Abstract
The mammalian dynamin-related guanosine triphosphatases Mfn1,2 and Opa1 are required for mitochondrial fusion. However, how their activities are controlled and coordinated is largely unknown. We present data that implicate the BCL-2–like protein CED-9 in the control of mitochondrial fusion in Caenorhabditis elegans. We demonstrate that CED-9 can promote complete mitochondrial fusion of both the outer and inner mitochondrial membrane. We also show that this fusion is dependent on the C. elegans Mfn1,2 homologue FZO-1 and the C. elegans Opa1 homologue EAT-3. Furthermore, we show that CED-9 physically interacts with FZO-1 in vivo and that the ability of CED-9 to interact with FZO-1 is important for its ability to cause mitochondrial fusion. CED-9–induced mitochondrial fusion is not required for the maintenance of mitochondrial morphology during embryogenesis or in muscle cells, at least under normal conditions and in the absence of stress. Therefore, we propose that the BCL-2–like CED-9 acts through FZO-1/Mfn1,2 and EAT-3/Opa1 to promote mitochondrial fusion in response to specific cellular signals.
Introduction
Mitochondria are essential organelles of eukaryotic cells that participate in several metabolic pathways, cell signaling, and apoptosis (Saraste, 1999; Suen et al., 2008; Szabadkai and Duchen, 2008). Mitochondria have a very plastic morphology and constantly fuse and divide. Genetic studies in Drosophila melanogaster and Saccharomyces cerevisiae have led to the identification of components of the enzymatic machineries that bring about mitochondrial fusion and fission (Okamoto and Shaw, 2005; Berman et al., 2008; Lackner and Nunnari, 2008; Westermann, 2008; Hoppins and Nunnari, 2009). Important components of these machineries are members of the family of dynamin-related GTPases (Praefcke and McMahon, 2004). It has been established that an inner mitochondrial membrane (IMM)– and outer mitochondrial membrane (OMM)–specific machinery acts during mitochondrial fusion. Specifically, the dynamin-related GTPases Fzo1/Mfn1,2 are required for OMM fusion (Hermann et al., 1998; Chen et al., 2003). Conversely, the dynamin-related GTPases Mgm1/Opa1 are required for IMM fusion (Sesaki et al., 2003; Misaka et al., 2006). Recent studies have demonstrated that the role of these proteins is conserved in Caenorhabditis elegans. Specifically, the Fzo1/Mfn1,2 homologue, FZO-1, and the Mgm1/Opa1 homologue, EAT-3, are required for mitochondrial fusion in C. elegans (Breckenridge et al., 2008; Ichishita et al., 2008; Kanazawa et al., 2008; Tan et al., 2008). EAT-3/Mgm1/Opa1 also plays a role in mitochondrial cristae structure maintenance (Frezza et al., 2006; Meeusen et al., 2006; Kanazawa et al., 2008). In addition, C. elegans EAT-3 has been shown to be required for resistance to oxidative stress caused by free radicals (Kanazawa et al., 2008).
The molecular mechanisms underlying mitochondrial fusion are still unclear. However, with the development of in vitro and in vivo mitochondrial fusion assays, these mechanisms are now being elucidated. Mitochondrial fusion seems to involve four distinct steps: (1) OMM tethering, (2) OMM fusion, (3) IMM tethering, and (4) IMM fusion (Hoppins and Nunnari, 2009). In addition, GTP binding and hydrolysis is required for mitochondrial fusion (Meeusen et al., 2004). IMM potential is also important for mitochondrial fusion; however, its mechanistic role in the fusion process remains elusive (Meeusen et al., 2004). The fusion of the OMM and IMM are temporally linked in vivo, indicating the existence of mechanisms to coordinate the OMM and IMM fusion machineries. It has been proposed that the S. cerevisiae protein Ugo1, which interacts with both Fzo1 and Mgm1, mediates the coordination of these two events (Sesaki and Jensen, 2004; Hoppins et al., 2009). To date, no structural or functional homologue of Ugo1 has been characterized in higher eukaryotes. Thus, how IMM and OMM fusion are coordinated in higher eukaryotes remains to be elucidated.
The mechanisms underlying the regulation of dynamin-related GTPases are starting to be elucidated (Cerveny et al., 2007). The activity of S. cerevisiae Fzo1 is in part controlled by protein degradation (Fritz et al., 2003; Neutzner and Youle, 2005), and mammalian Opa1 and yeast Mgm1 can be regulated through the generation of different isoforms and by proteolysis (Herlan et al., 2004; Cipolat et al., 2006; Ishihara et al., 2006). Recently, BCL-2–like proteins, which play critical roles during apoptosis (Youle and Strasser, 2008), have also been implicated in the control of mitochondrial morphology and have been proposed to do so by regulating the activity of dynamin-related GTPases (Jagasia et al., 2005; Delivani et al., 2006; Karbowski et al., 2006; Brooks et al., 2007; Li et al., 2008; Tan et al., 2008; Berman et al., 2009). All of these regulatory mechanisms are thought to induce appropriate mitochondrial morphology changes in response to certain cellular signals or events such as cell division (Cerveny et al., 2007).
In this study, we demonstrate that the BCL-2–like protein CED-9 of C. elegans can induce complete mitochondrial fusion in a manner that is dependent on the ability of CED-9 to physically interact with the dynamin-related GTPase FZO-1, the C. elegans orthologue of Fzo1/Mfn1,2. We propose that CED-9 acts to control and coordinate the fusion of the OMM and IMM in healthy cells in response to specific cellular signals.
Results
CED-9 can promote complete mitochondrial fusion in C. elegans
It has been reported that the expression of the C. elegans ced-9 gene (ced-9(wt)) in cultured mammalian cells can promote mitochondrial fusion (Delivani et al., 2006). To determine whether the expression of ced-9(wt) can also promote mitochondrial fusion in C. elegans, we analyzed the effect of ced-9(wt) expression on mitochondrial morphology in C. elegans embryos. To that end, we used transgenic lines expressing mitogfp and ced-9(wt) under the control of an inducible promoter and analyzed mitochondrial morphology in transgenic embryos during midgastrulation (∼150 min after the first cell division; we estimate that the level of CED-9 protein in transgenic embryos is about five times higher than the endogenous level). Similar to what we observed in wild-type embryos stained with tetramethyl rhodamine ethyl ester (TMRE; see Fig. 4 B, row 1), embryos expressing mitogfp alone had tubular mitochondria (82%, n = 14, four independent transgenic lines; Fig. 1 B, row 1). In contrast, the majority of embryos expressing mitogfp and ced-9(wt) had one or two highly globular mitochondria per cell (90%, n = 8, three independent transgenic lines; Fig. 1 B, row 2). A similar phenotype was observed in embryos expressing mitogfp and ced-9(n1950gf) (81%, n = 22, three independent transgenic lines; Fig. S1 B, row 2), indicating that this mutation, which enhances the ability of CED-9 to block apoptosis (Hengartner and Horvitz, 1994a), does not compromise the ability of CED-9 to promote mitochondrial fusion. In both cases, 3D reconstruction of z stacks confirmed the presence of one or two highly globular organelles per cell (Table S1 and Videos 1, 2, and 5).
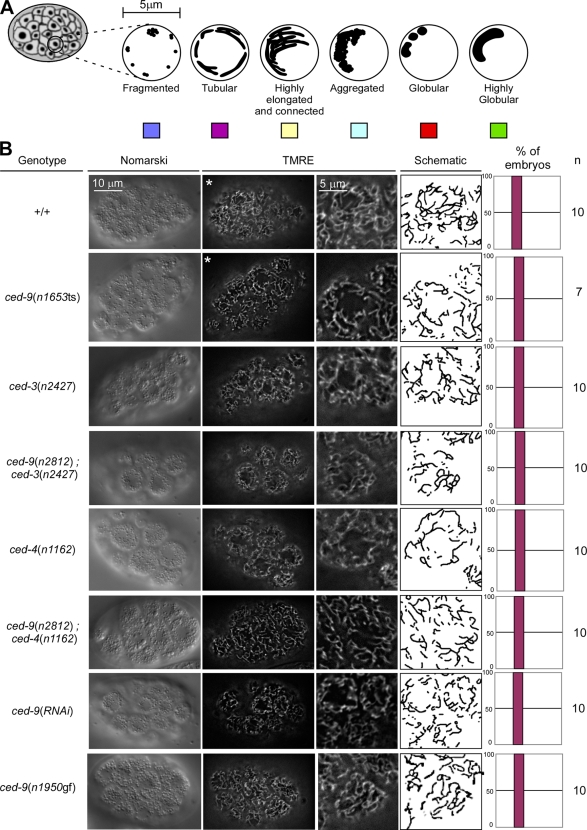
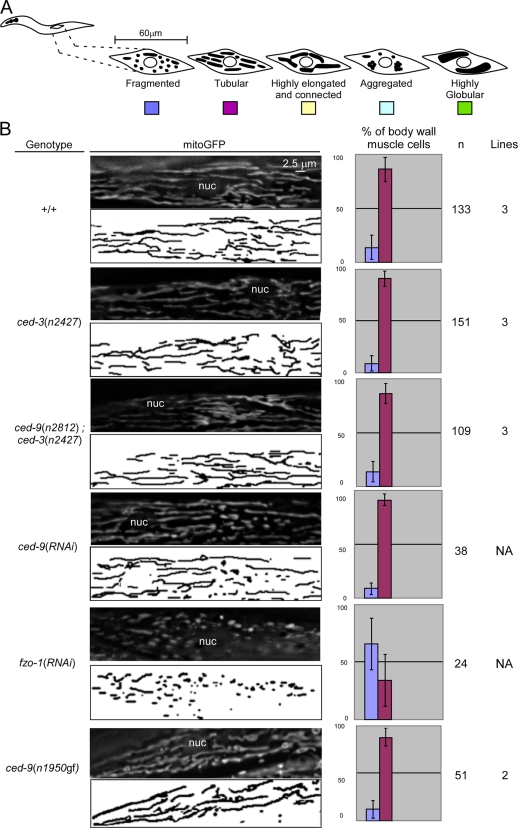
Figure 4.
