Abstract
Mucin-type protein O-glycosylation is initiated by the addition of α-GalNAc to Ser/Thr residues of a polypeptide chain. The addition of β-Gal to GalNAc by the UDP-Gal:glycoprotein-α-GalNAc β3 galactosyltransferase (T-synthase), forming the Core 1 structure (β-Gal(1-3)-α-GalNAc-O-Ser/Thr), is a common and biologically significant subsequent step in O-glycan biosynthesis. What dictates the sites of Core 1 glycosylation is poorly understood; however, the peptide sequence and neighboring glycosylation effects have been implicated. To systematically address the role of the peptide sequence on the specificity of T-synthase, we used the oriented random glycopeptide: GAGAXXXX(T-O-GalNAc)XXXXAGAG (where X = G, A, P, V, I, F, Y, S, N, D, E, H, R, and K) as a substrate. The Core 1 glycosylated product was isolated on immobilized PNA (Arachis hypogaea) lectin and its composition determined by Edman amino acid sequencing for comparison with the initial substrate composition, from which transferase preferences were obtained. From these studies, elevated preferences for Gly at the +1 position with moderately high preferences for Phe and Tyr in the +3 position relative to the acceptor Thr-O-GalNAc were found. A number of smaller Pro enhancements were also observed. Basic residues, i.e., Lys, Arg, and His, in any position were disfavored, suggesting electrostatic interactions as an additional important component modulating transferase specificity. This work suggests that there are indeed subtle specific and nonspecific protein-targeting sequence motifs for this transferase.
Keywords: Core 1 transferase, O-glycosylation, sequence motifs, T-synthase, UDP-Gal:glycoprotein-α-GalNAc β3 galactosyltransferase
Introduction
O-Glycosylation is a common post-translational modification found on many secreted and membrane-associated glycoproteins. Mucin-type O-glycans are categorized by their core structures, i.e., Cores 1–7, with the Core 1 structure, β-Gal (1-3) α-GalNAc-O-Ser/Thr (the T-antigen) being one of the most common (Brockhausen 1997). The Core 1 structure is formed by the transfer of galactose from UDP-Gal to α-GalNAc1-Ser/Thr catalyzed by the UDP-Gal:glycoprotein-α-GalNAc β3 galactosyltransferase (T-synthase). Because the Core 1 structure also serves as the precursor for further O-glycan elongation, including sialylation and fucosylation, it plays an important branchpoint role in mucin-type O-glycan biosynthesis (Brockhausen 1999; Ju et al. 2008). A decrease in the expression of T-synthase alters O-glycan elongation, resulting in abnormal truncated carbohydrate structures commonly leading to the exposure of the Tn antigen (GalNAc-O-Ser/Thr) as is found in several human diseases, i.e., Tn syndrome, IgA nephropathy, and cancers of the colon and breast (Springer 1997; Berger 1999; Freire et al. 2005; Ju and Cummings 2005; Smith et al. 2006; Li et al. 2007; Pinho et al. 2007). Overall, the biological importance of T-synthase is evidenced by its embryonic lethality when its expression is eliminated in the T-synthase knock-out mouse model (Xia et al. 2004).
The modulation of mucin-type O-glycan biosynthesis (initiation and elongation) is poorly understood. Recently, we demonstrated that the porcine salivary gland mucin (PSM) tandem repeat O-glycan structures, including the Core 1 structure, vary reproducibly with their position in the peptide sequence (Gerken et al. 2002). Based on our in vitro kinetic modeling of the porcine and canine salivary mucin tandem repeats glycosylated by ppGalNAc-T1 and -T2 (Gerken et al. 2004), we concluded that these transferases possess reproducible and specific sensitivities to the peptide sequence and neighboring glycosylation. To further characterize ppGalNAc-T peptide sequence preferences, we have recently developed an oriented random peptide substrate approach for obtaining the peptide preferences for the ppGalNAc-Ts, ±3 to 5 residues from the site of glycosylation (Gerken et al. 2006, 2008). From these latter studies, previously unknown features of the peptide substrate preferences of ppGalNAc-T1 and -T2 have been revealed.
Upon subsequent kinetic modeling of the in vivo PSM Core 1 glycosylation pattern, we could also show that the T-synthase was modulated by neighboring GalNAc glycosylation, similar to ppGalNAc-T1 and -T2 (Gerken 2004). However, in spite of earlier studies (Brockhausen et al. 1990; Granovsky et al. 1994), very little is known about the role of the peptide sequence on modulating the activity of the T-synthase, i.e., it is not known if there are optimal peptide sequences or motifs recognized by the transferase. In the present work, we have searched for such motifs using, as a T-synthase substrate, the oriented random glycopeptide GAGAXXXXT(O-GalNAc)XXXXAGAG (where X = random residues). From these studies, we have observed elevated preferences for Gly at the +1 position with moderately high preferences for Phe and Tyr in the +3 position relative to the acceptor Thr-O-GalNAc. Furthermore, we observed that basic residues in any position were disfavored suggesting electrostatic interactions as an additional important component modulating transferase specificity. This work represents the most thorough analysis of the Core 1 peptide substrate preferences performed to date and indeed suggests that there are subtle specific and nonspecific protein-targeting sequences (or motifs) for this transferase.
Results
We have recently utilized an oriented random peptide substrate approach for the determination of the peptide substrate preferences of the ppGalNAc-Ts (Gerken et al. 2006, 2008). Key to this approach was the use of lectin column chromatography for the isolation of the glycosylated product and the use of Edman amino acid sequencing for the determination of the positional enrichment factors. In the present work, we describe a similar approach for obtaining the T-synthase peptide preferences using the GP-II random glycopeptide GAGAXXXX(T-O-GalNAc)XXXXAGAG (where X = G, A, P, V, I, F, Y, S, N, D, E, H, R, and K) as an acceptor. Here, the Core 1 product was isolated on an immobilized lectin column specific for the Core 1 disaccharide, β-Gal (1-3) α-GalNAc-O-Ser/Thr, and its random residue composition was determined by Edman sequencing. From this data, the amino acid preferences were derived.
