Abstract
Genome-wide association studies have identified FGFR2 as a breast cancer (BC) susceptibility gene in populations of European and Asian descent, but a causative variant has not yet been conclusively identified. We hypothesized that the weaker linkage disequilibrium across this associated region in populations of African ancestry might help refine the set of candidate-causal single nucleotide polymorphisms (SNPs) previously identified by our group. Eight candidate-causal SNPs were evaluated in 1253 African American invasive BC cases and 1245 controls. A significant association with BC risk was found with SNP rs2981578 (unadjusted per-allele odds ratio = 1.20, 95% confidence interval 1.03–1.41, Ptrend = 0.02), with the odds ratio estimate similar to that reported in European and Asian subjects. To extend the fine-mapping, genotype data from the African American studies were analyzed jointly with data from European (n = 7196 cases, 7275 controls) and Asian (n = 3901 cases, 3205 controls) studies. In the combined analysis, SNP rs2981578 was the most strongly associated. Five other SNPs were too strongly correlated to be excluded at a likelihood ratio of < 1/100 relative to rs2981578. Analysis of DNase I hypersensitive sites indicated that only two of these map to highly accessible chromatin, one of which, SNP rs2981578, has previously been implicated in up-regulating FGFR2 expression. Our results demonstrate that the association of SNPs in FGFR2 with BC risk extends to women of African American ethnicity, and illustrate the utility of combining association analysis in datasets of diverse ethnic groups with functional experiments to identify disease susceptibility variants.
INTRODUCTION
Two recent genome-wide association studies (GWAS) have identified single nucleotide polymorphisms (SNPs) within the FGFR2 gene to be associated with breast cancer (BC) risk (1,2). This was the strongest susceptibility locus in both studies (and hence the strongest known common susceptibility locus for BC): the minor alleles of the associated SNPs confer a per-allele risk of ∼1.3-fold. FGFR2 is located on chromosome 10q and encodes fibroblast growth factor receptor 2, a receptor tyrosine kinase. It is amplified and overexpressed in breast tumors (3–5), and several lines of experimental evidence in humans and mice support that it is a BC oncogene (6–11).
While the GWAS provide overwhelming evidence that FGFR2 is a BC susceptibility locus, the associated SNPs may not be causally related to risk, as they are simply marker-SNPs correlated [in strong linkage disequilibrium (LD)] with other variants within the region. An important problem, therefore, is the identification of the causal variant or variants. Identification of the causal variant(s) will improve the accuracy of risk prediction (because the causal variants will be more strongly correlated with risk than other associated SNPs) and will constitute an important step towards understanding the functional basis of the disease susceptibility.
In this manuscript, we describe two parallel approaches to identifying causal variants in this intronic gene region: (i) a genetic epidemiological approach using fine-scale mapping with BC case–control studies from populations of European, Asian and African descent and (ii) biochemical probing to determine whether candidate SNPs reside in open chromatin indicating an increased probability of functionality. The first approach has previously been applied to this locus using case–control studies from populations of European and Asian ancestry (1). Here we extend that work to investigate candidate-causal variants in FGFR2 in studies of African American women, a population for which there was to-date no information on the association between FGFR2 variants and BC risk. Assuming that this association existed in African Americans, we aimed to utilize this population's different pattern of LD, which is typically weaker than in European or Asian populations (12–15), to refine further the set of possible causal variants.
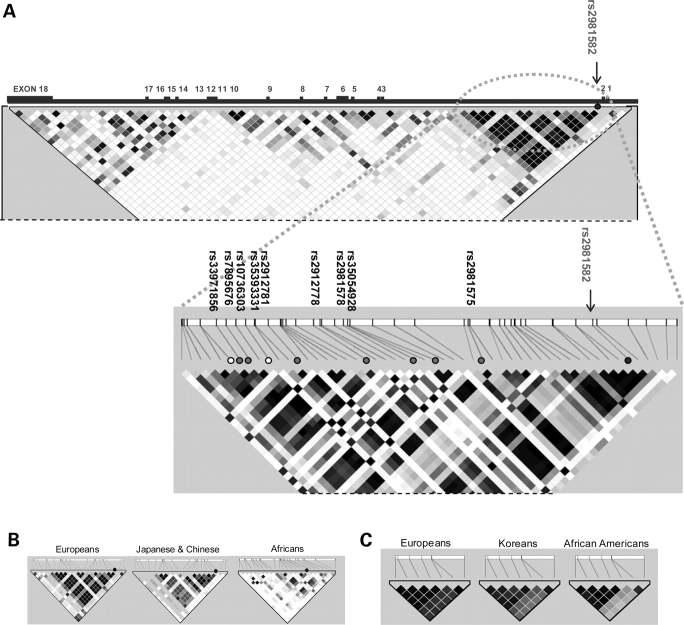
The initial marker-SNPs identified in both genome-wide studies fell in a 25 kb LD block almost entirely within intron 2 of FGFR2 (Fig. 1). The most strongly associated GWAS marker-SNP found by Easton et al. (1) was rs2981582. Haplotype analyses indicated that multiple haplotypes carrying the minor allele of rs2981582 were associated with risk, suggesting strongly that the causal variant was a common SNP strongly correlated with rs2981582. A catalog of all common variants in this region was generated by resequencing 45 subjects of European origin, allowing the detection of all variants with minor allele frequency ≥ 5% with > 97% probability. Of the 117 variants identified, 29 were highly correlated (r2 > 0.6) with rs2981582. In the previous analyses by Easton et al., 27 of the 29 SNPs were evaluated in multiple populations of European and Asian origin (two variants, rs35393331 and rs33971856, could not be assayed). In the combined analysis of European and Asian datasets, the most strongly associated SNP was rs7895676. Assuming a single causal variant and using the likelihood ratio test, 21 SNPs could be excluded as the causal variant at likelihood ratios <1/100 relative to rs7895676 (with 19 of these SNPs having ratios <1/1000) (1). Functional work on the eight remaining variants (including the two variants that could not be genotyped in the epidemiological studies) found that SNPs rs2981578 and rs7895676 affected binding of transcription factors Oct-1/Runx2 and C/EBPβ, respectively (16). Nevertheless, there remains uncertainty as to which variant(s) directly increases BC risk.
