Abstract
Age-related macular degeneration (AMD), a complex multigenic disorder and the most common cause of vision loss in the elderly, is associated with polymorphisms in the LOC387715/ARMS2 and HTRA1 genes on 10q26. Like humans, macaque monkeys possess a macula and develop age-related macular pathologies including drusen, the phenotypic hallmark of AMD. We genotyped a cohort of 137 unrelated rhesus macaques with and without macular drusen. As in humans, one variant within LOC387715/ARMS2 and one in HTRA1 were significantly associated with affected status. HTRA1 and the predicted LOC387715/ARMS2 gene were both transcribed in rhesus and human retina and retinal pigment epithelium. Among several primate species, orthologous exons for the human LOC387715/ARMS2 gene were present only in Old World monkeys and apes. In functional analyses, the disease-associated HTRA1 polymorphism resulted in a 2-fold increase in gene expression, supporting a role in pathogenesis. These results demonstrate that two genes associated with AMD in humans are also associated with macular disease in rhesus macaques and that one of these genes is specific to higher primates. This is the first evidence that humans and macaques share the same genetic susceptibility factors for a common complex disease.
INTRODUCTION
Age-related macular degeneration (AMD) has an incidence of almost 25% in those over 75 years of age (1). The hallmark of the condition is the accumulation of drusen, extracellular lipid and protein deposits under the central retina (macula). In the late stages of the disease, central blindness occurs due to invasion of new blood vessels from the choroid resulting in hemorrhage and gliosis (neovascularization or ‘wet AMD’) or degeneration of the retinal pigment epithelium (RPE) and photoreceptor cells (geographic atrophy or ‘dry AMD’) (2).
AMD is a complex disease with both genetic and environmental susceptibility factors, including several recently identified genetic variants. In particular, increased risk of the development and progression of AMD is associated with single nucleotide polymorphisms (cSNPs) in the complement factor H gene on chromosome 1q and a locus on chromosome 10q comprising LOC387715/ARMS2 (3–5), a transcribed sequence with as yet unknown function (6), and the adjacent promoter region of HTRA1, the gene for a serine protease (7–9). The two SNPs on 10q have been shown to confer similar increased AMD risk and to be in complete linkage disequilibrium in the Caucasian, Chinese and Japanese populations (7,9).
The rhesus macaque eye closely resembles the human eye in almost all respects. Most importantly, the macaque retina has a macula, a specialization shared only by monkeys, apes and man. The macula in both macaques and humans includes a foveal pit with several features underlying high spatial acuity, including the highest concentration of cone photoreceptors, displacement of inner retinal cell layers and vessels and a capillary-free zone; it is also marked by the presence of macular pigment. Furthermore, age-related drusen and pigmentary changes closely mimicking early to intermediate human AMD have been identified in a number of macaque colonies (10–13), including a population we phenotypically characterized at the Oregon National Primate Research Center.
In light of the identification of human genes conferring increased AMD risk, we hypothesized that the orthologous macaque genes would be similarly involved with age-related maculopathy and drusen formation in these animals. Furthermore, we hypothesized that the size of the genetic effect would be similar to that observed in humans, making it statistically feasible to detect an association in our sample. Currently, the regulators of complement activation (RCA) locus, a 388 kb interval which encompasses complement factor H and several related genes, is not well resolved in the rhesus genome. For this reason, we chose to focus here on potential genetic associations in the 10q region.
RESULTS
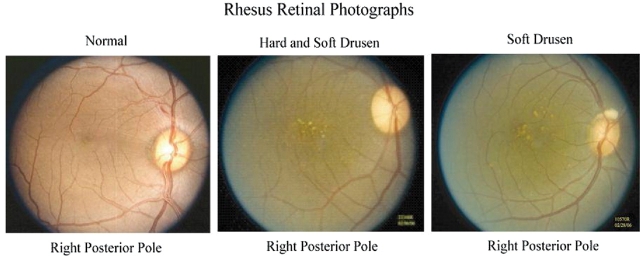
Representative examples of the spectrum of age-related maculopathy in the rhesus macaques are shown in Figure 1. Just as in human AMD, drusen accumulate over time, increase in size, become more indistinct or ‘soft’ and are accompanied by macular pigmentary changes. Two populations of rhesus monkeys, those originating from India and China, exhibited somewhat distinct phenotypes, with most rhesus of Chinese origin having the clinical and fluorescein angiographic characteristics of human ‘basal laminar drusen’.