The loss of ced-9 function does not cause detectable defects in mitochondrial morphology in embryos. (A) Schematic of the mitochondrial morphologies observed in embryos. (B) Mitochondria of wild-type (+/+), ced-3(n2427), ced-9(n2812); ced-3(n2427), ced-4(n1162), ced-9(n2812); ced-4(n1162), ced-9(n1653ts), ced-9(RNAi), and ced-9(n1950gf) embryos were stained using TMRE, as indicated in Materials and methods. Quantification of the different mitochondrial morphologies observed and the number of embryos analyzed are indicated. Videos 6 and 7 show 3D reconstructions of wild-type and ced-9(n1653ts) embryos marked with asterisks.
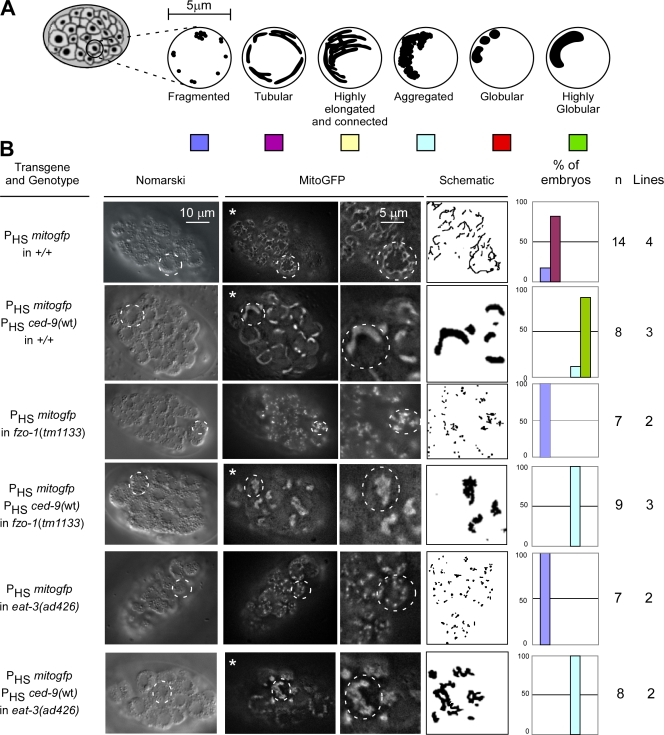
Figure 1.
CED-9 promotes FZO-1– and EAT-3–dependent mitochondrial fusion. (A) Schematic of the mitochondrial morphologies observed in embryos. (B) Wild-type (+/+), fzo-1(tm1133), or eat-3(ad426) transgenic lines expressing mitogfp alone (PHSmitogfp) or in the presence of ced-9(wt) (PHSmitogfp + PHSced-9(wt)) were analyzed by Nomarski optics and fluorescent microscopy. Quantification of the different mitochondrial morphologies observed and the number of embryos and transgenic lines analyzed are indicated. The outlines of representative cells are indicated with dashed circles. Videos 1–4 show 3D reconstructions of the embryos marked with asterisks.
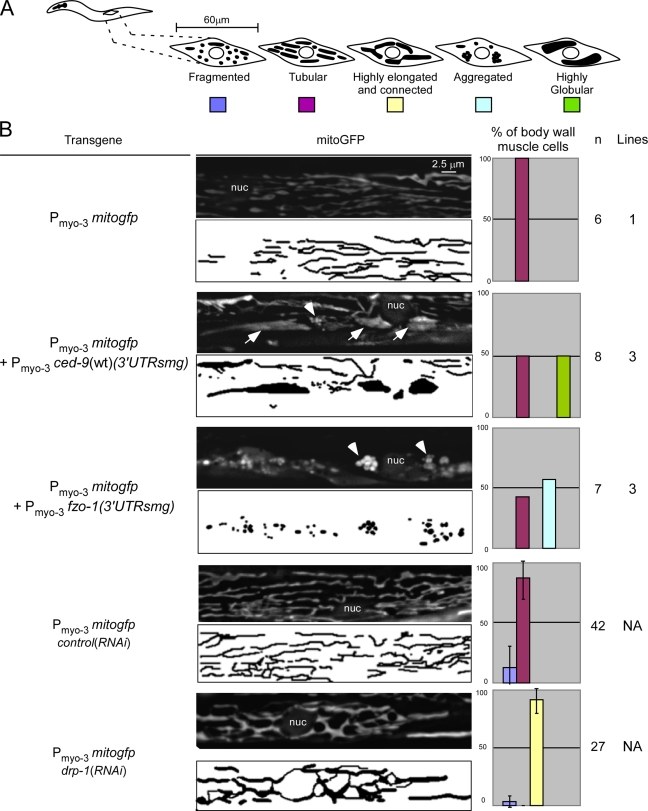
Unfortunately, the relatively small size of embryonic cells (∼5 μm) prevented us from analyzing the ultrastructure of these highly globular organelles. Therefore, we expressed ced-9(wt) in body wall muscle cells and analyzed mitochondrial morphology in these cells in larvae of the fourth larval stage (L4 larvae; at this stage of development, body wall muscle cells are ∼60-μm long and ∼8-μm wide). However, we failed to obtain stable transgenic lines expressing ced-9(wt) under the control of a body wall muscle–specific promoter (Pmyo-3), suggesting that the constitutive overexpression of ced-9(wt) in these cells might be toxic. For this reason, we conditionally expressed ced-9(wt) in body wall muscle cells using the Smg-inducible system (Pulak and Anderson, 1993). Body wall muscle cells of animals expressing mitogfp alone have tubular mitochondria (100%, n = 6, one line; Fig. 2 B, row 1). This result is consistent with previous studies (Ichishita et al., 2008; Kanazawa et al., 2008; Tan et al., 2008) and with the mitochondrial morphology we observed in wild-type embryos (Fig. 1). In contrast, after the induction of transgene expression, animals carrying a Pmyo-3 ced-9(wt) (3′untranslated region [UTR]smg) transgene have highly globular mitochondria (50%, n = 8, three independent lines; Fig. 2 B, row 2, arrows). This phenotype is reminiscent of the phenotype observed in embryos (Fig. 1) and is consistent with previous experiments (Tan et al., 2008). In some cells, we also noticed clusters of mitochondria (Fig. 2 B, row 3, arrowhead). These clusters, which are very similar to the aggregates induced by fzo-1 overexpression (57%, n = 7, one line; Fig. 2 B, row 5, arrowheads), could represent fusion intermediates.
Figure 2.
CED-9 promotes mitochondrial fusion, whereas FZO-1 promotes mitochondrial aggregation in body wall muscle cells. (A) Schematic of the mitochondrial morphologies observed in body wall muscle cells. (B) Transgenic lines expressing mitogfp alone (Pmyo-3 mitogfp), in the presence of ced-9(wt) (Pmyo-3 mitogfp + Pmyo-3 ced-9(wt) (3′UTRsmg)), or in the presence of fzo-1 (Pmyo-3 mitogfp + Pmyo-3 fzo-1 (3′UTRsmg)) were grown at 25°C to allow the expression of the smg-inducible transgene, as indicated in Materials and methods. These lines, as well as drp-1(RNAi) animals and lea-1(RNAi) animals (control(RNAi)) carrying a Pmyo-3 mitogfp transgene, were analyzed by fluorescent microscopy. Quantification of the different mitochondrial morphologies observed and the number of body wall muscle cells and transgenic lines analyzed are indicated. nuc, nucleus; arrows, fused mitochondria; arrowheads, aggregated mitochondria. Error bars indicate standard deviations.
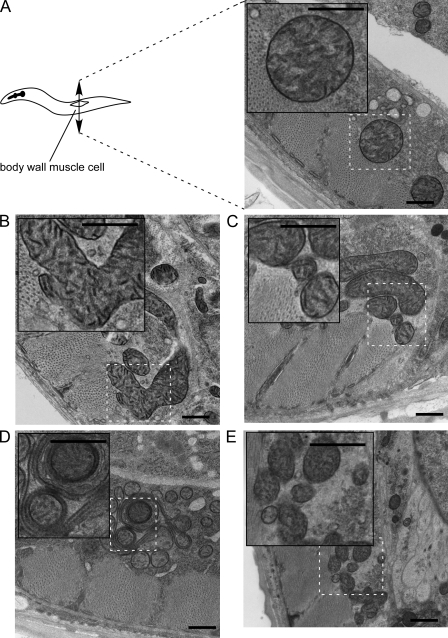
Analysis by EM of cross sections of wild-type body wall muscle cells of L4 larvae revealed mainly small and round mitochondria lying in the muscle belly just below the sarcomere. (12/12 muscle cells, four animals; Fig. 3 A). In contrast, we found that in a given cross section, ced-9(wt) overexpression can induce the formation of one large mitochondrion per cell (2/18 muscle cells, three animals; Fig. 3 B), whereas fzo-1 overexpression only induced the formation of clusters of mitochondria (3/14 muscle cells, three animals; Fig. 3 E). These phenotypes and their respective frequencies of occurrence are consistent with what we observed by fluorescent microscopy (Fig. 2). We estimate that ∼25% of the muscle cells are mitoGFP positive and therefore express the transgene. Because 50% and 57% of the mitoGFP-positive muscle cells show a phenotype after ced-9(wt) or fzo-1 overexpression, respectively (see previous paragraph), we expected to see alterations in mitochondrial morphology by EM in 12.5% and 14.5% of the cells, respectively. Using morphometric analysis, we confirmed that a subpopulation of mitochondria of ced-9(wt)–overexpressing animals have increased thickness (unpublished data). Interestingly, despite their abnormal size, CED-9–induced large mitochondria in general seem to have normal OMM, IMM, and cristae structures. In some cells overexpressing ced-9(wt), we observed mitochondrial clusters, which might constitute fusion intermediates (2/18 muscle cells, three animals; Fig. 3 C). Finally, in one case, we also observed mitochondria composed of multiple concentric layers as described previously by Tan et al. (2008; 1/18 muscle cells, three animals; Fig. 3 D). This phenotype could be caused by hyper fusion of the OMM. In summary, these results suggest that, as in cultured mammalian cells, the CED-9 protein can promote mitochondrial fusion in C. elegans.