Figure 1.
Diagram of FGFR2 and linkage disequilibrium blocks containing candidate SNPs. (A) The upper diagram is a map of the whole FGFR2 gene viewed relative to common (MAF >5%) SNPs in HapMap CEU. The lower diagram is a map of the 25 kb region of linkage derived from the resequence data. The most strongly associated tagSNP is rs2981582. The positions of remaining candidate SNPs are labeled with filled circles on the map with relative chromosomal positions indicated on the bar. The two variants that could not be genotyped are labeled with open circles. (B) 25 kb LD blocks in HapMap CEU, JPT+CHB and YRI populations. (C) LD block of the seven candidate-SNPs in study populations SEARCH, Seoul Breast Cancer Project and MEC-African Americans (MEC-AA).
In this study, we have extended the association studies of SNPs in FGFR2 to women of African American ethnicity. We have utilized case–control data from African American, Asian and European populations to identify the possible causal variants. Finally, we have used DNase I hypersensitivity assays to identify SNPs in accessible chromatin, as a method for identifying likely functional SNPs.
RESULTS
Association of FGFR2 SNPs in African Americans
The six SNPs identified as potentially causal for BC by Easton et al. (1) (rs7895676, rs10736303, rs2912781, rs2912778, rs2981578 and rs35054928) together with SNPs rs2981575 and rs2981582 were included in this study. SNP rs2981575 was not definitively excluded in the fine-scale mapping work in European and Asian populations performed by Easton et al., and it was one of the most strongly associated variants in Europeans (1). SNP rs2981582, the most significant GWAS marker-SNP, was excluded in the analyses by Easton et al. and is included here for comparison with other studies.
Of the eight SNPs considered in this analysis, five could be genotyped directly in the case–control studies using TaqMan® assays. From the four African American studies, a total of 1253 cases of invasive BC and 1245 controls were genotyped for the five SNPs (Table 1). The allele frequencies in African Americans differed markedly from those in Europeans and Asians; for four of the five SNPs, the minor (risk) allele in Europeans was the major allele in African Americans (Table 2). SNP allele frequencies did not differ by >15% across African American study centers (Supplementary Material, Table S1).
Table 1.
Summary of breast cancer case–control studies providing African American womena
| Study | US city | Number of subjects |
Age range | ||||
|---|---|---|---|---|---|---|---|
| Invasive cases | Controls | ER-pos/negb | PR-pos/negc | FH-cases/ctrlsd | |||
| Women's contraceptive and reproductive experiences study (CARE-AA) | Atlanta | 92 | 111 | 48/37 | 53/32 | 36/23 | 35–64 |
| Detroit | 80 | 94 | 25/27 | 25/27 | 18/14 | 35–64 | |
| Los Angeles | 173 | 148 | 102/54 | 78/55 | 40/29 | 35–64 | |
| Philadelphia | 75 | 56 | 26/24 | 21/29 | 33/13 | 35–64 | |
| Seattle | 32 | 26 | 19/13 | 17/15 | 9/6 | 35–64 | |
| Total | 452 | 435 | 220/155 | 194/158 | 136/85 | 35–64 | |
| Multiethnic cohort (MEC-AA) | Los Angeles | 436 | 653 | 242/93 | 159/110 | 88/78 | 45–83 |
| LIFE | Los Angeles | 140 | 0 | 61/55 | 58/53 | 19/0 | 23–49 |
| CARE-LA | Los Angeles | 225 | 157 | 116/86 | 88/76 | 4/3 | 35–64 |
| Total | 1253 | 1245 | 639/389 | 499/397 | 247/166 | ||
aCounts corrected for individuals present in more than one study (see Materials and Methods).
bEstrogen receptor positive/negative.
cProgesterone receptor positive/negative.
dCases/controls with positive family history, meaning a first-degree relative with breast cancer.
