Abstract
Eukaryotic cells possess a sophisticated membrane system to facilitate diverse functions. Whereas much is known about the nature of membrane systems in interphase, the organization and function of the mitotic membrane system are less well understood. In this study, we show that epsin, an endocytic adapter protein, regulates mitotic membrane morphology and spindle integrity in HeLa cells. Using epsin that harbors point mutations in the epsin NH2-terminal homology domain and spindle assembly assays in Xenopus laevis egg extracts, we show that epsin-induced membrane curvature is required for proper spindle morphogenesis, independent of its function in endocytosis during interphase. Although several other membrane-interacting proteins, including clathrin, AP2, autosomal recessive hypercholesterolemia, and GRASP65, are implicated in the regulation of mitosis, whether they participate through regulation of membrane organization is unclear. Our study of epsin provides evidence that mitotic membrane organization influences spindle integrity.
Introduction
Much progress has been made in understanding how the microtubule (MT)-based mitotic spindle captures and segregates chromosomes into daughter cells. However, it is not known whether membrane systems play a role in facilitating spindle organization in mitosis. In interphase, membrane organization and trafficking are regulated not only by membrane deformation, fusion, fission, and tethering systems but also by the MT cytoskeleton (McMahon and Gallop, 2005; Itoh and De Camilli, 2006). Interphase membranes also regulate MT organization. For example, the interphase trans-Golgi network functions as an MT-organizing center essential for both cell polarity and migration (Chabin-Brion et al., 2001; Efimov et al., 2007; Liu et al., 2007). Such an intimate relationship between MTs and membranes in interphase suggests that the two structures could regulate each other in mitosis to facilitate proper cell division.
Consistent with this idea, the ER and Golgi undergo dramatic reorganization during mitosis, with membrane tubules and vesicles being observed both within and surrounding mitotic spindles, suggesting a function for the spindle in the partitioning of mitotic membranes (Waterman-Storer et al., 1993; Axelsson and Warren, 2004; Altan-Bonnet et al., 2006; Lowe and Barr, 2007; Wei and Seemann, 2009). An increasing number of membrane-associated proteins have been shown to regulate different aspects of mitosis (Cao et al., 2003; Cao and Zheng, 2004; Royle et al., 2005; Sütterlin et al., 2005; Vong et al., 2005; Boucrot and Kirchhausen, 2007; Lehtonen et al., 2008). Moreover, a membranous mitotic spindle matrix has been isolated from Xenopus laevis egg extracts, which can stimulate MT assembly from pure tubulin in vitro (Tsai et al., 2006; Zheng and Tsai, 2006). These data suggest that proper organization of mitotic membranes could influence spindle assembly.
Endocytic proteins such as clathrin, AP2, and autosomal recessive hypercholesterolemia (ARH) induce membrane curvature either directly or indirectly by recruiting other proteins to drive endocytic vesicle formation. During mitosis, they localize to different places, with clathrin concentrating on the spindle, ARH localizing to centrosomes, and AP2 distributing throughout the cell (Royle et al., 2005; Boucrot and Kirchhausen, 2007; Lehtonen et al., 2008). Whereas clathrin appears to regulate kinetochore MT bundling independent of its role in membrane deformation and endocytosis, how AP2 and ARH regulate mitosis remains unclear.
In this study, we focus on epsin, an endocytic adapter protein which directly binds and deforms membranes through its epsin NH2-terminal homology (ENTH) domain (Ford et al., 2002). We provide evidence that the membrane deformation function of epsin affects spindle organization during mitosis.
Results and discussion
Reduction of epsin in human cells causes defects in spindle morphology
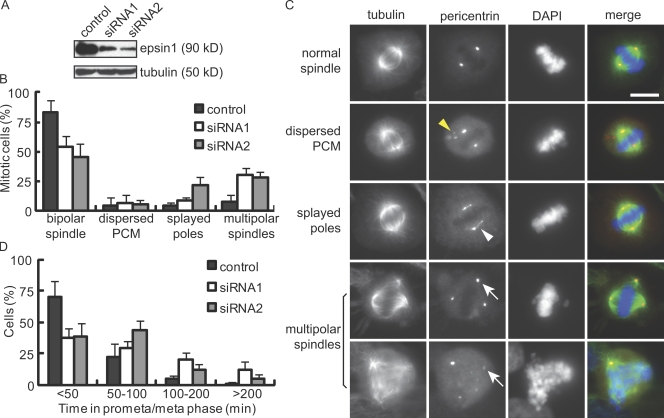
We used RNAi to determine whether endocytic adapter proteins might regulate mitosis in HeLa cells and found that epsin1 depletion caused a strong increase in mitotic cells with defective spindles (Fig. 1 C and Fig. S1, A and B). This was confirmed by two different siRNA oligonucleotides targeting different regions of epsin1, both of which reliably reduced epsin1 to 20–30% of the control levels in HeLa cells 48 h after siRNA transfection (Fig. 1 A). In contrast to control cells in which pericentrin staining was intense and focused at the spindle poles, epsin1 RNAi–treated cells showed a significant increase in splayed spindle poles and multipolar spindles (Fig. 1, B and C). There was no increase in the number of centrosomes in interphase cells (unpublished data). Although RNAi of epsin1 alone gave only a mild defect in spindle morphology in another human cell line, MCF-10A, codepletion of epsin1 and -2 resulted in a similar level of mitotic defects to those observed in HeLa cells (Fig. S2).
Figure 1.