Figure 3.
EM analysis of CED-9–induced mitochondrial fusion and FZO-1–induced mitochondrial aggregation. (A) Transgenic lines expressing mitogfp alone (Pmyo-3 mitogfp) show wild-type muscle mitochondria lying just below the sarcomere. Note its abundant internal cristae. (B–D) Transgenic lines expressing mitogfp in the presence of ced-9(wt) (Pmyo-3 mitogfp + Pmyo-3 ced-9(wt) (3′UTRsmg)) show mitochondria with abundant cristae. Three typical structures were observed. (B) Mitochondria form elongated and branched structures. These giant mitochondria seem fused. Nearby tubes may be part of these giant mitochondria; however, in one thin section, it is not possible to know for sure. (C) Mitochondria have smaller diameter and are closely clustered. In some cases, neighboring mitochondria may be physically attached to one another through the OMM. (D) Mitochondria lie closely together, and some show nested profiles in which one tube is completely enveloped by another. Cristae in some cases run circumferentially inside narrow outer tubes. (E) Transgenic lines expressing mitogfp in the presence of fzo-1 (Pmyo-3 mitogfp + Pmyo-3 fzo-1 (3′UTRsmg)) show mitochondria with abundant cristae, which are closely clustered and tend to have smaller diameters. In many cases, neighboring mitochondria may be physically attached to one another through the OMM. Insets show a magnified view of the indicated area. Bars, 500 nm.
CED-9–induced mitochondrial fusion is dependent on FZO-1 and EAT-3
The dynamin-related GTPases FZO-1 and EAT-3 are required for mitochondrial fusion in C. elegans (Breckenridge et al., 2008; Ichishita et al., 2008; Kanazawa et al., 2008; Tan et al., 2008). To investigate whether the ability of the CED-9 protein to promote mitochondrial fusion is dependent on a functional fzo-1 or eat-3 gene, we analyzed transgenic animals carrying loss of function mutations in the fzo-1 gene (fzo-1(tm1133)) or the eat-3 gene (eat-3(ad426)). Consistent with previous studies, transgenic animals homozygous for these loss of function mutations and expressing mitogfp have fragmented mitochondria (100%, n = 7, two independent lines; Fig. 1 B, rows 3 and 5).
We found that ced-9(wt) overexpression failed to promote mitochondrial fusion in fzo-1(tm1133) animals (0%, n = 9, three independent lines; Fig. 1 B, row 4). Instead, in these animals, mitochondria aggregated into one or two large clusters per cell, which is reminiscent of the phenotype caused by fzo-1 expression (Table S1 and Videos 3 and 8). We confirmed that the inability of ced-9(wt) expression to promote mitochondrial fusion in fzo-1(tm1133) embryos was not the result of lower levels of CED-9 protein (Fig. S2 A). Interestingly, we found that the level of bc1 protein (a protein of complex III of the electron transport chain) is reduced in fzo-1(tm1133) animals (Fig. S2 A). This result is consistent with the observation that fzo-1(tm1133) animals seem to have less active mitochondria when stained with TMRE (unpublished data).
Similarly, we found that ced-9(wt) expression failed to promote mitochondrial fusion in eat-3(ad426) animals (0%, n = 8, two independent lines; Fig. 1 B, row 6; Table S1; and Video 4). Instead, like in fzo-1(tm1133) animals, ced-9(wt) expression in eat-3(ad426) mutants induced mitochondrial aggregation. Again, the inability of ced-9(wt) expression to promote mitochondrial fusion in eat-3(ad426) embryos was not because of lower levels of CED-9 protein (Fig. S2 B).
We also observed that CED-9(n1950gf)–induced mitochondrial fusion was blocked by fzo-1(tm1133) and eat-3(ad426) (Fig. S1). Once again, the inability of ced-9(n1950gf) expression to promote mitochondrial fusion in the eat-3(ad426) or fzo-1(tm1133) mutant background was not because of lower levels of CED-9(n1950gf) protein (Fig. S2, D and E). Based on these results, we conclude that the CED-9 protein can promote mitochondrial fusion in a manner that is dependent on the dynamin-related GTPases FZO-1 and EAT-3.
ced-9(wt) expression might have a weak effect on mitochondrial mass in C. elegans
Overexpression of BCL-xL has been shown to increase mitochondrial mass in cultured mammalian cells 3–10-fold (Berman et al., 2009). Therefore, we investigated whether ced-9(wt) overexpression has the same effect in C. elegans. To that end, we measured the levels of three mitochondrial proteins by Western analysis: the bc1 protein, cytochrome oxidase I, and the α subunit of ATP synthase (these proteins are part of complexes III, IV, and V of the electron transport chain, respectively). No significant differences in the levels of these proteins were observed in transgenic lines overexpressing mitogfp and ced-9(wt) when compared with transgenic lines overexpressing mitogfp alone (Fig. S2 F). However, analysis of EM pictures revealed that the mitochondrial area is increased by ∼25% in transgenic lines overexpressing ced-9(wt) compared with control transgenic lines (Fig. S2 G). Therefore, we conclude that CED-9 might affect mitochondrial mass in C. elegans but to a much lesser extent than what has been observed for BCL-xL in mammalian cells.
ced-9 mutants do not display obvious defects in mitochondrial morphology
Based on MitoTracker staining, it has been reported that embryos homozygous for a temperature-sensitive partial loss of function mutation of ced-9, n1653ts, have fragmented mitochondria (Delivani et al., 2006). Based on this observation, it was proposed that ced-9 plays an important role in mitochondrial fusion. However, we found that in contrast to TMRE, MitoTracker does not label mitochondria in C. elegans embryos (Fig. S3). For this reason, we reexamined mitochondrial morphology in ced-9 mutants using TMRE. We were unable to detect a defect in mitochondrial morphology in ced-9(n1653ts) embryos at the nonpermissive temperature (100%, n = 7; Fig. 4 B, row 2). Furthermore, we failed to detect a defect in embryos homozygous for n2812, a null allele of the ced-9 gene (100%, n = 10; Fig. 4 B, rows 4 and 6). However, we cannot exclude that, in the strains analyzed, potential effects of ced-9 mutations on mitochondrial morphology are suppressed by the presence of secondary mutations. The occurrence of such compensatory, secondary mutations has previously been demonstrated in S. cerevisiae (Cheng et al., 2008). Therefore, to rule out the possibility that secondary mutations mask a mitochondrial phenotype in ced-9 mutants, we transiently inactivated the ced-9 gene by RNAi, which results in reduced CED-9 protein levels (Fig. S2 I). However, we failed to detect any mitochondrial morphology defects after ced-9 RNAi treatment (100%, n = 10; Fig. 4 B, row 7). We confirmed these results using 3D reconstruction of z stacks of wild-type and ced-9(n1653ts) embryos (Table S1 and Videos 6 and 7). Finally, we also found that the mitochondrial morphology in ced-9(n1950gf) embryos was indistinguishable from the morphology observed in wild-type embryos (100%, n = 10; Fig. 4 B, row 8).
To elucidate whether the CED-9 protein plays a role in mitochondrial fusion during later stages of development, we analyzed mitochondrial morphology in body wall muscle cells of L4 larvae. Animals homozygous for ced-9(n2812) do not display a significant fragmented mitochondrial morphology (+/+: 14%, n = 133; ced-3(n2427): 9%, n = 151; ced-9(n2812); ced-3(n2427): 14%, n = 109; Fig. 5). This result is consistent with previous experiments (Tan et al., 2008). In addition, we found that ced-9(RNAi) animals do not have a significant fragmented mitochondrial phenotype compared with control RNAi animals (control(RNAi): 12%, n = 42; ced-9(RNAi): 9%, n = 38). In contrast, we confirmed that, as previously shown (Ichishita et al., 2008), fzo-1(RNAi) animals have mainly fragmented mitochondria (66%, n = 24; Fig. 5). Western analysis confirmed that CED-9 and FZO-1 protein levels were reduced after ced-9 and fzo-1 RNAi treatment, respectively (Fig. S2 I). Using morphometric analysis, we found that ced-9(n2812); ced-3(n2427) animals have slightly shorter mitochondria compared with ced-3(n2427) animals (ced-3(n2427): 4.2 ± 1.6 μm, n = 21; ced-9(n2812); ced-3(n2427): 3.5 ± 1.3 μm, n = 38; Fig. S4). Similarly, ced-9(RNAi) animals have slightly smaller mitochondria compared with control(RNAi) animals. (control(RNAi): 4.5 ± 1.7 μm, n = 11; ced-9(RNAi): 4.3 ± 1.4 μm, n = 24; Fig. S4). Finally, ced-9(n1950gf) animals have slightly longer mitochondria compared with wild-type animals (+/+: 3.6 ± 1.1 μm, n = 25; ced-9(n1950gf): 4.2 ± 1.5 μm, n = 24; Fig. S4). However, in all three cases, the differences were not statistically significant (Fig. S4).
Figure 5.
The loss of ced-9 function does not cause detectable defects in mitochondrial morphology in body wall muscle cells. (A) Schematic of the mitochondrial morphologies observed in body wall muscle cells. (B) Wild-type (+/+), ced-3(n2427), ced-9(n2812); ced-3(n2427), ced-9(RNAi), fzo-1(RNAi), and ced-9(n1950gf) L4 larvae carrying a Pmyo-3 mitogfp transgene were analyzed by fluorescent microscopy. Quantification of the different mitochondrial morphologies observed and the number of body wall muscle cells and transgenic lines analyzed are indicated, and quantification of the mitochondrial length is shown in Fig. S4. nuc, nucleus. Error bars indicate standard deviations.
In summary, we conclude that, at least under normal growth conditions and in the absence of internal or external stress, ced-9 mutants do not display obvious defects in mitochondrial morphology during embryogenesis or in body wall muscle cells during larval development. However, we cannot rule out the idea that ced-9 mutants exhibit defects in the rate of mitochondrial fusion, which might not necessarily cause significant changes in mitochondrial morphology.