Method development
The T-synthase random glycopeptide substrate GP-II was designed based on our prior success using similar oriented random peptide substrates for characterizing ppGalNAc-T substrate specificity. Specifically, the substrate needed to be of sufficient size for separation from free UDP-Gal and Gal on Sephadex G10 gel filtration (Figure 1A) and contain flanking GAGA sequences for assessing product purity and integrity. Due to the expense of synthesizing glycopeptides, we also chose to have only one random glycopeptide synthesized rather than a series as was used for the ppGalNAc-Ts (Gerken et al. 2006, 2008). To compensate for this, the number of unique residues at each randomized position was increased to 14 residues (compared to 8–10 residues) while only lacking 6 residues (i.e., C, T, M, W, Q, and L), giving a statistical population of approximately 1.47 × 109 unique sequences. The glycosylation of GP-II by T-synthase was limited to less than 2–3% to ensure accurate preference determinations, as higher conversions would result in the loss of specificity.
Fig. 1.
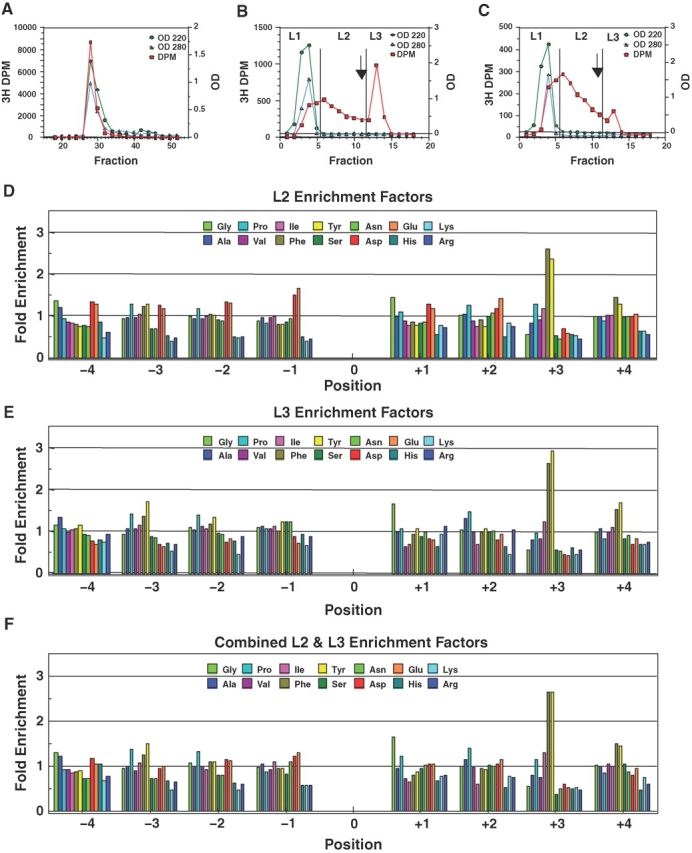
Isolation of the T-synthase Core 1 product of GP-II (GAGAXXXX(Thr-O-GalNAc)XXXXAGAG, X = G, A, P, V, I, F, Y, S, N, D, E, H, R, and K) by Sephadex G10 and PNA lectin chromatography (panels A–C) and the derived T-synthase amino acid substrate enhancement factors (D–F). (A) Sephadex G-10 gel filtration chromatography of a typical post dowex incubation mixture (determined to be ∼2% Core 1 glycosylated). Fractions 26–32 were pooled for the lectin chromatography in panel B. (B) PNA lectin affinity chromatography of the pool indicated in panel A. Arrow indicates the addition of 5 mM Gal. Fractions are labeled L1, L2, and L3 to represent the pass through, retarded, and bound fractions, respectively. (C) Lectin affinity chromatography of the L2 fraction from panel B. In panels A–C, circles represent the absorbance at 220 nm; diamonds absorbance at 280 nm, and squares 3H-Gal-R dpm. (D) Enhancement factors obtained for the L2 fraction in panel C, relative to the pre-lectin control. (E) Enhancement factors obtained for the L3 fraction in panel B. (F) 3H-weighted sum of the L2 and L3 enhancement values from panels D and E. In panels D–F, the “0” position represents Thr-O-GalNAc, the site of Core 1 glycosylation.
Isolation of the Core 1 random glycopeptide product of T-synthase on lectin affinity column chromatography
The PNA lectin possesses high specificity for the β-Gal residue of the Core 1 or T-antigen, and therefore was chosen initially for isolating the Core 1 random glycopeptide product of T-synthase (Lotan et al. 1975). A typical lectin column run of GP-II glycosylated with 3H-labeled UDP-Gal is shown in Figure 1B, where the fractions L1, L2, and L3 represent the pass through (predominantly nonmodified GP-II), retarded, and bound (Gal eluted) pools, respectively. In contrast to our previous experience with the use of lectin affinity chromatography (Gerken et al. 2006, 2008), a significant proportion of the apparent radiolabeled galactosylated GP-II product directly passes through the column with unmodified GP-II or was retarded, fractions L1 and L2, while only ∼1/3 of the radiolabeled product was eluted upon the addition of Gal, in the L3 fraction. Attempts to improve product binding by altering buffer composition were unsuccessful (data not shown) while the isolated pooled L1 fraction also failed to bind Jacalin, ABA, MPA, TKA, ACA, and APA lectin columns (data not shown). Rerunning the L2 fraction on the PNA column also gave a similar profile, Figure 1C. These unusual finding suggest that the profile observed on the PNA lectin column is not due to column overloading, but rather there is apparently a distribution of radiolabeled peptide products with different lectin binding affinities.
