Abstract
Glycosylation is among the most complex posttranslational modifications with an extremely high level of diversity that has made it refractory to high-throughput analyses. Despite its resistance to high-throughput techniques, glycosylation is important in many critical cellular processes that necessitate a productive approach to their analysis. To facilitate studies in glycosylation, we developed a high-throughput lectin microarray for defining mammalian cell surface glycan signatures. Using the lectin microarray we established a binary analysis of cell binding and hierarchical organization of 24 mammalian cell lines. The array was also used to document changes in cell surface glycosylation during cell development and differentiation of primary murine immune system cells. To establish the biological and clinical importance of glycan signatures, the lectin microarray was applied in two systems. First, we analyzed the cell surface glycan signatures and were able to predict mannose-dependent tropism using a model pathogen. Second, we used the glycan signatures to identify novel lectin biomarkers for cancer stem-like cells in a murine model. Thus, lectin microarrays are an effective tool for analyzing diverse cell processes including cell development and differentiation, cell–cell communication, pathogen-host recognition, and cell surface biomarker identification.
Keywords: biomaker, cell differentiation, glycan signature, lectin microarray, pathogen tropism
Introduction
The surfaces of all vertebrate cells are distinguished by a dense and diverse array of cell surface glycans, the “glycocalyx”. Glycosylation is a major form of posttranslational modification (Kukuruzinska et al. 1987; Lehle et al. 2006) and variation in glycosylation is associated with cell differentiation and malignant transformation (Hakomori 1985, 2002; Dennis et al. 1999; Dwek et al. 2001). This makes it a potentially valuable diagnostic indicator of cellular diversity and differentiation (Brooks and Leathem 1991). Specific surface glycosylation patterns are characteristic features of certain cell types, such as embryonic stem cells (Muramatsu 2002), and whole organs, such as the kidney and ABO blood group antigens (Palcic et al. 1990; Morgan and Watkins 2000). Recently, a growing body of evidence has indicated that changes in the expression of sugar-bearing molecules may actively direct a variety of cell biological processes (Haltiwanger and Lowe 2004; Van Dyken et al. 2007). The combinatorial possibilities inherent in a glycan structure far exceed DNA- and peptide-based structural diversity (Gabius et al. 2004). This enormous structural diversity coupled with accessibility at the cell surface makes glycosylation an important mechanism for regulating cellular interactions with the environment, neighboring cells, and pathogens.
The same structural diversity that provides a cell surface glycome with biological significance also makes it refractory to rapid detailed analysis. Nevertheless, lectins, which are glycan binding proteins, have long been used in techniques such as blots, flow cytometry, and immunohistochemistry to characterize serum or cells by focusing on individual glycans (Reisner et al. 1976; Yuan et al. 1986; Macartney 1987; Varki 1999; Ferguson et al. 2005). Lectin-based approaches are particularly advantageous because of their ability to discriminate sugar isomers (Hirabayashi 2004). Given the commercial availability of a large number of lectins with a variety of glycan specificities, their use in microarrays to provide more accurate and detailed “cell surface glycan signatures” is an inviting alternative to structural analyses, the latter of which are not amenable to rapid or high-throughput approaches. Although lectin microarrays have been used to profile glycoproteins and cell lysates (Kuno et al. 2005; Ebe et al. 2006; Lee et al. 2006; Pilobello et al. 2007), their use for profiling live cells has been limited to proof-of-principle studies (Zheng et al. 2005; Lee et al. 2006). For example, lectin microarrays accurately and predictably distinguish between glycosylation-defective CHO mutants (Tateno et al. 2007), and Hsu et al. (2006) used a 21-lectin microarray to distinguish O-antigens on lipopolysaccharides (LPS) of bacterial cell surfaces. Here, we demonstrate that a lectin microarray can be used to reveal biologically and clinically relevant cell surface glycan signatures and report the application of this technology to immune cell differentiation, pathogen-cell tropism, and cancer stem cell identification. These findings show that lectin microarrays are an excellent tool both for prediction of glycan-based cell–cell interactions and identification of novel cell surface biomarkers.
Results
Surface glycan signatures defined by lectin microarrays in mammalian cells
To probe the accessible surface glycans or cell surface glycan signatures, we collected a panel of 94 lectins with an extensive range of specificities (supplementary Table I). Fluorescently labeled live cells selectively interacted with a subset of lectins on the microarray (supplementary Figure 1A). Individual cells captured on 120-micron spots could be readily distinguished by fluorescent microscopy, indicating that their interactions with lectins immobilized on solid surfaces were due to the binding of intact cells (supplementary Figure 1A, inset). To determine if the binding signals observed resulted from direct interactions between the lectins and glycans on the cell surfaces, we tested the specificity using the simple sugar inhibitors, α-methyl-mannoside or the disaccharide lactose (Galβ1-4Glc). Labeled HeLa and 293 cells were incubated on lectin microarrays in the presence or absence of either α-methyl-mannoside (200 mM) or lactose (200 mM). The binding of both cell lines to the lectins LCA, specific for mannose, and TKA, specific for lactose or terminal galactose, was inhibited only by their respective glycan ligands (supplementary Figure 1B). Cell binding to SNA, which is not specific for either mannose or lactose, was unaffected by the presence of either soluble glycans. The lectin array demonstrates glycan-specific cell binding and is useful in probing accessible cell surface glycans.
Fig. 1.