Figure 1.
Age-related maculopathy in rhesus primates. Left image shows color photograph of retina with normal macula. Middle and right images show age-related macular drusen (yellow–white subretinal deposits).
Association analyses
Sequencing confirmed that in the macaque, as in humans, LOC387715/ARMS2 and the HTRA1 promoter region lie adjacent to each other. In rhesus monkeys of Indian origin (68 affected and 48 unaffected, Table 1), sequence analysis of this region identified eight previously undescribed polymorphisms, two in LOC387715/ARMS2 and six in the HTRA1 promoter region including a common deletion (Fig. 2). Adjusted unconditional logistic regression showed that homozygosity ‘CC’ for the 5′ rhesus LOC387715/ARMS2 SNP (low risk allele ‘T’) and homozygosity ‘AA’ for the fourth SNP (wild-type allele ‘C’) in the promoter region of HTRA1 were associated with the presence of drusen in the monkey population, P = 0.002 and P = 0.02, respectively (Table 2). Heterozygosity conferred no significant increased risk of age-related maculopathy. There were no significant deviations from Hardy–Weinberg equilibrium. The two maculopathy-associated SNPs were not in complete linkage disequilibrium (D′ 0.88). Figure 3 shows the alignment of the human and rhesus LOC387715/ARMS2 and HTRA1 sequences with the relative positions of the age-related maculopathy SNPs. In rhesus monkeys of Chinese origin, five of the same SNPs were found, including the drusen-associated SNP in LOC387715/ARMS2. The HTRA1 SNP and the deletion were not observed in these animals, but their numbers were not sufficient to permit meaningful estimations of association within this subpopulation alone.
Table 1.
Rhesus macaques included in this study
| Rhesus macaque origin | Age-related maculopathy | Unaffected controls | Total |
|---|---|---|---|
| Indian | 68 | 48 | 116 |
| Chinese | 13 | 8 | 21 |
| Total | 81 | 56 | 137 |
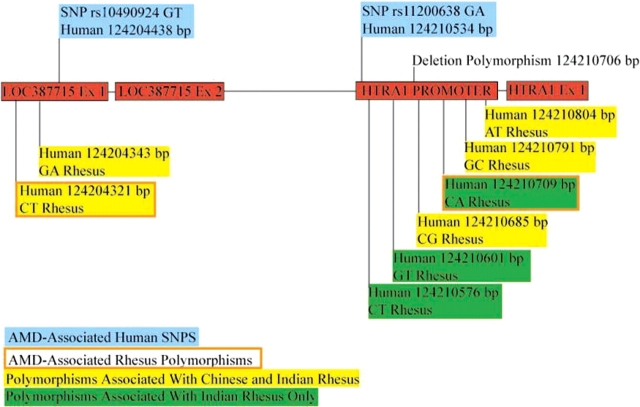
Figure 2.
A schematic of the LOC387715/ARMS2 locus and the HTRA1 promoter region, situated 6102 bp apart in humans. The positions of the AMD-associated human SNPs within exon 1 of LOC387715/ARMS2 (rs1049024) and within the HTRA1 promoter (rs11200638) are indicated with blue boxes. The position of the new SNPs identified in all rhesus (Chinese- and Indian origin), and in the Indian-origin population only, is indicated with yellow and green highlighted boxes, respectively. The two new SNPs that are associated with drusen development in rhesus are indicated with a highlighted box outlined with a brown line.
Table 2.