The loss of ced-9 function partially suppresses embryonic lethality caused by drp-1 RNAi
Animals treated with drp-1 RNAi have highly elongated and connected mitochondria (Labrousse et al., 1999). We tested whether drp-1 RNAi–induced changes in mitochondrial morphology are dependent on the presence of a functional ced-9 gene. To that end, we analyzed the effect of drp-1 RNAi on mitochondrial morphology in wild-type animals and ced-9(n2812) animals. In both cases, we observed highly elongated and connected mitochondria (unpublished data). However, although drp-1 RNAi induced 26% (± 6.2%, n = 309) of embryonic lethality on average in a wild-type background, it only induced 16% (± 2.8%, n = 278) of embryonic lethality on average in a ced-9(n2812) background. Interestingly, like the complete inactivation of drp-1 by RNAi in the ced-9(n2812) background, the partial inactivation of drp-1 by RNAi in a wild-type background also caused highly elongated and connected mitochondria and 9.8% (± 4.1%, n = 117) embryonic lethality on average. Thus, an effect on the rates of mitochondrial fusion and fission that does not cause detectable changes in steady-state mitochondrial morphology can cause a detectable increase in embryonic survival. Therefore, it is possible that the suppression of drp-1 RNAi–induced embryonic lethality by the loss of ced-9 function is caused by a reduction in the rate of mitochondrial fusion.
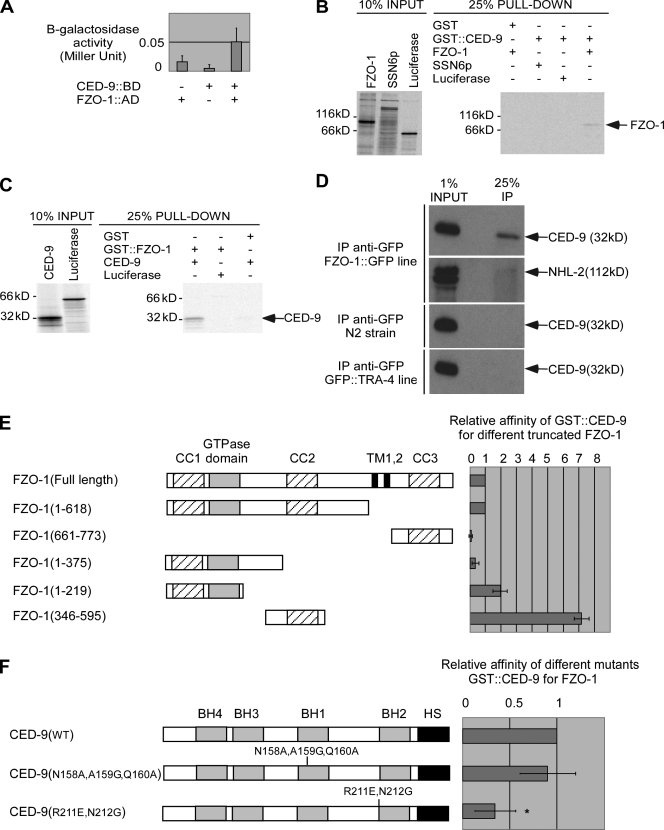
The CED-9 protein physically interacts with the FZO-1 protein
To determine whether CED-9 can physically interact with FZO-1 or EAT-3, we used the yeast two-hybrid system. We found that CED-9 can interact with FZO-1 (Fig. 6 A). We confirmed the interaction in vitro using GST pull-down experiments. Specifically, in vitro–translated, [35S]methionine-labeled FZO-1 protein copurified with GST-tagged CED-9 (GST∷CED-9) protein but not with GST alone. In vitro–translated, [35S]methionine-labeled control proteins (luciferase or the yeast protein SSN6p) did not copurify with GST∷CED-9, indicating that the interaction between GST∷CED-9 and FZO-1 is specific (Fig. 6 B). Similarly, in vitro–translated, [35S]methionine-labeled CED-9 copurified with GST-tagged FZO-1 (GST∷FZO-1) protein but not with GST alone (Fig. 6 C). To validate the interaction in vivo, using an anti-GFP antibody, we immunoprecipitated functional FZO-1∷GFP fusion protein or a control GFP fusion protein (GFP∷TRA-4) from lysates generated from transgenic animals expressing fzo-1∷gfp or gfp∷tra-4 under the control of an inducible promoter (PHSfzo-1∷gfp) or the tra-4 promoter (Ptra-4gfp∷tra-4). Using a CED-9–specific antibody (Fig. S2 H), we found that endogenous CED-9 coimmunoprecipitated with FZO-1∷GFP but not GFP∷TRA-4 (Fig. 6 D). Furthermore, endogenous CED-9 was not detected in precipitates from nontransgenic animals, and the anti-GFP antibody failed to coimmunoprecipitate an unrelated protein, NHL-2 (Hammell et al., 2009). Therefore, CED-9 can physically interact with FZO-1 in vivo, and this interaction is most probably direct.
Figure 6.
CED-9 physically interacts with FZO-1. (A) CED-9 interacts with FZO-1 in yeast. BD, Gal4p-binding domain; AD, Gal4p activation domain. (B and C) CED-9 interacts with FZO-1 in vitro. GST∷CED-9 and GST∷FZO-1 were used in pull-down experiments in the presence of in vitro–translated [35S]-labeled FZO-1 and CED-9, as described in Materials and methods. (D) CED-9 and FZO-1 interact in vivo. Embryonic lysates from wild-type (N2) animals or animals expressing PHS-fzo-1∷gfp or Ptra-4-gfp∷tra-4 were immunoprecipitated using anti-GFP antibodies, as described in Materials and methods. The precipitated proteins (IP) were analyzed for the presence of endogenous CED-9 or NHL-2 protein. (E) CED-9 interacts with the central domain of FZO-1 predicted to form a coiled-coil structure. The percentage of truncated FZO-1 protein pulled down was normalized to the percentage of full-length FZO-1 protein pulled down (n = 2). CC, coiled-coil regions; TM, transmembrane domain. (F) Mutations in the BH2 domain of CED-9 affect its ability to interact with FZO-1. The percentage of FZO-1 pulled down by the GST∷CED-9 mutant was normalized to the percentage of FZO-1 pulled down by GST∷CED-9(wt) (n = 6) . *, P < 0.0001 by Student's t test. Error bars indicate standard deviations.
To identify the domains of FZO-1 required for binding to CED-9, we tested fragments of the FZO-1 protein for their ability to bind to CED-9 in vitro. We found that FZO-1 binds to CED-9 predominantly through its central domain (aa 346–595), which is predicted to form a coiled-coil structure (Fig. 6 E). In addition, we found that FZO-1 can also bind to CED-9 through its N-terminal part (aa 1–219), which contains the GTPase domain and is predicted to form a coiled-coil structure (Fig. 6 E). We did not observe a significant interaction between aa 1–375 of the FZO-1 protein and CED-9, suggesting that aa 219–375 might form a domain that antagonizes the interaction between FZO-1 and CED-9. The presence of an inhibitory domain at aa 220–345 would also explain why aa 346–595 of the FZO-1 protein have a stronger affinity for CED-9 than the full-length FZO-1 protein (Fig. 6 E).
We also found that CED-9 interacts with EAT-3 in vitro and that this interaction is dependent on the N-terminal domain of the EAT-3 protein, which is predicted to form a coiled-coil structure (Fig. S5, A and B). However, we were not able to reproducibly confirm this interaction in vivo, indicating that this interaction might be very weak or transient.
CED-9(R211E, N212G) has reduced affinity for FZO-1
Two mutant CED-9 proteins, CED-9(N158A, A159G, Q160A) and CED-9(R211E, N212G), have been shown to have reduced affinity for the C. elegans CED-4 protein, a known interactor of CED-9 in healthy cells (Yan et al., 2004). We confirmed this using GST pull-down experiments (unpublished data). We then tested CED-9(N158A, A159G, Q160A) and CED-9(R211E, N212G) for their ability to bind to FZO-1 in vitro. We found that compared with wild-type CED-9 protein, the affinity of CED-9(R211E, N212G) for FZO-1 was three times lower. In contrast, the affinity of CED-9(N158A, A159G, Q160A) for FZO-1 was not significantly reduced (Fig. 6 F). aa 211 and 212 are located at the N terminus of the BH2 domain of CED-9. Therefore, the BH2 domain might play an important role in the interaction between CED-9 and FZO-1. Similarly, CED-9(R211E, N212G) but not CED-9(N158A, A159G, Q160A) has reduced affinity for EAT-3 in vitro (Fig. S5 C).
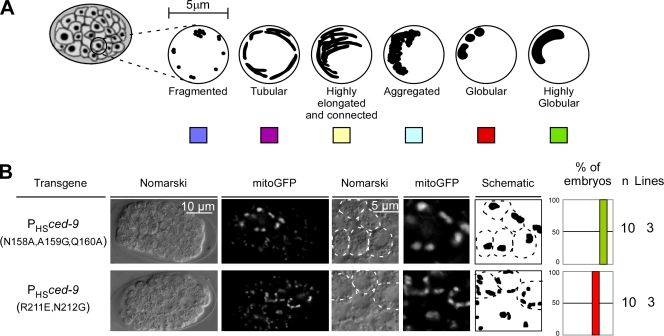
CED-9–induced mitochondrial fusion is dependent on the ability of CED-9 to interact with FZO-1
Because CED-9(R211E, N212G) has reduced affinity for FZO-1, we tested whether these mutations affect CED-9's ability to promote mitochondrial fusion. We used the protein CED-9(N158A, A159G, Q160A) as control protein because it has wild-type affinity with respect to FZO-1 binding but, like CED-9(R211E, N212G), reduced affinity with respect to CED-4 binding. We generated transgenic animals expressing mitogfp and either ced-9(N158A, A159G, Q160A) or ced-9(R211E, N212G) and analyzed mitochondrial morphology in transgenic embryos. We found that, like ced-9(wt) expression, the expression of ced-9(N158A, A159G, Q160A) caused the formation of one or two large, highly globular mitochondria per cell (100%, n = 10, three independent lines; Fig. 7 B, top). In contrast, ced-9(R211E, N212G) expression promoted mitochondrial fusion less efficiently. Specifically, it caused the formation of two to four smaller, globular mitochondria per cell (100%, n = 10, three independent lines; Fig. 7 B, bottom). The difference in mitochondrial fusion caused by ced-9(N158A, A159G, Q160A) or ced-9(R211E, N212G) expression was not caused by a difference in CED-9 protein level (Fig. S2 C). Therefore, we conclude that the ability of CED-9 to induce mitochondrial fusion is dependent on its ability to interact with FZO-1 but not CED-4.
