Abstract
O-Linked N-acetylglucosaminyltransferase (OGT) catalyzes the transfer of a single GlcNAc to the Ser or Thr of nucleocytoplasmic proteins. OGT activity, which may compete with that of kinases, is involved in signaling in animals and plants, and abnormalities in OGT activities have been associated with type 2 diabetes. Here, we show that ogt genes that predict enzymes with characteristic tetratricopeptide repeats and a spindly domain are present in some protists (Giardia, Cryptosporidium, Toxoplasma, and Dictyostelium) but are absent from the majority of protists examined (e.g., Plasmodium, Trypanosoma, Entamoeba, and Trichomonas). Similarly, ogt genes are present in some fungi but are absent from numerous other fungi, suggesting that secondary loss is an important contributor to the evolution of ogt genes. Nucleocytosolic extracts of Giardia and Cryptosporidium show OGT activity, and recombinant Giardia and Cryptosporidium OGTs are active in yeast and bacteria, respectively. These results suggest the possibility that O-GlcNAc modification of nucleocytosolic proteins also has function(s) in simple eukaryotes.
Keywords: Cryptosporidium, evolution, Giardia, O-GlcNAc transferase, recombinant expression
Introduction
O-Linked N-acetylglucosaminyltransferase (OGT) is a nucleocytoplasmic glycosyl transferase, which catalyzes the addition of a single β-O-linked N-acetylglucosamine (GlcNAc) to the serine or threonine of a polypeptide chain (reviewed in Love and Hanover (2005); Hart et al. (2007)). The C-terminus of the OGT contains the catalytic domain (also known as “spindly” or “spy” domain), which has a weak homology with glycogen phosphorylase (Roos and Hanover 2000; Wrabl and Grishin 2001). The N-terminus of the OGT contains a series of tetratricopeptide repeats (TPRs), which are involved in substrate recognition (Lubas and Hanover 2000; Martinez-Fleites et al. 2008).
In metazoa, OGT, which is essential, modifies transcription factors, nuclear pore proteins, kinases, and many other proteins (Hanover et al. 1987; Jackson and Tjian 1988; Shafi et al. 2000; Love and Hanover 2005; Hart et al. 2007). Numerous sites on nucleocytosolic proteins may be modified by either O-GlcNAc or O-phosphate, suggesting a possible role for the OGT in cellular signaling (Cheng et al. 2000; Hart et al. 2007). OGT activity, as well as O-GlcNAcase activity, has also been associated with hexosamine signaling in metazoa and has been implicated in human type 2 diabetes and in a Caenorhabditis elegans model of diabetes (Hanover et al. 2005; Lehman et al. 2005; Love and Hanover 2005; Forsythe et al. 2006; Whelan et al. 2008; Yang et al. 2008).
Plant OGTs, which are referred to as “spindly” and “secret agent,” negatively regulate the gibberellin signaling pathway and are important in gamete formation and embryogenesis (Hartweck et al. 2002; Silverstone et al. 2007). Recently, a Listeria OGT was shown to add O-GlcNAc to flagellar proteins, as well as down-regulate synthesis of flagellar proteins (Shen et al. 2006). OGT genes and activities are absent from Saccharomyces cerevisiae and Schizosaccharomyces pombe.
Giardia lamblia and Cryptosporidium parvum, which are the focus of the present study, are protists (single-cell eukaryotes) spread by the fecal-oral route that cause diarrhea (Steiner et al. 1997). However, Giardia and Cryptosporidium are unrelated in their morphology and phylogeny. Giardia, which has two similar nuclei, eight flagellae, and an adherence disc, is a deeply divergent and minimal protist (Morrison et al. 2007). For example, Giardia makes an N-glycan precursor containing just two GlcNAc and has a single nucleotide-sugar transporter (NST) that transports UDP-GlcNAc from the cytosol to secretory compartments (Samuelson et al. 2005; Banerjee et al. 2008). In contrast, metazoa have N-glycan precursors with 14 sugars and 18–22 NSTs.
Cryptosporidium has a single nucleus and is related in its appearance and phylogeny to Plasmodium and Toxoplasma (Abrahamsen et al. 2004). Cryptosporidium makes an N-glycan precursor containing eight sugars and has numerous NSTs that transport activated Gal, GalNAc, Man, and Fuc (Samuelson et al. 2005 and our unpublished data).
The first goal here was to determine whether any protists have a predicted OGT, and if so, use phylogenetic methods to determine the ancestry of the protist OGTs. The second goal was to determine whether cytosolic extracts of Giardia and Cryptosporidium, which have ogt genes, have OGT activity. The third goal was to determine whether recombinant Giardia and Cryptosporidium OGTs have the predicted activities.
