Abstract
Notch signaling plays an important role in developmental processes and adult tissue homeostasis. Altered Notch signaling has been associated with various diseases including cancer. While the importance of altered Notch signaling in cancers of hematopoietic and epithelial origins has been established, its role in tumors of mesenchymal origin is less clear. Here, we report that human osteosarcoma cell lines and primary human osteosarcoma tumor samples show significant up-regulation of Notch, its target genes and Osterix. Notch inhibition by γ-secretase inhibitors or by using lentiviral mediated expression of dominant negative Mastermind-like protein (DN-MAML) decreases osteosarcoma cell proliferation in vitro. In vivo, established human tumor xenografts in nude mice show decreased tumor growth after chemical or genetic inhibition of Notch signaling. Finally, transcriptional profiling of osteosarcomas from p53 mutant mice confirmed up-regulation of Notch1 target genes Hes1, Hey1 and its ligand Dll4. Our data suggest that activation of Notch signaling contributes to the pathogenesis of human osteosarcomas and its inhibition may be a therapeutic approach for the treatment of this mesenchymal tumor.
INTRODUCTION
Notch signaling plays an important role in developmental processes and adult tissue homeostasis by regulating cell fate determination, proliferation, differentiation and apoptosis (1). Notch signaling is triggered upon interaction of the receptor with its transmembrane ligand. Upon this ligand-receptor interaction, Notch receptor undergoes to proteolytic cleavage at the membrane by Presenilins. This cleavage releases Notch intra cellular domain (NICD), which is then translocated to the nucleus where it can interact with a transcriptional activator Mastermind-like protein (MAML) and a transcription factor RBP-jK to activate the transcription of downstream target genes. Altered Notch signaling has been associated with many different diseases including cancer (2–5). Experimental evidence supports that Notch can act both as an oncogene and tumor suppressor gene depending on its expression levels and timing in a cell-type and context-dependent manner (6,7). Aberrant Notch signaling is involved in solid and hematological tumor formation, and activating mutations of Notch1 in humans promotes T-cell acute lymphatic leukemia (T-ALL) (2,8,9). Osteosarcoma is the most common primary malignant bone tumor affecting predominantly teenagers and young adults. These mesenchymally derived high-grade tumors most frequently affect the metaphyseal regions of the distal femur, proximal tibia and proximal humerus. Although current understanding of molecular pathogenesis of osteosarcomas is limited, alterations of tumor suppressor gene expression and deregulation of major signaling pathways including the Wnt, TGF-β and Shh have been reported (10). Emerging data strongly support that osteosarcomas may result from genetic and epigenetic disruptions of osteoblast terminal differentiation (10). The role of Notch signaling in osteoblast differentiation, proliferation and lineage commitment was recently identified (11,12). Because of the importance of altered Notch signaling in tumorigenesis of hematopoietic (leukemia) and epithelial (gastrointestinal, breast and pancreas) origins, and because our previously published data support both pro-proliferative and anti-differentiation effects for pathological gain of Notch signaling, we investigated the role of Notch in osteosarcomas (8,11,13–16).
RESULTS
Notch and notch target gene expression are up-regulated in human osteosarcomas
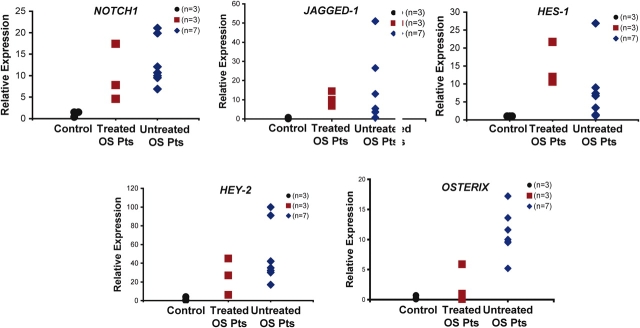
By quantitative RT–PCR, we demonstrated that expression of NOTCH1, the Notch ligand JAGGED-1, and the transcription factors HES-1, and HEY-2, which are direct targets of NOTCH1 signaling, were significantly up-regulated in seven primary untreated osteosarcoma samples and three post-treatment osteosarcoma samples, when compared with wild-type osteoblasts (n = 3) (Fig. 1 and Table 1). Interestingly, we also observed significant up-regulation of the Osterix transcription factor. This is consistent with our previous observation that gain of Notch function in committed osteoblasts of transgenic mice leads to proliferation of an immature osteoblastic population, arrest of osteoblast maturation and up-regulation of the Osterix transcription factor (11).