Therefore, to determine whether the different chromatographic behaviors are reflected by differences in their composition, the L2 and L3 fractions were separately pooled, passed over Sephadex G10, and submitted to Edman sequencing. The resulting L2 and L3 preferences, plotted in Figure 1D and E, are quite similar, both dominated by Tyr and Phe enhancements at the +3 position (discussed below). However, a closer inspection reveals that the L2 fraction is increased in Asp and Glu compared to the L3 fraction at nearly all random positions, especially at the −1 position. Likewise, the basic residues (Arg, Lys, His) content is decreased in the L2 fraction compared to L3. These observations suggest that the PNA lectin (and the other lectin columns) poorly binds highly acidic (negatively charged) peptides. Regardless, the general features of the preferences for the L2 and L3 fractions are very similar and together account for ∼77% of the radiolabeled material applied to the PNA column. The weighted average preferences (by 3H content) of the combined L2 and L3 fractions are given in Figure 1F. Since we have only accounted for ∼ 77% of the radiolabeled peptide, attempts were made to further characterize the nature of the radiolabeled product contained in the L1 fraction.
What is the origin of the 3H unbound L1 peak?
To determine whether the radiolabel in the L1 fraction was due to residual UDP-3H-Gal, an identical incubation was performed lacking enzyme. After passing over dowex and isolation of the peptide fraction (based on OD 220) via G10 chromatography, the PNA lectin column profile shown in Figure 2E was obtained. As shown in the figure, a small peak of 3H label indeed appears in the L1 fraction with identical chromatographic behavior as directly added UDP-3H-Gal (data not shown). Based on these results, we conclude that some, but not all, of the radioactivity in the L1 fraction is due to UDP-3H-Gal that was not bound to the dowex and which also does not bind to the PNA lectin. To determine the optimal amount of dowex necessary to remove unreacted UDP-3H-Gal, two different incubations were tested, one containing all reagents and another just lacking the glycopeptide acceptor (data not shown). From these two experiments, it was determined that two dowex column runs (2 × 3 mL) were sufficient to remove the majority (∼90%) of the UDP-3H-Gal, while additional dowex runs began to remove 3H-labeled random glycopeptide. Thus, the remaining UDP-3H-Gal could only be removed at the cost of removing the glycosylated product.
Fig. 2.
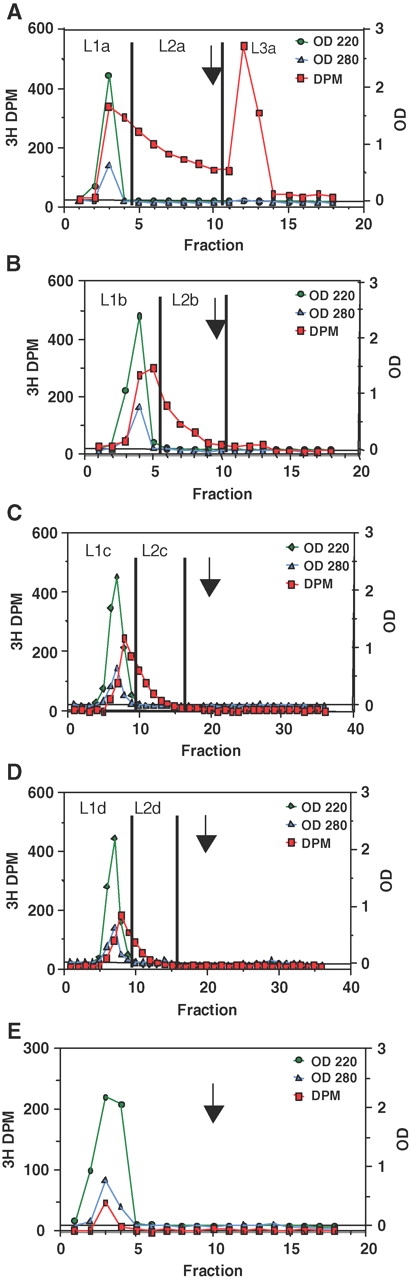
Isolation of the T-synthase retarded L2 glycopeptide product by multiple lectin chromatography. (A) Initial PNA lectin chromatography of a GP-II incubation with T-synthase (determined to be ∼0.8% Core 1 glycosylated). (B–D) Subsequent PNA lectin chromatography of the L1 fraction in panel A. (E) Lectin affinity chromatography of an incubation lacking only enzyme, demonstrating the trace presence of UDP-3H-Gal. (Note: In panels C and D fraction volumes were ½ of those collected in panels A, B, and E.)
Next, to confirm that the 3H-radiolabeled product collected from the L1 fraction was indeed a Core 1 product, the L1 fraction, as well as L2 and L3 fractions, was each submitted to β-galactosidase treatment and their products separated on G10 chromatography (supplementary Figure S1). From these experiments, ∼86% of the 3H label in the L1 fraction was digested while 100% of the L2 and L3 fractions were digested and freed of 3H-Gal. Together these results demonstrate that the L1 fraction contains a small amount of UDP-3H-Gal and a significant amount of the weakly binding 3H-Core 1 product.