Results and discussion
Distinct origin of the Giardia OGT
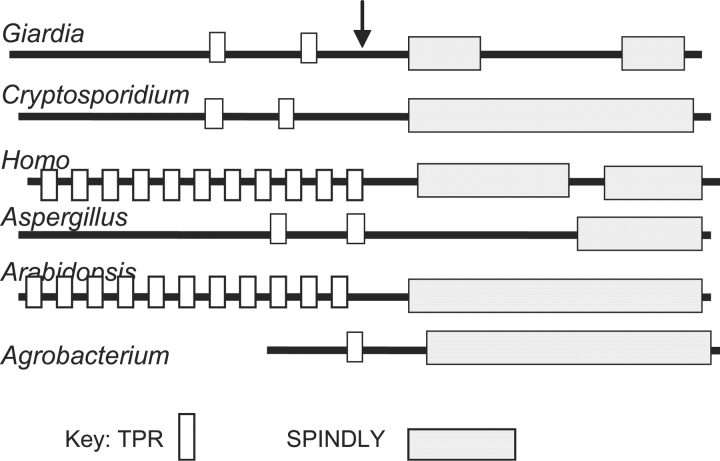
The Giardia genome predicts a single 1480-aa OGT, which contains just two TPRs at its N-terminus and a “spy” (catalytic) domain that has a large insert in the C-terminus (Figure 1 and supplementary Figure 1). The Cryptosporidium genome predicts a single 1032-aa long OGT, which also contains just two N-terminal TPRs but has an uninterrupted C-terminal “spy” domain. In contrast, human OGT, which is 1046-aa long, has 11 TPRs.
Fig. 1.
The OGTs of protists, fungus, and bacterium have fewer N-terminal TPRs than previously characterized OGTs of metazoa and plants. Diagrams of selected OGTs show distribution of TPRs and C-terminal, catalytic “spy” domains. Arrow indicates start of recombinant Giardia OGT expressed in Saccharomyces (see Figure 3). Please see supplementary Figures 1 and 2 for alignment of the catalytic domains of Giardia and Cryptosporidium OGTs with those of the most similar metazoan and plant, respectively.
Toxoplasma and Dictyostelium have predicted OGTs, which are absent from other protists examined (Eimeria, Entamoeba, Leishmania, Plasmodium, Spironucleus, Theileria, Trichomonas, and Trypanosoma). Predicted OGTs, which are present in all metazoa and plants examined, are also present in Yarrowia, Aspergillus, Magnaporthea, and Neurospora but are absent from Saccharomyces, Schizosaccharomyces, Candida, and Cryptococcus.
We looked for but did not find protist homologs of the O-GlcNAcase, which removes O-linked GlcNAc from nucleocytosolic proteins of metazoan (Gao et al. 2001). Homologs of this O-GlcNAcase are present in numerous bacteria but are absent from plants and fungi (with the exception of Histoplasma), which contain OGTs.
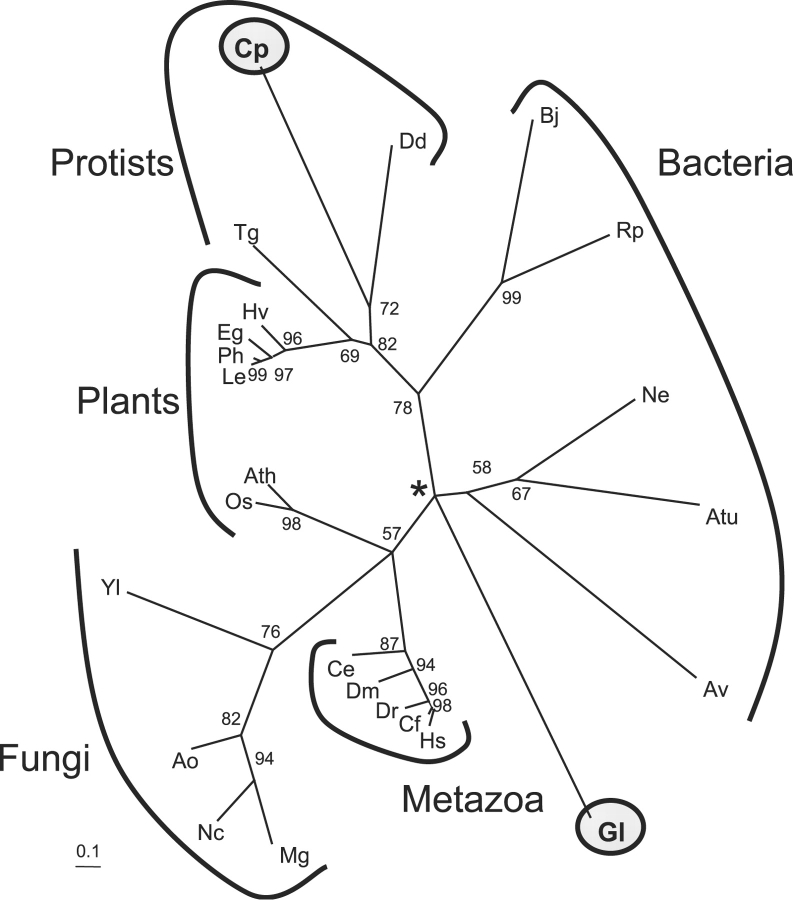
Phylogenetic analyses were performed to determine the origins of the Giardia and Cryptosporidium OGTs with the following results (Figure 2). First, the OGT tree, which was drawn by maximum likelihood methods, is star shaped, so that it is not possible to determine the ancestry of the Giardia OGT. Second, the OGTs of the other protists (Cryptosporidium, Toxoplasma, and Dictyostelium) appear to share a common ancestor with each other and more distant common ancestry with some plants (e.g., Lycopersicon and Petunia) and some bacteria (Bradyrhizobium and Rhodopseudomonas). Third, metazoan OGTs are very tightly clustered and appear to share more distant common ancestry with fungal OGTs and OGTs of some plants (Arabidopsis and Oryza).
