanoseconds to ensure that the total potential energies did not undergo any systematic changes. The structure of the wt KLP10A monomer remains the same as the original crystal structure except for residues whose coordinates were generated in the simulation, whereas the mutant KLP10A monomer undergoes significant conformational changes in several distinct parts of the molecule.
To simulate the kinesin–tubulin complex, we first dock the structures of the wt and mutant KLP10A monomer obtained from previous simulations into the crystal structure of the complex (Protein Data Bank accession no. 3EDL) by minimizing the root mean square difference between our KLP10A molecule and the kinesin molecule in the crystal structure. Coordinates for residues missing in the original crystal structure of the tubulin heterodimer (Protein Data Bank accession no. 1SA0; α-tubulin, 37–46 and 279–284; β-tubulin, 276–283) were constructed from idealized internal coordinates. The resulting structures were subjected to first energy minimization and then molecular dynamics simulations as described for the monomers.
Casein kinase inhibition in S2 cells
CK1 inhibition by RNAi was performed as described in Cell culture and RNAi. For chemical inhibition of CK1, all drugs were diluted into an equal final volume of DMSO and were added to S2 cells that had been plated on concanavalin A dishes for 2 h before treatment. The concentration of kinase inhibitors (EMD) was selected based on their Ki values (Table S1). S2 cells were fixed in 100% methanol (−20°C) and stained with anti–α-tubulin antibody (1:200; Sigma-Aldrich) and antiphospho–histone H3 antibody (1:500; Millipore). Fluorescent secondary antibodies (Cy2-conjugated anti–mouse and rhodamine Red-X–conjugated anti–rabbit; Jackson ImmunoResearch Laboratories).
Statistics
Means of different experimental treatments were compared by one-way analysis of variance and the Bonferroni posttest. To compare frequencies of phenotypes (Fig. 3 a), the statistical test was performed using contingency tables and the Fisher exact test using Prism software. In figures, statistical significance is indicated with asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001), and error bars indicate SD.
Online supplemental material
Fig. S1 shows MS/MS sequencing spectrum of KLP10A peptide 572–579 and a distribution histogram of the fluorescence intensities of cells expressing the mRFP fusion KLP10A constructs. Fig. S2 shows images of live S2 cells expressing EGFP-KLP10A wt and S573E protein. Fig. S3 shows poleward flux rates in cells expressing KLP10A wt and S573E, an electron micrograph showing that removal of the neck restores the ability of KLP10A S573E to form rings, and the effects of CK1 inhibition by IC261. Video 1 shows a live S2 cell stably transfected with EGFP-tubulin and mRFP-KLP10A wt in interphase. Video 2 shows the same but in a mitotic cell. Video 3 is a molecular dynamics simulation of the effect of S573 phosphorylation on the structure of the motor/neck domain. Video 4 shows the effects of CK1 inhibition on MTs in live S2 cells. Table S1 outlines characteristics of the kinase inhibitors used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200902113/DC1.
Acknowledgments
We thank Laura B. Granell-Ortiz for her assistance with the molecular dynamics simulations.
This work was supported by the National Institutes of Health (grant R01 GM65940) to D.J. Sharp. D.J. Sharp is a Scholar of the Leukemia and Lymphoma Society.