Unconditional logistic regression results for the novel SNPs identified in this study
| Rhesus gene and SNP position | Genotypes | Odds ratio | P -value |
|---|---|---|---|
| LOC387715 10682 | TT | 1.00 | |
| CT | 1.66 (0.49–6.10) | 0.55 | |
| CC | 8.76 (1.96–47.38) | 0.002 | |
| LOC387715 10704 | AA | 1.00 | |
| AG | 0.40 (0.05–1.91) | 0.29 | |
| GG | 1.10 (0.11–7.99) | 1.00 | |
| HTRA1 16943 | CC | 1.00 | |
| CT | 1.01 (0.44–2.32) | 1.00 | |
| TT | 2.17 (0.45–17.23) | 0.46 | |
| HTRA1 16968 | GG | 1.00 | |
| GT | 1.35 (0.60–3.09) | 0.542 | |
| TT | 0.83 (0.08–8.30) | 1.00 | |
| HTRA1 17052 | CC | 1.00 | |
| CG | 1.07 (0.40–2.81) | 1.00 | |
| GG | 0.67 (0.22–1.99) | 0.59 | |
| HTRA1 17076 | CC | 1.00 | |
| AC | 0.94 (0.42–2.11) | 1.00 | |
| AA | ∞ | 0.02 | |
| HTRA1 17158 | CC | 1.00 | |
| CG | 1.05 (0.45–2.47) | 1.00 | |
| GG | 0.95 (0.33–2.78) | 1.00 | |
| HTRA1 17171 | AA | 1.00 | |
| AT | 1.01 (0.17–5.27) | 1.00 | |
| TT | 1.56 (0.25–8.54) | 0.68 |
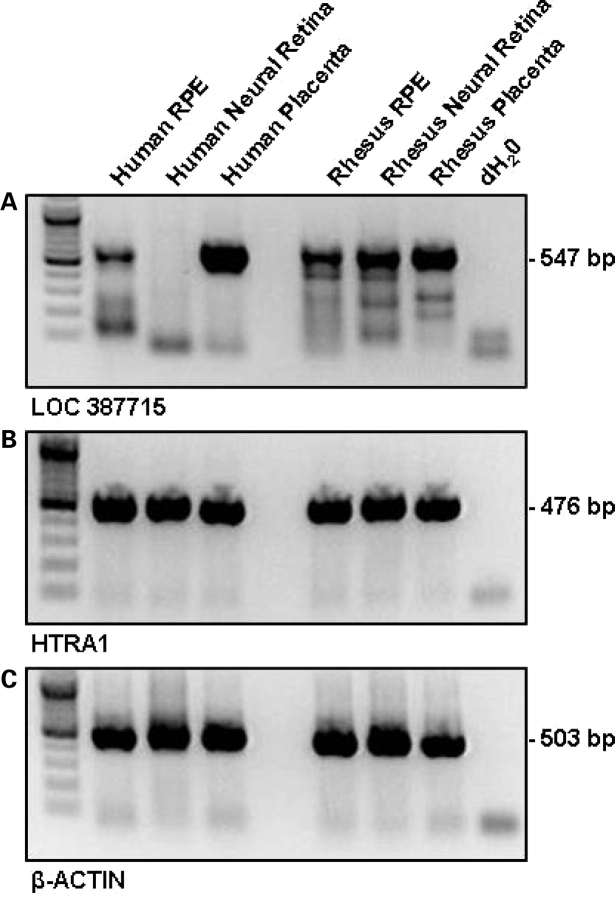
Figure 3.
LOC387715/ARMS2 and HTRA1 transcripts are present within human and rhesus tissue. RT–PCR was performed using intron-spanning primers designed to consensus human and rhesus (A) LOC387715/ARMS2 and (B) HTRA1 exonic sequence; (C) β-actin was used as a positive control.
The two specific SNPs (rs10490924 and rs11200638) associated with human AMD were not polymorphic in Indian and Chinese rhesus macaques; the low-risk sequences were conserved in each case. As in humans, it is unknown whether the SNPs associated with maculopathy are themselves pathological or are in linkage disequilibrium with other functional variants.