Table 2.
Association of FGFR2 SNPs with breast cancer risk in African American women
| SNP | Chr10 position | Risk allele | Risk allele frequencya |
Per-allele OR (95% CI) | P-trend | Homogeneity testb | ||
|---|---|---|---|---|---|---|---|---|
| Europeans | East Asians | African Americans | ||||||
| rs10736303 | 123324447 | G | 0.49 | 0.55 | 0.84 | 1.17 (1.00–1.37) | 0.046 | 0.57 |
| 1.21 (1.02–1.44)c | 0.025c | |||||||
| rs2981578 | 123330301 | G | 0.49 | 0.55 | 0.83 | 1.20 (1.03–1.41) | 0.021 | 0.64 |
| 1.24 (1.04–1.47)c | 0.014c | |||||||
| rs35054928 | 123330423 | C | 0.43 | 0.44 | 0.60 | 1.05 (0.93–1.18) | 0.44 | 0.91 |
| 1.02 (0.90–1.16)c | 0.71c | |||||||
| rs2981575 | 123336106 | G | 0.42 | 0.43 | 0.57 | 1.11 (0.99–1.25) | 0.075 | 0.46 |
| 1.09 (0.97–1.24)c | 0.15c | |||||||
| rs2981582d | 123342307 | A | 0.41 | 0.32 | 0.45 | 1.03 (0.89–1.19) | 0.67 | 0.24 |
| 1.01 (0.86–1.18)c | 0.94c | |||||||
aAllele frequency based on pooling studies from each ethnic group.
bHomogeneity test across African American study centers.
cAnalysis adjusted for age at diagnosis and family history.
dAnalysis excluded data from study CARE-AA without genotype data for this SNP.
Significant associations with BC risk were observed for two of the five SNPs: rs10736303 and rs2981578 [unadjusted per-allele odds ratios (ORs) 95% confidence intervals (CIs) and P-values 1.17 (1.00–1.37) P = 0.046 and 1.20 (1.03–1.41) P = 0.021, respectively]. For all SNPs, the risk allele in African Americans was the same as in European and Asian populations. The two significant associations remained after adjusting for age at diagnosis and family history of BC (Table 2). The per-allele ORs for SNPs rs10736303 or rs2981578 did not vary significantly by age at diagnosis (data not shown).
SNPs rs10736303 and rs2981578 were analyzed further to see if the associations varied by tumor characteristics. Estrogen receptor (ER) status was available for 82% of cases, with 639 ER-positive and 389 ER-negative tumors. Progesterone receptor (PR) status was available for 72% of cases, with 499 PR-positive and 397 PR-negative tumors. Both SNPs rs10736303 and rs2981578 demonstrated stronger associations with ER-positive tumors [unadjusted per-allele ORs = 1.24 (95% CI 1.02–1.51) and 1.26 (95% CI 1.03–1.52), respectively] than ER-negative tumors [ORs = 1.04 (95% CI 0.83–1.31) and 1.07 (95% CI 0.85–1.35) (Table 3)]. However, the association with ER-positive tumors was not significantly different from that with ER-negative tumors. Similarly, a stronger association was observed for both rs10736303 and rs2981578 with PR-positive tumors [ORs = 1.28 (95% CI 1.03–1.58) and 1.30 (95% CI 1.05–1.61), respectively] versus PR-negative tumors [ORs = 1.02 (95% CI 0.82–1.28) and 1.04 (95% CI 0.87–1.24)], but again these differences were not significant.
Table 3.