Fractionation of L1 fraction by repeated lectin chromatography
As shown above, the 3H content of the L1 fraction contained the significant weakly binding Core 1 product. Therefore, in a separate experiment, we attempted to further separate the L1 fraction into additional L1 and L2 fractions, by repeated lectin chromatography. This is shown in Figure 2A–D, where each subsequent panel represents the rerunning on the lectin column of the prior L1 fraction (labeled L1a–d). As can be seen in Figure 2B and C, running the L1 fraction over additional lectin columns results in the further separation into additional L1 and L2 fractions. At the end of the fractionation process, the combined L2a + L3a fractions represent 76% of the initial 3H label, the combined L2b + L2c + L2d fractions ∼14%, and the final L1d fraction ∼10% of the initial loaded 3H label. Based on our β-galactosidase incubations and our UDP-3H-Gal control studies, we estimate that ∼2/3 of the final L1d fraction is nonbinding Core 1 product and ∼1/3 UDP-3H-Gal. Thus, all but ∼7% of the Core 1 product could be separated into L2 and L3 fractions.
The pooled L2a + L3a and pooled L2b + L2c + L2d fractions (Figure 2A–D) were Edman sequenced and their substrate preferences obtained as plotted in Figure 3A and B, respectively. As expected, the L2a + L3a pool (Figure 3A) gave nearly identical preferences as obtained in the previous experiment, where the individual L2 and L3 preferences were summed (Figure 1F). The L2b + L2c + L2d pool (Figure 3B) gave enhanced acidic residue preferences and reduced basic residue enhancements, while maintaining the elevated Tyr and Phe preferences at the +3 position. By further combining the L2 + L3 and L2b + L2c + L2d preferences, weighted according to their 3H content, the preferences given in Figure 3C were obtained, representing ∼93% of the total Core 1 product of T-synthase.
Fig. 3.
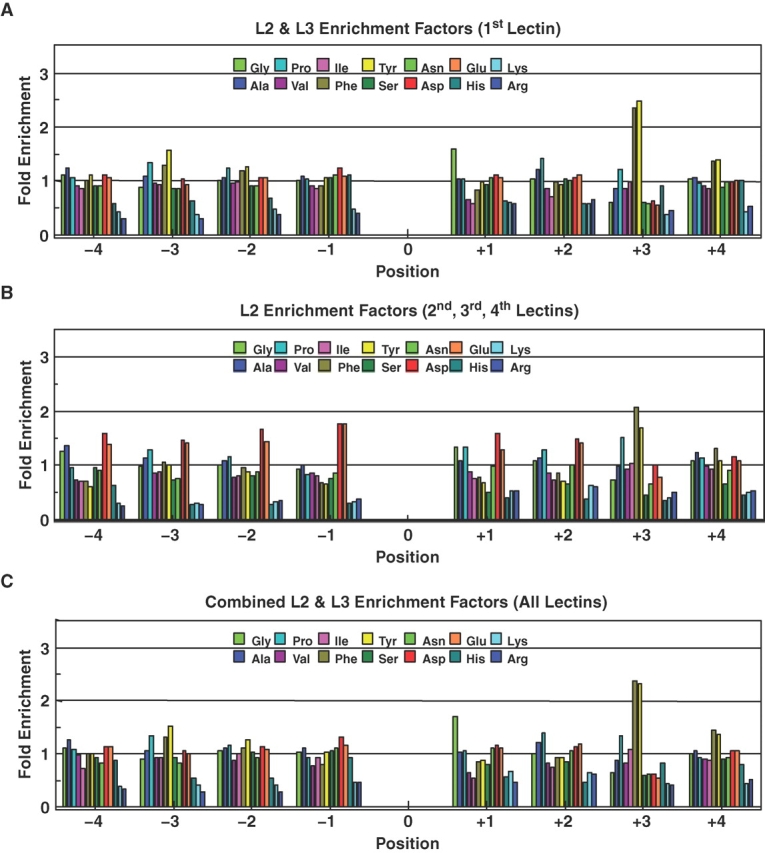
T-synthase amino acid residue enhancement factors for the pooled GP-II fractions in Figure 2. (A) Enhancement factors obtained for the combined L2a + L3a pool from Figure 2A. (B) Enhancement factors obtained for the L2b + L2c + L2d pool from Figures 2B–D. (C) Summed (3H weighted) enhancement factors from panels A and B, accounting for ∼93% of the T-synthase GP-II product.
As a final control for the nonspecific binding of non-Core 1 glycosylated GP-II to the PNA column, we applied the same amount of GP-II to the lectin column as in a standard reaction and collected the equivalent L1, L2, and L3 fractions (data not shown). Subsequent Edman sequencing of the L2 and L3 fractions revealed the absence of detectable GP-II; therefore, the observed preferences shown in Figures 1 and 3 are not artifacts of unmodified GP-II substrate binding to the PNA column. On this basis, we take the preferences given in 3C to nearly fully represent the peptide residue preferences for the human T-synthase.
Discussion
Using an oriented random glycopeptide substrate acceptor, GP-II, we have performed the most comprehensive analysis to date of the peptide substrate preferences of the T-synthase. These studies were unexpectedly more difficult than our previous random substrate studies on the ppGalNAc transferases due to the partial binding of the Core 1 product to the PNA and other lectin columns. Nevertheless, after repeated fractionation on a PNA column, over 90% of the Core 1 random glycopeptide product could be isolated for analysis (Figure 2). We concluded that much of the unbound radiolabel represented UDP-3H-Gal and nonbinding 3H-labeled Core 1 product. In Figure 3C, we have plotted the finally obtained enhancement values, ±4 residues of the acceptor site, GalNAc-O-Thr, for the 14-amino-acid residues present in GP-II. The plot clearly shows that the T-synthase is uniquely influenced by the nature of the acceptor substrate's flanking peptide residues.