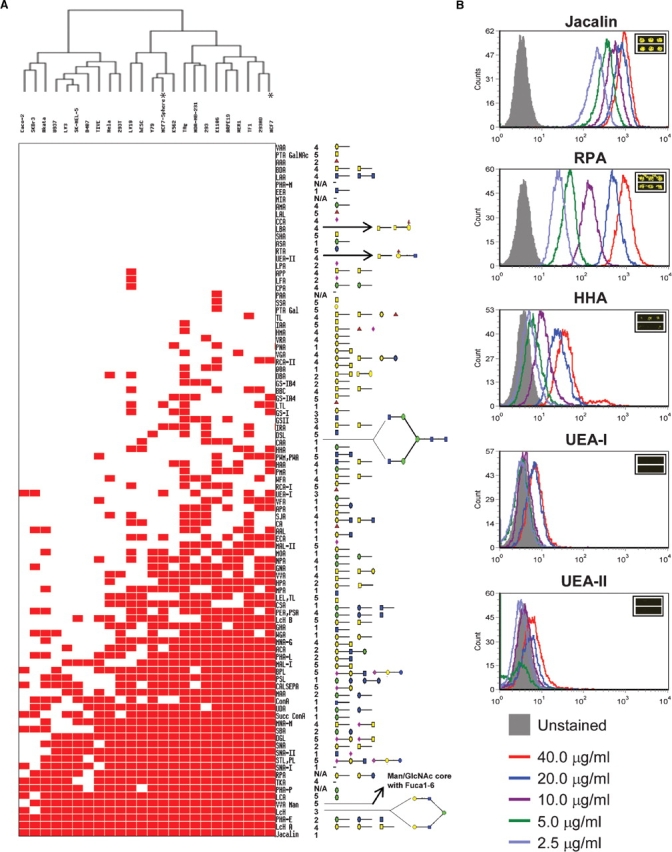
Binary cell binding signature. (A) Thumbnail overview of the one-way hierarchical cluster of 24 human cell lines (rows) and 94 lectins (columns). The corresponding carbohydrate specificities were depicted to the right as cartoons on the basis of information provided from five different sources. (B) Validation of cell-lectin binding by FACS. Cells were gated on FCS versus side scatter and all analysis was done on viable gated cells. Histogram overlays showing lectin staining of several indicated lectins at varying concentrations bound to K562 cells for Jacalin, RPA, HHA, UEA-I, and UEA-II which show a range of lectin binding from highly bound lectins (Jacalin), to moderately bound lectins (RPA and HHA), to non-binding lectins (UEA-I and UEA-II). Insets are the microarray images for the respective lectins. Glycan legend: Galactose ◯, GalNAc □, Glucose  , GlcNAc
, GlcNAc  , Mannose
, Mannose  , Fucose
, Fucose  , Sialic acid
, Sialic acid  . *MCF7 cells under normal and sphere culture conditions showing different lectin binding profiles used to demonstrate glycan signature-based biomarker discovery.
. *MCF7 cells under normal and sphere culture conditions showing different lectin binding profiles used to demonstrate glycan signature-based biomarker discovery.
A lectin binding signature for mammalian cell lines
To demonstrate the capability of the lectin microarrays to distinguish among the cell surface glycan repertoires of mammalian cells, 24 different types of human cell lines were tested (supplementary Table II). To account for significant variations in size, morphology, and labeling efficiency of cells, absolute quantification and cross-cell line normalization were not performed, but instead, a binary analysis was used to process the data (supplementary Figures 2 and 3). Each lectin was designated either bound or unbound to every cell line using a two-step method (see Materials and methods for details).
Fig. 2.
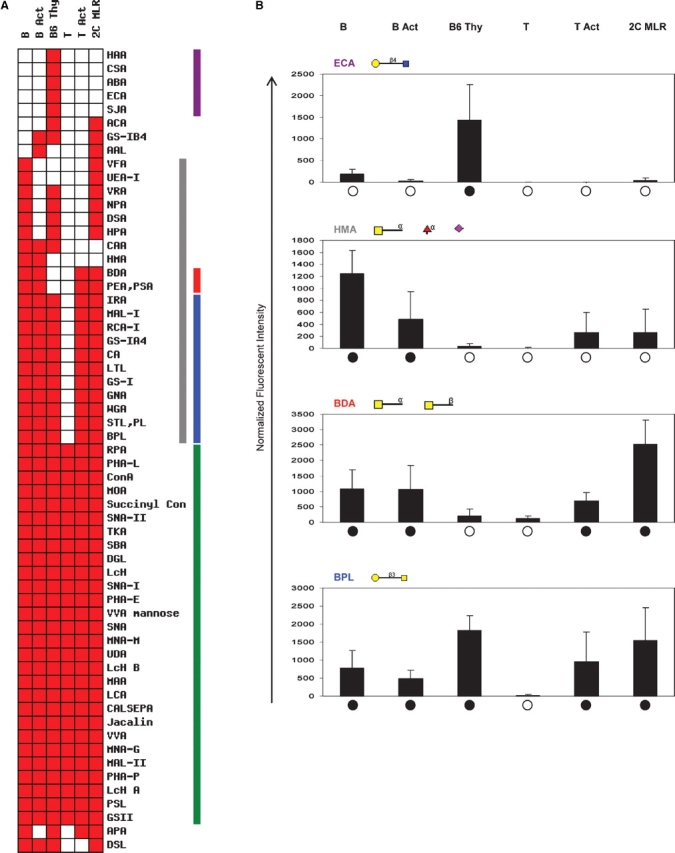
Differential cell binding in lymphocytes. (A) Cell binding signature showing the binding profile of B cells (B), activated B cells (B Act), thymocytes (B6 Thy), T cells (T), PMA/Ionomycin-activated T cells (T Act), and allogeneically activated T cells (2C MLR). Purple bar represents lectins bound to thymocytes only. Gray bar represents lectins which bound to B cells but not T cells. Red bar highlights two lectins which bound to all cell types except thymocytes and naïve T cells. Blue bar indicates binding to all cells except naïve T cells. Green bar shows lectins which bound all cell types. (B) Normalized spot intensities for selected lectins. Spot intensities are the average of six values, each consisting of the spot fluorescence minus the background. Values for each cell type are normalized to cell labeling fluorescence intensity as assessed by flow cytometry. Filled black circles or open circles indicate binding or no binding shown in the heat map, respectively.
Fig. 3.
E. coli-mammalian cell binding assay. (A) Schematic diagram. The freshly cultured cells were seeded to 24-well plates and cultured until a cell monolayer formed. E. coli strains were incubated with or without d-mannose at room temperature and loaded to the 24-well plates with or without d-mannose. The plates were incubated and washed with FBS-free cell culture media and PBS to remove the unbound E. coli. The mammalian cells were disrupted to release the bound E. coli. The released E. coli cells were plated onto a LB agar plate followed by incubation at 37°C overnight. The single colonies were counted next day. (B) Wildtype E. coli K12 and FimH mutant (fimhΔ) binding with three cell lines. The number of the bound mannose-specific lectin/total lectin is 16/52, 14/44, and 6/17 for MCF7, MDA-MB-231, and SkBr3, respectively. The ratios of (K12+Man)/K12 and (Mutant+Man)/Mutant, which represent the nonmannose inhibitable E. coli-mammalian cell binding of K12 and FimH mutant, were calculated. The error bar represents the standard error of the mean (SEM) of three independent experiments.