Evolution of the coding region of the primate LOC387715/ARMS2 gene
Consensus primers were designed to human and chimp gDNA and tested for amplification in a variety of apes (Pan paniscus, Pan troglodytes, Gorilla gorilla, Pongo pygmaeus), Old World monkeys (Macaca mulatta, Macaca nemestrina), New World monkeys (Lagothrix lagotricha, Ateles geoffroyi, Saguinus labiatus) and a prosimian (Lemur catta). Primers designed to exon 1 of LOC387715/ARMS2 amplified products of the expected size from all apes and monkeys, but no product was obtained from the prosimian. Comparison of all primate exon 1 products indicated 77.2% sequence identity to each other. Pan paniscus, P. troglodytes, G. gorilla and the New World monkeys have the same predicted ATG start site as human. Pongo pygmaeus has a CTG codon, whereas both macaques possess a GTG codon at the equivalent site. Primers designed to exon 2 of LOC387715/ARMS2 amplified products of the expected size from apes and Old World monkeys only, and the predicted TGA stop codon at codon 108 within exon 2 is only present in humans and apes. The human and P. paniscus LOC387715/ARMS2 sequences are predicted to code for 107 and 106 amino acid proteins, respectively. The Gorilla sequence has a premature TGA stop at codon 38. The macaques have a CGA at codon 108 and an alternative in-frame TAA stop codon 24 bp downstream (Table 3). At the human AMD susceptibility SNP locus, the wild-type codon is conserved in both Old and New World lineages.
Table 3.
Summary of sequencing results of LOC387715/ARMS2 exons 1 and 2 and the HTRA1 promoter region from human and 10 non-human primate species, showing sequences necessary for initiation of protein translation, termination and optimal exon–intron splicing and the nucleotides occupying the human-AMD SNP positions
| Primate | ATG start codon? | Codon 69 | Exon 1 donor-site? | Exon 2 acceptor-site? | TGA stop codon? | HTRA1 promoter rs11200638 | Other comments |
|---|---|---|---|---|---|---|---|
| Human: Homo sapiens | Yes:ATG | GCT/TCT Ala/Ser | Yes:GT | Yes:AG | Yes:TGA | GG/AA | |
| Chimpanzee: P. troglodyte | Yes:ATG | GCT | Yes:GT | Yes:AG | Yes:TGA | GG | |
| Bonobo: P. paniscus | Yes:ATG | GCT | Yes:GT | Yes:AG | Yes:TGA | GG | |
| Gorilla: G. gorilla | Yes:ATG | GCT50 | Yes:GT | Yes:AG | Yes:TGA | GG | Codon 38: TGA (stop) |
| Sumatran orangutan: P. pygmaeus | NO:CTG | GCT | Yes:GT | Yes:AG | Yes:TGA | GG | |
| Rhesus monkey: M. mulatta | NO:GTG | GCT | Yes:GT | NO:AA | NO:CGA | GG | Alternative TAA stop? Missing 1 bp at codon 37 |
| Pigtailed macaque: M. nemestrina | NO:GTG | GCT | Yes:GT | N/A | NO:CGA | GG | Alternative TAA stop? Missing 1 bp at codon 37 |
| Common woolly monkey: L. lagotricha | Yes:ATG | GCT | Yes:GT | N/A | N/A | CC | |
| Black-handed spider monkey: A. geoffroyi | Yes:ATG | GCT | Yes:GT | N/A | N/A | CC | |
| Red-chested moustached tamarin: S. Labiatus | Yes:ATG | GCT | Yes:GT | N/A | N/A | CC | |
| Ring-tailed lemur: L. catta | N/A | N/A | N/A | N/A | N/A | N/A |
Characterization of the LOC387715/ARMS2 transcript in rhesus monkeys
Reverse transcriptase (RT)–PCR using exon-spanning primers yielded products of the predicted size for a transcribed and spliced gene from rhesus RPE and placenta (Fig. 3A). However, in repeated experiments, the product from neural retina could not be amplified consistently. This contrasts with our consistently negative findings for human neural retina (Fig. 3). Sequence analysis confirmed the presence of the exon 1 donor splice position identical to human. However, the rhesus monkey utilizes an alternative exon 2 acceptor splice site situated 18 bp further downstream, equivalent to codon 106 within the human sequence. Thus, the first two codons of exon 2 in the rhesus monkey (codons 100 and 101) align to codons 106 and 107 in the human sequence. The rhesus monkey also uses an alternative termination codon situated 21 bp downstream relative to the human stop codon. The human and rhesus predicted coding region of exon 1 share 90% identity (Fig. 4A). Exon 1 has an in-frame deletion resulting in loss of codon 37. This corresponds to a 330 bp open-reading frame (ORF), from the predicted GTG start codon to the TAA termination codon. The 32 bp ORF of rhesus exon 2 aligns with 93% identity, corresponding to codons 106–108 and 24 bp of the 3′-UTR of human LOC387715/ARMS2. The human and rhesus predicted coding region of exon 1 share 90% identity (Fig. 4A).