Figure 7.
CED-9(R211E, N212G) is less effective in the promotion of mitochondrial fusion. (A) Schematic of the mitochondrial morphologies observed in embryos. (B) Transgenic lines overexpressing mitogfp in combination with ced-9(N158A, A159G, Q160A) (PHSmitogfp + PHSced-9(N158A, A159G, Q160A)) or ced-9(R211E, N212G) (PHSmitogfp + PHSced-9(R211E, N212G)) were analyzed by Nomarski optics and fluorescent microscopy. The outline of cells is indicated in the schematic of the mitochondrial network. Quantification of the different mitochondrial morphologies observed and the number of embryos and transgenic lines analyzed are indicated.
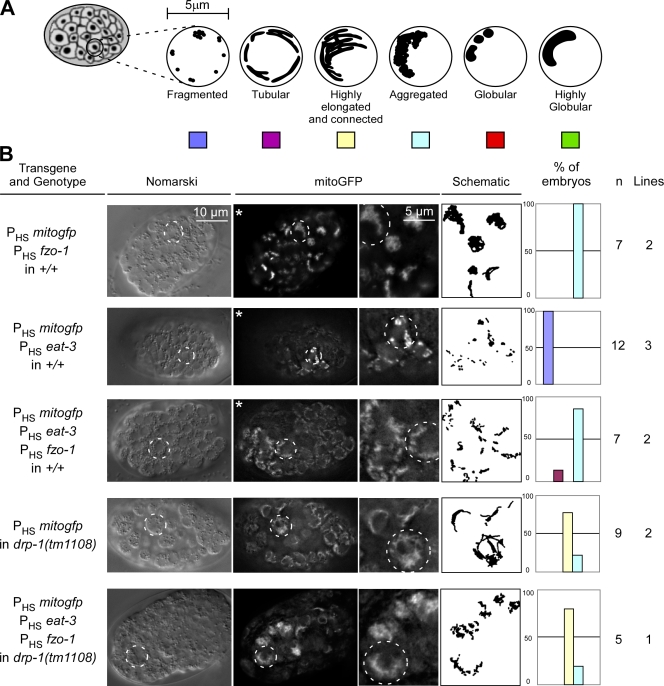
The expression of fzo-1 and eat-3 is not sufficient to cause mitochondrial fusion
To determine whether fzo-1 or eat-3 expression causes excessive mitochondrial fusion as observed in animals expressing ced-9(wt), we generated transgenic animals expressing mitogfp with fzo-1, eat-3, or fzo-1 and eat-3 and analyzed mitochondrial morphology in transgenic embryos. Unlike ced-9(wt) expression, the expression of fzo-1 caused the tubular mitochondria to aggregate into one or two large clusters per cell (100%, n = 7, two independent lines; Fig. 8 B, row 1; Table S1; and Video 8). This result was confirmed in body wall muscle cells. Although ced-9(wt) overexpression caused a complete mitochondrial fusion (Figs. 2 and 3), fzo-1 overexpression caused only mitochondrial aggregation (Figs. 2 and 3). Surprisingly, the expression of eat-3 caused mitochondria to fragment (100%, n = 12, three independent lines; Fig. 8 B, row 2; Table S1; and Video 9). Furthermore, like the expression of fzo-1 alone, the expression of both fzo-1 and eat-3 caused mitochondria to aggregate into one or two large clusters per cell (85%, n = 7, two independent lines; Fig. 8 B, row 3; Table S1; and Video 10). Finally, the expression of both fzo-1 and eat-3 in the absence of a functional drp-1 gene (i.e., in the drp-1(tm1108) background) caused many of the transgenic animals to be nonviable (unpublished data). The few embryos that we were able to analyze exhibited a highly elongated and connected mitochondrial morphology similar to the morphology observed in drp-1(tm1108) embryos expressing mitogfp alone (100%, n = 5, one independent line; Fig. 8 B, row 5).
Figure 8.
Expression of fzo-1 and eat-3 does not promote complete mitochondrial fusion. (A) Schematic of the mitochondrial morphologies observed in embryos. (B) Wild-type (+/+) or drp-1(tm1108) transgenic lines expressing mitogfp in the presence of fzo-1 (PHSmitogfp + PHSfzo-1), eat-3 (PHSmitogfp + PHSeat-3), or both fzo-1 and eat-3 (PHSmitogfp + PHSfzo-1 + PHSeat-3) were analyzed by Nomarski optics and fluorescent microscopy. Quantification of the different mitochondrial morphologies observed and the number of embryos and transgenic lines analyzed are indicated. The outlines of representative cells are indicated with dashed circles. Videos 8–10 show 3D reconstructions of the embryos indicated by asterisks.
In summary, we conclude that overexpression of the mitochondrial fusion machinery is not sufficient to cause mitochondrial fusion in either a wild-type or drp-1–deficient background. However, we cannot completely rule out the possibility that the lethality induced by the expression of fzo-1 and eat-3 in the drp-1(tm1108) background masks a potential fusion phenotype.
Discussion
ced-9(wt) expression in C. elegans is sufficient to induce FZO-1– and EAT-3–dependent mitochondrial fusion
Members of the family of BCL-2–like proteins have recently been implicated in the regulation of mitochondrial fusion (Cerveny et al., 2007). For example, the BCL-2–like proteins Bax and Bak have been shown to be required for mitochondrial fusion in mammals (Karbowski et al., 2006). Furthermore, the C. elegans BCL-2–like protein CED-9 has been shown to promote mitochondrial fusion when expressed in cultured mammalian cells (Delivani et al., 2006). Recently, CED-9 has also been shown to promote mitochondrial fusion in C. elegans body wall muscle cells (Tan et al., 2008). In this study, we confirm that CED-9 can promote mitochondrial fusion in C. elegans body wall muscle cells, and we demonstrated that it can also promote mitochondrial fusion in C. elegans embryos. In addition, we show that CED-9–induced mitochondrial fusion generates giant mitochondria, which in general have normal OMM, IMM, and cristae structure. Therefore, CED-9 can promote the complete fusion of the OMM and IMM. Furthermore, we demonstrate that CED-9's ability to promote mitochondrial fusion is dependent on the genes fzo-1 and eat-3. Specifically, ced-9(wt) expression in embryos carrying loss of function mutations in the fzo-1 or eat-3 gene failed to cause complete mitochondrial fusion and, instead, caused mitochondrial aggregation.
In contrast, fzo-1, eat-3, or fzo-1 and eat-3 overexpression did not result in the generation of giant mitochondria and therefore presumably did not cause complete fusion of the OMM and IMM. Instead, fzo-1 overexpression promoted mitochondrial aggregation, which is reminiscent of the phenotype caused by Mfn1,2 expression in cultured mammalian cells (Santel et al., 2003). It has been proposed that by promoting the tethering of mitochondria and the fusion of the OMM, Mfn1,2 acts at an early step during mitochondrial fusion (Koshiba et al., 2004). Although CED-9 promotes a complete fusion of both membranes, FZO-1 might therefore only cause hyper tethering and/or fusion of the OMM. Surprisingly, both the loss of eat-3 function and eat-3 overexpression caused mitochondrial fragmentation. Thus, EAT-3 might have dual roles in fusion and fission in C. elegans, as it has been proposed for Opa1 in mammalian cells (Cipolat et al., 2004; Griparic et al., 2004). The mechanism through which Opa1 overexpression causes fission remains elusive. However, it does not seem to be caused by a block in fusion but by Drp1-dependent fission (Chen et al., 2005).
The CED-9 protein promotes mitochondrial fusion by directly interacting with FZO-1
We demonstrate that CED-9 can interact with FZO-1 in yeast, in vitro, and in vivo. In addition, mutations that affect the BH2 domain of CED-9 reduce the affinity of CED-9 for FZO-1. The analysis of the CED-9(R211E, N212G) mutant revealed that the ability of CED-9 to promote mitochondrial fusion is not only dependent on the presence of a functional FZO-1 protein but on the ability of CED-9 to physically interact with it. Furthermore, the identification of the domains of FZO-1 required for the interaction of FZO-1 with CED-9 revealed a domain predicted to form a coiled-coil structure. The function of this domain appears to be evolutionary conserved because Bax and Bak have been shown to interact with Mfn2, and the corresponding domain of Mfn2 has been shown to be required for this interaction (Karbowski et al., 2006; Brooks et al., 2007). The identification of a physical interaction between CED-9 and FZO-1 supports the notion that CED-9 plays a role in mitochondrial fusion. However, several questions remain to be addressed. For example, it is currently unclear how CED-9 regulates the activity of FZO-1. Recent studies have shown that CED-9 in C. elegans and BCL-xL in mammals can enhance the GTPase activity of the fission protein DRP-1/Drp1 (Li et al., 2008; Tan et al., 2008). Therefore, it is possible that CED-9 regulates FZO-1 function and promotes mitochondrial fusion by enhancing the GTPase activity of FZO-1. Alternatively, CED-9 could affect the ability of FZO-1 to dimerize. We found that CED-9 can also interact with EAT-3 in vitro. However, we were not able to reproducibly observe this interaction in vivo, which suggests that it might be very weak or transient.
Based on our results, we speculate that CED-9–induced mitochondrial fusion proceeds through the following sequential steps: (1) CED-9 interacts with FZO-1 to promote mitochondrial tethering and/or OMM fusion, and (2) CED-9–dependent activation of FZO-1 results in the FZO-1–dependent activation of EAT-3 through a mechanism that remains to be elucidated. Alternatively, CED-9 might insert into the OMM and interact with EAT-3 in the intermembrane space to directly promote tethering and/or IMM fusion.