Per-allele odds ratios (ORs) and 95% confidence intervals (CIs) for the association between FGFR2 SNPs rs10736303 and rs2981578 and breast cancer risk by tumor receptor status in African American women
| SNP | Unadjusted analysisa |
Adjusted analysisb |
||||||
|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P-value | OR | 95% CI | P-value | |||
| ER-positive cases versus controls | rs10736303 | 1.24 | 1.02–1.51 | 0.028 | 1.23 | 1.00–1.52 | 0.050 | |
| rs2981578 | 1.26 | 1.03–1.52 | 0.021 | 1.23 | 1.00–1.52 | 0.049 | ||
| ER-negative cases versus controls | rs10736303 | 1.04 | 0.83–0.31 | 0.72 | 1.12 | 0.87–1.44 | 0.37 | |
| rs2981578 | 1.07 | 0.85–1.35 | 0.54 | 1.14 | 0.89–1.46 | 0.31 | ||
| ER-positive cases versus ER-negative cases | rs10736303 | 1.23 | 0.96–1.59 | 0.11 | 1.19 | 0.90–1.58 | 0.22 | |
| rs2981578 | 1.21 | 0.94–1.55 | 0.13 | 1.18 | 0.89–1.56 | 0.25 | ||
| PR-positive cases versus controls | rs10736303 | 1.28 | 1.03–1.58 | 0.025 | 1.28 | 1.02–1.61 | 0.037 | |
| rs2981578 | 1.30 | 1.05–1.61 | 0.016 | 1.28 | 1.02–1.62 | 0.032 | ||
| PR-negative cases versus controls | rs10736303 | 1.02 | 0.82–1.28 | 0.85 | 1.10 | 0.86–1.41 | 0.47 | |
| rs2981578 | 1.03 | 0.83–1.29 | 0.77 | 1.10 | 0.86–1.41 | 0.45 | ||
| PR-positive cases versus PR-negative cases | rs10736303 | 1.20 | 0.93–1.57 | 0.17 | 1.10 | 0.83–1.47 | 0.51 | |
| rs2981578 | 1.21 | 0.94–1.57 | 0.15 | 1.11 | 0.83–1.48 | 0.48 | ||
aStratified by study center.
bAdjusted for age at diagnosis and family history of breast cancer.
Four of the five SNPs were genotyped in all four African American studies (rs10736303, rs2981578, rs35054928 and rs2981575). Haplotype analysis based on these SNPs identified three haplotypes significantly associated with increased risk of BC, including one unique to African Americans (Table 4). All three haplotypes contained the risk alleles of the two associated SNPs rs10736303 and rs2981578. A fourth rare haplotype also carrying these two risk alleles was not significantly associated with risk, but the 95% CI included the ORs of the other risk haplotypes. The three risk haplotypes were associated with risk in all three populations, although only one achieved statistical significance in Europeans.
Table 4.
Haplotype analysis of four candidate SNPs genotyped by TaqMan® in African American, UK and Korean studies
| Haplotype | Frequencya | ORb | 95% CI | P-value |
|---|---|---|---|---|
| African Americans | ||||
| h0000 | 0.15 | 1 | ||
| h1111 | 0.54 | 1.22 | 1.03–1.46 | 0.025 |
| h1100 | 0.21 | 1.26 | 1.02–1.54 | 0.032 |
| h1110 | 0.07 | 1.15 | 0.18–7.17 | 0.350 |
| h1101 | 0.03 | 1.97 | 1.26–3.09 | 0.003 |
| UK | ||||
| h0000 | 0.50 | 1 | ||
| h1111 | 0.41 | 1.25 | 1.18–1.31 | 1.14 × 10−17 |
| h1100 | 0.06 | 1.08 | 0.97–1.21 | 0.14 |
| h1110 | 0.02 | 1.08 | 0.88–1.31 | 0.47 |
| Koreans | ||||
| h0000 | 0.44 | 1 | ||
| h1111 | 0.40 | 1.22 | 1.10–1.36 | 1.89 × 10−4 |
| h1100 | 0.09 | 1.24 | 1.03–1.49 | 0.02 |
| h1110 | 0.03 | 1.51 | 1.07–2.14 | 0.02 |
| SNP order | base 0 | base 1 (risk allele) | ||
| rs10736303 | A | G | ||
| rs2981578 | A | G | ||
| rs35054928 | — | C | ||
| rs2981575 | A | G | ||
aHaplotype frequency of cases and controls combined.
bAdjusted for study centre, age and family history.
Evaluation of the candidate causative variants utilizing data from multiple populations
In the previous fine-scale mapping analysis (1), 27 of the 29 variants were evaluated in European and Asian case–control studies (Supplementary Material, Table S2a). The other two variants (rs35393331 and rs33971856) could not be genotyped by high-throughput methods and so were not considered further in that study nor this one; thus they remain on the list of candidate-causal variants. Following the previous analysis, 21 SNPs could be excluded as causal variants based on likelihood ratios <1/100 relative to the most significantly associated SNP, rs7895676; likelihood ratios were <1/1000 for 19 of the 21 SNPs (1). One SNP, rs2981575, was only marginally excluded by this analysis (likelihood ratio = 1/224) and was one of the most significant variants in Europeans. This SNP together with the six remaining candidate SNPs (rs7895676, rs10736303, rs2912781, rs2912778, rs2981578 and rs35054928) were evaluated further in this study as seven candidate-causal SNPs. Additionally, we included the GWAS marker-SNP rs2981582 for reference.
The SNPs amenable to TaqMan® analyses (rs10736303, rs2981578, rs35054928, rs2981575 and rs2981582) were genotyped in the African American studies (as described previously) together with additional European and Asian cases and controls (Table 5). The genotypes of an additional three SNPs which were not amenable to high-throughput genotyping methods (rs7895676, rs2912781 and rs2912778) were determined in 95 samples from the CARE-AA study by bi-directional sequencing. These data were used to impute genotypes for all African American women (see Materials and Methods).