Using this analysis, a cell binding map was generated (Figure 1A). Significant differences in lectin binding patterns were observed among different cell lines. For example, less than 20 lectins could capture the hESC, Caco-2, D407, and U937 cells, while more than 50 lectins captured the TAg, 293, K1106, and MCF7 cells. These data indicate that the accessible cell surface glycans of mammalian cells vary significantly from one cell type to another. Accordingly, the cell lines were clustered into two major groups. Some related cell types clustered tightly, for example, the retina-derived cell lines, ARPE19, WER1, and Y79. In contrast, three breast cancer cell lines, MCF7, MDA-BA-231, and SkBr3, were not tightly clustered. This observation suggests that similar cell lines still possess variations in their cell surface glycan signatures and can be captured by the lectin microarray.
On the basis of their interactions with cell types, the 94 lectins could be grouped into four categories, i.e., zero binding (no cell line), low binding (1–9 cell lines), medium binding (10–19 cell lines), and high binding (20–24 cell lines). Three lectins recognized all of the 24 cell lines tested and 17 lectins did not show any binding activity. Therefore, the other 74 lectins were informative in terms of cell-type classification in this study. With the exception of two lectins of unknown saccharide specificities, all high binding lectins recognize simple sugars, such as mannose, galactose, and fucose, which are generally found on all mammalian cell surfaces. More complex glycan structures are recognized by lectins in the low or medium binding categories. For those lectins that showed no binding activity to the tested cells, it is possible that they either have relatively low affinity or are of low quality.
To further validate the lectin binding profile, we analyzed representative lectin-cell binding using flow cytometry. Lectins that were selected were representative of four categories: Jacalin bound to all cell types; RPA bound to most cell types; HHA and UEA-I bound to only a few cell types; and UEA-II bound to no cell types (Figure 1B). The flow cytometry data for each lectin correlated well with microarray binding (compare Figure 1B with the respective insets). High cell binding on the microarray was reflected as a large shift in the mean channel fluorescence (MCF) by flow cytometry, whereas low cell binding to the microarray was verified by negligible shifts in MCF over unstained control.
The analysis also showed that there is additional information that we are currently not harvesting in the binary lectin binding profile. For example, HHA lectin had intermediate binding capacity in the microarray analysis (Figure 1B, inset). In our ranking system this appeared only as a positive binding event. Flow cytometry experiments confirm the intermediate nature of the interaction of K562 cells with the HHA lectin (Figure 1B). As microarray technology matures, one could extract additional information through quantitative analysis of lectin binding.
A lectin binding signature for differentiating lymphocytes
The lectin microarray discriminated between closely related primary murine cells and revealed glycosylation changes in differentiating immune system cells (Rudd et al. 1999). We probed the lectin microarrays with naïve and activated B cells, thymocytes (immature T cells that are lineage committed but not fully differentiated), and naïve and activated T cells (Figure 2A). Freshly isolated thymocytes were captured by the largest number of lectins, indicative of the diversity of surface glycans. In contrast, naïve T cells bound to the lowest number of lectins. Two lectins BDA and PEA/PSA bound to B cells, activated B cells, and activated T cells (Figure 2A) but not thymocytes or naïve T cells. Interestingly, activation shifted the lectin profile of T cells from naïve toward one more similar to thymocytes. Lectin microarray analysis also showed that T cells activated with either phorbol myristyl acetate (PMA)/ionomycin or a mixed lymphocyte reaction (MLR) displayed different binding profiles. The lectins that bound MLR-activated but not PMA/ionomycin-activated T cells are specific for terminal Gal, terminal GalNAc, Man, and Fuc saccharides. This finding implies that even though both cells are “activated,” they represent phenotypically different cells. When examining B cells, we see that changes in lectin binding produced by activating B cells and by activating T cells overlap significantly with each other. Glycosylation changes in activating B cells and T cells are quite similar and may be the result of their common lymphocyte lineage.
Certain differential patterns of lectin binding to immune B and T cells were observed (Figure 2). Twelve of the seventeen surface lectin binding motifs (supplementary Figure 4) for lectin binding in immature, mature, and activated T lymphocytes were identical (data not shown). Despite the similarities, there are also clear differences among the lymphocyte populations. The largest segment of differentially bound lectins, such as the Gal-GalNAc specific BPL, bound all cells with the exception of naïve T cells (Figure 2A). Lectins which captured thymocytes and activated T cells but not naïve T cells have relatively narrow sugar specificities usually consisting of Gal, GalNAc, and GlcNAc. Differential recognition of Gal-GalNAc, the core-1 O-glycan, reflects known changes in glycan expression when transitioning from immature thymocyte cells to naïve T cells to activated T cells (Daniels et al. 2001, 2002; Walzel et al. 2006). Interestingly, there were no lectins that bound exclusively to naïve T cells. The lymphocytes also provide a reminder that cell binding events are not simply binary (Figure 2B). After normalizing fluorescent intensities to cell labeling efficiency, as assessed by flow cytometry, there are still significant variations in fluorescence intensity between cell types represented in our reductionist binary analysis. Overall, the lectin signatures across lymphocytes show fewer differences than the 24 human cell lines. The general similarity in surface glycan expression was to be expected as the cells share a common developmental lineage.
Fig. 4.
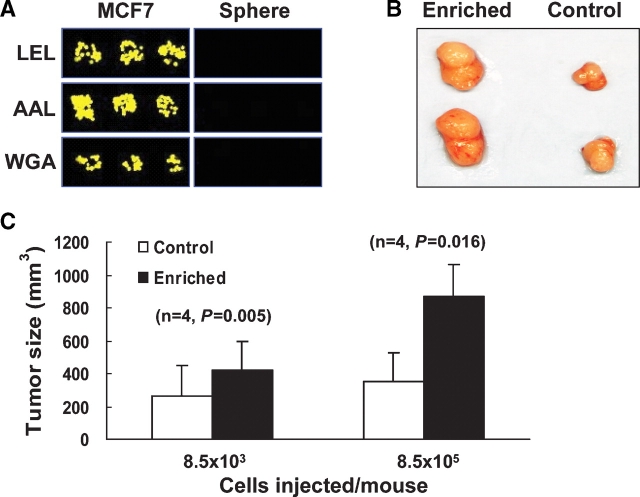
Enhanced tumorigenicity in mice injected with LEL depleted MCF7 cells. (A) Microarray image of LEL, AAL, and WGA capture of normal or sphere cultured MCF7 cells. (B) Representative tumors in two mice at the enriched cell injection sites (left) and at the control cell injection sites (right), the top two tumors are from one mouse and the bottom two are from another mouse. (C) Average tumor sizes (n = 4). P-values are calculated using a one-tailed pair-wise t-test.