The most striking feature of the enhancements plotted in Figure 3C is the very pronounced (∼2.5-fold) enhancement for both Phe and Tyr at the +3 position, and the smaller, but reproducible (∼1.5-fold), enhancements for these residues at the −3 and +4 positions (Figures 1E, F and 3A and C). In addition, a large, nearly 2-fold, enhancement for Gly at the +1 position is found, while much smaller (∼1.3-fold) enhancements for Pro at the −3, +2, and +3 positions are also clearly apparent. These latter more subtle enhancements (and those discussed below) are reproducibly seen across multiple experiments (e.g., compare Figures 1E and 3A), and therefore may provide additional important insight into the specificity of the transferase. In general, the hydrophobic residues (Ala, Pro, Val, Ile, Phe, and Tyr) are well tolerated at all positions (i.e., values ∼1), except at the +1 position where Val and Ile appear to be moderately disfavored (∼0.6) relative to Phe and Tyr. Another striking feature is the relatively uniform neutral to slightly positive (∼1.0–1.3) enhancements for the acidic Asp and Glu residues at all (except +3) positions compared to the uniformly disfavored (∼0.3–0.8) enhancement values found at all positions for the basic residues His, Lys, and Arg. It is noteworthy that the acidic residues (Asp and Glu) display essentially identical enhancement values at each position across the entire substrate sequence, and that nearly the same is observed for the basic residues, particularly for the highly basic Lys and Arg residues. (Note this selection of acidic charge residues over basic residues cannot be an artifact of the dowex ion exchange column as it would have enriched the basic residues over the acidic residues).
These common charged residue enhancement patterns strongly suggest that a large surface of the T-synthase, in the vicinity of the acceptor binding/catalytic site, is highly basic and that electrostatic interactions play a significant role in modulating the transferase's peptide acceptor preferences. Finally, consistent with the above, the uncharged hydrophylic residues, Ser and Asn, show essentially identical enhancement patterns which are in the neutral range (∼1), except at the +3 position where they are less favored than Tyr, Phe, and Pro. Taken together these observations suggest that the T-synthase's acceptor binding site does not discriminate between hydrophylic residues, except by their charge. While the transferase possesses specific sites for the interactions of the hydrophobic residues Phe and Tyr and perhaps Pro, as previously suggested by Granovsky et al. (1994), the observed +1 Gly preference may reflect the need for conformational flexibility or reduced steric interactions rather than a specific interaction of Gly on the transferase. Unfortunately, not until a three-dimensional structure of the T-synthase is reported can we further speculate on the nature of the interactions of the transferase with its substrate peptide acceptor.
Several years ago Brockhausen et al. (1990) and Granovsky et al. (1994) reported the characterization of the effects of the peptide sequence and glycosylation on the activity of partially purified preparations of the porcine and rat T-synthase. These workers using a series of short (3–8 residue) glycopeptides, based on the human MUC2 mucin tandem repeat (PTT(T-O-GalNAc)PIST), obtained kinetic data characterizing the effects of amino acid substitution of the residues directly N- and C-terminal of the acceptor Thr-O-GalNAc. They observed that Glu, Pro, and Gly gave the highest Vmax/Km values at the −1 site while Lys, Arg, and Thr gave the lowest values. At the +1 site, substitutions of Gly, Val, and Met gave the largest Vmax/Km values while Cys, Pro, Phe, and Leu gave the lowest values. Furthermore, these workers observed that Glu and Asp both gave low Km values while unblocked (i.e., charged) N- and C-terminal residues seemed to be inhibitory in the shorter glycopeptides. Our observations with the GP-II random glycopeptide substrate are in general, but not complete, agreement with these earlier findings. We indeed observe the Glu and Gly enhancements at the −1 and +1 sites, respectively, and the inhibition of Lys and Arg, but we do not observe significant enhancements for Pro/Gly or Val at the −1 or +1 sites, respectively (Figure 3). Inhibition by Pro and Phe at the +1 site is also not observed by us. It is likely that these earlier studies may have been biased due to the peptide length or because of the fixed nature of residues flanking MUC2 sequence. Furthermore, since our data are obtained from a single random glycopeptide substrate, the comparisons of the reactivity of different amino acid residues are expected to be relatively free of methodological and experimental errors, as compared to the earlier work utilizing a large series of individual glycopeptide substrates. Nevertheless, our present studies with the GP-II random glycopeptide substrate significantly expand on this earlier work by obtaining the peptide residue preferences of the human T-synthase over the range of four residues N- and C-terminal of the acceptor site.
Since Core 1 glycosylation requires the prior glycosylation at a given Ser or Thr by a ppGalNAc-T, one might expect that the preferences we obtained for T-synthase would parallel those of one or more of the ppGalNAc-Ts. However, this does not presently appear to be the case for at least those ppGalNAc-Ts which have had their preferences characterized to date (i.e., ppGalNAc-T1 and -T2 (Gerken et al. 2006, 2008) and ppGalNAc-T5, -T10, and -T12 (unpublished)). Most common to these ppGalNAc-Ts (except for T10) is a high (3- to 6-fold) enhancement for Pro at the +3 position, with variable Pro enhancements observed at the −3, −1, and +1 sites depending on the transferase isoform. In contrast, T-synthase shows only a slight Pro enhancement at +3 and neutral-to-weak Pro enhancements at the other sites. Interestingly, ppGalNAc-T1 does have an elevated Tyr (but not Phe) enhancement at +3 position while ppGalNAc-T12 displays weak Tyr enhancements at the −2 and −3 positions (unpublished). We presently do not know whether T-synthase utilizes its unusually high Tyr/Phe +3 preferences for specifically glycosylating any in vivo protein targets, but as Tyr and Phe are comparatively rare in mucin domain sequences, the motif is obviously not required for mucin Core 1 glycosylation. However, it is possible that one or more proteins lacking classical mucin domain residues or characteristics may utilize the +3 Tyr/Phe motif for efficient Core 1 glycosylation.