Figure 4.

An alignment of human and rhesus (A) exon 1 LOC387715/ARMS2 sequence and (B) HTRA1 promoter. The position of maculopathy-associated SNPs in rhesus monkeys (↓) compared with humans (⇓) is shown.
Rhesus HTRA1 tissue expression and pSEAP reporter assay of promoter activity
Using primers that spanned an intron, designed to the consensus human and rhesus HTRA1 exonic sequence, RT–PCR confirmed the presence of HTRA1 expression within rhesus neural, RPE/choroid and placental tissue (Fig. 3B). An alignment of the relevant human and rhesus promoter region is shown in Figure 4B.
The low- and high-risk alleles of the associated HTRA1 promoter SNP were then assayed using the pSEAP:luciferase reporter method to evaluate the functional effects on gene expression in 293T human microvascular endothelial cells. Each experiment was repeated four times. Clones with the ‘A’ high-risk allele consistently showed double the expression level of pSEAP when compared with wild-type ‘C’ clones (P = 0.001).
DISCUSSION
Over several years, we have prospectively ascertained and serially examined the eyes of a large cohort of aged rhesus monkeys. We have observed that a large proportion over the age of 18 years (equivalent to age 54 in humans) develop drusen and retinal pigmentary changes clinically identical to those seen in intermediate AMD in humans. Drusen typically start as small ‘hard’ yellow subretinal deposits that become more numerous and increase in size over time. The borders of larger drusen become indistinct or ‘soft’ in appearance. Interestingly, differences were found between rhesus originating from India and China, two populations that exhibit distinctive morphological, physiological and genetic characteristics (14).
In humans, increased risk of the development and progression of AMD is associated with cSNPs in the complement factor H gene on chromosome 1q and a locus on chromosome 10q26 in which reside LOC387715/ARMS2 (3,4,6) and HTRA1 (7–9). We hypothesized that the same genes may be involved in determining age-related maculopathy in our rhesus cohort. Currently, the RCA locus, a 388 kb interval which encompasses the complement factor H gene, is not well resolved in the rhesus genome. For this reason, we chose to evaluate the potential genetic associations in the 10q region.
Sequencing confirmed that in the macaque, as in humans, the LOC387715/ARMS2 and HTRA1 genes lie adjacent to each other. Direct sequencing revealed several novel SNPs, two of which were associated with the presence of age-related maculopathy in the rhesus macaques of Indian origin: one in the coding region of LOC387715/ARMS2 and one in the promoter region of HTRA1. These associated SNPs were not in complete linkage disequilibrium (D′ 0.88), unlike the situation in humans. The relatively small numbers of animals available for analysis in this study precluded further analysis that might identify which of the two SNPs conferred the risk of AMD. Furthermore, the two SNPs (rs10490924 and rs11200638) associated with human AMD were not polymorphic in rhesus macaques; the low-risk sequences were conserved in each case.
In humans, it is as yet unknown whether the SNPs associated with maculopathy are themselves pathological or are in linkage disequilibrium with other functional variants. To begin to address this question of allelic causality in the rhesus, we first examined the evolution of the two-exon LOC387715/ARMS2 gene, which shows no homology to other non-primate mammalian genomic sequences and is reportedly not present in rodents (3). Our data show that the LOC387715/ARMS2 sequence appeared in the genome after the split of prosimians and simians 60–65 million years ago and therefore generally corresponds with the evolution of the macula. The second exon, which is conserved in apes and Old World monkeys only, appears to have evolved after the division of Old and New World monkeys 40–50 million years ago (Fig. 5).