Figure 1.
Notch Signaling is up regulated in human osteosarcoma. (A) Expression of Notch1, Jagged-1, Osterix and known Notch1 target genes Hey-2 and Hes-1 by Q-RT–PCR. Tumor RNA was obtained from (n = 7) untreated, and (n = 3) treated osteosarcoma patients (OS Patients), and (n = 3) control human osteoblasts.
Table 1.
Clinical characteristics of osteosarcoma samples
| BPC no. | Diagnosis | Chemotherapy status | Site | Age | Sex | Race |
|---|---|---|---|---|---|---|
| T5-2000-03-P2048 | Osteosarcoma, metastatic | Relapse post | Lung | 21 | F | W |
| T7-2000-05-P4021 | Osteosarcoma, fibroblastic | Post | Femur, right distal | 16 | M | W |
| T16-2001-09-P4021 | Osteosarcoma | Post-chemo-DS | Femur, right distal | 16 | F | W |
| T20-2002-02-P1074 | Osteosarcoma, chrondroblastic | Pre-IB | Femur, right distal | 9 | M | W |
| T21-2002 02 P1195 | Osteosarcoma | Pre-IB | Femur, left distal | 9 | F | W |
| T29-2003-02-P2097 | Osteosarcoma | Pre-IB | Femur, left distal biopsy | 11 | M | W |
| T31-2003-03-P2000 | Osteosarcoma with telangiectatic features | Pre-IB | Tibia, left proximal | 6 | M | B |
| T35-2003-10-P1062 | Osteosarcoma | Pre-IB | Femur, right | 10 | F | Other |
| T36-2003-11-P1025 | Osteosarcoma | Pre-IB | Tibia, right proximal | 15 | F | W |
| T38-2004-04-P0223 | Osteosarcoma, conventional | Pre-IB | Femur, left | 17 | M | W |
IB, initial biopsy; DS, definitive surgery.
Inhibition of notch signaling with γ-secretase inhibitors decreases the proliferation osteosarcoma cells in vitro
If up-regulation of Notch signaling is pathogenic in human osteosarcomas, inhibition of Notch activity might reduce the proliferation of osteosarcoma cells. To test this, we incubated human osteosarcoma SaOs2 cells with two different γ-secretase inhibitors to inhibit Notch signaling (DAPT and Compound E). This pharmacological inhibition of Notch signaling resulted in a significant reduction in cell proliferation (Fig. 2A). Additionally, two other human osteosarcoma cell lines namely SJSA-1 and Crl-1423 showed the similarly reduced proliferation after treatment with DAPT (Fig. 2B). Importantly, cell viability as assessed by Trypan blue staining was unaffected, indicating absence of an effect on cell death (apoptosis or necrosis) by these compounds (Fig. 2C). While inhibition of γ-secretase would be expected to abolish all Notch signaling, other Presenilin-dependent, Notch-independent functions may be affected. Hence, we determined the specificity of Notch inhibition on this cellular phenotype by transducing osteosarcoma cells with a lentiviral vector expressing a dominant negative Mastermind-like 1 protein (DN-MAML1). DN-MAML1 would be expected to directly inhibit Notch at the level of transcriptional complex assembly. We observed a similarly significant inhibition on cellular proliferation suggesting that chemical inhibition was at least in part Notch-specific (Fig. 2D).
Figure 2.
Chemical inhibition of Notch signaling reduces proliferation of human osteosarcoma cell lines. (A) Proliferation of human SaOs2 osteosarcoma cells were measured by BrdU incorporation after treatment of these cells either with γ-secretase inhibitor (DAPT) for 24 h with the indicated doses or another γ-secretase inhibitor Compound E (500 nM), *P < 0.05. (B) Proliferation of two additional human osteosarcoma cell lines i.e. SJSA and CRL-1423 were measured by BrdU incorporation after treatment of these cells with 10 uM DAPT for 24 hours. (C) Cell viability of human SaOs2 osteosarcoma cells was measured by counting Trypan blue positive cells after adding increasing dosages of DAPT at indicated time periods. (D) SaOs2 cells were transduced either with a control vector or a lentivirus expressing dominant negative Mastermind like-1 (DN-MAML). The cells were sorted for YFP and the positive cells were re-plated for the measurement of BrdU incorporation, *P < 0.05.