Using glycan signatures to predict Escherichia coli tropism
To further explore the potential of the glycan signatures described above, an E. coli-mammalian cell-binding system was employed. E. coli bind to a variety of mammalian cells, such as the pathogenic E. coli strain K1, which binds to brain microvascular endothelial cells (BMEC) with high affinity (Kim 2006) and causes meningitis. Type 1 fimbriae-based mannose-specific E. coli binding is one of the major contributors for E. coli-mammalian cell binding (Teng et al. 2005) and the FimH subunit determines the mannose-specific binding of type I fimbriae (Hung et al. 2002). Clinically, human urinary tract and bladder infections caused by mannose-specific E. coli-mammalian cell binding are treated with oral administration of mannose (Svanborg and Godaly 1997).
The glycan signatures revealed that mannose-specific lectin binding is significantly different among different cell lines. For example, of the 16 mannose-specific lectins that bound to MCF7 only 6 bound to SkBr3. If such differences reflect a higher density of accessible mannose residues on MCF7 cells, the mannose-specific E. coli binding could be proportionally greater to those cells. To test this hypothesis, we performed E. coli-mammalian cell binding assays (Figure 3A) using three breast cancer cell lines MCF7, MDA-MB-231, and SkBr3, which bound to 16, 14, and 6 mannose-specific lectins, respectively. We calculated a mannose inhibition index of the bacterial binding of wildtype K12 E. coli (WT) and the FimH knockout mutant (fimHΔ) by evaluating the ratio of bacterial binding (colony counts) in the presence and absence of soluble d-mannose (Figure 3B). The mannose inhibition index values of the WT for MCF7, MDA-MB-231, and SkBr3 are 0.48, 0.58, and 0.76, respectively, which are statistically different among these three cell lines (P < 0.05) except MCF7 versus MDA-MB-231 (P = 0.078). These ratios are inversely proportional to the number of the mannose-specific lectins bound to them. In contrast, the mannose inhibition index values of fimHΔ cells for MCF7, MDA-MB-231, and SkBr3 are 0.96, 1, and 1.03, respectively, which are close to 1 and not significantly different (P ≥ 0.4). These results indicate that, as expected, the mannose-independent fimHΔ-mammalian cell binding is not inhibited by the addition of mannose. Taken together, the data indicate that the mannose-specific E. coli-mammalian cell binding is predicted by the mannose-specific lectin chip binding. Thus, the glycan signatures can be used to predict bacterial tropisms for mammalian cells.
Biomarker discovery based on glycan signatures
To examine the power of the microarray platform for biomarker discovery, we analyzed lectin binding in a model cancer stem-like cell system by comparing the cell surface glycan signatures of all 24 cell types including MCF7 cells under standard and sphere culture conditions. Cancer stem-like cells were chosen for analysis due to the dearth of cell surface markers to differentiate between cancer stem-like cells and cancer cell lines. Because of this issue, cancer stem-like cells are often differentiated from other cells functionally by their ability to grow as spheres when cultured, as a side population (SP) which reflects expression of drug transporters seen by flow cytometry, or by their tumorigenicity in vivo in NOD/SCID models. Ultimately, the limited ability to define unique cell surface markers has limited the phenotypic characterization of these important cells.
The model system analyzed was the breast cancer cell line, MCF7, which can be grown under both standard and sphere culture conditions. When grown as a sphere culture, MCF7 cells express the phenotypes associated with known breast cancer stem-like cells (Ponti et al. 2005; Phillips et al. 2006). By contrast, only a fraction of MCF7 cells grown under standard conditions express those markers or phenotypically could be seen as SP cells (Ponti et al. 2005; Phillips et al. 2006; Zhou et al. 2007). Interestingly, the glycan signatures demonstrated that MCF7 cells grown under two different conditions have distinct lectin binding profiles and are not clustered together (Figure 1A marked by asterisks). The lectins LEL, AAL, and WGA showed the largest difference in fluorescent intensities. Each of these lectins effectively captured MCF7 cells but failed to capture MCF7 sphere cells (Figure 4A). The most significant difference was seen for LEL. This indicates that the LEL lectin may be used to distinguish these cancer cell subpopulations.
To demonstrate the usefulness of LEL as a potential biomarker, we looked for enhanced in vivo tumorigenicity in NOD/SCID mice injected with LEL-depleted MCF7 cells compared to those injected with regular MCF7 cells. A marked difference was seen in tumorigenicity between the two different groups (Figure 4B and C). The average tumor size of mice injected with the LEL-depleted cancer stem-like cell enriched cultures (8.5 × 105 cells/mouse) was 870 mm3 versus 350 mm3 for the control group injected with the similar number of regular MCF7 cells (P = 0.016), and average tumor sizes of animals injected with a 100-fold fewer LEL-depleted cancer stem-like cell enriched cultures were 420 mm3 versus 260 mm3 for the control group, respectively (P = 0.005), which was seen after two additional weeks of growth. Thus, the lectin microarray identified novel cell surface markers on cancer stem-like cells which were useful in enriching and studying cancer stem-like cells.
Discussion
Traditional technologies to study cell glycosylation include mass spectrometry, flow cytometry (Batisse et al. 2004; Venable et al. 2005), immunocytochemistry (Wearne et al. 2006), and histochemistry (Carter and Brooks 2006), which are not conducive for high-throughput analyses. Recently, as a proof of concept, lectin microarrays were used to capture selected glycosylation-defective cell lines (Tateno et al. 2007). We created a lectin microarray and exploited the ability of lectins to differentiate the accessible cell surface glycans on live human cells. We found a unique cell binding signature for each cell type analyzed. Additionally, we have shown that differentiation of immune cells with distinct biological characteristics shows differential binding to the lectin microarray. Therefore, we not only present a high-throughput assessment of the accessible surface glycans of mammalian cells, but also demonstrate potential biological applications, including prediction of E. coli tropism and cancer stem cell biomarker discovery.
A key feature of infectious pathogens is their ability to directly and preferentially bind cell surface receptors via adhesins. The vast majority of adhesins are lectins, which recognize carbohydrate moieties on cell surfaces. For example, type I fimbriae expressed by various pathogenic strains of E. coli preferentially recognize mannose residues. As these residues are found on numerous glycoproteins, these types of interactions are not usually considered to be specific for a particular receptor. Interestingly, they appear to determine the tropism of a microorganism for a specific host tissue, due to preferential expression and/or accessibility of certain carbohydrate moieties in some tissues (Van Nhieu et al. 2005). We successfully applied the glycan signatures generated in this study to predict E. coli mannose-specific binding to mammalian cells. To our knowledge, this is the first example that reveals the potential use of lectin microarray cell profiling for the study of infectious pathogen tropism and pathogen–host interactions.