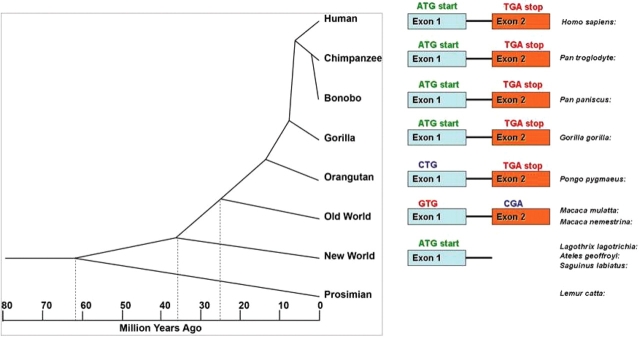
Figure 5.
A schematic indicating the evolution and appearance of LOC387715/ARMS2 exons within the primate lineage. Exon 1 sequence with a putative ATG start codon arose about 43 million years ago after the split from prosimians; it was maintained in apes but not in Old World monkeys. Exon 2 appeared within Old World monkeys after the split from New World monkeys; a putative TGA termination codon in exon 2 arose within the hominid group, i.e. it is present only in apes and humans.
We observed that the rhesus sequence contains an alternative acceptor splice site that is equivalent in position to human codons 99 and 100 (data not shown). The rhesus sequence includes a CGA arginine codon at the predicted TGA termination codon position observed in human. An alternative TAA stop codon present in rhesus, 21 bp downstream of the CGA codon, may be used for protein termination.
Whether the LOC387715/ARMS2 transcript is translated in vivo in rhesus macaques is still unknown. mRNA transcript has been detected in human placental, neural retinal tissue and retinal pigment epithelial (RPE) cells, and we found similar tissue expression in rhesus macaque. Although orangutan and rhesus monkeys possess CUG and GUG codons, respectively, instead of the classic AUG start codon, the CUG and GUG codons are used in other mammalian genes (including humans) as alternative non-AUG initiation codons (15). Thus, of the species examined, only humans, the two species of chimpanzees, orangutan and rhesus monkeys possess a sequence that is predicted to be transcribed and translated into a protein. An alternative explanation is that the ARMS2 region in non-human primates is a pseudo-gene. It is also possible that the LOC387715/ARMS2 gDNA sequence acts as a distant enhancer or cis-regulatory element for the HTRA1 gene and therefore that polymorphisms within the LOC387715/ARMS2 region influence transcription of HTRA1.
Dewan et al. (7) reported that homozygosity for the minor allele in the human HTRA1 SNP resulted in a small but significant increase in promoter activity. Similarly, in the pSEAP reporter assay used to evaluate the effect of the ‘A’ allele in the associated rhesus SNP, we found a consistent doubling of promoter activity.
In summary, we conclude that LOC387715/ARMS2 may be a newly evolved and transcribed gene found only in higher primates, a pseudogene, or a recent HTRA1 promoter development. Our analysis shows that novel SNPs in both the LOC387715/ARMS2 and HTRA1 genes are associated with the development of drusenoid maculopathy in monkeys and that the HTRA1 variant has functional consequences for gene expression. A hypothesis consistent with our promoter functional analyses and the human studies of Dewan et al. (7) is that elevated expression of HTRA1 increases the risk in both humans and macaque monkeys.
Our findings demonstrate for the first time that macaque monkeys and humans share the same susceptibility genes for a common complex disorder. Thus, this study strengthens the value of an important non-human primate model to investigate the genetic factors that lead to AMD and to develop potential therapies for this blinding disorder. Since LOC387715/ARMS2 appears to be present only in animals with a macula and its polymorphisms are associated with disease in this part of the retina, we speculate that this transcribed locus may have a role in macular function.
MATERIALS AND METHODS
All studies were approved by the Institutional Animal Care and Use Committee of the Oregon National Primate Research Center/Oregon Health and Science University and conformed to NIH guidelines.