Chemical and genetic inhibition of notch signaling reduces tumor burden in vivo
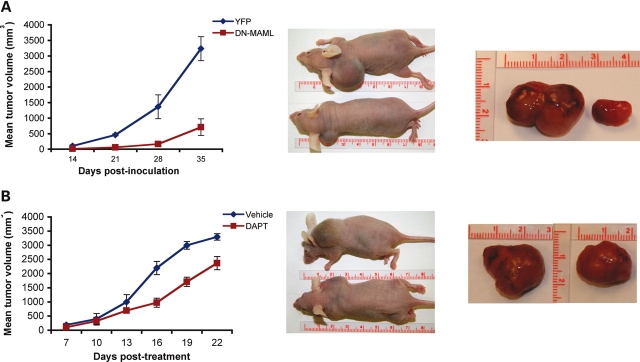
We investigated how disruption of Notch signaling could influence the response of established human tumor xenografts in nude mice. First, we implanted SJSA-1 cells, transduced with either a lentivirus expressing YFP (SJSA-1-YFP) or dominant negative Mastermind-like (SJSA-1-DN-MAML), into nude mice. Nude mice bearing SJSA-1-DN-MAML cells showed markedly reduced tumor growth in comparison to the mice implanted with SJSA-1-YFP (Fig. 3A). To determine, if the tumors could similarly be treated by pharmacological inhibition of Notch/Presenilin signaling with γ-secretase inhibitors, nude mice bearing SJSA-1 xenografts were treated with intraperitoneal injections with 125 mg/kg DAPT for 2 weeks. No weight loss or significant pathological changes were observed during the course of the study (data not shown). However, DAPT-treated mice also showed substantial tumor retention in comparison to vehicle-treated control group (Fig. 3B). Interestingly, no metastasis was observed in these mice as it was reported in a recent paper (17).
Figure 3.
Genetic and chemical inhibition of Notch signaling reduces tumor burden in nude mice. (A) Mean tumor volume in nude mice after inoculated with osteosacoma cells expressing either lentivirus-mediated YFP or DN-MAML1. (B) Pharmacological inhibition of Notch signaling via DAPT. Mice were injected with 125 mg/kg DAPT daily for 22 days.
Mutations in the p53 gene have been associated with a wide range of human tumors, including osteosarcomas (18,19). Since p53 heterozygous mice develop osteosarcomas, we investigated the expression of Notch pathway components in the tumors obtained from p53 heterozygous mice using a microarray analysis. Transcriptional profiling of osteosarcomas showed that majority of Notch-signaling pathway components were significantly up-regulated in comparison to both an osteoblastic cell line MC3T3 and primary calvarial osteoblasts (Fig. 4A and B). These included the ligands Jagged2, Delta-like 4, Hes and Hey genes suggesting that increased Notch expression in p53 heterozygous mice might contribute the formation of osteosarcomas.
Figure 4.
Osteosarcomas from p53 mutant mice, exhibit up-regulation of Notch and its target genes. (A) mRNA expression of Notch and Notch target genes as measured by RNA microarray analysis. Expression levels are relative to MC3T3 osteoblastic cells (left most lane). Red indicates high expression relative to MC3T3 cells and green represents low expression (see logarithmic scale at upper right). OS: osteosarcoma. Each lane represents a different osteosarcoma sample, and sample names are labeled above the lanes. Some genes have more than one row of data, which indicate that there is more than one probe for the same gene on the microarray chip. (B) Expression of Notch ligand Dll4 and known Notch1 target genes Hes-1 and Hey-1 in osteosarcomas of p53+/− mice by Q-RT–PCR. Tumor RNA was obtained from p53+/− mice (n = 3), and calvarial RNA obtained from wt mice (n = 3) was used as a control, *P < 0.05.