Table 5.
Summary of additional European and Asian data generated for fine-scale mapping
| Study | Abbreviation | Number of subjects |
SNPs genotyped | |
|---|---|---|---|---|
| Invasive cases | Controls | |||
| Studies of Epidemiology and Risk Factors in Cancer Heredity Sets 1 and 2 | SEARCH | 4470 | 4560 | rs35054928 |
| SEARCH set 3 | 2301 | 2280 | rs10736303 | |
| rs2981578 | ||||
| rs35054928 | ||||
| Seoul Breast Cancer Project | SBCP | 2442 | 1780 | rs35054928 |
Haplotype frequencies were calculated for the seven candidate-causal SNPs (Table 6). One haplotype not present in Europeans and Asians at frequency ≥1% was identified in African Americans. As expected, the correlations between the SNPs in African Americans were weaker than those observed in Asians or Europeans (Supplementary Material, Table S4).
Table 6.
Estimated-haplotype frequencies in 80 UK, 84 Korean, 84 CARE African American samples, for seven candidate-SNPs in FGFR2
| Haplotypea | rs7895676 | rs10736303 | rs2912781 | rs2912778 | rs2981578 | rs35054928 | rs2981575 | UKb | Koreansc | AAd |
|---|---|---|---|---|---|---|---|---|---|---|
| h1111111 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0.49 | 0.39 | 0.60 |
| h0000000 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.43 | 0.39 | 0.16 |
| h1111100 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0.06 | 0.10 | 0.17 |
| h1111110 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | n/a | 0.03 | 0.04 |
| h0010000 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | n/a | 0.02 | 0.04 |
| h0001100 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | n/a | 0.02 | n/a |
| h0110100 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | n/a | 0.02 | n/a |
| h0111101 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | n/a | n/a | 0.02 |
| SNP order | base 0 | base 1 (risk allele) | ||||||||
| rs7895676 | T | C | ||||||||
| rs10736303 | A | G | ||||||||
| rs2912781 | A | G | ||||||||
| rs2912778 | T | C | ||||||||
| rs2981578 | A | G | ||||||||
| rs35054928 | — | C | ||||||||
| rs2981575 | A | G |
aAllele 0 for all SNPs is allele in LD with commoner allele G in rs2981582.
bSEARCH study population.
cSeoul Breast Cancer Project.
dCARE-AA.
Using these new data in combination with the existing European and Asian data, likelihoods were calculated for each variant, based on the assumption that there is one single disease-causing allele. The most strongly associated SNP was rs2981578, and likelihood ratios were calculated for the other SNPs relative to it (Table 7). As in the previous analysis in European and Asian women (1), rs2981582, the GWAS marker-SNP included in this study for reference purposes, could be definitively excluded as the causal variant at a likelihood ratio of <1/100 relative to rs2981578. Of the seven candidate-causal SNPs, rs35054928 had a likelihood ratio of <1/100 relative to rs2981578, indicating that it was unlikely to be the causal variant. SNP rs2981575, however, which had been tentatively excluded (likelihood ratio of 1/224) by the previous analysis (1), had a likelihood ratio of 1/30 in this extended analysis and could no longer be excluded at this level. The other five SNPs, including rs7895676 which was the most strongly associated SNP in the previous analysis, were too highly correlated with each other in the three populations and could not be excluded from being the causative variant (based on a likelihood ratio of >1/100 relative to rs2981578).
Table 7.
Likelihoods ratios of breast cancer risk for candidate SNPs compared with most significant SNP rs2981578 by race and overall
| Log10 (likelihood ratio) relative to rs2981578 |
|||||||
|---|---|---|---|---|---|---|---|
| SNP | Chr10 position | Allelesa | Europeansb | Asiansc | AAsd | Total | Likelihood ratio SNP/rs2981578 |
| rs7895676 | 123323987 | T/C | 0.68 | −0.54 | 0.43 | 0.57 | 1/3.7 |
| rs10736303 | 123324447 | A/G | 1.54 | 0.43 | −0.01 | 1.95 | 1/89.1 |
| rs2912781 | 123326484 | A/G | 0.66 | 0.96 | −0.23 | 1.39 | 1/24.5 |
| rs2912778 | 123328644 | T/C | 1.33 | −1.00 | −0.26 | 0.07 | 1/1.2 |
| rs2981578 | 123330301 | A/G | 0.00 | 0.00 | 0.00 | 0.00 | 1/1.0 |
| rs35054928 | 123330423 | −/C | −1.37 | 2.75 | 0.94 | 2.32 | 1/208.9 |
| rs2981575 | 123336106 | A/G | −2.17 | 3.29 | 0.36 | 1.48 | 1/30.2 |
| rs2981582 | 123342307 | G/A | −1.21 | 5.22 | 0.58 | 4.59 | 1/38904.5 |
aAllele in LD with allele G in rs2981582 listed first.
bSEARCH sets 1, 2, 3 and MEC-W studies.
cMEC-J, TBCS, SBCP, TWBCS studies.
dAfrican Americans: CARE-AA, MEC-AA, CARE-LA, LIFE.