Effects of epsin1 reduction on mitosis in HeLa cells. (A) Western blotting of epsin1 48 h after siRNA transfection. Each oligonucleotide targeting different regions of human epsin1 caused an ∼80% reduction of epsin1 at 48 h. α-Tubulin was used as a loading control. (B) Quantification of spindle defects in HeLa cells treated with control or epsin1 siRNA oligonucleotides. Results from five independent experiments were averaged. Spindles were analyzed based on α-tubulin and pericentrin staining, with >100 prometaphase and metaphase cells counted in each condition per experiment. (C) Examples of normal and abnormal spindles visualized by α-tubulin (green), pericentrin (red), and DAPI (blue) staining. Yellow and white arrowheads indicate cytoplasmic pericentrin staining and stretched pericentrin staining along the unfocused spindle poles, respectively. White arrows point to multiple spindle poles. (D) Reduction of epsin1 affects mitotic progression. HeLa cells stably expressing histone H2B–GFP were treated with control or epsin1 siRNA. Live imaging was acquired at 3-min intervals during 32–48 h after transfection (Fig. S1 C). Quantification of the time between nuclear envelope breakdown and chromosome segregation is shown. Results from three independent experiments were averaged, with >100 mitotic cells analyzed in each condition. (B and D) Error bars show standard deviation. PCM, pericentriolar material. Bar, 10 μm.
Live imaging of HeLa cells expressing histone H2B–GFP showed that the time cells spent in prometaphase and metaphase, which was determined from the first sign of nuclear envelope breakdown to the first sign of anaphase chromosome separation (Fig. S1 C), was significantly increased in epsin1 RNAi–treated cells (Fig. 1 D). There was no obvious defect in chromosome segregation, as a majority of the cells proceeded through mitosis after a delay. Interestingly, a recent study also showed that deletion of NuMA (nuclear/mitotic apparatus protein) in mouse embryonic fibroblasts prevented spindle pole focusing without rendering obvious chromosome segregation defects (Silk et al., 2009).
Reduction of epsin alters membrane organization in mitosis
Next, we performed immunofluorescence staining for epsin1. Although clathrin was enriched on mitotic spindles relative to cytoplasmic staining, as previously reported (Royle et al., 2005), we found that epsin1 was diffusely distributed in the mitotic cytoplasm, including the spindle region, with no obvious localization to centrosomes or spindle poles (Fig. S1 D). In addition, although epsin1 has been shown previously to interact with tubulin and MTs in rat brain lysate (Hussain et al., 2003), we could not detect any interaction between epsin1 and MTs or tubulin in mitotic Xenopus egg extracts (Fig. S3). Therefore, it is unlikely that epsin1 contributes toward spindle organization by directly regulating MT assembly dynamics in mitosis.
Epsin1 is known to induce membrane curvature through its ENTH domain in vitro (Ford et al., 2002). Thus, epsin1 may organize mitotic membranes, which in turn facilitates spindle morphogenesis. We used a membrane-permeable fluorescent dye, DiOC6(3), to visualize mitotic membranes in live cells by confocal microscopy (Waterman-Storer et al., 1993). To identify mitotic cells, chromosomes were labeled with Hoechst 33258. In control RNAi–treated cells, the membrane network uniformly surrounded mitotic chromosomes, whereas epsin1 RNAi–treated cells often showed distorted and uneven membrane distribution (Fig. 2 A and Videos 1–4). The percentage of cells with abnormal membrane morphology was significantly higher with epsin1 siRNA treatment than with control siRNA treatment (control, 9.2 ± 3.8%; epsin1 siRNA, 45.8 ± 8.9%; Student's two-tailed t test, P = 0.0016; n > 100 in each condition).
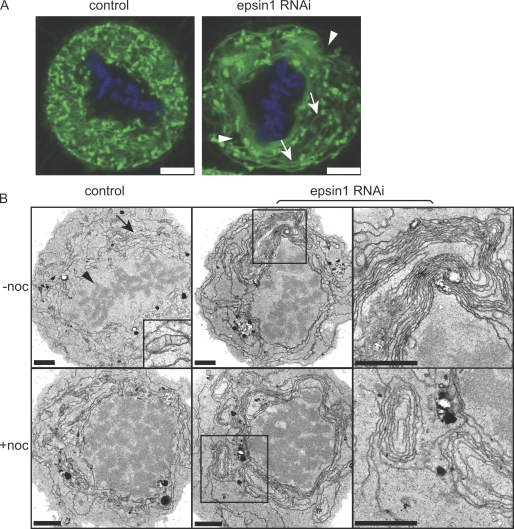
Figure 2.
Effects of epsin1 reduction on membrane organization in mitotic HeLa cells. (A) Control or epsin1-depleted cells were labeled with DiOC6(3) and Hoechst 33258 to visualize membranes and chromosomes, respectively. Confocal images of mitotic cells at one z section are shown. Epsin1 depletion caused aberrant membrane morphologies such as areas without fluorescent signal (arrowheads) or long-running membrane tubules or sheets (arrows), which were not seen in normal control cells. (B) EM images of control siRNA– or epsin1 siRNA2–treated cells are shown. Control mitotic cells have even membrane structures (arrow) surrounding the condensed chromosomes (arrowhead). An area containing mitochondria is enlarged in the inset (top left). Abnormal membrane structures such as whorls with multiple layers of membranes are present in epsin1 siRNA2–treated mitotic cells. Cells were analyzed 3 d after RNAi treatment with or without nocodazole arrest (+noc). Similar results were obtained with an additional epsin1 siRNA oligonucleotide (siRNA1). Boxed areas in the middle panels are enlarged in the right panels. Bars: (A) 5 μm; (B) 2 μm.