Ascertainment and phenotyping
Older members of the colony of 4000 rhesus monkeys (M. mulatta) at the Oregon National Primate Research Center were surveyed for age-related eye disease. Stereoscopic color retinal fundus photographs were obtained from over 700 colony members, including 245 of those 18 years of age and older (equivalent to human age 54 years using the generally accepted 3:1 ratio for human:rhesus age). Selected cases also were evaluated by fluorescein angiography. From these, we identified a cohort of 81 unrelated monkeys with macular drusen (Table 1), a phenotype indistinguishable from early to intermediate human AMD (Fig. 1) and occurring with similar prevalence. The retinal photographs were formally graded for the number and size of macular drusen using classifications based on the AREDS AMD grading system (16), using a scaling factor of 7% to adjust for differences in eye size and photograph magnification. Monkeys were classified as affected when six or more drusen <108 µm in diameter (equivalent to <125 µm in humans) or more than one druse >108 µm were present. Phenotype was determined in all cases by three independent observers (MLK, NL and PJF). For comparison, we selected a control population of 56 similarly evaluated, unrelated rhesus macaques, all ≥18 years of age, with no drusen, including 48 of Indian origin (39 females, nine males) and eight of Chinese origin (five females, three males).
Genotyping and sequencing
DNA was extracted from venous blood drawn from all affected and unaffected subjects. As the rhesus sequence at the pertinent genomic interval was unknown, consensus human and chimp primers were designed to successfully amplify this region and were tested for amplification in a variety of apes, Old World Monkeys, New World Monkeys and a prosimian. Sequencing was performed at the Oregon Health and Science University Sequencing Core Facility.
Statistical analyses
Adjusted unconditional logistic regression analysis was performed to determine the relationship between the genotype and phenotype. Estimates of Hardy–Weinberg equilibrium and linkage disequilibrium were also calculated.
RT–PCR of rhesus LOC387715 and HTRA1
Cultured RPE cells harvested from adult human donors and rhesus macaques were homogenized, and total RNA was isolated from neural retina, RPE and placenta using the RNAqueous kit (Ambion Inc., Austin, TX, USA). RT–PCR with oligo-dT primers was then performed. First-strand cDNA was amplified using forward primers situated in exon 1 and reverse primers within exon 2. PCR products were visualized in an agarose gel under UV light, excised, purified and sequenced. Full-length cDNA spanning the region from the predicted initiation codon to the termination codon was cloned into an expression vector.
pSEAP reporter assay of rhesus HTRA1 promoter activity
A 350 bp fragment of the rhesus HTRA1 promoter region with the low-risk ‘C’ allele at position 17076 (Fig. 2) and a similar fragment with the high-risk allele ‘A’ at the same position were cloned into a pMetLuc-Reporter vector (Clontech, cat. #631729) using XhoI and BamHI restriction enzyme sites. The vector uses a dual secreted reporter system with luciferase as a control for transfection efficiency and SEAP for the promoter under study.
In four separate experiments, 293T cells were nucleofected (AMAXA, cat. #VCA-1003) with clones containing either the wild-type or risk allele. Media were collected from each assay at 24 h. The pMetLuc-Control vector provided with the kit was used as an internal positive control, and blank wells were assayed for background signal. pSEAP signal was assayed according to manufacturer’s instructions (Clontech protocol #PT3940-2) and normalized using the corresponding luciferase signal in each well.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by grants from The Foundation Fighting Blindness, Owing Mills, MD (M.N., P.J.F.); the National Institutes of Health (NIH) National Eye Institute R01-EY12203 (M.K.) and National Center for Research Resources RR-00163 (M.N., N.L., B.F.); the Clayton Foundation (J.T.S.); the Macular Degeneration Center Research Fund of the Casey Eye Institute (M.K., P.J.F.), Research to Prevent Blindness, New York, NY (P.J.F.) and China NSF grant no. 30730057 (J.O.).