DNase I hypersensitivity of intron 2 of the FGFR2 gene
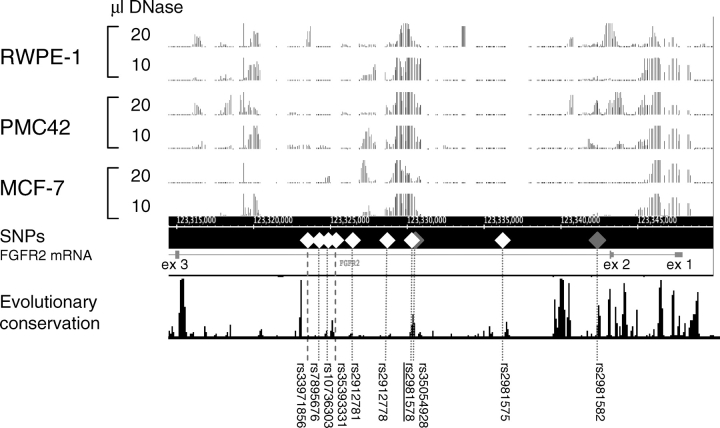
Functional assays may offer additional insight about SNPs that are too strongly correlated to be distinguished by epidemiological approaches. We surmised that any SNP functioning in gene regulation would lie in a region of the chromatin that is accessible to regulatory factors. By probing the chromatin with DNase I, we were able to identify regions of the chromatin that are in an open configuration without having to make assumptions about the nature of the proteins involved (17,18). To this end, we used two BC cell lines PMC42 and MCF-7 (both expressing the ER) and the prostate cell line RWPE-1, and generated DNase I hypersensitivity profiles for the region of interest using two different concentrations of DNase I (Fig. 2). As expected, we found the FGFR2 promoter to be in an open configuration, serving as a positive control in our experiment. Of the remaining possible eight causative SNPs, only two SNPs mapped to a region of the FGFR2 intron that is highly accessible in breast cell lines, including the most highly associated SNP rs2981578. SNP rs35393331 lies in a 1 kb region of repetitive sequence that is not covered by probes on our microarray, and its status is therefore undetermined. The current analysis suggests that the remaining SNPs lie in predominantly closed chromatin. Interestingly, the two breast cell lines as well as the prostate cell lines display very similar regions of open chromatin, with the most open stretch overlapping regions of sequence conservation, further strengthening the supposition that this region contains regulatory elements.
Figure 2.
DNase I hypersensitivity profiles of the genomic region of FGFR2 covering exons 1–3. Profiles for the two breast (PMC42 and MCF-7) and one prostate (RWPE-1) cell line were generated using two different concentrations of DNase I. The ACME (45) algorithm was used to generate the plots with a 95% cut-off and a sliding window of 500 bp. Grey diamonds indicate SNPs that were excluded on the basis of the likelihood ratio test. Two SNPs that could not be genotyped are indicated by the thick dotted line. Mammalian conservation of 17 species was taken from the UCSC genome browser (28).
DISCUSSION
SNPs in intron two of FGFR2 have been reported to be significantly associated with risk of BC in European (1,2), Asian (1,19), Ashkenazi Jewish (20) and Israeli populations (21). This present study extends this association to women of African American ethnicity. Of the SNPs genotyped in all African American subjects, SNPs rs10736303 and rs2981578 showed the most significant associations, and their estimated per-allele ORs were similar to those found in European and Asian subjects (1). A recent meta-analysis found that SNP rs2981582 was more strongly associated with ER-positive than ER-negative tumors and PR-positive than PR-negative tumors (22). A similar, although not significant, pattern was observed in the African American studies presented here. These results suggest strongly that the same causal variant(s) are present in all three populations and confer similar risks of BC in all populations.
Women from Africa and Eastern Asia have lower age-adjusted world incidence rates of BC (<40 per 100 000 in most countries) than non-Hispanic White women in USA (98.5 per 100 000 woman). The age-adjusted world incidence rate for black women in USA (81.7 per 100 000 women) is, however, closer to that of US non-Hispanic Whites than Africans (23). The frequencies of the risk-association alleles in FGFR2 are similar in European and Asian populations, but substantially higher in African Americans. On the basis of these frequencies, the BC risk in African Americans would be predicted to be 16% higher than in Europeans. The fact that risk is lower is consistent with the extensive evidence that the international variation in the risk of BC is driven mainly by different lifestyle and environmental exposures rather than differing genetic factors (24–26).