A major goal of high-throughput analyses can often relate to identification of novel biomarkers that have therapeutic and/or diagnostic values. We successfully identified novel and unique lectin binding patterns that can differentiate cancer stem-like cells in a model human breast cancer cell line. In vivo tumorigenicity experiments showed that indeed these lectins could be used to enrich cancer stem-like cells. Thus, combined with other cancer stem cell enrichment methods, lectin microarray technology promises to be an important tool for identifying cell surface markers in tumors, and thus may facilitate therapies targeting cancer stem-like cells.
The current plant-based lectin microarray platform analyzes surface-accessible glycans that are often responsible for cell–cell or cell–pathogen interactions and have been shown to be targets of subtle variations (Gabius et al. 2004). While these have been very useful, we believe the addition of human and animal lectins will likely further empower the lectin microarray technology as they can reveal subtleties in the glycan structures on cell surfaces, which are their natural ligands. However, as evidenced by the biomarker study, even the current plant-based lectin microarrays are useful in analysis of systems that have been refractory to cell surface biomarker definition. In summary, here we showed the power of a high-throughput lectin microarray to profile cells. These studies indicate that lectin microarrays can be leveraged in future applications including monitoring cell differentiation, clinical diagnostics, and biomarker discovery.
Materials and methods
Lectins and slides
Ninety-four lectins were collected from several sources listed in supplementary Table I. The Schott NexterionR slides were purchased from Schott North America, Inc. (Louisville, KY). The FullMoon slides were purchased from FullMoon BioSystems, Inc. (Sunnyvale, CA). The FAST slides were purchased from Whatman, Inc. (Florham Park, NJ). The nickel slides were obtained from Xenopore, Inc. (Saddlebrook, NJ). Aldehyde and SuperProtein slides were ordered from TeleChem International, Inc. (Sunnyvale, CA).
Lectin microarray fabrication and quality control
Lectin proteins were resuspended in a phosphate buffered saline (PBS) buffer with 0.02% Tween20 and 25% glycerol to a final concentration of 1 μg/μL. Bovine serum albumin (BSA, 0.05 μg/μL) was also added to the buffer to improve spot morphology. The lectins were printed on different slides using the ChipWriter Pro (Bio-Rad, Hercules, CA) microarrayer. Three spots per lectin were printed in each block and 12 blocks were printed per slide. After printing, slides were covered with aluminum foil and stored at 4°C for future use. To monitor the quality of the lectin microarrays, they were stained with 549 NHS Ester (DyLight) in 100-fold dilution at room temperature for 1 h. The stained slides were washed twice with TBST followed by one wash with water. The dried slides were scanned with a GenePix 4100B (Axon, Sunnyvale, CA) scanner at 10 μm resolution (supplementary Figure 5). The scanning conditions were 600 mV laser power and 33% PMT value at the Cy3 channel.
Culture of human cell lines and cell harvest
Twenty-three human cell lines and the sphere culture of MCF7 were collected (supplementary Table II). Suspension cells were collected with three PBS washes. Attached cells were dissociated enzymatically (0.05% trypsin in 0.5 mM EDTA (Invitrogen)) and mechanically by three PBS washes using a glass Pasteur pipette.
Cell probing, microarray scanning, and data extraction
Lectin microarrays were blocked for 1 h at room temperature in 50 mM ethanolamine in a borate buffer (pH 8.0). Blocked slides were washed once in PBS with 0.5% Tween20, followed by two washes in PBS. Cells were labeled with CFSE cell tracking dye (Invitrogen, Carlsbad, CA) at 10 μM concentrations for 5 min at 4° C. The labeling efficiency was determined by flow cytometry. Each lectin block on the chip was seeded with 5 × 105 CFSE-labeled cells in PBS with 0.5 mM CaCl2 and 1% BSA; 200 mM lactose or 200 mM α-methyl-mannoside was also added in the inhibitor experiments. Cells were allowed to bind on lectin microarrays at room temperature for 1 h and the excess and/or unbound cells were gently removed by submerging and inverting the slides in PBS. The slides were continuously washed on a shaker for 5 min, air-dried, and scanned with a microarray scanner (Axon) at 5 μm resolution. The scanning condition was set to 300 mV laser power and 40% PMT value for Cy5 channel and 200 mV laser power and 10% PMT value for FITC channel, unless otherwise stated. GenePix5.0 (Axon) was used for extracting binding signals from the scanned images.
Mouse T cell and B cells
C57BL/6, 2C TCR transgenic, and Balb/c mice were maintained in specific pathogen-free conditions. Single cell suspension was created by mashing harvested spleens over 70 μm nylon cell strainers (BD Biosciences, San Jose, CA) with PBS (GIBCO, Carlsbad, CA). The cell suspension was centrifuged and resuspended with an ACK Lysing buffer (Quality Biological, Inc.) for 3 min to remove red blood cells. Cells are washed with PBS and resuspended in RPMI-1640 complete media (RPMI-1640 (GIBCO), 10% FCS (Atlanta Biologicals), Mem nonessential amino acids (GIBCO), Mem vitamin solution (GIBCO), β-Mercaptoethanol (GIBCO), 10 μg/mL Cipro (Sigma, St. Louis, MO). Cells were further enriched (as indicated) for B or T cells using MACS kits for either mouse CD19+ or pan T cell (Miltenyi Biotech, Auburn, CA).
In vitro lymphocyte activation
T cells isolated from C57BL/6 mice as described above were stimulated nonspecifically using 2 ng/mL PMA and 400 ng/mL ionomycin in complete RPMI-1640 media supplemented with T-cell growth factor. B cells isolated from C57BL/6 mice as described above were stimulated nonspecifically using 100 ng/mL LPS in complete RPMI-1640 media supplemented with T-cell growth factor (Mackensen et al. 1994). Allogeneic stimulation of T cells was achieved by culturing splenocytes from 2C TCR transgenic mice with irradiated splenocytes from Balb/c mice at a 1:1.4 ratio in RPMI-1640 complete media supplemented with T-cell growth factor.
Lectin flow cytometry
Lectins were labeled with fluorescein-5-isothiocyanate ‘Isomer-I’ (Invitrogen) at 50 molar excess in PBS for 1 h at room temperature. The excess free FITC was removed using PD-10 Sephadex Columns (GE Healthcare, Piscataway, NJ). Labeled lectins were concentrated with VivaSpin centrifugal concentrators (Sartorius AG, Goettingen, Germany). The lectin concentration was measured using a UV–visible spectrophotometer (Shimadzu Corp., Columbia, MD). Staining with labeled lectins was performed at 4°C for 1 h with indicated concentrations of labeled lectin. Cells were washed with a FACS wash buffer (PBS with 2% FCS and 0.05% Sodium Azide) prior to reading flow cytometry samples on FACSCalibur (BD Biosciences).