We have also shown that T-synthase may utilize charge–charge interactions to modulate its activity, rejecting basic peptides over neutral and acidic peptide acceptors. These electrostatic interactions may serve to help drive Core 1 glycosylation to completion at one site or to inhibit or minimize Core 1 glycosylation at another–solely based on the peptide substrate net charge. Two examples where we can speculate that this may indeed be the case can be found in the cell surface mucin, P-selectin glycoprotein ligand-1 (PSGL1) (Liu et al. 1998), and in the phosphaturic factor fibroblast growth factor 23 (FGF23) (Kato et al. 2006). The specific O-glycosylation of Thr 57 of human PSGL1 by a Core 2 sLex structure is an integral component of PSGL1's binding to P-selectin (Liu et al. 1998). One would expect the peptide sequence at this site to be optimized for highly efficient glycosylation and elongation relative to the remaining ∼70 or so Ser and Thr residues of the PSGL1 mucin domain. Since the formation of the Core 1 structure is the second step in the biosynthesis of the Core 2 sLex structure, a rapid and efficient Core 1 glycosylation of Thr 57 would drive the biosynthesis of the complete sLex structure at Thr 57 relative to other potential glycosylation sites. Indeed, the peptide sequence surrounding Thr 57 in h-PSGL1 (42QATEYEYLDYDFLPET*57EPPEML63) is highly acidic consistent with this site being optimized for Core 1 glycosylation by T-synthase. Furthermore, the acidic charge density at Thr 57 is the highest among all of the potential glycosylation sites in the PSGL1 mucin domain.
In FGF23, the specific glycosylation of Thr 178 by ppGalNAc-T3 and its further sialylation are necessary to prevent furin protease cleavage at Arg 179 (Kato et al. 2006). The in vitro Core 1 glycosylation of Thr 178 fails to inhibit furin cleavage at Arg 179 (Kato et al. 2006). Therefore, one could expect this regulatory site to have evolved for efficient sialylation and poor Core 1 glycosylation. The peptide sequence around Thr 178 (171TPIPRRHT*178RSAEDDS*185ERDP189) supports this conclusion as Thr 178 is surrounded by four basic residues, which would be expected to significantly reduce Core 1 glycosylation at this site by T-synthase. Interestingly, the residues surrounding Ser 185, C-terminal of the cleavage site, are highly acidic, suggesting that this site may be targeted for efficient Core 1 glycosylation. Thus, we propose for at least T-synthase and perhaps the sialyl transferases, ST6 GalNAc I and II, that their activities may be differently modulated by the nature of the charge of the acceptor peptide core. Confirmation of these predictions with native and charge mutant PSGL1 and FGF23 glycopeptides with T-synthase and the ST6 GalNAc sialyl transferases will be the focus of future studies.
In summary, in addition to being modulated by neighboring glycosylation, our results clearly indicate that T-synthase recognizes specific features of the peptide portion of the substrate, including overall charge and the presence of Gly, Phe, Tyr, and perhaps Pro at specific positions, up to four residues from the site of glycosylation, and indeed may use these features to modulate sites of Core 1 glycosylation.
Materials and methods
Materials and reagents
Human recombinant T-synthase was HPC4-epitope tagged and bound to HPC4-AffiGel-10 beads, ∼2.4 units/mL suspended resin (1 unit = 1 μmole/h transferred to benzyl-O-GalNAc) (Ju et al. 2002). Random glycopeptide substrate, GP-II, GAGAXXXX(Thr-O-GalANAc)XXXXAGAG, where X = G, A, P, V, I, F, Y, S, N, D, E, H, R, and K was custom synthesized by Bachem Bioscience, Inc. (King of Prussa, PA), pH adjusted to approximately 7.5 with dilute NaOH and/or HCl and lyophilized from water before making a pH 7.5 stock solution. Its sequence and composition was confirmed by Edman sequencing. UDP-3H-Gal was purchased from ARC Inc. (St. Louis, MO). PNA (Arachis hypogaea), AIA (Jacalin, Artocarpus integrifolia), ABA (Agaricus bisporus), MPA (Maclura pomifera), TKA (Trichosanthes kirilowii), ACA (Amaranthus caudatus), and APA (Abrus precatorious) lectins were obtained from EY Laboratories (San Mateo, CA). Sephadex G10 and Dowex (1 × 8) ion exchange resin (100–200 mesh) were supplied from Pharmacia (Peapack, NJ) and Acros Organics (Morris Plains, NJ), respectively. Liquid scintillation counting was performed on a Beckman Model LS5801 (Fullerton, CA).
Glycosylation of random glycopeptide by human T-synthase
Random glycopeptide (GP-II) incubations consisted of 100 mM MES (pH 7.2), 20 mM MnCl2, 0.1 mM CaCl2, a 1/100 dilution protease inhibitor cocktails P8340 and P8849 (Sigma-Aldrich, St. Louis), 0.2 mM UDP-galactose ([3H]-labeled, 0.32 μCi/μL), 15–50 μL T-synthase (∼0.04–0.12 units), and 5 mM GP-II. Reactions were incubated ∼1–5 h at 37°C in a TAITEC shaking MicroIncubator M-36 (San Jose, CA) at a speed sufficient to maintain a bead suspension. Reactions were stopped by the addition of a 2× volume of 250 mM EDTA. UDP and nonhydrolyzed UDP-Gal were removed by two passages over an ∼3 mL Dowex (1 × 8) anion-exchange columns (see Results). 3H-Gal incorporation was determined via scintillation counting 1/100 of the sample before and after the dowex columns. Core 1 glycosylation of GP-II ranged from 0.8% to 2% based on 3H-Gal radiolabel incorporation. The eluant was lyophilized and chromatographed on a Sephadex G10 column (0.7 × 113 cm) in a 50 mM acetic acid buffer, pH 4.5, with NH4OH. The fractions were monitored for absorbance at 220 and 280 nm and incorporation of radioactivity by scintillation counting. Core 1 glycosylated GP-II fractions were pooled based on 3H radioactivity and lyophilized.