African Americans represent a population of mixed ethnicity, and it has not been possible to assess the ancestral composition of study subjects. However, the allele frequencies and risk estimates were similar in the four component studies, suggesting that the risk estimates should be broadly applicable to all African American women.
The confirmed disease association combined with the pattern of LD within the African American population indicates that African American case–control studies may help eliminate variants which cannot be eliminated in Europeans due to strong SNP correlation. We were indeed able to eliminate one candidate FGFR2 SNP, rs35054928, by combining African American case–control data with data from European and Asian studies. SNP rs2981575 could not be excluded in the present analysis; however, assuming it is not the causal variant, it should be possible to exclude this SNP with larger studies in African or African American women, as it is weakly correlated with the other candidate SNPs. The remaining five candidate-causal SNPs (rs7895676, rs10736303, rs2912781, rs2912778, rs2981578), however, are strongly correlated (all r2 > 0.81) in all populations examined so far. It will therefore be difficult (and perhaps impossible) to separate their effects using epidemiological studies, even with much larger sample sizes. Additionally, the two variants (rs35393331 and rs33971856) that could not be directly evaluated in any study remain candidate-causal variants, leaving in total eight candidate variants.
Given the location of the associated SNPs, in intron two of the gene, the most plausible hypothesis is that the association with risk is mediated through differential expression of FGFR2. Recent analyses by Meyer et al. indicate that the rare allele of the marker-SNP rs2981582 in Europeans is associated with increased FGFR2 expression in breast tissue. They also report that candidate SNP rs2981578, the most strongly associated SNP in the present analysis, alters binding of transcription factors Oct-1/Runx2. Furthermore, Meyer et al. (16) report that luciferase assays in transfected cell lines showed that the minor allele of rs2981578 increased transcription 2–5-fold over the common allele. A second SNP, rs7895676, was found to alter binding of the C/EBPβ transcription factor in vitro, on naked DNA. However, when assaying occupancy at this site in vivo using chromatin immunoprecipitation, only a very slight difference was seen between the two alleles.
On the assumption that susceptibility is mediated through binding of transcription factors, a causative variant would be expected to reside in a region of open chromatin. Of the eight remaining candidate variants, we have shown that only two (rs2912778 and rs2981578) lie in a region of highly accessible chromatin. Five SNPs lie in chromatin that is in a more closed conformation, whereas one SNP is in a region that is not covered by our probe set due to the repetitive nature of the surrounding sequence, and therefore cannot be assessed (rs35393331). The region of open chromatin includes SNP rs2981578, the most strongly associated variant in this present study, consistent with its association with increased transcription noted above. Conversely rs7895676, the other SNP suggested as a potentially important transcription factor binding site by Meyer et al., lies in a region of less open chromatin. Our current analyses, therefore, point to the region surrounding rs2981578 as the most critical. However, it is important to note that DNase hypersensitive sites are dynamic and can change in response to cell signaling or during development (27).
Evolutionary conservation of the sequence surrounding each SNP may also give an indication of the relative evolutionary importance of the eight remaining variants, all of which are non-coding. In a previous evaluation of the conservation across six placental mammalian species and the opossum, SNPs rs10736303 and rs2981578 were the only of the eight remaining variants conserved across all six placental mammals (1). Using the University of California Santa Cruz (UCSC) human genome browser (http://genome.ucsc.edu/), we also considered conservation scores from the alignment of 28 vertebrate species and the subset of 17 placental mammals (28). SNP rs2981578 (the most strongly associated variant in this present study) had a conservation score of >0.75 within 50 bp on either side using both species alignment measurements, whereas none of the other seven SNPs reached this score using either measurement. Thus for SNP rs2981578 conclusions from the genetic, biochemical and conservation analyses all converge, increasing the probability that this is a causal variant for BC.
It should be noted that the present analysis has focused on common SNPs with minor allele frequency >5%. The possibility remains that these SNPs also tag other types of variants (such as multi-allelic repeats) or rarer variants, which were not specifically targeted during the resequencing effort.
For the four SNPs that were not genotyped in all studies, the imputation of genotypes also introduces a source of uncertainty. To evaluate the accuracy of genotype imputation, we calculated the estimated r2 between the imputed and actual SNP (see Materials and Methods). In all three ethnic groups, all imputed SNPs had estimated r2 values >0.90 with the exception of two SNPs in Asians studies (rs2912781 with r2 = 0.84 and rs2912778 with r2 = 0.87). The imputation approach assumes that the reference panel of genotyped SNPs provides haplotype frequencies representative of those in the populations in which SNPs are being imputed. In our study, SNPs genotyped in the Seoul Breast Cancer Project were used as a reference panel for the imputation in the Asian studies, which could bias the results should the Korean haplotype frequencies differ greatly from the other Asian populations. In fact, we found that SNP allele frequencies were similar within this broad ethnic group, suggesting that the use of Korean haplotypes for the imputation was reasonable.