Fig. 2.
Phylogenetic analyses suggest a unique origin of the Giardia (Gl) OGT, while OGTs of Cryptosporidium (Cp), Toxoplasma (Tg), and Dictyostelium (Dd) share a common origin with each other. The phylogenetic tree of selected protist, fungal, plant, metazoan, and bacterial OGTs was constructed using the maximum likelihood method. OGTs of Giardia and Cryptosporidium, which were tested here, are highlighted by shaded circles. Lengths of branches are proportional to differences between sequences, while numbers at nodes refer to bootstrap values for 100 trees. Asterisk indicates unresolved central node in this star-shaped tree, so that it is not possible to determine the ancestry of the Giardia OGT. Metazoa include Caenorhabditis elegans (Ce), Canis familiaris (Cf), Drosophila melanogaster (Dm), Danio rerio (Dr), and Homo sapiens (Hs). Fungi include Aspergillus oryzae (Ao), Neurospora crassa (Nc), Magnaporthea grisea (Mg), and Yarrowia lipolytica (Yl). Plants include Arabidopsis thaliana (Ath), Eustoma grandiflorum (Eg), Hordeum vulgare (Hv), Lycopersicon esculentum (Le), Oryxa sativa (Os), and Petunia hybrida (Ph). Bacteria include Anabaena variabilis (Av), Agrobacterium tumefacience (Atu), Bradyrhizobium japonicum (Bj), Nitrosomonas europaea (Ne), and Rhodopseudomonas palustris (Rp).
The absence of ogt genes in other protists and fungi strongly suggests secondary loss as an explanation for the present diversity of OGTs. In particular, OGT is present in Dictyostelium but absent from the closely related ameba Entamoeba; OGT is present in Giardia but absent from the closely related diplomonad Spironucleus; and OGTs are present in Toxoplasma and Cryptosporidium but are absent from the closely related apicomplexans Plasmodium and Theileria. The present diversity of N-glycan precursors has also been explained by secondary loss of Alg enzymes, which synthesize the N-glycan precursors (Samuelson et al. 2005).
The presence of OGTs in a mixed clade composed of protists, bacteria, and selected plants or in a mixed clade composed of metazoa, fungi, and selected plants suggests the possibility of lateral gene transfer of ogt genes. The possible common ancestry of plant and Rhodobacter OGTs, subsequent to lateral gene transfer, has previously been proposed (Roos and Hanover 2000). While lateral gene transfer is an important contributor to evolution of bacteria and some protists (Giardia, Entamoeba, and Trichomonas), it is not usually an important contributor to evolution of plants or metazoa (De Koning et al. 2000; Loftus et al. 2005; Morrison et al. 2007).