Footnotes
Abbreviations used in this paper:
- CK1
- casein kinase 1
- dsRNA
- double-stranded RNA
- LC
- liquid chromatography
- MALDI
- matrix-assisted laser desorption/ionization
- mRFP
- monomeric RFP
- MS
- mass spectrometry
- MT
- microtubule
- TOF
- time of flight
- UTR
- untranslated region
- wt
- wild type
References
- Andrews P.D., Ovechkina Y., Morrice N., Wagenbach M., Duncan K., Wordeman L., Swedlow J.R. 2004. Aurora B regulates MCAK at the mitotic centromere.Dev. Cell. 6:253–268 [DOI] [PubMed] [Google Scholar]
- Blom N., Sicheritz-Pontén T., Gupta R., Gammeltoft S., Brunak S. 2004. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence.Proteomics. 4:1633–1649 [DOI] [PubMed] [Google Scholar]
- Brooks B.R., Bruccoleri R.E., Olafson B.D., States D.J., Swaminathan S., Karplus M. 1983. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations.J. Comput. Chem. 4:187–217 [Google Scholar]
- Desai A., Verma S., Mitchison T.J., Walczak C.E. 1999. Kin I kinesins are microtubule-destabilizing enzymes.Cell. 96:69–78 [DOI] [PubMed] [Google Scholar]
- Goshima G., Vale R.D. 2005. Cell cycle-dependent dynamics and regulation of mitotic kinesins in Drosophila S2 cells.Mol. Biol. Cell. 16:3896–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzer K.M., Ems-McClung S.C., Kline-Smith S.L., Lipkin T.G., Gilbert S.P., Walczak C.E. 2006. Full-length dimeric MCAK is a more efficient microtubule depolymerase than minimal domain monomeric MCAK.Mol. Biol. Cell. 17:700–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa M., Hirokawa N. 2006. High-resolution cryo-EM maps show the nucleotide binding pocket of KIF1A in open and closed conformations.EMBO J. 25:4187–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W., Zhang X., Kline-Smith S.L., Rosasco S.E., Barrett-Wilt G.A., Shabanowitz J., Hunt D.F., Walczak C.E., Stukenberg P.T. 2004. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity.Curr. Biol. 14:273–286 [DOI] [PubMed] [Google Scholar]
- Mennella V., Rogers G.C., Rogers S.L., Buster D.W., Vale R.D., Sharp D.J. 2005. Functionally distinct kinesin-13 family members cooperate to regulate microtubule dynamics during interphase.Nat. Cell Biol. 7:235–245 [DOI] [PubMed] [Google Scholar]
- Mishima M., Pavicic V., Grüneberg U., Nigg E.A., Glotzer M. 2004. Cell cycle regulation of central spindle assembly.Nature. 430:908–913 [DOI] [PubMed] [Google Scholar]
- Moore A.T., Rankin K.E., von Dassow G., Peris L., Wagenbach M., Ovechkina Y., Andrieux A., Job D., Wordeman L. 2005. MCAK associates with the tips of polymerizing microtubules.J. Cell Biol. 169:391–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moores C.A., Cooper J., Wagenbach M., Ovechkina Y., Wordeman L., Milligan R.A. 2006. The role of the kinesin-13 neck in microtubule depolymerization.Cell Cycle. 5:1812–1815 [DOI] [PubMed] [Google Scholar]
- Ogawa T., Nitta R., Okada Y., Hirokawa N. 2004. A common mechanism for microtubule destabilizers-M type kinesins stabilize curling of the protofilament using the class-specific neck and loops.Cell. 116:591–602 [DOI] [PubMed] [Google Scholar]
- Ostap E.M., Lin T., Rosenfeld S.S., Tang N. 2002. Mechanism of regulation of Acanthamoeba myosin-IC by heavy-chain phosphorylation.Biochemistry. 41:12450–12456 [DOI] [PubMed] [Google Scholar]
- Ovechkina Y., Wagenbach M., Wordeman L. 2002. K-loop insertion restores microtubule depolymerizing activity of a “neckless” MCAK mutant.J. Cell Biol. 159:557–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers G.C., Rogers S.L., Schwimmer T.A., Ems-McClung S.C., Walczak C.E., Vale R.D., Scholey J.M., Sharp D.J. 2004. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase.Nature. 427:364–370 [DOI] [PubMed] [Google Scholar]
- Schaefer M., Bartels C., Karplus M. 1998. Solution conformations and thermodynamics of structured peptides: molecular dynamics simulation with an implicit solvation model.J. Mol. Biol. 284:835–848 [DOI] [PubMed] [Google Scholar]
- Schaefer M., Bartels C., Leclerc F., Karplus M. 2001. Effective atom volumes for implicit solvent models: comparison between Voronoi volumes and minimum fluctuation volumes.J. Comput. Chem. 22:1857–1879 [DOI] [PubMed] [Google Scholar]
- Tan D., Asenjo A.B., Mennella V., Sharp D.J., Sosa H. 2006. Kinesin-13s form rings around microtubules.J. Cell Biol. 175:25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D., Rice W.J., Sosa H. 2008. Structure of the kinesin13-microtubule ring complex.Structure. 16:1732–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale R.D., Fletterick R.J. 1997. The design plan of kinesin motors.Annu. Rev. Cell Dev. Biol. 13:745–777 [DOI] [PubMed] [Google Scholar]
- Woehlke G., Ruby A.K., Hart C.L., Ly B., Hom-Booher N., Vale R.D. 1997. Microtubule interaction site of the kinesin motor.Cell. 90:207–216 [DOI] [PubMed] [Google Scholar]
- Wordeman L. 2005. Microtubule-depolymerizing kinesins.Curr. Opin. Cell Biol. 17:82–88 [DOI] [PubMed] [Google Scholar]