DISCUSSION
Unfortunately, little is known about the molecular basis of osteosarcoma, the most common primary malignant tumor of bone and the second highest cause of cancer-related death in children. It is likely that at least in a subset of osteosarcomas, the initiating events have features of a committed osteoprogenitor cell. Our data together with previous reports suggest that in a pathological context, gain of Notch function in committed osteoblast progenitors may be implicated in the pathogenesis of osteosarcomas in part by expanding an immature osteoblastic population via up-regulation of Cyclins and the Osterix transcription factor (11). Our previous observation that significant up-regulation of CYCLIN D1 in the osteoblast-specific Notch1 transgenic mice correlates with the observation in humans where 10% of osteosarcomas show amplification of the chromosomal region encoding CYCLIN D1 (20).
Here, primary human osteosarcomas show transcriptional up-regulation of NOTCH, NOTCH ligand (JAGGED-1) and Notch target transcription factors (HES-1 and HEY-2). Expression of Notch in a case of human osteosarcoma was reported previously (21). However, function of Notch in osteosarcoma still remains unknown. A recent study demonstrated that expression of HES1 was associated with invasive and metastatic potential of osteosarcomas, and inhibition of Notch pathway by a γ-secretase inhibitor eliminated invasion in Matrigel without affecting cell proliferation, survival or anchorage-independent growth (17). Hence, Notch effects may contribute to both proliferative and metastatic potential in osteosarcomas. While our data support a proliferative effect of Notch in osteosarcomas, we were unable to detect any obvious metastasis of the tumors generated in nude mice. Different phenotypic effects likely depend on the context dependence of specific cell lines used in each study.
Published reports support a role for dysregulation of the master osteoblast transcription factor Runx2 in osteosarcomas via functions related to growth-related G1 transition in osteoblastic cells and direct regulation of BAX in a pro-apoptotic program (22,23). Interestingly, our and others' data show that Notch can directly inhibit Runx2 function in vivo supporting dysregulation of Runx2 as another consequence of gain of Notch in osteosarcoma. Finally, our data suggest that increased Notch signaling occurs downstream of p53 loss of function in one of the few mouse models of osteosarcoma pathogenesis. Together our and others' data suggest that loss of p53 in a committed osteoprogenitor population leads to gain of function of Notch signaling with consequences on Cyclins and Osterix upregulation and Runx2 inhibition to account for proliferative and metastatic potential of osteosarcomas. While it is very likely that this pathway accounts for only one aspect of the complex pathogenesis of osteosarcoma, it provides a potential therapeutic target in what is until now a poorly treatable cancer.
MATERIALS AND METHODS
Plasmids
For lentivirus vector production, plasmid encoding dominant-negative (DN) form of Mastermind-Like 1 (MAML) fused to eGFP was obtained from Warren Pear. The DN-MAML portion was excised and placed upstream of eYFP and downstream of an IRES. This 1.6 kb IRES-DNMAML-eYFP cassette replaced IRES-eYFP in pHIV-IRES-eYFP to make pHIV-IRES-DNMAML-eYFP. VSV G-pseudo-typed vector supernatants were produced as previously described (24). After 72 h, cell culture supernatants were harvested and clarified. Typical titers after concentration by ultracentrifugation were in excess of 108 IU/ml, for the two SIN vectors as assessed on HOS cells by epifluorescence microscopy. Titers of unconcentrated non-SIN vectors were in excess of 107 IU/ml.
Cell culture, BrDU incorporation and cell viability assay
SJSA1, SaOs2 and CRL1423 cells were purchased from American Type Culture Collection. Cells were treated with BrdU labeling reagent according to manufacturer's instructions for 6 h, washed with PBS and fixed with 70% ethanol for 25 min at 4°C (Zymed). Three to five areas for each genotype (n = 3 slides) were counted by two independent observers blinded to genotype. BrdU positive cells over total cells were scored visually and with Automeasure software (Zeiss Axiovision). Cells treated either with 1−5 µM DAPT or 500 nM Compound E (Calbiochem) versus carrier (DMSO) for 24 or 48 h. For the cell viability assay 5 × 104 SaOs2 cells were seeded into 12-well plates. Twenty-four hours after seeding, cells were treated with 1 and 5 µM DAPT (Calbiochem) for 8 and 24 h. Cell viability was measured via counting the Trypan blue positive cells by Beckman Coulter Vi-Cell XR 2.03.