Molecular characterization of the Giardia OGT
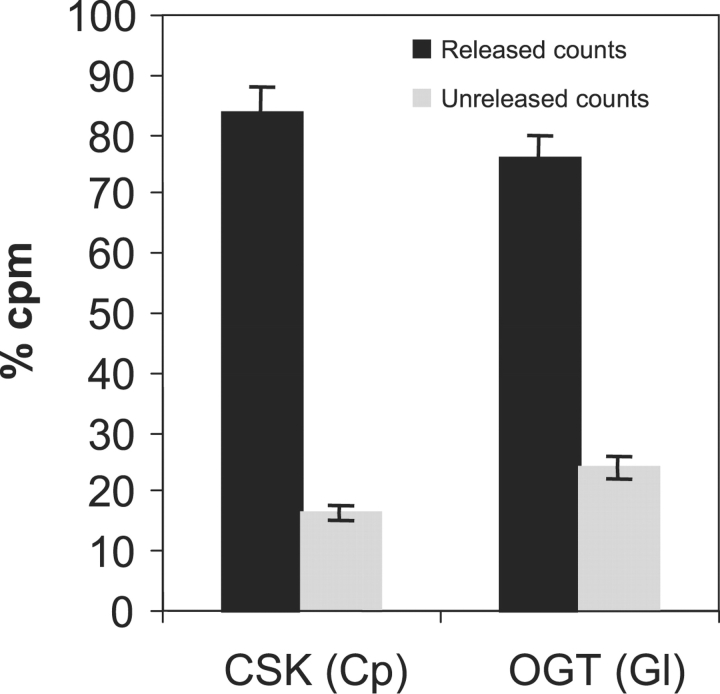
Messenger RNAs encoding Giardia lamblia OGT (GlOGT) are present in Giardia trophozoites and increase by ∼4-fold when Giardia encyst in vitro (Banerjee et al. 2008). Nucleocytosolic extracts of Giardia have OGT activity in the presence of radiolabeled UDP-GlcNAc (Figure 3A), and this result was confirmed by the release of a single GlcNAcitol from nucleocytosolic glycoproteins by reductive β-elimination (data not shown). A recombinant His-tagged catalytic domain of GlOGT, which was expressed in Saccharomyces and purified on Ni-NTA beads, failed to glycosylate casein kinase (a test substrate for OGTs) but autoglycosylated with kinetics similar to that of the OGT in Giardia nucleocytosolic extracts (Figure 3B). A single GlcNAcitol was released by reductive β-elimination of the glycosylated protein (Figure 3C). In addition, β-hexosaminidase released ∼75% of the radiolabeled GlcNAc from the autoglycosylated GlOGT, confirming the expected β-O-GlcNAc linkage (Figure 4). A recombinant human OGT, which is missing in the TPRs, also autoglycosylates but does not glycosylate casein kinase (Lubas and Hanover 2000).
Fig. 3.
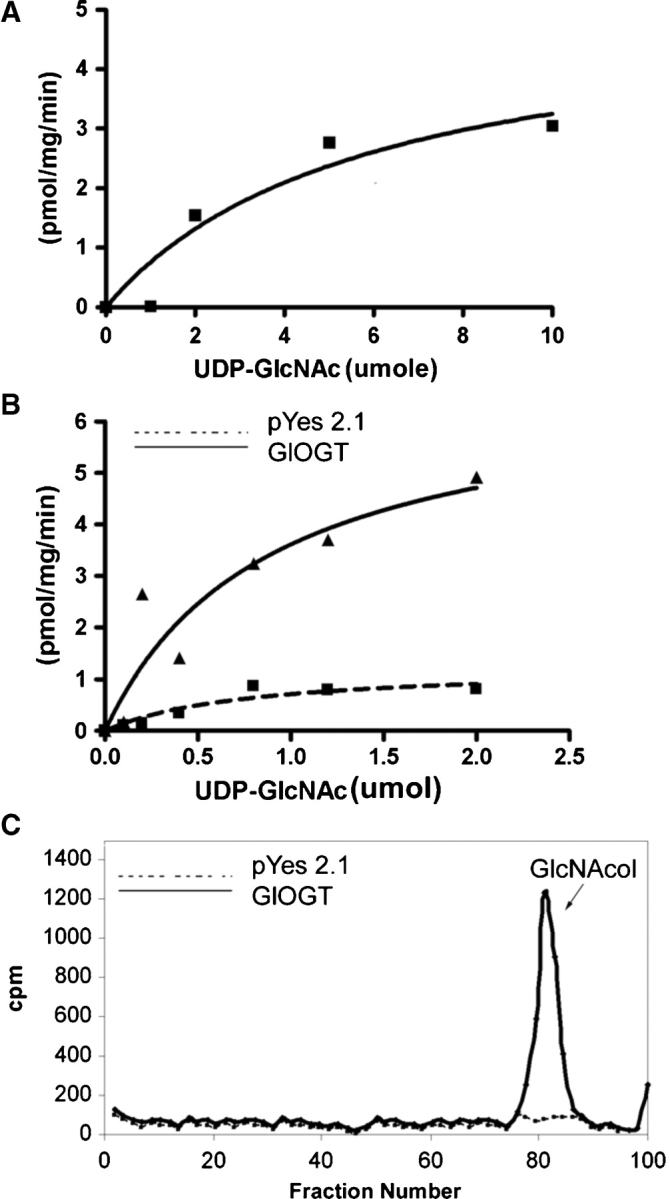
OGT activities of Giardia nucleocytosolic extract and of the recombinant GlOGT expressed in yeast. (A) The OGT activity of the Giardia nucleocytosolic extract with itself showed a Km of 5.8 μM UDP-GlcNAc and a Vmax of 5.1 pgm/min/mg protein. (B) The activity of recombinant GlOGT (catalytic domain only), again with itself, showed a Km of 0.9 μM UDP- GlcNAc and a Vmax of 5.6 pgm/min/mg protein. (C) Transfer of a single, O-linked GlcNAc to recombinant GlOGT was confirmed by performing a reductive β-elimination on the precipitated pellet following the OGT assay. Analysis of the products on a Bio-Gel P2 showed a single GlcNAcitol. The negative control in B and C is the empty vector. In each case, representative data are shown from three experiments performed on separate days.