Next, we performed EM on control or epsin1 RNAi–treated cells (Fig. 2 B). Abundant membrane tubules and fragments were observed in control metaphase cells. These membrane structures usually surrounded condensed chromosomes in an even distribution (Fig. 2 B). However, in epsin1-depleted cells, uniform short membrane tubules were replaced by abnormally long membrane tubules or sheets frequently associated with layers of membrane whorls (Fig. 2 B). Cells with abnormal membrane organization were as follows: control, 2.8 ± 3.9% (n = 30); epsin1 siRNA2, 37.2 ± 2.7% (n = 57); Student's two-tailed t test, P = 0.014. The distorted membrane morphology was not caused by prolonged arrest in mitosis, as nocodazole-arrested mitotic cells with control RNAi treatment did not show such defects (Fig. 2 B). In addition, a significant increase of cells with aberrant membrane organization was observed in epsin1-depleted cells arrested in mitosis with nocodazole (Fig. 2 B). Cells with abnormal membrane organization were as follows: control, 6.3 ± 8.8% (n = 39); epsin1 siRNA2, 49.2 ± 4.6% (n = 79); Student's two-tailed t test, P = 0.049. Similar defects in membrane organization were also observed with a different siRNA oligonucleotide (siRNA1; unpublished data). There was no apparent membrane morphology change in interphase cells with either siRNA oligonucleotide (unpublished data), suggesting epsin1 specifically mediates mitotic membrane organization, which in turn modulates spindle assembly.
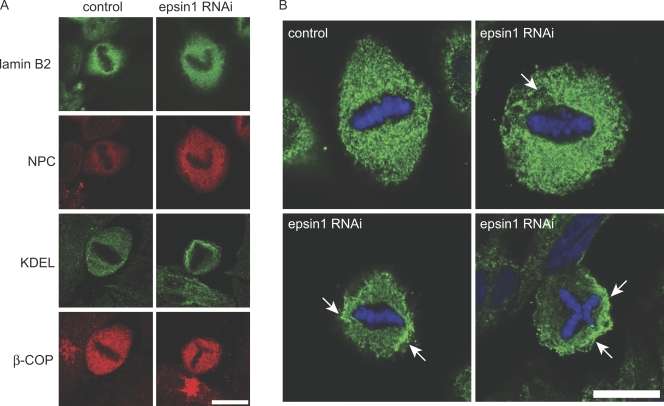
We found that epsin1 reduction did not cause obvious changes in the distribution of markers for Golgi (β-Cop), nuclear pores (nucleoporin; nuclear pore complex; detected by MAb414), or nuclear lamina (lamin B2) in mitosis (Fig. 3 A). However, the evenly distributed mitotic ER network around the spindle (Puhka et al., 2007), which is revealed by the KDEL antibody in control cells, became disorganized upon epsin1 knockdown by RNAi (Fig. 3, A and B). Cells with uneven KDEL signal were as follows: control, 15.7 ± 8% (n = 56); epsin1 siRNA2, 52.5 ± 2.3% (n = 64); Student's two-tailed t test, P = 0.011. This is reminiscent of the membrane morphology defects observed in live cells by membrane dye labeling (Fig. 2 A). Therefore, epsin regulates the organization of the mitotic membrane network that contains ER membranes, which may in turn influence spindle integrity.
Figure 3.
Mitotic ER network marked by the KDEL antibody is affected by epsin1 reduction in HeLa cells. (A) Confocal images of markers for subcellular compartments in mitosis in control or epsin1 knockdown cells. The maximum projection of images obtained for nuclear lamina (lamin B2), nucleoporins (nuclear pore complex [NPC]; labeled by MAb414), ER (KDEL), and Golgi (β-COP) is shown. (B) Confocal section of mitotic cells stained by the KDEL antibody, which marks the ER network. Arrows indicate areas of uneven ER membrane distribution around condensed chromosomes in mitotic cells after epsin1 knockdown by RNAi, which are not seen in normal control cells. Bars, 10 μm.
The membrane deformation function of epsin is required for proper mitotic spindle morphology
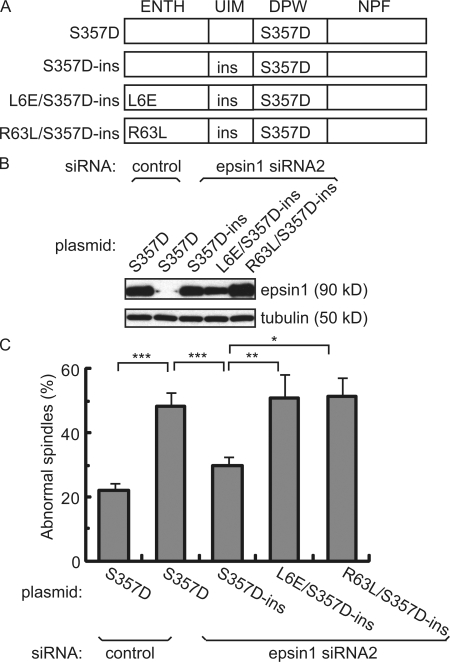
To further elucidate whether the membrane-bending activity of epsin1 is required for proper spindle morphology, we designed various epsin1 expression constructs for siRNA rescue. By introducing point mutations into the wobble codons of siRNA-targeting sequences, we generated RNAi-insensitive versions of epsin1 expression constructs that have the wild-type amino acid sequence. Because epsin1 is known to be phosphorylated by Cdc2 kinase on residue Ser357 during mitosis (Chen et al., 1999; Kariya et al., 2000; Rossé et al., 2003), we further introduced a single mutation at Ser357 to produce the phosphomimic and nonphosphorylatable forms Ser357D and Ser357A, respectively. All three constructs were able to rescue the defects caused by epsin1 depletion when cotransfected with the siRNA (Fig. 4 and Fig. S1, E and F), suggesting that the mitotic function of epsin1 does not depend on its phosphorylation by Cdc2. However, as a previous study showed that overexpression of wild type and Ser357A but not Ser357D interfered with clathrin-mediated endocytosis (Kariya et al., 2000), we used the mitotic phosphomimic form Ser357D to further investigate whether the membrane-bending function of epsin is involved in spindle morphogenesis.
Figure 4.