The role of CED-9 in mitochondrial fusion
Our results show that CED-9 can promote complete mitochondrial fusion in C. elegans by interacting with the mitochondrial fusion machinery. These results are consistent with previous work by Delivani et al. (2006) in cultured mammalian cells and by Tan et al. (2008) in C. elegans. However, our results are in apparent contradiction with recent work from Breckenridge et al. (2009), who proposed that CED-9 plays no role in mitochondrial dynamics in C. elegans. The discrepancy between their results and our results can possibly be explained by the higher concentration of the PHSced-9(wt) transgene used by Breckenridge et al. (2009) to generate transgenic lines (25 ng/μl instead of 5 ng/μl in our study). Based on our analyses of body wall muscle cells, the resulting high levels of CED-9 protein might be toxic and therefore cause transgenic animals to be nonviable. Breckenridge et al. (2009) selected for transgenic animals that carry stable, integrated transgenes in their genomes. During this process, CED-9–dependent toxicity might have led to the selection of lines generating very low levels of CED-9 protein from the ced-9(wt) transgene. This low level of CED-9 might be sufficient to block apoptosis but not to promote mitochondrial fusion. Because Breckenridge et al. (2009) did not analyze CED-9 protein levels in their transgenic lines, it remains to be determined how their results relate to the results presented in this study.
Our results also indicate that, unlike FZO-1 and EAT-3, CED-9 does not play a major role in mitochondrial fusion during embryogenesis or in body wall muscle cells during larval development. Indeed, even though ced-9 loss of function animals have slightly shorter mitochondria and ced-9(n1950gf) animals have slightly longer mitochondria when compared with control animals, the differences are not statistically significant. Similarly, we found that ced-9(n2812) can suppress drp-1 RNAi–induced embryonic lethality, possibly by reducing the rate of mitochondrial fusion. However, we could not detect an effect of ced-9(n2812) on the mitochondrial morphology phenotype caused by drp-1 RNAi.
If ced-9 does not have a major role in mitochondria fusion, what could its physiological role be? We are considering the following four possibilities: (1) the role of ced-9 in mitochondrial fusion might be redundant with that of one or more additional genes. Thus, the inactivation of ced-9 alone might not be sufficient to cause a defect in mitochondrial fusion. (2) ced-9 might play a minor role in mitochondrial fusion during embryogenesis and in body wall muscle cells during larval development. In that case, the loss of ced-9 function might not cause a detectable phenotype when analyzing steady-state mitochondrial morphology. (3) ced-9 has previously been shown to promote mitochondrial fission during apoptosis (Jagasia et al., 2005). Therefore, ced-9 has both profusion and profission activities. As a consequence, the loss of ced-9 function might affect both fusion and fission rates, resulting in no net change in steady-state mitochondrial morphology. (4) ced-9 might play a role in mitochondrial fusion only during certain developmental stages, in specific cell types, or under conditions of stress. Therefore, the loss of ced-9 function might cause a detectable phenotype only in the presence of such a specific cellular signal. We favor the last two possibilities. Specifically, we propose that in the absence of a specific cellular signal, the CED-9 protein promotes both mitochondrial fusion and fission and that in healthy cells, these opposing activities are balanced. Furthermore, we propose that in response to a specific cellular signal, CED-9 profusion and/or profission activity changes, which affects the balance between fusion and fission and thus mitochondrial morphology. The goal of future studies is to further test this model experimentally and to fully elucidate the physiological role of ced-9 in mitochondrial dynamics.
C. elegans CED-9 is not the only BCL-2–like protein with an apparent role in both mitochondrial fusion and fission. For example, although BCL-xL expression promotes mitochondrial fusion in cultured mammalian cells, it has been shown to be required for mitochondrial fragmentation in neurons (Delivani et al., 2006; Li et al., 2008; Berman et al., 2009). Similarly, depending on the cellular context, Bax and Bak can either promote mitochondrial fusion or fission (Desagher and Martinou, 2000; Karbowski et al., 2002, 2006; Brooks et al., 2007). Therefore, analyzing the regulation of the opposing roles of BCL-2–like proteins in mitochondrial dynamics represents another future challenge.
Materials and methods
General methods and strains
C. elegans strains were cultured as described previously (Brenner, 1974). Bristol N2 was used as the wild-type strain. Mutations used in this study are listed below and were described previously by Riddle et al. (1997) except where noted otherwise: LG I, smg-1(cc546ts) (Pulak and Anderson, 1993); LG II, fzo-1(tm1133) (National BioResource Project), dpy-10(e128), and eat-3(ad426); LG III, ced-4(n1162), ced-9(n1653ts), ced-9(n1950gf), and ced-9(n2812) (Hengartner and Horvitz, 1994b); LG IV, drp-1(tm1108) (National BioResource Project), ced-3(n717), and ced-3(n2427) (Hengartner and Horvitz, 1994b); and LG X, lin-15(n765ts).
Transgenic animals
For the analysis of mitochondrial morphology in embryos, all plasmids were injected into lin-15(n765ts) animals using 50 ng/μl pL15EK, which contains the lin-15–rescuing fragment, as a coinjection marker. PHSmitogfp was injected at 1 ng/μl, PHSced-9(wt) (pBC876/877), PHSced-9(N158A, A159G, Q160A) (pBC636/637), PHSced-9(R211E, N212G) (pBC638/639), PHSdrp-1 (pBC401/403), and PHSfzo-1 (pBC647/648) at 5 ng/μl, PHSeat-3 (pBC673/674) at 20 ng/μl, and PHSfzo-1∷gfp (pBC683/684) at 10 ng/μl.
For the analysis of mitochondrial morphology in body wall muscle cells, all plasmids were coinjected with pRF4, which contains the rol-6(su1006dm) allele, which causes a dominant Rol phenotype (80 ng/μl; Kramer et al., 1990). Pmyo-3 mitogfp (provided by A. Van der Bliek, University of California, Los Angeles, Los Angeles, CA) was injected at 5 ng/μl, Pmyo-3 ced-9(wt) (3′UTRsmg) (pBC862) at 20 ng/μl, and Pmyo-3 fzo-1 (3′UTRsmg) (pBC863) at 20 ng/μl. For a summary of all the transgenic lines generated for this study, see Table S2.
Analysis of mitochondrial morphology in embryos using TMRE or MitoTracker staining
Adult animals were grown at 20°C in the presence of 30 μM TMRE (Invitrogen) for 16 h as described previously (Jagasia et al., 2005) or in the presence of 0.4 μg/ml MitoTracker-CMXRos as described previously (Delivani et al., 2006). For ced-9(n1653ts), adults were grown at 20°C in the presence of TMRE for 12 h and then shifted to 25°C for 4 h.
RNA-mediated interference
RNAi was performed as described previously (Timmons et al., 2001). For ced-9 RNAi, rrf-3(pk1426) L4 larvae were grown at 15°C on ced-9 RNAi plates for 24 h and transferred for 16 h onto regular TMRE plates without RNAi. Adults were cut open, and embryos were mounted in M9 medium and analyzed by Nomarski optics and fluorescent microscopy.
Partial inactivation of drp-1 by RNAi was achieved by mixing the Escherichia coli strain producing drp-1 dsRNA with an E. coli strain carrying a mock RNAi plasmid.
Analysis of mitochondrial morphology in body wall muscle cells
Adult Rol animals (transgenic animals) were allowed to lay eggs for 2 h at 20°C and were then removed. Plates were incubated for 2–3 d at 20°C, and Rol L4 larvae of the next (F1) generation were analyzed by Nomarski optics and fluorescent microscopy. For RNAi experiments, Rol L4 larvae were grown on RNAi plates for 24 h at 20°C and transferred to fresh RNAi plates for 2–3 d. Rol L4 larvae of the F1 generation were analyzed by Nomarski optics and fluorescent microscopy. Muscle cells with >50% of fragmented mitochondria were scored as muscle cells with a mitochondrial fragmentation phenotype. Scoring was performed blind.
Overexpression experiments
L4 larvae from transgenic lines were picked and grown at 20°C for 24 h. Nonmultivulval adults (transgenic animals) were transferred to new plates, allowed to lay eggs for 2 h at 20°C, heat shocked at 32°C for 45 min, and incubated at 20°C for 1 h and 15 min before being removed from the plates. Eggs laid during the 4-h period were mounted in M9 medium and analyzed by Nomarski optics and fluorescent microscopy. Throughout this study, transgenic animals carrying extrachromosomal arrays were used. Those animals are mosaic for the presence of the transgenes and thus the expression of the transgenes. However, the mitochondrial morphology observed in individual mitoGFP-positive cells in a given embryo was in general homogeneous. For this reason, the data are presented as the percentage of embryos showing a certain phenotype.
Pmyo-3 smg–inducible system
This system takes advantage of a temperature-sensitive allele of a component of the Smg RNA surveillance machinery (smg-1(cc546ts)). In a smg-1(cc546ts) mutant background, an mRNA with an abnormally long 3′ UTR (3′UTRsmg) is degraded at the permissive temperature (15°C) but translated at the nonpermissive temperature (25°C). Full-length ced-9 and fzo-1 cDNAs were cloned into pPD118.60 (provided by A. Fire, Stanford School of Medicine, Stanford, CA) to construct Pmyo-3 ced-9(wt) (3′UTRsmg) and Pmyo-3 fzo-1 (3′UTRsmg). smg-1(cc546ts) transgenic animals carrying Pmyo-3 ced-9(wt) (3′UTRsmg) or Pmyo-3 fzo-1(3′UTRsmg) were maintained at 15°C and transferred at 25°C to induce the expression of the transgene. L4 larvae of the F1 generation were analyzed by fluorescent microscopy or transmission EM.