Fig. 4.
β-Hexaminidase release of radiolabeled O-GlcNAc from autoglycosylated GlOGT (Figure 3B and C) and from casein kinase (CSK), which was glycosylated by the recombinant CpOGT (Figure 5B and C). In each case, the vast majority of the radiolabeled GlcNAc was released by β-hexosaminidase, confirming the expected β-O-GlcNAc linkage.
Molecular characterization of the Cryptosporidium OGT
Messenger RNAs encoding CpOGT are present in Cryptosporidium sporozoites (data not shown), and nucleocytosolic extracts of Cryptosporidia have OGT activity in the presence of radiolabeled UDP-GlcNAc (Figure 5A). A recombinant full-length CpOGT, which was expressed as a GST-fusion enzyme in Escherichia coli and purified on glutathione-agarose, glycosylated casein kinase (a test target for OGTs) with kinetics similar to that of the OGT in nucleocytosolic extracts of Cryptosporidia (Figure 5B). A single GlcNAcitol was released by reductive β-elimination of the glycosylated proteins (Figure 5C). In addition, β-hexosaminidase released ∼83% of the radiolabeled GlcNAc from the glycosylated casein kinase, confirming the expected β-O-GlcNAc linkage (Figure 4).
Fig. 5.
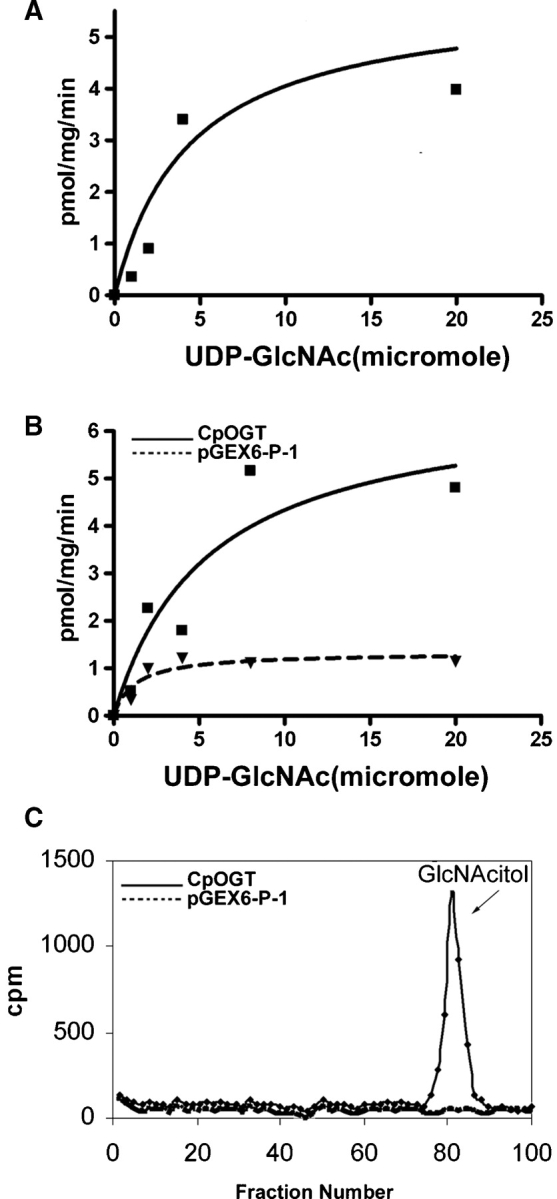
OGT activities of Cryptosporidium nucleocytosolic extract and of the recombinant CpOGT expressed as a GST-fusion protein in bacteria. (A) The OGT activity of the nucleocytosolic extract with itself showed a Km of 4.3 μM UDP-GlcNAc and a Vmax of 5.8 pgm/min/mg protein. (B) The activity of recombinant full-length CpOGT with casein kinase showed a Km of 5.4 μM UDP-GlcNAc and a Vmax of 6.7 pgm/min/mg protein. (C) Transfer of O-linked GlcNAc to casein kinase was confirmed by performing a reductive β-elimination on the precipitated pellet following the OGT assay. Analysis of the products on a Bio-Gel P2 showed a single GlcNAcitol. The negative control in B and C is GST alone. In each case, representative data are shown from three experiments performed on separate days.
Major conclusions and unresolved questions
To our knowledge, the presence of putative ogt genes in some protists and fungi has not previously been shown, and the OGT activities of protist membranes and of recombinant enzymes have not previously been demonstrated. These results suggest the possibility that O-GlcNAc modification of nucleocytosolic proteins also has function(s) in simple eukaryotes. Conversely, the absence of ogt genes (likely by secondary loss) in so many protists and fungi indicates that these organisms have alternative methods for regulating activities of nucleocytosolic proteins.