Epsin1 regulates spindle integrity through its membrane-bending ENTH domain. (A) Domain structure of epsin1 expression constructs with the indicated mutations. The ENTH domain interacts with membrane and has membrane-bending activity. The ubiquitin-interacting motif (UIM) binds to ubiquitin. DPW motifs interact with AP2. The NPF motif interacts with proteins such as Eps15. The mutant epsin1Ser357D (S357D) mimics the mitotic phosphorylated form. The RNAi-insensitive rescue construct epsin1Ser3573D-ins (S357D-ins) contains mutations in wobble codons in the sequence corresponding to siRNA2, which do not affect protein sequences. Mutant epsins epsin1L6E/Ser357D-ins (L6E/S357D-ins) and epsin1R63L/Ser357D-ins (R63L/S357D-ins) are derived from epsin1Ser357D-ins but contain an additional point mutation in the ENTH domain that abolishes the membrane-bending activity of epsin1. (B) Western blot analysis of epsin1 expression in HeLa cells. Epsin1Ser357D expression was inhibited by siRNA2 but not by control siRNA, whereas expression of epsin1Ser357D-ins, epsin1L6E/Ser357D-ins, and epsinR63L/Ser357D-ins was not affected by treatment with siRNA2. (C) Rescue of spindle morphology defects in HeLa cells by epsin1Ser357D-ins but not by epsin1L6E/Ser357D-ins or epsin1R63L/Ser357D-ins. The percentage of defective spindles, as judged by α-tubulin and pericentrin staining, was quantified in HeLa cells cotransfected with the siRNA oligonucleotide and each expression construct. The percentages of cells with abnormal spindle morphology from at least three independent experiments are shown, with >100 prometaphase and metaphase cells counted in each condition per experiment. Error bars show standard deviation. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The membrane-bending activity of epsin requires its ENTH domain; therefore, we generated epsin1 expression constructs epsin1L6E/Ser357D-ins and epsin1R63L/Ser357D-ins, which are known to be defective in membrane deformation and lipid binding, respectively (Fig. 4 A; Itoh et al., 2001; Ford et al., 2002). We found that although both mutants were expressed at similar levels as epsin1Ser357D-ins (Fig. 4 B), they failed to rescue the mitotic defects caused by epsin1 RNAi (Fig. 4 C). This suggests that the ability of epsin1 to induce membrane curvature is required for spindle organization in mitosis.
Epsin1 facilitates spindle organization independent of its endocytic function
To formally establish that epsin1 has a mitosis-specific role in spindle organization independent of its endocytic function in interphase, we used the cell-free spindle assembly assay using Xenopus egg extracts (Murray, 1991). Because spindle assembly in egg extracts is not confined within a cell cortex, we may not observe the same types of spindle defects observed in whole cells. Nonetheless, if epsin regulates spindle assembly independent of its role in endocytosis, we should observe spindle defects when its activity is inhibited.
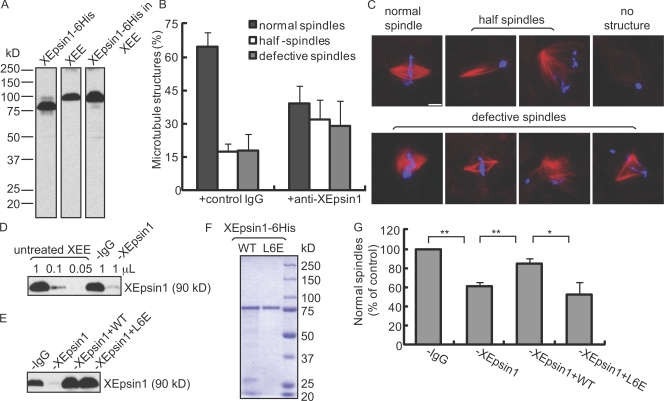
We generated a polyclonal antibody against the ENTH domain of Xenopus epsin1 (XEpsin1; also known as MP90), which recognized the purified bacterially expressed XEpsin1-6His and a single band in egg extracts (Fig. 5 A). The retarded migration of the endogenous XEpsin1 on SDS-PAGE is most likely caused by posttranslational modifications such as phosphorylation and ubiquitination (Oldham et al., 2002; Polo et al., 2002; Horvath et al., 2007). Consistent with this idea, when purified XEpsin1-6His was incubated with egg extracts depleted of endogenous XEpsin1 (Fig. 5, D and E), it migrated to the same position as endogenous XEpsin1 (Fig. 5 A).
Figure 5.
Epsin1 affects spindle assembly in Xenopus egg extracts. (A) Characterization of XEpsin1 antibody. Rabbit polyclonal antibodies against the ENTH domain of XEpsin1 recognized a single band in Xenopus egg extract (XEE) by Western blot analysis. 6His-tagged XEpsin1 purified from bacteria showed an apparent smaller molecular mass than the endogenous protein. After incubating with the Xenopus egg extract that was immunodepleted of endogenous XEpsin1, the recombinant XEpsin1 migrated as a similar size as the endogenous protein (XEpsin1-6His in XEE). (B) XEpsin1 antibody inhibits spindle assembly in cycled Xenopus egg extracts. Quantifications of percentages of MT structures formed in cycled extract after the addition of control antibody (+control rabbit IgG) or XEpsin1 antibody (+anti-XEpsin1) were performed by analyzing structures associated with >300 sperm in each reaction. Results from four independent experiments were averaged. (C) Examples of Xenopus sperm associated with normal spindle, abnormal MT structures, or no MT structures. MT structures were visualized by rhodamine-tubulin, and chromosomes were stained with DAPI. (D) Western blot analysis of XEpsin1 in extracts after immunodepletion. Different amounts of untreated egg extracts were loaded as standards. Mock-depleted extract (−IgG) had a similar level of XEpsin1 as nondepleted extract, whereas ∼90% of XEpsin1 was depleted by XEpsin1 antibody (−XEpsin1). (E) Western blot analysis of XEpsin1 after depletion (−XEpsin1) and add back of recombinant wild-type or mutant XEpsin1 (−XEpsin1 + WT or −XEpsin1 + L6E) compared with mock-depleted extract. (F) Coomassie blue staining of SDS-PAGE to show the quality of purified wild-type (WT) and mutant (L6E) XEpsin1 proteins. (G) Effects of depletion/add back of XEpsin1 on spindle assembly in CSF extracts. Mock-depleted extract, XEpsin1-depleted extract (−XEpsin1), or XEpsin1-depleted extract with the addition of wild type or the L6E mutant of the XEpsin1-6His protein was allowed to form spindles by incubation with sperm DNA. The number of normal spindles was quantified and normalized against mock-depleted extract. Results from three independent experiments, each containing structures associated with >300 sperms per reaction, were averaged. *, P < 0.05; **, P < 0.01. (B and G) Error bars show standard deviation. Bar, 10 μm.