Isolation of Core 1 glycosylated GP-II glycopeptide product of T-synthase
The Core 1 GP-II product was bound to a PNA (Arachis hypogaea) lectin column (10 mL, 22 × 0.8 cm) equilibrated at 4°C in a buffer (15 mM NaCl, 2.5 mM Tris, pH 7.3, and 0.002% NaN3) with protease inhibitors added to the loaded sample (10 μL each P8340 and P8849). Prior loading, 1/100 of the sample was removed for Edman amino acid sequencing for use as pre-lectin control. The column was washed with 30 mL of buffer at a flow rate of ∼8 mL/h, and 5 mL of 5 mM Gal in the buffer was used to release bound Core 1 glycopeptide. Pass through, retarded, and bound fractions (L1, L2, and L3, respectively) were monitored and pooled based on 3H radioactivity and lyophilized. The 3H-labeled Core 1 product was further separated from free Gal and buffers on Sephadex G10 as described above. As described in the Results, multiple lectin columns were performed to fully separate bound from unbound product.
Lectin column trials for unbound L1 fraction from PNA lectin columns
Samples from the L1 fraction of the PNA lectin column were passed over each of the following 1 mL lectin columns, i.e., AIA, ABA, MPA, TKA, ACA, and APA, which were equilibrated as described above for the PNA lectin column. The unbound protein was eluted with the 3 × 1 mL buffer (15 mM NaCl, 2.5 mM Tris, pH 7.3, and 0.002% NaN3) and 1 mL of 5 mM Gal in the buffer was added to each column to release any bound Core 1 3H-Gal glycosylated product. Each fraction was collected separately and 3H radioactivity was monitored by scintillation counting.
β-Galactosidase treatments of lectin column fractions L1, L2, and L3
In one experiment, fractions from a lectin column chromatography run were incubated with β-galactosidase to confirm Core 1 product formation. Incubations consisted of the 50 μL sodium citrate buffer (100 mM, pH 4.5), 1/100 dilution of protease inhibitor cocktails P8340 and P8849, and 50 μg (0.01 units) bovine testes β-galactosidase (Sigma). Reactions were incubated overnight at 33°C and chromatographed on a Sephadex G10 column as described above.
Edman amino acid sequencing
Pulsed liquid phase Edman amino acid sequencing was performed on an Applied Biosystems Procise 494 protein sequencer (Applied Biosystems, Foster City, CA) at a C18 PTH column temperature of 55°C. Data analysis was performed as described earlier (Gerken et al. 2004, 2006). Enrichment factors (preferences) were obtained by dividing the mole fractions of the lectin-isolated product by the mole fractions obtained from sequencing a control pre-lectin aliquot. Values greater than 1 represent enhanced preferences while values less than 1 represent reduced preferences.
Supplementary Data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Funding
The National Institutes of Health Grants R01-GM068559 (to R.D.C.) and R01-CA78834 (to T.A.G.).
Acknowledgments
We wish to thank Oliver Jamison for his technical support and for his reading the manuscript and offering helpful suggestions. Undergraduate student assistance of Brian Sauer, Mansi Patel, and Andrew Holpuch are also acknowledged.
Conflict of interest statement
None declared.
Abbreviations
- Core 1 structure
β-Gal (1-3) α-GalNAc-O-Ser/Thr
- FGF23
phosphaturic factor fibroblast growth factor 23
- GP-II
random glycopeptide substrate GAGAXXXX(Thr-O-GalNAc)XXXXAGAG, where X = G, A, P, V, I, F, Y, S, N, D, E, H, R, and K
- PNA
lectin from Arachis hypogaea; ppGalNAc-T, UDP-α-GalNAc:polypeptide α-N-acetylgalactosaminyltransferase
- PSGL1
P-selectin glycoprotein ligand
- PSM
porcine salivary gland mucin
- T-synthase
UDP-Gal:glycoprotein-α-GalNAc β3 galactosyltransferase
References
- Berger EG. Tn-syndrome. Biochimica et Biophysica Acta. 1999;1455:255–268. doi: 10.1016/s0925-4439(99)00069-1. [DOI] [PubMed] [Google Scholar]
- Brockhausen I. Biosynthesis and functions of O-glycans and regulation of mucin antigen expression in cancer. Biochem Soc Trans. 1997;25:871–874. doi: 10.1042/bst0250871. [DOI] [PubMed] [Google Scholar]
- Brockhausen I. Pathways of O-glycan biosynthesis in cancer cells. Biochim Biophys Acta. 1999;1473:67–95. doi: 10.1016/s0304-4165(99)00170-1. [DOI] [PubMed] [Google Scholar]
- Brockhausen I, Moller G, Merz G, Adermann K, Paulsen H. Control of mucin synthesis: The peptide portion of synthetic O-glycopeptide substrates influences the activity of O-glycan core 1 UDP-galactose:N-acetyl-α-galactosaminyl-R β 3-galactosyltransferase. Biochemistry. 1990;29:10206–10212. doi: 10.1021/bi00496a008. [DOI] [PubMed] [Google Scholar]
- Freire T, Bay S, Mensdorff-Pouilly S, Osinaga E. Molecular basis of incomplete O-glycan synthesis in MCF-7 breast cancer cells: Putative role of MUC6 in Tn antigen expression. Cancer Res. 2005;65:7880–7887. doi: 10.1158/0008-5472.CAN-04-3746. [DOI] [PubMed] [Google Scholar]