REFERENCES
- 1.Klein R., Klein B.E., Knudtson M.D., Meuer S.M., Swift M., Gangnon R.E. Fifteen-year cumulative incidence of age-related macular degeneration: the Beaver Dam Eye Study. Ophthalmology. 2007;114:253–262. doi: 10.1016/j.ophtha.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 2.Klein M.L., Francis P.J. Genetics of age-related macular degeneration. Ophthalmol. Clin. North Am. 2003;16:567–574. doi: 10.1016/s0896-1549(03)00063-4. [DOI] [PubMed] [Google Scholar]
- 3.Jakobsdottir J., Conley Y.P., Weeks D.E., Mah T.S., Ferrell R.E., Gorin M.B. Susceptibility genes for age-related maculopathy on chromosome 10q26. Am. J. Hum. Genet. 2005;77:389–407. doi: 10.1086/444437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maller J., George S., Purcell S., Fagerness J., Altshuler D., Daly M.J., Seddon J.M. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nat. Genet. 2006;38:1055–1059. doi: 10.1038/ng1873. [DOI] [PubMed] [Google Scholar]
- 5.Francis P.J., George S., Schultz D.W., Rosner B., Hamon S., Ott J., Weleber R.G., Klein M.L., Seddon J.M. The LOC387715 gene, smoking, body mass index, environmental associations with advanced age-related macular degeneration. Hum. Hered. 2007;63:212–218. doi: 10.1159/000100046. [DOI] [PubMed] [Google Scholar]
- 6.Kanda A., Chen W., Othman M., Branham K.E., Brooks M., Khanna R., He S., Lyons R., Abecasis G.R., Swaroop A. A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proc. Natl Acad. Sci. USA. 2007;104:16227–16232. doi: 10.1073/pnas.0703933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dewan A., Liu M., Hartman S., Zhang S.S., Liu D.T., Zhao C., Tam P.O., Chan W.M., Lam D.S., Snyder M., et al. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science. 2006;314:989–992. doi: 10.1126/science.1133807. [DOI] [PubMed] [Google Scholar]
- 8.Seddon J.M., Francis P.J., George S., Schultz D.W., Rosner B., Klein M.L. Association of CFH Y402H and LOC387715 A69S with progression of age-related macular degeneration. JAMA. 2007;297:1793–1800. doi: 10.1001/jama.297.16.1793. [DOI] [PubMed] [Google Scholar]
- 9.Yang Z., Camp N.J., Sun H., Tong Z., Gibbs D., Cameron D.J., Chen H., Zhao Y., Pearson E., Li X., et al. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science. 2006;314:992–993. doi: 10.1126/science.1133811. [DOI] [PubMed] [Google Scholar]
- 10.El-Mofty A., Gouras P., Eisner G., Balazs E.A. Macular degeneration in rhesus monkey (Macaca mulatta) Exp. Eye Res. 1978;27:499–502. doi: 10.1016/0014-4835(78)90027-1. [DOI] [PubMed] [Google Scholar]
- 11.Hope G.M., Dawson W.W., Engel H.M., Ulshafer R.J., Kessler M.J., Sherwood M.B. A primate model for age related macular drusen. Br. J. Ophthalmol. 1992;76:11–16. doi: 10.1136/bjo.76.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stafford T.J., Anness S.H., Fine B.S. Spontaneous degenerative maculopathy in the monkey. Ophthalmology. 1984;91:513–521. doi: 10.1016/s0161-6420(84)34275-0. [DOI] [PubMed] [Google Scholar]
- 13.Umeda S., Suzuki M.T., Okamoto H., Ono F., Mizota A., Terao K., Yoshikawa Y., Tanaka Y., Iwata T. Molecular composition of drusen and possible involvement of anti-retinal auto-immunity in two different forms of macular degeneration in cynomolgus monkey (Macaca fasicularis) FASEB J. 2005;12:1683–1685. doi: 10.1096/fj.04-3525fje. [DOI] [PubMed] [Google Scholar]
- 14.Ferguson B., Street S.L., Wright H., Pearson C., Jia Y., Thompson S.L., Allibone P., Dubay C.J., Spindel E., Norgren R.B., Jr Single nucleotide polymorphisms (SNPs) distinguish Indian-origin and Chinese-origin rhesus macaques (Macaca mulatta) BMC Genomics. 2007;8 doi: 10.1186/1471-2164-8-43. 43.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Touriol C., Bornes S., Bonnal S., Audigier S., Prats H., Prats A.C., Vagner S. Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biol. Cell. 2003;95:169–178. doi: 10.1016/s0248-4900(03)00033-9. [DOI] [PubMed] [Google Scholar]
- 16.Age-related Eye Disease Study Group. The Age-Related Eye Disease Study (AREDS): design implications. AREDS report no. 1. Control. Clin. Trials. 1999;20:573–600. doi: 10.1016/s0197-2456(99)00031-8. [DOI] [PMC free article] [PubMed] [Google Scholar]