We estimated the endogenous XEpsin1 concentration to be ∼70 nM in egg extracts. Because the ENTH domain of XEpsin1 appeared to be important for spindle organization (Fig. 4), the ENTH domain antibody might inhibit assembly of spindles when added to the egg extracts. We first incubated either the ENTH domain antibody or rabbit IgG purified from nonimmunized rabbits with the cycled egg extract at a final IgG concentration of 0.15 mg/ml and then initiated spindle assembly using Xenopus sperm. We observed a reduction of normal spindle assembly and a corresponding increase in sperm associating with half spindles that were unable to proceed into bipolar spindles, defective spindles with distorted or reduced MT fibers, or no MT structures at all (Fig. 5, B and C).
Next, we immunodepleted XEpsin1 from cytostatic factor (CSF)–arrested egg extracts. Approximately 90% of the endogenous XEpsin1 was depleted, as judged by Western blot analysis (Fig. 5, D and E). Similar to ENTH domain antibody addition, we found that depletion of XEpsin1 also resulted in a significant reduction of normal spindle assembly compared with mock-depleted egg extracts and a corresponding increase of sperm associated with either no spindles or with defective spindles (Fig. 5, C and G).
Finally, we expressed and purified wild-type XEpsin1-6His and mutant XEpsin1L6E-6His (defective in membrane bending), and used the proteins to rescue the XEpsin1-depleted egg extracts (Fig. 5, E and F). The addition of wild-type XEpsin1 but not XEpsin1L6E-6His resulted in a significant rescue of normal spindle formation in XEpsin1-depleted egg extracts (Fig. 5 G). Similarly, XEpsin1R63L-6His, which is defective in lipid binding, also failed to rescue the spindle defects caused by XEpsin1 depletion (unpublished data). Therefore, the spindle organization function of XEpsin1 requires its membrane-bending activity but is independent of its role in endocytosis during interphase.
We show that epsin regulates spindle morphology indirectly through its ability to regulate mitotic membrane organization. Although most studies of spindle assembly have focused on structures and forces produced within the spindle MTs, both our study (Fig. 2 and Videos 1 and 3) and previous work (Waterman-Storer et al., 1993; Axelsson and Warren, 2004; Altan-Bonnet et al., 2006) have shown that the mitotic spindle is built inside a highly interconnected membrane network, which could form an elastic module to provide support for MTs during spindle assembly. A disorganized membrane network such as those caused by epsin reduction would exhibit uneven elasticity and deformability when subjected to forces from MTs, which could affect spindle morphology. We suggest that the membrane elasticity could be provided in part by the intermediate filament protein lamin B (Tsai et al., 2006; Ma et al., 2009). Consistent with this, we have found that lamin B balances motor activities during spindle assembly in Xenopus egg extracts (unpublished data).
Besides epsin, other proteins may also contribute to the organization of various membrane compartments in the mitotic cytosol. This may explain the relatively mild spindle defects we observed in epsin reduction. Proteins such as AP2, ARH, and GRASP65 have functions in mitosis (Sütterlin et al., 2005; Boucrot and Kirchhausen, 2007; Lehtonen et al., 2008). It will be interesting to determine whether they have a role in mitotic membrane organization as well. Other proteins such as BAR (Bin, amphiphysin, and Rvs) or F-BAR (FCH-BAR) domain–containing proteins, coat proteins such as COPI and COPII, and enzymes involved in lipid metabolism may also be involved in regulating mitotic membrane organization (McMahon and Gallop, 2005; Itoh and De Camilli, 2006). Our study of epsin and mitotic membrane should stimulate further investigation into how structures outside the spindle proper could affect MT organization within the spindle.
Materials and methods
siRNAs, plasmids, proteins, and antibodies
siRNAs for human epsin1 (siRNA1, 5′-GGAAGACGCCGGAGTCATT-3′; siRNA2, 5′-GGACCTTGCTGACGTCTTC-3′; siRNA3, 5′-GAACTGGCGTCACGTTTAC-3′), clathrin heavy chain (5′-GCAATGAGCTGTTTGAAGA-3′), Eps15 (EGF receptor pathway substrate 15; 5′-AAACGGAGCTACAGATTAT-3′), Eps15R (Eps15 related; 5′-GCACTTGGATCGAGATGAG-3′; Huang et al., 2004), and control siRNA (5′-CGTACGCGGAATACTTCGA-3′) as well as SMARTpool siRNA for epsin2 and -3 were purchased from Thermo Fisher Scientific. Full-length cDNA clones encoding human epsin1 (IMAGE clone 5244118) and XEpsin1 (IMAGE clone 3405036) were obtained from Open Biosystems. Wild-type human epsin1 was cloned into the pEF6/V5-His vector (Invitrogen). The phosphomimic and nonphosphorylatable forms of epsin1, Ser357D and Ser357A, respectively, were generated by site-directed mutagenesis (TCA to GAT or GCA, respectively). Five wobble codon mutations in the central siRNA-binding region were introduced by overlapping PCR (5′-GGACCTTGCTGACGTCTTC-3′ to 5′-GGATCTCGCCGATGTTTTC-3′; underlining indicates the nucleotides mutated) to produce the rescue construct epsin1Ser357D-ins. The rescue construct was further mutated to give rise to epsin1L6E/Ser357D-ins (TTG to GAG) and epsin1R63L/Ser357D-ins (CGG to CTG) mutants.