- Gerken TA. Kinetic modeling confirms the biosynthesis of mucin Core 1 (β-Gal(1-3) α-GalNAc-O-Ser/Thr) O-glycan structures are modulated by neighboring glycosylation effects. Biochemistry. 2004;43:4137–4142. doi: 10.1021/bi036306a. [DOI] [PubMed] [Google Scholar]
- Gerken TA, Gilmore M, Zhang J. Determination of the site-specific oligosaccharide distribution of the O-glycans attached to the porcine submaxillary mucin tandem repeat: Further evidence for the modulation of O-glycan side chain structures by peptide sequence. J Biol Chem. 2002;277:7736–7751. doi: 10.1074/jbc.M111690200. [DOI] [PubMed] [Google Scholar]
- Gerken TA, Ten Hagen KG, Jamison O. Conservation of peptide acceptor preferences between Drosophila and mammalian polypeptide-GalNAc transferase orthologue pairs. Glycobiology. 2008;18:861–870. doi: 10.1093/glycob/cwn073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken TA, Raman J, Fritz TA, Jamison O. Identification of common and unique peptide substrate preferences for the UDP-GalNAc:polypeptide α-N-acetylgalactosaminyltransferases T1 & T2 (ppGalNAc T1 & T2) derived from oriented random peptide substrates. J Biol Chem. 2006;281:32403–32416. doi: 10.1074/jbc.M605149200. [DOI] [PubMed] [Google Scholar]
- Gerken TA, Tep C, Rarick J. Role of peptide sequence and neighboring residue glycosylation on the substrate specificity of the uridine 5′-diphosphate-α-N-acetylgalactosamine:polypeptide N-acetylgalactosaminyl transferases T1 and T2: Kinetic modeling of the porcine and canine submaxillary gland mucin tandem repeats. Biochemistry. 2004;43:9888–9900. doi: 10.1021/bi049178e. [DOI] [PubMed] [Google Scholar]
- Granovsky M, Bielfeldt T, Peters S, Paulsen H, Meldal M, Brockhausen J, Brockhausen I. UDPgalactose:glycoprotein-N-acetyl-d-galacto- samine 3-β-d-galactosyltransferase activity synthesizing O-glycan core 1 is controlled by the amino acid sequence and glycosylation of glycopeptide substrates. Eur J Biochem. 1994;221:1039–1046. doi: 10.1111/j.1432-1033.1994.tb18822.x. [DOI] [PubMed] [Google Scholar]
- Ju T, Aryal RP, Stowell CJ, Cummings RD. Regulation of protein O-glycosylation by the endoplasmic reticulum-localized molecular chaperone Cosmc. J Cell Biol. 2008;182:531–542. doi: 10.1083/jcb.200711151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju T, Brewer K, D’Souza A, Cummings RD, Canfield WM. Cloning and expression of human Core 1 β 1,3-galactosyltransferase. J Biol Chem. 2002;277:178–186. doi: 10.1074/jbc.M109060200. [DOI] [PubMed] [Google Scholar]
- Ju T, Cummings RD. Protein glycosylation: Chaperone mutation in Tn syndrome. Nature. 2005;437:1252. doi: 10.1038/4371252a. [DOI] [PubMed] [Google Scholar]
- Kato K, Jeanneau C, Tarp MA, Benet-Pages A, Lorenz-Depiereux B, Bennett EP, Mandel U, Strom TM, Clausen H. Polypeptide GaINAc-transferase T3 and familial tumoral calcinosis: Secretion of FGF23 requires O-glycosylation. J Biol Chem. 2006;281:18370–18377. doi: 10.1074/jbc.M602469200. [DOI] [PubMed] [Google Scholar]
- Li GS, Zhang H, Lv JC, Shen Y, Wang HY. Variants of C1GALT1 gene are associated with the genetic susceptibility to IgA nephropathy. Kidney Int. 2007;71:448–453. doi: 10.1038/sj.ki.5002088. [DOI] [PubMed] [Google Scholar]
- Liu W, Ramachandran V, Kang J, Kishimoto TK, Cummings RD, McEver RP. Identification of N-terminal residues on P-selectin glycoprotein ligand-1 required for binding to P-selectin. J Biol Chem. 1998;273:7078–7087. doi: 10.1074/jbc.273.12.7078. [DOI] [PubMed] [Google Scholar]
- Lotan R, Skutelsky E, Danon D, Sharon N. The purification, composition, and specificity of the anti-T lectin from peanut (Arachis hypogaea) J Biol Chem. 1975;250:8518–8523. [PubMed] [Google Scholar]
- Pinho S, Marcos NT, Ferreira B, Carvalho AS, Oliveira MJ, Santos-Silva F, Harduin-Lepers A, Reis CA. Biological significance of cancer-associated sialyl-Tn antigen: Modulation of malignant phenotype in gastric carcinoma cells. Cancer Lett. 2007;249:157–170. doi: 10.1016/j.canlet.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Smith AC, de Wolff JF, Molyneux K, Feehally J, Barratt J. O-Glycosylation of serum IgD in IgA nephropathy. J Am Soc Nephrol. 2006;17:1192–1199. doi: 10.1681/ASN.2005101115. [DOI] [PubMed] [Google Scholar]
- Springer GF. Immunoreactive T and Tn epitopes in cancer diagnosis, prognosis, and immunotherapy. J Mol Med. 1997;75:594–602. doi: 10.1007/s001090050144. [DOI] [PubMed] [Google Scholar]
- Xia L, Ju T, Westmuckett A, An G, Ivanciu L, McDaniel JM, Lupu F, Cummings RD, McEver RP. Defective angiogenesis and fatal embryonic hemorrhage in mice lacking core 1-derived O-glycans. J Cell Biol. 2004;164:451–459. doi: 10.1083/jcb.200311112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