RNA extraction and Q-RT–PCR analysis
Total RNA was extracted using TRIzol reagent (Invitrogen). cDNAs were synthesized from extracted RNA by using Superscript III First Strand RT–PCR kit (Invitrogen). Real-time quantitative PCR amplifications were performed on LightCycler (Roche) and with TaqMan assay (Applied Biosystems probe HS00172878-M1). β-actin and β2-microglobulin genes were used as internal controls for the quantity and quality of the cDNAs in real time PCR assays.
In vivo studies
Athymic female nude mice (6–8-week-old BALB/cAnNCr-nu/nu) were purchased from National Cancer Institute (NCI). SJSA-1 (2 × 106) cells were resuspended in HBSS and injected subcutaneously. Once tumors reached to 40–70 mm3, animals were treated with daily intraperitoneal subcutaneous injections of DAPT in corn oil 125 mg/kg/day for 22 days. Tumor volume was measured with a caliper and calculated by the formula of 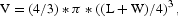 L being the longest cross-section and W being the shortest.
L being the longest cross-section and W being the shortest.
Gene expression microarray analysis
RNA was extracted from rhabdomyosarcoma or osteosarcomas (about 3 mm3) or Mc3T3 cell (60 mm dish) with RNeasy Mini Kit (Qiagen). Following quality check of the RNA, microarray analysis was performed at Baylor Microarray Core Facility (www.bcm.edu/mcfweb/) with Affymetrix Mouse Genome 430 2.0 Chips. Microarray data were analyzed with Genesifter software (www.genesifter.net).
Patient samples
Osteosarcoma tumor samples were obtained from Pediatric Cooperative Human Tissue Network Biopathology Center (Columbus, OH, USA). The tissue samples were cut into 2–3 mm wrapped in the foil, snap frozen in liquid nitrogen or cold isopentane. Samples are stored at 70°C or colder until they are shipped to the investigator. The RNA from the tissues was extracted as described above and the relative expression of each tumor sample was calculated separately. Control human osteoblast RNAs were purchased from Cell Applications Inc. (see Table 1 for clinical characteristics of the samples).
Statistical analyses
Data are expressed as mean values±standard deviation (SD). Statistical significance was computed using Student's paired t-test. A P-value <0.05 was considered statistically significant.
FUNDING
This work was supported by NIH grants ES11253 (B.L.), HD22657 (B.L.), DE016990 (B.L.), the Rolanette and Berdon Lawrence Bone Disease Program and the Baylor College of Medicine (BCM) Intellectual and Developmental Disabilities Research Center (IDDRC).
ACKNOWLEDGEMENTS
We thank Van Nguyen and Jianning Tao for their technical assistance.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Bray S.J. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell. Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 2.Koch U., Radtke F. Notch and cancer: a double-edged sword. Cell Mol. Life Sci. 2007;64:2746–2762. doi: 10.1007/s00018-007-7164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miele L., Golde T., Osborne B. Notch signaling in cancer. Curr. Mol. Med. 2006;6:905–918. doi: 10.2174/156652406779010830. [DOI] [PubMed] [Google Scholar]
- 4.Roy M., Pear W.S., Aster J.C. The multifaceted role of Notch in cancer. Curr. Opin. Genet. Dev. 2007;17:52–59. doi: 10.1016/j.gde.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Gridley T. Notch signaling and inherited disease syndromes. Hum. Mol. Genet. 2003;12(Spec no. 1):R9–R13. doi: 10.1093/hmg/ddg052. [DOI] [PubMed] [Google Scholar]
- 6.Weng A.P., Aster J.C. Multiple niches for Notch in cancer: context is everything. Curr. Opin. Genet. Dev. 2004;14:48–54. doi: 10.1016/j.gde.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Radtke F., Raj K. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat. Rev. Cancer. 2003;3:756–767. doi: 10.1038/nrc1186. [DOI] [PubMed] [Google Scholar]