Full-length XEpsin1 (aa 1–594), ENTH domain (aa 1–171), and a fragment of the ENTH domain (aa 10–171) were cloned into the bacterial expression vector pET21a (EMD) in frame with a C-terminal His tag. L6E (TTG to GAG) and R63L (CGG to CTG) in full-length or the ENTH domain of XEpsin1 were generated by site-directed mutagenesis.
His-tagged proteins of XEpsin1 variants were purified from BL21(DE3)pLysS. Bacteria were grown in Luria-Bertani broth or 2× yeast extract/tryptone bacterial medium and induced with IPTG for 2 h at 37°C or for 5 h at 25°C. Cells were collected and sonicated in lysis buffer (20 mM Hepes, pH 7.7, 200 mM KCl, 1% Triton X-100, and 10 mM imidazole). The soluble fraction was then incubated with Ni–nitrilotriacetic acid agarose resin (QIAGEN) at 4°C for 1 h, washed with wash buffer (20 mM Hepes, pH 7.7, 200 mM KCl, and 40 mM imidazole), and eluted in elution buffer (20 mM Hepe, pH 7.7, 200 mM KCl, and 250 mM imidazole). The buffer was exchanged to extraction buffer (10 mM Hepes, pH 7.7, 100 mM KCl, 2 mM MgCl2, 0.1 mM CaCl2, and 5 mM EGTA) using PD-10 desalting columns (GE Healthcare). Aliquots were snap frozen in liquid nitrogen and stored at −80°C. Protein concentrations were determined by Coomassie blue staining of SDS-PAGE and compared with BSA standards.
Rabbit polyclonal antibodies against XEpsin1 were generated using the His-tagged fragment of the ENTH domain (aa 10–171) as antigen. Removal of the first 9 aa greatly increased the solubility and yield of the recombinant protein. Pericentrin antibodies were raised against a fragment of pericentrin (M8; Doxsey et al., 1994). Chicken anti–lamin B2 antibody was generated against the C-terminal region of mouse lamin B2 (aa 383–589). Rabbit polyclonal antibodies against human epsin1 (H-130; Santa Cruz Biotechnology, Inc.), mouse monoclonal antibody against clathrin heavy chain (BD) and Eps15 (BD), and rabbit monoclonal antibody against Eps15R (Epitomics, Inc.) were used at 1:200, 1:1,000, 1:250, and 1:2,500, respectively, for Western blot analyses. For immunostaining, mouse anti–α-tubulin (DM1-α; Sigma-Aldrich) and mouse anti–γ-tubulin (GTU-88; Sigma-Aldrich) antibodies were diluted at 1:1,000, and Mab414 (Covance), KDEL (Abcam), and β-COP (Thermo Fisher Scientific) were diluted at 1:200, whereas anti-epsin1, anti-Eps15, and anti-Eps15R were diluted at 1:100.
Cell culture and transfection
HeLa cells were maintained in DME supplemented with 10% FBS using standard tissue culture procedures. MCF-10A (human breast cell line) cells were maintained in DME/F12 supplemented with 5% horse serum, 20 ng/ml EGF, 500 ng/ml hydrocortisone, 100 ng/ml cholera toxin, and 10 μg/ml insulin. For transfection, cells were seeded in 6-well plates with or without coverslips (coated with poly-l-Lys for MCF-10A cells) and incubated overnight. The next day, the cells were transfected with 10 nM siRNA (unless otherwise indicated) using Lipofectamine RNAiMax (Invitrogen) according to the manufacturer's instructions. For RNAi rescue experiments in HeLa cells, 50 nM siRNA and 1 μg of plasmid were cotransfected by incubating separately with 1 μl Lipofectamine 2000 (Invitrogen) in Opti-MEM (Invitrogen) and then mixed together before adding to the cells. HeLa cells stably expressing histone H2B–GFP were maintained and transfected as above.
Immunofluorescence and live cell imaging
Cells grown on coverslips were fixed 48 h after transfection with 4% paraformaldehyde for 15 min and permeabilized in TBS containing 0.1% Triton X-100 for 15 min. After blocking with 3% BSA in TBS, coverslips were incubated with DM1-α and M8 diluted in the blocking solution, followed by Alexa Fluor 488– and Alexa Fluor 594–conjugated secondary antibodies (Invitrogen) together with DAPI, and mounted. Images were acquired by MetaMorph (MDS Analytical Technologies) through a charge-coupled device camera (CoolSNAP HQ; Photometrics) on an inverted microscope (Eclipse TE200; Nikon) with a Plan-Apochromat 60× NA 1.40 oil lens (Nikon). Confocal images were obtained by a confocal laser-scanning microscope (TCS SP5 DM6000B; Leica) with an HCX Plan-Apochromat 63× NA 1.4 oil lens.
For mitotic timing measurements, cells maintained in a LiveCell chamber (Pathology Devices) were imaged by IPLab (BD) through a CoolSNAP HQ charge-coupled device camera on an inverted microscope (TE2000; Nikon) with a Hoffman modulation contrast ELWD Plan-Fluor 10× NA 0.3 lens 32–48 h after siRNA transfection of HeLa cells stably expressing histone H2B–GFP. To visualize live mitotic membranes, cells grown on the coverslip were labeled with Hoechst 33258 and DiOC6(3) (Invitrogen) as previously described (Waterman-Storer et al., 1993) 2 d after siRNA transfection. Mitotic cells with condensed chromosomes were picked randomly and imaged live on a TCS SP5 DM6000B confocal laser-scanning microscope with an HCX Plan-Apochromat 63× NA 1.4 oil lens at 1-μm intervals (z section). Cells were scored based on membrane morphologies. For 3D reconstruction, images were obtained at 0.118-μm intervals and assembled with Imaris imaging software (Bitplane).