Image acquisition and processing
Imaging was performed at 20°C using a microscope equipped with a 100× 1.3 NA oil lens (Axioskop 2; Carl Zeiss, Inc.) and a charge-coupled device camera (1300; Micromax). A Nomarski and fluorescent z stack of each embryo (0.5 μm/slice) was acquired using MetaMorph software (version 7.1; MDS Analytical Technologies). The same acquisition time was used for each experiment (10 ms for Nomarski, 100 ms for TMRE staining, and 200 ms for mitoGFP). The fluorescent images were deconvoluted (10 iterations) blind using AutoDeblur/AutoVisualize software (version 1.4.1; Media Cybernetics). Images shown in the figures correspond to single planes. Images were inverted, and their contrast was enhanced using Paint Shop Pro X software (Corel). The resulting images were used to generate the schematics of the mitochondrial network. Analysis of mitochondrial length in body wall muscle cells was performed on deconvoluted images using MetaMorph.
Transmission EM
Wild-type and mutant nematodes were prepared for EM by high pressure freezing (HPM 010; Bal-Tec) and freeze substitution (RMC FS-7500; Boeckeler Instruments) into 1% osmium tetroxide in acetone (2% water added) following a standard protocol (Weimer, 2006) and embedded in plastic resin (Embed 812; Electron Microscopy Sciences). Semithin sections (0.5–0.75 μm) were cut (MT6000; RMC) and stained with Toluidine blue to determine the location within the fixed animal. When the desired area was located, thin sections (70–90 nm) were collected on 400 mesh 8 U thin-bar grids (EMS400H-Cu). Finally, the grids were stained with 2% methanolic uranyl acetate for 20 min followed by Reynold's lead citrate for a maximum of 5 min. Samples were viewed on a transmission electron microscope (1010; JEOL) at 100 kV.
Yeast two-hybrid interactions
The full-length fzo-1 cDNA was cloned into the vector pACT2 to create a GAL4 DNA transactivation domain fusion. The full-length ced-9 cDNA was cloned into the vector pBridge to create a GAL4-binding domain fusion (Bai and Elledge, 1996). Yeast strain Y190 was cotransformed with these plasmids by using standard protocols and transformants selected on SD medium lacking tryptophane and leucine. β-Galactosidase activity was assayed on filters as described previously by Bai and Elledge (1996). Three independent experiments were performed using three independent clones of each strain.
In vitro interactions
In vitro interactions were performed by GST pull-down essentially as described previously (Conradt and Horvitz, 1998). In brief, GST∷CED-9 was produced in BL21(DE3) cells, extracted with 1 ml of NETN buffer (20 mM Tris-HCl, pH 8, 0.5% NP-40, 100 mM NaCl, and a cocktail of protease inhibitor [Roche]) by six sonication pulses of 10 s at 30% power (digital sonifier; Branson) and purified using glutathione agarose beads (GE Healthcare). FZO-1 and EAT-3 were in vitro translated in the presence of [35S]methionine using the TNT Rabbit Reticulocyte Lysate System (Promega). The binding of [35S]methionine-labeled FZO-1 or EAT-3 with purified GST∷CED-9 was performed for 4 h at 4°C in 500 μl of binding buffer (30 mM Tris-HCl, pH 7.5, 0.5% Triton X-100, 100 mM NaCl, 2 mM MgCl2, 1 mM DTT, 0.5% BSA, and a cocktail of protease inhibitor). Agarose beads were washed three times with 1 ml of binding buffer, one time with 1 ml of binding buffer without BSA, resuspended in Laemmli buffer, and boiled for 5 min. The input and pull-down were analyzed by SDS-PAGE and autoradiography using a Storm Imager (GE Healthcare). For GST∷FZO-1, the fzo-1 cDNA was cloned into the pGEX-4T-3, expressed in BL21(DE3) cells, and purified using the FZO-1 extraction buffer (30 mM Tris-HCl, pH 7.5, 1% Triton X-100, 100 mM NaCl, 2 mM MgCl2, 1 mM DTT, and a cocktail of protease inhibitor).The binding of [35S]methionine-labeled CED-9 with purified GST∷FZO-1 was performed as described for the binding of [35S]methionine-labeled Fzo1 with GST∷CED-9.
In vivo interactions
In vivo interactions were performed by coimmunoprecipitation essentially as described previously (Grote and Conradt, 2006). In brief, embryos from N2 animals and transgenic animals expressing fzo-1∷gfp or gfp∷tra-4 were resuspended in 1 vol of 2× coimmunoprecipitation buffer (100 mM Tris-HCl, pH 7.5, 2% Triton X-100, 200 mM NaCl, 4 mM MgCl2, 2 mM DTT, and a cocktail of protease inhibitor) and homogenized by sonicating six times for 10 s at 30% power (digital sonifier; Branson). Immunoprecipitations were performed with monoclonal anti-GFP antibody (anti-AFP; mAb 3E6; MP Biomedicals) for 2 h at 4°C. Immunoprecipitates were captured using 20 μl protein A–Sepharose beads (GE Healthcare) for 1 h at 4°C, washed three times with 1 ml of coimmunoprecipitation buffer, eluted with 20 μl of Laemmli buffer, and boiled for 5 min. The immunoprecipitates and the input were analyzed by SDS-PAGE. To detect CED-9, we used a rabbit polyclonal anti–CED-9 antibody (1:2,000). To detect NHL-2, we used rabbit polyclonal anti–NHL-2 antibody (1:5,000; Hammell et al., 2009).
Online supplemental material
Fig. S1 shows that ced-9(n1950gf) promotes FZO-1– and EAT-3–dependent mitochondrial fusion. Fig. S2 shows Western analyses of the different transgenic lines used in this study. Fig. S3 shows MitoTracker versus TMRE mitochondrial staining of embryos. Fig. S4 shows the analysis of mitochondrial length in body wall muscle cells of different mutant strains. Fig. S5 shows that CED-9 interacts with EAT-3 in vitro. Videos 1–10 were generated from the z stacks of the embryos after 3D reconstruction using AutoDeblur/AutoVisualize software. A description of the transgenic embryos shown in the videos is indicated in Table S1. A description of the transgenic lines used in this study is indicated in Table S2. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200905070/DC1.
Acknowledgments
We thank E. Lambie (Dartmouth College, Hanover, NH), D. Hall (Albert Einstein College of Medicine, New York, NY), and members of the Conradt laboratory for comments on the manuscript, D. Mayka for excellent technical support, D. Hall for help and training in electron microscopy (D. Hall is supported by National Institutes of Health grant RR12596), L. Gunther and K. Nguyen (Albert Einstein College of Medicine) for their help with high pressure freezing runs, L. Howard (Dartmouth College) for excellent technical support for electron microscopy analysis, E. Lambie for use of the microinjection set up, A. Fire for plasmids, A. van der Bliek for plasmids and unpublished data, B. Trumpower (Dartmouth College) for antibodies, C. Hammell and V. Ambros (University of Massachusetts Medical School, Worcester, MA) for antibodies, S. Mitani (National BioResource Project, Tokyo, Japan) for fzo-1(tm1133) and drp-1(tm1108), and the C. elegans Genetics Center for strains (supported by the National Center for Research Resources).
This work was supported by funding from the Howard Hughes Medical Institute Award (76200-560801) to Dartmouth Medical School under the Biomedical Research Support Program for Medical Schools, a W.P. and D.N. Harris Distinguished Visiting Professorship (to C.N. David), an American Cancer Society grant (RSG-06-110-1-CCG), and a National Institutes of Health grant (GM076651 to B. Conradt).