Future studies, which might help determine the function of the protist OGTs, include the following. First, knockout studies of the OGTs, which are not presently possible in Giardia or Cryptosporidium, may be performed in Dictyostelium and Toxoplasma, where these technologies are readily available. Second, identification of the “OGTome” (nucleocytosolic proteins modified by O-linked GlcNAc) may suggest numerous possible functions for O-GlcNAc modification in these protists. Third, in the context of the hexosamine hypothesis (the idea that OGT and O-GlcNAcase are involved in glucose sensing and signaling) (Love and Hanover 2005), it would be interesting to examine the role of O-GlcNAc modification of nucleocytosolic proteins during encystation by Giardia. This is because OGT mRNAs are increased 4-fold during encystation and UDP-GlcNAc (the limiting substrate for the OGT) is also increased during encystation (Sener et al. 2004; Banerjee et al. 2008).
Material and methods
Bioinformatic methods
The predicted proteins of Giardia lamblia and Cryptosporidium parvum, which are present at GiardiaDB and CryptoDB, respectively, were searched with PSI-BLAST using the OGT and O-GlcNAcase (also known as MGEA5) protein sequences from Homo sapiens (Altschul et al. 1997; Gao et al. 2001). Similar methods were used to search the predicted proteins of representative protists (Dictyostelium discoideum, Eimeria tenella, Entamoeba histolytica, Leishmania major, Plasmodium falciparum, Spironucleus vortens, Theileria annulata, Toxoplasma gondii, Trypanosoma brucei, and Trypanosoma cruzi), metazoa, fungi, and plants in the NR database at the NCBI or at specific databases (e.g., PlasmoDB or GeneDB). Trichomonas vaginalis proteins were searched at websites maintained by the J. Craig Venter Institute, Rockville, MD. Predicted OGTs were examined for conserved domains using the CD search at NCBI (Marchler-Bauer et al. 2007). OGTs were aligned using multiple sequence comparison by log-expectation (MUSCLE) (Edgar 2004). The alignment was manually refined, and gaps were removed using BioEdit. The finished alignment was used to construct the phylogenetic tree using TREE-PUZZLE, a program to reconstruct phylogenetic trees from molecular sequence data by the maximum likelihood method (Schmidt et al. 2002).
Parasite manipulations
Trophozoites of the genome project WB strain of Giardia were grown axenically in TYI-S media supplemented with 10% serum and 1 mg/mL bile. Giardia were chilled on ice for 20 min and concentrated at 2000 rpm for 5 min, and genomic DNA and total RNA were isolated using Promega and Invitrogen kits. Alternatively, Giardia cells were lysed by sonication in 10 mM Hepes, pH 7, 10 mM MgCl2, and 25 mM NaCl, supplemented with protease inhibitor cocktail (Sigma). The lysate was centrifuged at 6000 rpm for 5 min to remove unbroken cells and nuclei, and the supernatant was centrifuged at 37,000 rpm in a SW60 rotor (Beckman) for 1 h. The residual pellet was discarded, and the supernatant (nucleocytosolic extract) was used for assays of the endogenous OGT.
Cryptosporidium parvum (Iowa strain) oocysts, which were passaged through newborn calves, were obtained from Bunch Grass Farm (Dury, Idaho) and stored at 4°C. Contaminating fecal bacteria were lysed by 5–10 pulses of sonication in phosphate buffered saline (PBS) and intact oocysts were concentrated by centrifugation at 1100 × g. Cryptosporidium sporozoites were induced to excyst by incubating oocysts in a RPMI medium with 0.75% Na-taurocholate, pH 7.5, for 2 h at 37°C. DNA, RNA, and nucleocytosol of Cryptosporidia were isolated as described for Giardia.
Cloning and expression of Giardia and Cryptosporidium OGTs
The 3′ end of the Giardia ogt gene, which encodes the catalytic “spy” domain, was amplified from the genomic DNA by PCR (forward primer: GAATTCATGCCCTACCACTGTTATCTTTA and reverse primer: CTCGAGCTGTGCCCCCGT ACTCTTTA) and cloned into in the pYES2.1/V5-His-TOPO vector (Invitrogen), which makes polyHis-tagged proteins under the Saccharomyces Gal1 promoter. The GlOGT construct was transformed into Saccharomyces cerevisiae strain BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), and yeast were induced with 2% Gal for 16 h at 30°C. Yeast were lysed with glass-beads, and the recombinant GlOGT was purified using Ni-NTA beads (Invitrogen). Expression and purification of GlOGT was confirmed on Western blots of purified GlOGT, using a horse-radish peroxidase-labeled antibody to the polyHis epitope-tag.