EM imaging
HeLa cells in 35-mm dishes were treated with siRNA for 72 h, followed by fixation in 3% glutaraldehyde, 1% formaldehyde, and 2 mM CaCl2 in 0.1 M cacodylate, pH 7.4, for 1 h, and subjected to uranyl acetate staining. For mitotic arrest, cells were fed with fresh medium containing 100 ng/ml nocodazole 14 h before fixation. Mitotic cells with condensed chromosomes were observed on an electron microscope (Tecnai 12; Philips). Images of mitotic cells were taken from control and epsin1 RNAi–treated cells. To quantify the membrane defects, all images were scrambled together and then analyzed blindly based on membrane morphology. Two siRNA oligonucleotides for epsin1 (siRNA1 and -2) produced essentially the same phenotype; results from siRNA2-treated samples are shown.
Xenopus egg extract manipulations
CSF-arrested extracts were prepared from Xenopus eggs collected and dejellied (Murray, 1991). The egg extracts were made by a crushing spin in an ultracentrifuge (L5-50; Beckman Coulter). For antibody addition experiments, extracts were driven into interphase by incubating with 0.4 mM Ca2+ and 300 demembranated sperm/μl, energy mix (3.8 mM creatine phosphate, 0.5 mM ATP, 0.5 mM MgCl2, and 0.05 mM EGTA, pH 7.7), and 20 μg/ml rhodamine-tubulin at 20°C for 60–90 min and monitored by nuclei formation. An equal volume of fresh CSF-arrested extract was added to the interphase egg extract along with purified XEpsin1 antibody or control IgG (0.15 mg/ml final concentration) to cycle the egg extracts back into mitosis. The reactions were incubated at 20°C for another 50–70 min to allow spindle formation before they were diluted into BRB80 (80 mM K-Pipes, pH 6.8, 1 mM MgCl2, and 1 mM EGTA) containing 30% glycerol and spun onto 12-mm round coverslips through BRB80 containing 40% glycerol. After fixation by cold methanol, the coverslips were stained with DAPI and mounted. Alternatively, fresh CSF-arrested extract was incubated with Dynabeads protein A (Invitrogen) coated with either control antibody or XEpsin1 antibody for 1 h at 4°C. After retrieval of the magnetic beads, mock- or XEpsin1-depleted extract was used for spindle assembly with sperm nuclei, energy mix, and rhodamine-tubulin. For rescue experiments, purified wild-type XEpsin1-6His, mutant XEpsin1L6E-6His, or XEpsin1R63L-6His protein was added to the extract after depleting the endogenous protein and incubated on ice for 20–30 min before starting the spindle assembly reaction.
Online supplemental material
Fig. S1 shows mitosis in normal or siRNA-transfected HeLa cells. Fig. S2 shows mitotic defects in MCF-10A cells depleted of epsin family members. Fig. S3 shows a lack of interaction between epsin1 and MTs or epsin1 and tubulin in mitotic Xenopus egg extract. Videos 1 and 2 show membrane organization during normal mitosis and during mitosis in epsin1-depleted cells, respectively. Videos 3 and 4 show 3D reconstructions of membrane organization during normal mitosis in a HeLa cell and during mitosis in an epsin1-depleted HeLa cell, respectively. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200902071/DC1.
Acknowledgments
We thank Dr. Liang Liang for wild-type, Ser357A, and Ser357D human epsin1 expression constructs, Mike Sepanski for EM and Michael McCaffery for advice on the EM images, Ona Martin and Rong Chen for technical support, and the members of Zheng laboratory for critical comments.
This work is supported by the National Institutes of Health (grant GM56312 to Y. Zheng). Y. Zheng is an investigator of the Howard Hughes Medical Institute.
Footnotes
Abbreviations used in this paper:
- ARH
- autosomal recessive hypercholesterolemia
- CSF
- cytostatic factor
- ENTH
- epsin NH2-terminal homology
- MT
- microtubule
- XEpsin1
- Xenopus epsin1
References
- Altan-Bonnet N., Sougrat R., Liu W., Snapp E.L., Ward T., Lippincott-Schwartz J. 2006. Golgi inheritance in mammalian cells is mediated through endoplasmic reticulum export activities.Mol. Biol. Cell. 17:990–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson M.A., Warren G. 2004. Rapid, endoplasmic reticulum-independent diffusion of the mitotic Golgi haze.Mol. Biol. Cell. 15:1843–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucrot E., Kirchhausen T. 2007. Endosomal recycling controls plasma membrane area during mitosis.Proc. Natl. Acad. Sci. USA. 104:7939–7944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao K., Zheng Y. 2004. The Cdc48/p97-Ufd1-Npl4 complex: its potential role in coordinating cellular morphogenesis during the M-G1 transition.Cell Cycle. 3:422–424 [PubMed] [Google Scholar]
- Cao K., Nakajima R., Meyer H.H., Zheng Y. 2003. The AAA-ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis.Cell. 115:355–367 [DOI] [PubMed] [Google Scholar]
- Chabin-Brion K., Marceiller J., Perez F., Settegrana C., Drechou A., Durand G., Poüs C. 2001. The Golgi complex is a microtubule-organizing organelle.Mol. Biol. Cell. 12:2047–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Slepnev V.I., Di Fiore P.P., De Camilli P. 1999. The interaction of epsin and Eps15 with the clathrin adaptor AP-2 is inhibited by mitotic phosphorylation and enhanced by stimulation-dependent dephosphorylation in nerve terminals.J. Biol. Chem. 274:3257–3260 [DOI] [PubMed] [Google Scholar]