MCF7 sphere cell culture
Human breast cancer cell line MCF7 cells were obtained from American Type Culture Collection (ATCC, Manassas, VA). Cells were grown in a DMEM medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), 100 units/mL penicillin, and 100 μg/mL streptomycin (Invitrogen), in a 37°C incubator containing 5% CO2. Sphere cell culture was performed according to published protocol with modifications (Dontu et al. 2003; Ponti et al. 2005). Briefly, single cells were plated in ultralow attachment plates (Corning, Corning, NY) at a density of 20 000 viable cells/mL in primary culture and 1000 cells/mL in passages. MCF7 sphere cells were grown in a serum-free mammary epithelial growth medium without bovine pituitary extract (MEGM, BioWhittaker, Walkersville, MD), but supplemented with B27 (Invitrogen), 20 ng/mL EGF, and 20 ng/mL bFGF (BD Biosciences). In order to passage sphere cells (SP), spheres were collected into a 15 mL conical tube and allowed to settle for 15 min. Supernatant was removed. Sphere cells were dissociated enzymatically with 0.05% trypsin and 0.5 mM EDTA (Invitrogen) and mechanically by a glass Pasteur pipette. The cells obtained from dissociation were passed through a 40 μm sieve and analyzed microscopically for single cells and subjected to the following experiments.
Lectin staining and SP analysis
For SP analysis, cells were collected and resuspended in 37°C DMEM containing 2% FBS at 1 × 106 cells/mL. Hoechst 33342 cell staining was performed according to protocol originally developed by Goodell (1996) with slight modification. Briefly, the cells were incubated with Hoechst 33342 (Sigma) at 5 μg/mL for 90 min at 37°C. Following staining, the cells were spun down and further incubated with FITC-conjugated lectin LEL (EY labs, San Mateo, CA) at 40 μg/mL for 30 min on ice. Then, cells were washed and resuspended in HBSS. Following the staining, the cells were spun down and resuspended in HBSS (Invitrogen) containing 1 μg/mL propidium iodide and maintained at 4°C for flow cytometry analysis. Cell analysis was performed on a Moflo cytometer (Dako, Carpinteria, CA) equipped with a Coherent Enterprise II laser emitting MLUV at 351 nm and blue 488 nm lines. The Hoechst 33342 emission was first split using a 610-dsp filter and then the red and the blue emissions were collected through 670/630 and 450/465 nm bandpass filters, respectively.
E. coli-mammalian cell binding assay
Wildtype E. coli strain K12 (CGSC strain: 4401/EMG2) and FimH mutant (fimhΔ) (CGSC strain: 11068/JW4283) were provided by the Yale E. coli Genetic Stock Center. The protocol was modified from Teng et al. (2005). Briefly, the freshly cultured cells were seeded to collagen-coated 24-well plates at ∼200 000 cells/well and cultured at 37°C, 5% CO2 for about 48 h until a monolayer was formed. E. coli K12 cells were cultured in liquid LB broth overnight, then adjusted OD620 to 0.2 and incubated with or without 100 mM D-mannose at room temperature for 1 h. Following the incubation period, 100 μL of the E. coli cells (∼107) was added to each well of the 24-well plates with or without 100 mM d-mannose. The plates were incubated at 37°C, 5% CO2 for 90 min followed by two washes with FBS-free DMEM (4.5 g/L glucose) media and two washes with PBS to remove the unbound E. coli cells. The mammalian cells were disrupted to release the bound E. coli by treatment with 0.025% Triton X-100 at room temperature for 20 min. The suspensions were properly diluted and plated onto an LB agar plate, followed by incubation at 37°C overnight. The numbers of single colonies for each experiment were counted the next day.
LEL+ Cell depletion and tumorigenicity assay
The M450 epoxy magnetic beads (Invitrogen) were coupled with lectin LEL (EY labs) according to the manufacturer's instruction at a ratio of 200 μg of LEL to 1 mL of magnetic beads. Over 108 MCF7 cells were washed and resuspended in the lectin-cell binding buffer (1 × PBS with 1% BSA and 0.5 mM CaCl2). LEL-coupled magnetic beads were added to the resuspended cells at a bead-to-cell ratio of 2:1. The cells and beads were incubated for 30 min at 4°C with gentle tilting and rotation. Bead-bound cells were removed magnetically (1.5 min). The same step of cell depletion was repeated once and ≥99% of the parental cells was estimated to be removed. The remaining cells were resuspended in the solution of medium:Matrigel (BD Biosciences) (1:1) for injection. Female NOD/SCID mice (The Jackson Laboratory) were given subcutaneous injections of β-estradiol (Sigma) dissolved in pure sesame oil (0.1 mg per 0.05 mL sesame oil per mouse) 1 day before tumor inoculation and at weekly intervals (Hardman et al. 1999; Kasukabe et al. 2005). The enriched and control cells were injected into the same mouse into the upper right and left mammary pads, respectively, by using 22-gauge needles. An amount equivalent to 0.2 mL of cells was injected. Tumor growth was monitored thereafter. Five weeks after inoculation the mice were sacrificed, and the tumor tissues were analyzed by Hematoxylin and Eosin staining (H & E).
Data analysis
To determine cell binding to lectin spots on the microarray, the local background intensity was subtracted from each lectin spot. A histogram of the subtracted background intensities for each microarray was plotted, centered on zero, and fitted to a normal distribution. Positive intensity (above background) appeared as a long tail in the histogram, representing a range of positive binding intensities (supplementary Figure 2). If the intensity of a spot was 6 standard deviations (SD) above 0, it was scored positive.
For each lectin, the scores of the six replicate spots (duplicate microarrays and triplicate spots on each microarray) were used in a voting scheme to determine whether cells bound to that lectin. If more than four spots of a lectin were scored positive, it was defined as positive. Most of the lectins (80%) had either zero or six spots scored positive (supplementary Figure 3).
Supplementary Data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Funding
National Institutes of Health (U54RR020839-01 to H.Z., AI29575, AI44129, and CA108835 to J.P.S., EY015684 to J.Q., GM 28470 to I.J.G., NS037096 to R.L.S., AI44063 to Y.Z., 5 T32AI07247-25 to Y.L.); Ho Ching Yang Fellowship to J.Z.; WW Smith Charitable Trust to H.Z.