The full-length Cryptosporidium ogt gene was amplified from the genomic DNA by PCR (forward primer: ATGCTGAAAGATGGTGGAGTT and reverse primer: CTCGAGTTATTTGCATGCAGATAAA) and cloned into the pGEX-6P-1 vector as a glutathione-S-transferase (GST) fusion protein. The recombinant CpOGT was induced by IPTG and purified using glutathione agarose (GE Life Sciences). Expression and purification of CpOGT in bacteria was confirmed by Western blots, using an anti-GST antibody (GE Life Sciences).
OGT assays
The OGT activities were determined for Giardia and Cryptosporidium nucleocytosolic extracts, as well as for recombinant GlOGT and CpOGT, which were expressed in yeast or bacteria, respectively. For assaying endogenous OGT activity, 25–50 μg of protist nucleocytosolic proteins were incubated with 0.1 μCi of tritiated UDP-GlcNAc in the OGT assay buffer (50 mM Tris–HCl, pH 7.5, 12.5 mM MgCl2 and 1 mM DTT) for 30 min. The reaction was stopped with an equal volume of ice cold 20% TCA, and precipitated proteins were washed three times in water, resuspended in the Eoscint scintillation fluid, and counted in a liquid scintillation counter. Each assay was performed on 3 separate days.
The radiolabeled product was further characterized by reductive β-elimination and chromatography on a Bio-Gel P2 column. Briefly, the TCA pellets were resuspended in 50 mM NaOH containing 1 M NaBH4 at room temperature for 16 h. The reaction was stopped with acetic acid and freed of boric acid by repeated evaporation with methanol containing 1% acetic acid in a rotary evaporator. The residue was passed through a mixed bed AG501-X8 resin (BioRad), and the flow-through was dried, resuspended in water, chromatographed, and counted.
For assaying recombinant GlOGT, 1–2 μg of the purified recombinant protein was used, and the transfer of O-GlcNAc to the enzyme itself was assayed (Lubas and Hanover 2000). For assaying recombinant CpOGT, 1–2 μg of purified recombinant protein was used, and the transfer of O-GlcNAc to casein kinase was determined. The radiolabeled product was characterized by β-elimination, as described above. In addition, 3 μg each of radiolabeled GlOGT and casein kinase was digested for 1 h at 37°C with 1 unit of β-hexosaminidase (New England Biolabs) in 100 μL of buffer that was supplied by the manufacturer. The reaction was stopped by adding an equal volume of chilled methanol, and the released products were cleaned using the GlycocleanR (Prozyme) and counted, as described above.
Supplementary Data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Funding
National Institutes of Health grants AI048082 (to J.S.) and GM31318 (to P.W.R.).
Conflict of interest statement
None declared.
Glossary
Abbreviations
- CpOGT
Cryptosporidium parvum OGT
- GlOGT
Giardia lamblia OGT
- GST
glutathione-S-transferase
- NST
nucleotide-sugar transporter
- OGT
O-linked N-acetylglucosaminyltransferase
- TPR
tetratricopeptide repeat
References
- Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA, Deng M, Liu C, Widmer G, Tzipori S, et al. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004;304:441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Cui J, Robbins PW, Samuelson J. Use of Giardia, which appears to have a single nucleotide-sugar transporter for UDP-GlcNAc, to identify the UDP-Glc transporter of Entamoeba. Mol Biochem Parasitol. 2008;159:44–53. doi: 10.1016/j.molbiopara.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Cole RN, Zaia J, Hart GW. Alternative O-glycosylation/O-phosphorylation of the murine estrogen receptor beta. Biochemistry. 2000;39:11609–11620. doi: 10.1021/bi000755i. [DOI] [PubMed] [Google Scholar]
- de Koning AP, Brinkman FS, Jones SJ, Keeling PJ. Lateral gene transfer and metabolic adaptation in the human parasite Trichomonas vaginalis. Mol Biol Evol. 2000;17:1769–1773. doi: 10.1093/oxfordjournals.molbev.a026275. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsythe ME, Love DC, Lazarus BD, Kim EJ, Prinz WA, Ashwell G, Krause MW, Hanover JA. Caenorhabditis elegans ortholog of a diabetes susceptibility locus: Oga-1(O-GlcNAcase) knockout impacts O-GlcNAc cycling, metabolism, and dauer. Proc Natl Acad Sci USA. 2006;103:11952–11957. doi: 10.1073/pnas.0601931103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Wells L, Comer FI, Parker GJ, Hart GW. Dynamic O-glycosylation of nuclear and cytosolic proteins: Cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain. J Biol Chem. 2001;276:9838–9845. doi: 10.1074/jbc.M010420200. [DOI] [PubMed] [Google Scholar]
- Hanover JA, Cohen CK, Willingham MC, Park MK. O-Linked N-acetylglucosamine is attached to proteins of the nuclear pore. Evidence for cytoplasmic and nucleoplasmic glycoproteins. J Biol Chem. 1987;262:9887–9894. [PubMed] [Google Scholar]