- Doxsey S.J., Stein P., Evans L., Calarco P.D., Kirschner M. 1994. Pericentrin, a highly conserved centrosome protein involved in microtubule organization.Cell. 76:639–650 [DOI] [PubMed] [Google Scholar]
- Efimov A., Kharitonov A., Efimova N., Loncarek J., Miller P.M., Andreyeva N., Gleeson P., Galjart N., Maia A.R., McLeod I.X., et al. 2007. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network.Dev. Cell. 12:917–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford M.G., Mills I.G., Peter B.J., Vallis Y., Praefcke G.J., Evans P.R., McMahon H.T. 2002. Curvature of clathrin-coated pits driven by epsin.Nature. 419:361–366 [DOI] [PubMed] [Google Scholar]
- Horvath C.A., Vanden Broeck D., Boulet G.A., Bogers J., De Wolf M.J. 2007. Epsin: inducing membrane curvature.Int. J. Biochem. Cell Biol. 39:1765–1770 [DOI] [PubMed] [Google Scholar]
- Huang F., Khvorova A., Marshall W., Sorkin A. 2004. Analysis of clathrin-mediated endocytosis of epidermal growth factor receptor by RNA interference.J. Biol. Chem. 279:16657–16661 [DOI] [PubMed] [Google Scholar]
- Hussain N.K., Yamabhai M., Bhakar A.L., Metzler M., Ferguson S.S., Hayden M.R., McPherson P.S., Kay B.K. 2003. A role for epsin N-terminal homology/AP180 N-terminal homology (ENTH/ANTH) domains in tubulin binding.J. Biol. Chem. 278:28823–28830 [DOI] [PubMed] [Google Scholar]
- Itoh T., De Camilli P. 2006. BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature.Biochim. Biophys. Acta. 1761:897–912 [DOI] [PubMed] [Google Scholar]
- Itoh T., Koshiba S., Kigawa T., Kikuchi A., Yokoyama S., Takenawa T. 2001. Role of the ENTH domain in phosphatidylinositol-4,5-bisphosphate binding and endocytosis.Science. 291:1047–1051 [DOI] [PubMed] [Google Scholar]
- Kariya K., Koyama S., Nakashima S., Oshiro T., Morinaka K., Kikuchi A. 2000. Regulation of complex formation of POB1/epsin/adaptor protein complex 2 by mitotic phosphorylation.J. Biol. Chem. 275:18399–18406 [DOI] [PubMed] [Google Scholar]
- Lehtonen S., Shah M., Nielsen R., Iino N., Ryan J.J., Zhou H., Farquhar M.G. 2008. The endocytic adaptor protein ARH associates with motor and centrosomal proteins and is involved in centrosome assembly and cytokinesis.Mol. Biol. Cell. 19:2949–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Vong Q.P., Zheng Y. 2007. CLASPing microtubules at the trans-Golgi network.Dev. Cell. 12:839–840 [DOI] [PubMed] [Google Scholar]
- Lowe M., Barr F.A. 2007. Inheritance and biogenesis of organelles in the secretory pathway.Nat. Rev. Mol. Cell Biol. 8:429–439 [DOI] [PubMed] [Google Scholar]
- Ma L., Tsai M.Y., Wang S., Lu B., Chen R., Iii J.R., Zhu X., Zheng Y. 2009. Requirement for Nudel and dynein for assembly of the lamin B spindle matrix.Nat. Cell Biol. 11:247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon H.T., Gallop J.L. 2005. Membrane curvature and mechanisms of dynamic cell membrane remodelling.Nature. 438:590–596 [DOI] [PubMed] [Google Scholar]
- Murray A.W. 1991. Cell cycle extracts.Methods Cell Biol. 36:581–605 [PubMed] [Google Scholar]
- Oldham C.E., Mohney R.P., Miller S.L., Hanes R.N., O'Bryan J.P. 2002. The ubiquitin-interacting motifs target the endocytic adaptor protein epsin for ubiquitination.Curr. Biol. 12:1112–1116 [DOI] [PubMed] [Google Scholar]
- Polo S., Sigismund S., Faretta M., Guidi M., Capua M.R., Bossi G., Chen H., De Camilli P., Di Fiore P.P. 2002. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins.Nature. 416:451–455 [DOI] [PubMed] [Google Scholar]
- Puhka M., Vihinen H., Joensuu M., Jokitalo E. 2007. Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells.J. Cell Biol. 179:895–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossé C., L'Hoste S., Offner N., Picard A., Camonis J. 2003. RLIP, an effector of the Ral GTPases, is a platform for Cdk1 to phosphorylate epsin during the switch off of endocytosis in mitosis.J. Biol. Chem. 278:30597–30604 [DOI] [PubMed] [Google Scholar]
- Royle S.J., Bright N.A., Lagnado L. 2005. Clathrin is required for the function of the mitotic spindle.Nature. 434:1152–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silk A.D., Holland A.J., Cleveland D.W. 2009. Requirements for NuMA in maintenance and establishment of mammalian spindle poles.J. Cell Biol. 184:677–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sütterlin C., Polishchuk R., Pecot M., Malhotra V. 2005. The Golgi-associated protein GRASP65 regulates spindle dynamics and is essential for cell division.Mol. Biol. Cell. 16:3211–3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M.Y., Wang S., Heidinger J.M., Shumaker D.K., Adam S.A., Goldman R.D., Zheng Y. 2006. A mitotic lamin B matrix induced by RanGTP required for spindle assembly.Science. 311:1887–1893 [DOI] [PubMed] [Google Scholar]
- Vong Q.P., Cao K., Li H.Y., Iglesias P.A., Zheng Y. 2005. Chromosome alignment and segregation regulated by ubiquitination of survivin.Science. 310:1499–1504 [DOI] [PubMed] [Google Scholar]
- Waterman-Storer C.M., Sanger J.W., Sanger J.M. 1993. Dynamics of organelles in the mitotic spindles of living cells: membrane and microtubule interactions.Cell Motil. Cytoskeleton. 26:19–39 [DOI] [PubMed] [Google Scholar]
- Wei J.H., Seemann J. 2009. The mitotic spindle mediates inheritance of the Golgi ribbon structure.J. Cell Biol. 184:391–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Tsai M.Y. 2006. The mitotic spindle matrix: a fibro-membranous lamin connection.Cell Cycle. 5:2345–2347 [DOI] [PubMed] [Google Scholar]