Acknowledgments
We thank Dr. Y. Zhou for his help on the summarizing of the lectin carbohydrate specificity and Hao Zhang for flow cytometry analysis of cancer stem-like cells. We also thank Drs. J. Zhang, L. Cheng, G. Chen, X. Li, D.J. Zack, M. Li, J.O. Liu, F. Pan, and S.D. Hayward for providing cell lines; Dr. K.S. Kim for his help on E. coli-mammalian cell binding assay; and Dr. Jef Boeke for critical comments and helpful discussion.
Conflict of interest statement
None declared.
References
- Batisse C, Marquet J, Greffard A, Fleury-Feith J, Jaurand MC, Pilatte Y. Lectin-based three-color flow cytometric approach for studying cell surface glycosylation changes that occur during apoptosis. Cytometry A. 2004;62:81–88. doi: 10.1002/cyto.a.20094. [DOI] [PubMed] [Google Scholar]
- Brooks SA, Leathem AJ. Prediction of lymph node involvement in breast cancer by detection of altered glycosylation in the primary tumour. Lancet. 1991;338:71–74. doi: 10.1016/0140-6736(91)90071-v. [DOI] [PubMed] [Google Scholar]
- Carter TM, Brooks SA. Detection of aberrant glycosylation in breast cancer using lectin histochemistry. Methods Mol Med. 2006;120:201–216. doi: 10.1385/1-59259-969-9:201. [DOI] [PubMed] [Google Scholar]
- Daniels MA, Devine L, Miller JD, Moser JM, Lukacher AE, Altman JD, Kavathas P, Hogquist KA, Jameson SC. CD8 binding to MHC class I molecules is influenced by T cell maturation and glycosylation. Immunity. 2001;15:1051–1061. doi: 10.1016/s1074-7613(01)00252-7. [DOI] [PubMed] [Google Scholar]
- Daniels MA, Hogquist KA, Jameson SC. Sweet ‘n’ sour: The impact of differential glycosylation on T cell responses. Nat Immunol. 2002;3:903–910. doi: 10.1038/ni1002-903. [DOI] [PubMed] [Google Scholar]
- Dennis JW, Granovsky M, Warren CE. Glycoprotein glycosylation and cancer progression. Biochim Biophys Acta. 1999;1473:21–34. doi: 10.1016/s0304-4165(99)00167-1. [DOI] [PubMed] [Google Scholar]
- Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwek MV, Ross HA, Leathem AJ. Proteome and glycosylation mapping identifies post-translational modifications associated with aggressive breast cancer. Proteomics. 2001;1:756–762. doi: 10.1002/1615-9861(200106)1:6<756::AID-PROT756>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Ebe Y, Kuno A, Uchiyama N, Koseki-Kuno S, Yamada M, Sato T, Narimatsu H, Hirabayashi J. Application of lectin microarray to crude samples: Differential glycan profiling of lec mutants. J Biochem (Tokyo) 2006;139:323–327. doi: 10.1093/jb/mvj070. [DOI] [PubMed] [Google Scholar]
- Ferguson RE, Jackson DH, Hutson R, Wilkinson N, Harnden P, Selby P, Banks RE. Detection of glycosylation changes in serum and tissue proteins in cancer by lectin blotting. Adv Exp Med Biol. 2005;564:113–114. doi: 10.1007/0-387-25515-X_19. [DOI] [PubMed] [Google Scholar]
- Gabius HJ, Siebert HC, Andre S, Jimenez-Barbero J, Rudiger H. Chemical biology of the sugar code. Chembiochem. 2004;5:740–764. doi: 10.1002/cbic.200300753. [DOI] [PubMed] [Google Scholar]
- Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakomori S. Aberrant glycosylation in cancer cell membranes as focused on glycolipids: Overview and perspectives. Cancer Res. 1985;45:2405–2414. [PubMed] [Google Scholar]
- Hakomori S. Glycosylation defining cancer malignancy: New wine in an old bottle. Proc Natl Acad Sci USA. 2002;99:10231–10233. doi: 10.1073/pnas.172380699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haltiwanger RS, Lowe JB. Role of glycosylation in development. Annu Rev Biochem. 2004;73:491–537. doi: 10.1146/annurev.biochem.73.011303.074043. [DOI] [PubMed] [Google Scholar]
- Hardman WE, Moyer MP, Cameron IL. Fish oil supplementation enhanced CPT-11 (irinotecan) efficacy against MCF7 breast carcinoma xenografts and ameliorated intestinal side-effects. Br J Cancer. 1999;81:440–448. doi: 10.1038/sj.bjc.6690713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi J. Lectin-based structural glycomics: Glycoproteomics and glycan profiling. Glycoconj J. 2004;21:35–40. doi: 10.1023/B:GLYC.0000043745.18988.a1. [DOI] [PubMed] [Google Scholar]
- Hsu KL, Pilobello KT, Mahal LK. Analyzing the dynamic bacterial glycome with a lectin microarray approach. Nat Chem Biol. 2006;2:153–157. doi: 10.1038/nchembio767. [DOI] [PubMed] [Google Scholar]
- Hung CS, Bouckaert J, Hung D, Pinkner J, Widberg C, DeFusco A, Auguste CG, Strouse R, Langermann S, Waksman G. Structural basis of tropism of Escherichia coli to the bladder during urinary tract infection. Mol Microbiol. 2002;44:903–915. doi: 10.1046/j.1365-2958.2002.02915.x. [DOI] [PubMed] [Google Scholar]
- Kasukabe T, Okabe-Kado J, Kato N, Sassa T, Honma Y. Effects of combined treatment with rapamycin and cotylenin A, a novel differentiation-inducing agent, on human breast carcinoma MCF-7 cells and xenografts. Breast Cancer Res. 2005;7:R1097–R1110. doi: 10.1186/bcr1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KS. Microbial translocation of the blood-brain barrier. Int J Parasitol. 2006;36:607–614. doi: 10.1016/j.ijpara.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Kukuruzinska MA, Bergh ML, Jackson BJ. Protein glycosylation in yeast. Annu Rev Biochem. 1987;56:915–944. doi: 10.1146/annurev.bi.56.070187.004411. [DOI] [PubMed] [Google Scholar]
- Kuno A, Uchiyama N, Koseki-Kuno S, Ebe Y, Takashima S, Yamada M, Hirabayashi J. Evanescent-field fluorescence-assisted lectin microarray: A new strategy for glycan profiling. Nat Methods. 2005;2:851–856. doi: 10.1038/nmeth803. [DOI] [PubMed] [Google Scholar]