- Hanover JA, Forsythe ME, Hennessey PT, Brodigan TM, Love DC, Ashwell G, Krause M. A Caenorhabditis elegans model of insulin resistance: Altered macronutrient storage and dauer formation in an OGT-1 knockout. Proc Natl Acad Sci USA. 2005;102:11266–11271. doi: 10.1073/pnas.0408771102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- Hartweck LM, Scott CL, Olszewski NE. Two O-linked N-acetylglucosamine transferase genes of Arabidopsis thaliana L. Heynh. have overlapping functions necessary for gamete and seed development. Genetics. 2002;161:1279–1291. doi: 10.1093/genetics/161.3.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Tjian R. O-Glycosylation of eukaryotic transcription factors: Implications for mechanisms of transcriptional regulation. Cell. 1988;55:125–133. doi: 10.1016/0092-8674(88)90015-3. [DOI] [PubMed] [Google Scholar]
- Lehman DM, Fu DJ, Freeman AB, Hunt KJ, Leach RJ, Johnson-Pais T, Hamlington J, Dyer TD, Arya R, Abboud H, et al. A single nucleotide polymorphism in MGEA5 encoding O-GlcNAc-selective N-acetyl-beta-d glucosaminidase is associated with type 2 diabetes in Mexican Americans. Diabetes. 2005;54:1214–1221. doi: 10.2337/diabetes.54.4.1214. [DOI] [PubMed] [Google Scholar]
- Loftus B, Anderson I, Davies R, Alsmark UC, Samuelson J, Amedeo P, Roncaglia P, Berriman M, et al. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433:865–868. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- Love DC, Hanover JA. The hexosamine signaling pathway: Deciphering the “O-GlcNAc code”. Sci STKE. 2005:re13. doi: 10.1126/stke.3122005re13. [DOI] [PubMed] [Google Scholar]
- Lubas WA, Hanover JA. Functional expression of O-linked GlcNAc transferase. Domain structure and substrate specificity. J Biol Chem. 2000;275:10983–10988. doi: 10.1074/jbc.275.15.10983. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, et al. CDD: A conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Fleites C, Macauley MS, He Y, Shen DL, Vocadlo DJ, Davies GJ. Structure of an O-GlcNAc transferase homolog provides insight into intracellular glycosylation. Nat Struct Mol Biol. 2008;15:764–765. doi: 10.1038/nsmb.1443. [DOI] [PubMed] [Google Scholar]
- Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, Best AA, Cande WZ, Chen F, Cipriano MJ, et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;317:1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- Roos MD, Hanover JA. Structure of O-linked GlcNAc transferase: Mediator of glycan-dependent signaling. Biochem Biophys Res Commun. 2000;271:275–280. doi: 10.1006/bbrc.2000.2600. [DOI] [PubMed] [Google Scholar]
- Samuelson J, Banerjee S, Magnelli P, Cui J, Kelleher DJ, Gilmore R, Robbins PW. The diversity of dolichol-linked precursors to Asn-linked glycans likely results from secondary loss of sets of glycosyltransferases. Proc Natl Acad Sci USA. 2005;102:1548–1553. doi: 10.1073/pnas.0409460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: Maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Sener K, Shen Z, Newburg DS, Jarroll EL. Amino sugar phosphate levels in Giardia change during cyst wall formation. Microbiology. 2004;150:1225–1230. doi: 10.1099/mic.0.26898-0. [DOI] [PubMed] [Google Scholar]
- Shafi R, Iyer SP, Ellies LG, O’Donnell N, Marek KW, Chui D, Hart GW, Marth JD. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci USA. 2000;97:5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen A, Kamp HD, Gründling A, Higgins DE. A bifunctional O-GlcNAc transferase governs flagellar motility through anti-repression. Genes Dev. 2006;20:3283–3295. doi: 10.1101/gad.1492606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Tseng TS, Swain SM, Dill A, Jeong SY, Olszewski NE, Sun TP. Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol. 2007;143:987–1000. doi: 10.1104/pp.106.091025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner TS, Thielman NM, Guerrant RL. Protozoal agents: What are the dangers for the public water supply? Annu Rev Med. 1997;48:329–340. doi: 10.1146/annurev.med.48.1.329. [DOI] [PubMed] [Google Scholar]
- Whelan SA, Lane MD, Hart GW. Regulation of the O-linked beta-N-acetylglucosamine transferase by insulin signaling. J Biol Chem. 2008;283:21411–21417. doi: 10.1074/jbc.M800677200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrabl JO, Grishin NV. Homology between O-linked GlcNAc transferases and proteins of the glycogen phosphorylase superfamily. J Mol Biol. 2001;314:365–374. doi: 10.1006/jmbi.2001.5151. [DOI] [PubMed] [Google Scholar]
- Yang X, Ongusaha PP, Miles PD, Havstad JC, Zhang F, So WV, Kudlow JE, Michell RH, Olefsky JM, Field SJ, et al. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature. 2008;451:964–969. doi: 10.1038/nature06668. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.