Footnotes
Abbreviations used in this paper:
- IMM
- inner mitochondrial membrane
- OMM
- outer mitochondrial membrane
- TMRE
- tetramethyl rhodamine ethyl ester
- UTR
- untranslated region
References
- Bai C., Elledge S.J. 1996. Gene identification using the yeast two-hybrid system.Methods Enzymol. 273:331–347 [DOI] [PubMed] [Google Scholar]
- Berman S.B., Pineda F.J., Hardwick J.M. 2008. Mitochondrial fission and fusion dynamics: the long and short of it.Cell Death Differ. 15:1147–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman S.B., Chen Y.B., Qi B., McCaffery J.M., Rucker E.B., III, Goebbels S., Nave K.A., Arnold B.A., Jonas E.A., Pineda F.J., Hardwick J.M. 2009. Bcl-xL increases mitochondrial fission, fusion, and biomass in neurons.J. Cell Biol. 184:707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breckenridge D.G., Kang B.H., Kokel D., Mitani S., Staehelin L.A., Xue D. 2008. Caenorhabditis elegans drp-1 and fis-2 regulate distinct cell-death execution pathways downstream of ced-3 and independent of ced-9.Mol. Cell. 31:586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breckenridge D.G., Kang B.H., Xue D. 2009. Bcl-2 proteins EGL-1 and CED-9 do not regulate mitochondrial fission or fusion in Caenorhabditis elegans.Curr. Biol. 19:768–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans.Genetics. 77:71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks C., Wei Q., Feng L., Dong G., Tao Y., Mei L., Xie Z.J., Dong Z. 2007. Bak regulates mitochondrial morphology and pathology during apoptosis by interacting with mitofusins.Proc. Natl. Acad. Sci. USA. 104:11649–11654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny K.L., Tamura Y., Zhang Z., Jensen R.E., Sesaki H. 2007. Regulation of mitochondrial fusion and division.Trends Cell Biol. 17:563–569 [DOI] [PubMed] [Google Scholar]
- Chen H., Detmer S.A., Ewald A.J., Griffin E.E., Fraser S.E., Chan D.C. 2003. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development.J. Cell Biol. 160:189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Chomyn A., Chan D.C. 2005. Disruption of fusion results in mitochondrial heterogeneity and dysfunction.J. Biol. Chem. 280:26185–26192 [DOI] [PubMed] [Google Scholar]
- Cheng W.C., Teng X., Park H.K., Tucker C.M., Dunham M.J., Hardwick J.M. 2008. Fis1 deficiency selects for compensatory mutations responsible for cell death and growth control defects.Cell Death Differ. 15:1838–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolat S., Martins de Brito O., Dal Zilio B., Scorrano L. 2004. OPA1 requires mitofusin 1 to promote mitochondrial fusion.Proc. Natl. Acad. Sci. USA. 101:15927–15932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolat S., Rudka T., Hartmann D., Costa V., Serneels L., Craessaerts K., Metzger K., Frezza C., Annaert W., D'Adamio L., et al. 2006. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling.Cell. 126:163–175 [DOI] [PubMed] [Google Scholar]
- Conradt B., Horvitz H.R. 1998. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9.Cell. 93:519–529 [DOI] [PubMed] [Google Scholar]
- Delivani P., Adrain C., Taylor R.C., Duriez P.J., Martin S.J. 2006. Role for CED-9 and Egl-1 as regulators of mitochondrial fission and fusion dynamics.Mol. Cell. 21:761–773 [DOI] [PubMed] [Google Scholar]
- Desagher S., Martinou J.C. 2000. Mitochondria as the central control point of apoptosis.Trends Cell Biol. 10:369–377 [DOI] [PubMed] [Google Scholar]
- Frezza C., Cipolat S., Martins de Brito O., Micaroni M., Beznoussenko G.V., Rudka T., Bartoli D., Polishuck R.S., Danial N.N., De Strooper B., Scorrano L. 2006. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion.Cell. 126:177–189 [DOI] [PubMed] [Google Scholar]
- Fritz S., Weinbach N., Westermann B. 2003. Mdm30 is an F-box protein required for maintenance of fusion-competent mitochondria in yeast.Mol. Biol. Cell. 14:2303–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griparic L., van der Wel N.N., Orozco I.J., Peters P.J., van der Bliek A.M. 2004. Loss of the intermembrane space protein Mgm1/OPA1 induces swelling and localized constrictions along the lengths of mitochondria.J. Biol. Chem. 279:18792–18798 [DOI] [PubMed] [Google Scholar]
- Grote P., Conradt B. 2006. The PLZF-like protein TRA-4 cooperates with the Gli-like transcription factor TRA-1 to promote female development in C. elegans.Dev. Cell. 11:561–573 [DOI] [PubMed] [Google Scholar]
- Hammell C.M., Lubin I., Boag P.R., Blackwell T.K., Ambros V. 2009. nhl-2 Modulates microRNA activity in Caenorhabditis elegans.Cell. 136:926–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengartner M.O., Horvitz H.R. 1994a. Activation of C. elegans cell death protein CED-9 by an amino-acid substitution in a domain conserved in Bcl-2.Nature. 369:318–320 [DOI] [PubMed] [Google Scholar]
- Hengartner M.O., Horvitz H.R. 1994b. C. elegans cell survival gene ced-9 encodes a functional homolog of the mammalian proto-oncogene bcl-2.Cell. 76:665–676 [DOI] [PubMed] [Google Scholar]
- Herlan M., Bornhövd C., Hell K., Neupert W., Reichert A.S. 2004. Alternative topogenesis of Mgm1 and mitochondrial morphology depend on ATP and a functional import motor.J. Cell Biol. 165:167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann G.J., Thatcher J.W., Mills J.P., Hales K.G., Fuller M.T., Nunnari J., Shaw J.M. 1998. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p.J. Cell Biol. 143:359–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S., Nunnari J. 2009. The molecular mechanism of mitochondrial fusion.Biochim. Biophys. Acta. 1793:20–26 [DOI] [PubMed] [Google Scholar]
- Hoppins S., Horner J., Song C., McCaffery J.M., Nunnari J. 2009. Mitochondrial outer and inner membrane fusion requires a modified carrier protein.J. Cell Biol. 184:569–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichishita R., Tanaka K., Sugiura Y., Sayano T., Mihara K., Oka T. 2008. An RNAi screen for mitochondrial proteins required to maintain the morphology of the organelle in Caenorhabditis elegans.J. Biochem. 143:449–454 [DOI] [PubMed] [Google Scholar]
- Ishihara N., Fujita Y., Oka T., Mihara K. 2006. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1.EMBO J. 25:2966–2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagasia R., Grote P., Westermann B., Conradt B. 2005. DRP-1-mediated mitochondrial fragmentation during EGL-1-induced cell death in C. elegans.Nature. 433:754–760 [DOI] [PubMed] [Google Scholar]
- Kanazawa T., Zappaterra M.D., Hasegawa A., Wright A.P., Newman-Smith E.D., Buttle K.F., McDonald K., Mannella C.A., van der Bliek A.M. 2008. The C. elegans Opa1 homologue EAT-3 is essential for resistance to free radicals.PLoS Genet. 4:e1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M., Lee Y.J., Gaume B., Jeong S.Y., Frank S., Nechushtan A., Santel A., Fuller M., Smith C.L., Youle R.J. 2002. Spatial and temporal association of Bax with mitochondrial fission sites, Drp1, and Mfn2 during apoptosis.J. Cell Biol. 159:931–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M., Norris K.L., Cleland M.M., Jeong S.Y., Youle R.J. 2006. Role of Bax and Bak in mitochondrial morphogenesis.Nature. 443:658–662 [DOI] [PubMed] [Google Scholar]
- Koshiba T., Detmer S.A., Kaiser J.T., Chen H., McCaffery J.M., Chan D.C. 2004. Structural basis of mitochondrial tethering by mitofusin complexes.Science. 305:858–862 [DOI] [PubMed] [Google Scholar]
- Kramer J.M., French R.P., Park E.C., Johnson J.J. 1990. The Caenorhabditis elegans rol-6 gene, which interacts with the sqt-1 collagen gene to determine organismal morphology, encodes a collagen.Mol. Cell. Biol. 10:2081–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrousse A.M., Zappaterra M.D., Rube D.A., van der Bliek A.M. 1999. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane.Mol. Cell. 4:815–826 [DOI] [PubMed] [Google Scholar]
- Lackner L.L., Nunnari J.M. 2008. The molecular mechanism and cellular functions of mitochondrial division.Biochim. Biophys. Acta. 10.1016/j.bbadis.2008.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Chen Y., Jones A.F., Sanger R.H., Collis L.P., Flannery R., McNay E.C., Yu T., Schwarzenbacher R., Bossy B., et al. 2008. Bcl-xL induces Drp1-dependent synapse formation in cultured hippocampal neurons.Proc. Natl. Acad. Sci. USA. 105:2169–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S., McCaffery J.M., Nunnari J. 2004. Mitochondrial fusion intermediates revealed in vitro.Science. 305:1747–1752 [DOI] [PubMed] [Google Scholar]
- Meeusen S., DeVay R., Block J., Cassidy-Stone A., Wayson S., McCaffery J.M., Nunnari J. 2006. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1.Cell. 127:383–395 [DOI] [PubMed] [Google Scholar]
- Misaka T., Murate M., Fujimoto K., Kubo Y. 2006. The dynamin-related mouse mitochondrial GTPase OPA1 alters the structure of the mitochondrial inner membrane when exogenously introduced into COS-7 cells.Neurosci. Res. 55:123–133 [DOI] [PubMed] [Google Scholar]
- Neutzner A., Youle R.J. 2005. Instability of the mitofusin Fzo1 regulates mitochondrial morphology during the mating response of the yeast Saccharomyces cerevisiae.J. Biol. Chem. 280:18598–18603 [DOI] [PubMed] [Google Scholar]
- Okamoto K., Shaw J.M. 2005. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes.Annu. Rev. Genet. 39:503–536 [DOI] [PubMed] [Google Scholar]
- Praefcke G.J., McMahon H.T. 2004. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 5:133–147 [DOI] [PubMed] [Google Scholar]
- Pulak R., Anderson P. 1993. mRNA surveillance by the Caenorhabditis elegans smg genes.Genes Dev. 7:1885–1897 [DOI] [PubMed] [Google Scholar]
- Riddle D.L. 1997. C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 1222 pp [PubMed] [Google Scholar]
- Santel A., Frank S., Gaume B., Herrler M., Youle R.J., Fuller M.T. 2003. Mitofusin-1 protein is a generally expressed mediator of mitochondrial fusion in mammalian cells.J. Cell Sci. 116:2763–2774 [DOI] [PubMed] [Google Scholar]
- Saraste M. 1999. Oxidative phosphorylation at the fin de siècle.Science. 283:1488–1493 [DOI] [PubMed] [Google Scholar]
- Sesaki H., Jensen R.E. 2004. Ugo1p links the Fzo1p and Mgm1p GTPases for mitochondrial fusion.J. Biol. Chem. 279:28298–28303 [DOI] [PubMed] [Google Scholar]
- Sesaki H., Southard S.M., Yaffe M.P., Jensen R.E. 2003. Mgm1p, a dynamin-related GTPase, is essential for fusion of the mitochondrial outer membrane.Mol. Biol. Cell. 14:2342–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen D.F., Norris K.L., Youle R.J. 2008. Mitochondrial dynamics and apoptosis.Genes Dev. 22:1577–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabadkai G., Duchen M.R. 2008. Mitochondria: the hub of cellular Ca2+ signaling.Physiology (Bethesda). 23:84–94 [DOI] [PubMed] [Google Scholar]
- Tan F.J., Husain M., Manlandro C.M., Koppenol M., Fire A.Z., Hill R.B. 2008. CED-9 and mitochondrial homeostasis in C. elegans muscle.J. Cell Sci. 121:3373–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., Court D.L., Fire A. 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans.Gene. 263:103–112 [DOI] [PubMed] [Google Scholar]
- Weimer R.M. 2006. Preservation of C. elegans tissue via high-pressure freezing and freeze-substitution for ultrastructural analysis and immunocytochemistry.Methods Mol. Biol. 351:203–221 [DOI] [PubMed] [Google Scholar]
- Westermann B. 2008. Molecular machinery of mitochondrial fusion and fission.J. Biol. Chem. 283:13501–13505 [DOI] [PubMed] [Google Scholar]
- Yan N., Gu L., Kokel D., Chai J., Li W., Han A., Chen L., Xue D., Shi Y. 2004. Structural, biochemical, and functional analyses of CED-9 recognition by the proapoptotic proteins EGL-1 and CED-4.Mol. Cell. 15:999–1006 [DOI] [PubMed] [Google Scholar]
- Youle R.J., Strasser A. 2008. The BCL-2 protein family: opposing activities that mediate cell death.Nat. Rev. Mol. Cell Biol. 9:47–59 [DOI] [PubMed] [Google Scholar]