- Lee MR, Park S, Shin I. Protein microarrays to study carbohydrate-recognition events. Bioorg Med Chem Lett. 2006;16:5132–5135. doi: 10.1016/j.bmcl.2006.07.028. [DOI] [PubMed] [Google Scholar]
- Lehle L, Strahl S, Tanner W. Protein glycosylation, conserved from yeast to man: A model organism helps elucidate congenital human diseases. Angew Chem Int Ed Engl. 2006;45:6802–6818. doi: 10.1002/anie.200601645. [DOI] [PubMed] [Google Scholar]
- Macartney JC. Fucose-containing antigens in normal and neoplastic human gastric mucosa: A comparative study using lectin histochemistry and blood group immunohistochemistry. J Pathol. 1987;152:23–30. doi: 10.1002/path.1711520104. [DOI] [PubMed] [Google Scholar]
- Mackensen A, Carcelain G, Viel S, Raynal MC, Michalaki H, Triebel F, Bosq J, Hercend T. Direct evidence to support the immunosurveillance concept in a human regressive melanoma. J Clin Invest. 1994;93:1397–1402. doi: 10.1172/JCI117116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan WT, Watkins WM. Unravelling the biochemical basis of blood group ABO and Lewis antigenic specificity. Glycoconj J. 2000;17:501–530. doi: 10.1023/a:1011014307683. [DOI] [PubMed] [Google Scholar]
- Muramatsu T. Development: Carbohydrate recognition in spermatogenesis. Science. 2002;295:53–54. doi: 10.1126/science.1068156. [DOI] [PubMed] [Google Scholar]
- Palcic MM, Ratcliffe RM, Lamontagne LR, Good AH, Alton G, Hindsgaul O. An enzyme-linked immunosorbent assay for the measurement of Lewis blood-group alpha-(1-4)-fucosyltransferase activity. Carbohydr Res. 1990;196:133–140. doi: 10.1016/0008-6215(90)84112-8. [DOI] [PubMed] [Google Scholar]
- Phillips TM, McBride WH, Pajonk F. The response of CD24(−/low)/CD44 + breast cancer-initiating cells to radiation. J Natl Cancer Inst. 2006;98:1777–1785. doi: 10.1093/jnci/djj495. [DOI] [PubMed] [Google Scholar]
- Pilobello KT, Slawek DE, Mahal LK. A ratiometric lectin microarray approach to analysis of the dynamic mammalian glycome. Proc Natl Acad Sci. 2007;104:11534. doi: 10.1073/pnas.0704954104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D, Pilotti S, Pierotti MA, Daidone MG. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65:5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- Reisner Y, Linker-Israeli M, Sharon N. Separation of mouse thymocytes into two subpopulations by the use of peanut agglutinin. Cell Immunol. 1976;25:129–134. doi: 10.1016/0008-8749(76)90103-9. [DOI] [PubMed] [Google Scholar]
- Rudd PM, Wormald MR, Stanfield RL, Huang M, Mattsson N, Speir JA, DiGennaro JA, Fetrow JS, Dwek RA, Wilson IA. Roles for glycosylation of cell surface receptors involved in cellular immune recognition. J Mol Biol. 1999;293:351–366. doi: 10.1006/jmbi.1999.3104. [DOI] [PubMed] [Google Scholar]
- Svanborg C, Godaly G. Bacterial virulence in urinary tract infection. Infect Dis Clin North Am. 1997;11:513–529. doi: 10.1016/s0891-5520(05)70371-8. [DOI] [PubMed] [Google Scholar]
- Tateno H, Uchiyama N, Kuno A, Togayachi A, Sato T, Narimatsu H, Hirabayashi J. A novel strategy for mammalian cell surface glycome profiling using lectin microarray. Glycobiology. 2007 doi: 10.1093/glycob/cwm084. [DOI] [PubMed] [Google Scholar]
- Teng CH, Cai M, Shin S, Xie Y, Kim KJ, Khan NA, Di Cello F, Kim KS. Escherichia coli K 1 RS 218 interacts with human brain microvascular endothelial cells via type 1 fimbria bacteria in the fimbriated state. Infect Immun. 2005;73:2923–2931. doi: 10.1128/IAI.73.5.2923-2931.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyken SJ, Green RS, Marth JD. Structural and mechanistic features of protein O glycosylation linked to CD8+ T-cell apoptosis. Mol Cell Biol. 2007;27:1096–1111. doi: 10.1128/MCB.01750-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nhieu GT, Sansonetti PJ, Lafont F. Signaling via adhesion molecules and bacterial internalization. In: Cossart P, editor. Cellular Microbiology. Herndon, VA: ASM Press; 2005. [Google Scholar]
- Varki A. Essentials of Glycobiology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. [PubMed] [Google Scholar]
- Venable A, Mitalipova M, Lyons I, Jones K, Shin S, Pierce M, Stice S. Lectin binding profiles of SSEA-4 enriched, pluripotent human embryonic stem cell surfaces. BMC Dev Biol. 2005;5:15. doi: 10.1186/1471-213X-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walzel H, Fahmi AA, Eldesouky MA, Abou-Eladab EF, Waitz G, Brock J, Tiedge M. Effects of N-glycan processing inhibitors on signaling events and induction of apoptosis in galectin-1-stimulated Jurkat T lymphocytes. Glycobiology. 2006;16:1262–1271. doi: 10.1093/glycob/cwl037. [DOI] [PubMed] [Google Scholar]
- Wearne KA, Winter HC, O′Shea K, Goldstein IJ. Use of lectins for probing differentiated human embryonic stem cells for carbohydrates. Glycobiology. 2006;16:981–990. doi: 10.1093/glycob/cwl019. [DOI] [PubMed] [Google Scholar]
- Yuan M, Itzkowitz SH, Boland CR, Kim YD, Tomita JT, Palekar A, Bennington JL, Trump BF, Kim YS. Comparison of T-antigen expression in normal, premalignant, and malignant human colonic tissue using lectin and antibody immunohistochemistry. Cancer Res. 1986;46:4841–4847. [PubMed] [Google Scholar]
- Zheng T, Peelen D, Smith LM. Lectin arrays for profiling cell surface carbohydrate expression. J Am Chem Soc. 2005;127:9982–9983. doi: 10.1021/ja0505550. [DOI] [PubMed] [Google Scholar]
- Zhou J, Wulfkuhle J, Zhang H, Gu P, Yang Y, JD, Margolick JB, Liotta LA, Petricoin E, III, Zhang Y. Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc Natl Acad Sci USA. 2007;104:16158–16163. doi: 10.1073/pnas.0702596104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.