- 8.Weng A.P., Ferrando A.A., Lee W., Morris J.P.t., Silverman L.B., Sanchez-Irizarry C., Blacklow S.C., Look A.T., Aster J.C. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 9.Bolos V., Grego-Bessa J., de la Pompa J.L. Notch signaling in development and cancer. Endocrinol. Rev. 2007;28:339–363. doi: 10.1210/er.2006-0046. [DOI] [PubMed] [Google Scholar]
- 10.Tang N., Song W.X., Luo J., Haydon R.C., He T.C. Osteosarcoma development and stem cell differentiation. Clin. Orthop. Relat. Res. 2008;466:2114–2130. doi: 10.1007/s11999-008-0335-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engin F., Yao Z., Yang T., Zhou G., Bertin T., Jiang M.M., Chen Y., Wang L., Zheng H., Sutton R.E., et al. Dimorphic effects of Notch signaling in bone homeostasis. Nat. Med. 2008;14:299–305. doi: 10.1038/nm1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hilton M.J., Tu X., Wu X., Bai S., Zhao H., Kobayashi T., Kronenberg H.M., Teitelbaum S.L., Ross F.P., Kopan R., et al. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat. Med. 2008;14:306–314. doi: 10.1038/nm1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyamoto Y., Maitra A., Ghosh B., Zechner U., Argani P., Iacobuzio-Donahue C.A., Sriuranpong V., Iso T., Meszoely I.M., Wolfe M.S., et al. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell. 2003;3:565–576. doi: 10.1016/s1535-6108(03)00140-5. [DOI] [PubMed] [Google Scholar]
- 14.van Es J.H., van Gijn M.E., Riccio O., van den Born M., Vooijs M., Begthel H., Cozijnsen M., Robine S., Winton D.J., Radtke F., et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 15.Hu C., Dievart A., Lupien M., Calvo E., Tremblay G., Jolicoeur P. Overexpression of activated murine Notch1 and Notch3 in transgenic mice blocks mammary gland development and induces mammary tumors. Am. J. Pathol. 2006;168:973–990. doi: 10.2353/ajpath.2006.050416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klinakis A., Szabolcs M., Politi K., Kiaris H., Artavanis-Tsakonas S., Efstratiadis A. Myc is a Notch1 transcriptional target and a requisite for Notch1-induced mammary tumorigenesis in mice. Proc. Natl Acad. Sci. USA. 2006;103:9262–9267. doi: 10.1073/pnas.0603371103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang P., Yang Y., Zweidler-McKay P.A., Hughes D.P. Critical role of notch signaling in osteosarcoma invasion and metastasis. Clin. Cancer Res. 2008;14:2962–2969. doi: 10.1158/1078-0432.CCR-07-1992. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Lang G.A., Iwakuma T., Suh Y.A., Liu G., Rao V.A., Parant J.M., Valentin-Vega Y.A., Terzian T., Caldwell L.C., Strong L.C., et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 19.Olive K.P., Tuveson D.A., Ruhe Z.C., Yin B., Willis N.A., Bronson R.T., Crowley D., Jacks T. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Wang L.L. Biology of osteogenic sarcoma. Cancer J. 2005;11:294–305. doi: 10.1097/00130404-200507000-00005. [DOI] [PubMed] [Google Scholar]
- 21.Kawakami T., Siar C.H., Ng K.H., Shimizu T., Okafuji N., Kurihara S., Hasegawa H., Tsujigiwa H., Nagatsuka H., Nagai N. Expression of Notch in a case of osteosarcoma of the maxilla. Eur. J. Med. Res. 2004;9:533–535. [PubMed] [Google Scholar]
- 22.Eliseev R.A., Dong Y.F., Sampson E., Zuscik M.J., Schwarz E.M., O'Keefe R.J., Rosier R.N., Drissi M.H. Runx2-mediated activation of the Bax gene increases osteosarcoma cell sensitivity to apoptosis. Oncogene. 2008;27:3605–3614. doi: 10.1038/sj.onc.1211020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galindo M., Pratap J., Young D.W., Hovhannisyan H., Im H.J., Choi J.Y., Lian J.B., Stein J.L., Stein G.S., van Wijnen A.J. The bone-specific expression of Runx2 oscillates during the cell cycle to support a G1-related antiproliferative function in osteoblasts. J. Biol. Chem. 2005;280:20274–20285. doi: 10.1074/jbc.M413665200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sutton R.E., Wu H.T., Rigg R., Bohnlein E., Brown P.O. Human immunodeficiency virus type 1 vectors efficiently transduce human hematopoietic stem cells. J. Virol. 1998;72:5781–5788. doi: 10.1128/jvi.72.7.5781-5788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]