This fine-scale mapping work highlights the complexity involved in pinpointing causal variants within loci identified by GWAS, and the utility of combining genetic mapping with data on sequence context, biochemical and functional studies. Although the identification of causative variants, particularly in non-coding regions, poses a great challenge, such efforts are critical for furthering our understanding of the genetic architecture of complex genetic diseases.
MATERIALS AND METHODS
Study populations
African American study subjects were participants in four population-based studies. The first study, abbreviated CARE-AA, includes African American participants from a case–control study conducted in five metropolitan areas of USA (Atlanta, Detroit, Los Angeles, Philadelphia and Seattle) as part of the National Institute of Child Health and Human Development's Women's Contraceptive and Reproductive Experiences (CARE) Study (29,30). Briefly, cases aged 35–64 years and newly diagnosed with invasive BC between July 1994 and April 1998 were ascertained by the Surveillance, Epidemiology and End Results (SEER) population-based cancer registries at four sites and by field staff monitoring hospital records at the fifth site. Both younger cases and African American cases were intentionally oversampled. Controls were identified through centralized random-digit dialing and matched on study center location, race and age group. An in-person interview on BC risk factors was completed by 4575 cases (76.5% of those eligible) and 4682 controls (78.6% of those eligible). Funding allowances permitted the collection of blood from 33% of interviewed women. Hence, blood collection was requested from all cases and controls with a first-degree family history of BC, plus a random sample of those without a first-degree family history. Of those asked, 1644 (80.2%) cases and 1451 (74.3%) controls provided blood. African American individuals represented 29.7% of cases and 31.6% of controls. A total of 435 African American cases and 452 African American controls age 35–64 years were available for the present analysis.
The second study included the African American participants in Los Angeles (CARE-LA) component of the CARE (29). As is described below, only individuals whose blood samples were not collected in the CARE-AA study were included in these analyses to ensure no duplicate observations. Participation rates for the CARE-LA cases and controls were >70%. Blood specimens were collected from 82% of cases (n = 397) and 80% of controls (n = 236).
Only cases were available from the third study, the LIFE Los Angeles-based case–control study (31). This study was comprised incident invasive BC cases age 20–49 years diagnosed between 2000 and 2003 identified from Los Angeles SEER registry. The participation rate for African American cases in LIFE was 60%. Further details about the CARE-LA and LIFE studies have been previously described (31–33). On the basis of shared geography and similarity in age of participants as well as allele frequencies of genotyped FGFR2 SNPs, individuals from these studies were combined into one stratum [as has previously been done i.e. (33)].
The fourth study, abbreviated MEC-AA, included African American cases and controls primarily from the Multiethnic Cohort Study (MEC) (34). Blood sample collection began in 1994, with participation rates ≥65% for cases and controls. Incident BC cases were identified by cohort linkage to SEER registries and controls were a randomly selected sample of MEC participants frequency-matched on 5-year age group. The present analysis includes 439 invasive BC cases aged 45–83 years diagnosed up to December 2004 and 654 controls free of cancer up to December 2004.
All cases and controls from Los Angeles were assessed to ensure no overlap. A common identification number was used to identify 237 pairs of duplicate samples in CARE-AA and CARE-LA; these were removed from CARE-LA in the analysis. CARE-AA maintained records identifying any participants in the MEC-AA study by comparing the information on name, birth date and address while the studies were in the field; four pairs in CARE-AA that overlapped with MEC-AA were removed from the MEC-AA study.
The two populations of European ethnicity and four of Asian ethnicity that were genotyped for the fine-scale mapping work have been described previously (1). Briefly, SEARCH is a population-based study of cancer in East Anglia involving a three-stage design with a total of 6771 cases of invasive BC and 6840 controls (35). Within the Multiethnic Cohort, described above, the White and Japanese populations were included in this study with 425/435 White cases/controls and 448/394 Japanese cases/controls (34). Finally, three Asian studies, the Seoul Breast Cancer Project (36), the Taiwan Breast Cancer Study (37,38) and the IARC Thai Breast cancer study (39) are each hospital-based case–control studies with 2442/1780, 921/940 and 474/390 cases/controls, respectively. All studies had the relevant institutional review board approval.
Genotyping
SNP genotyping was conducted at individual study centers with the exceptions of Seoul Breast Cancer Project study samples, which were genotyped at Strangeways Research Laboratory in Cambridge, UK and the CARE-AA samples, which were genotyped at Ostrander Laboratory, NHGRI, Bethesda, MD, USA. Genotyping of five SNPs (rs10736303, rs2981578, rs35054928, rs2981575, rs2981582) was carried out using a fluorescent 5' exonuclease assay (TaqMan®) and the ABI PRISM 7900 Sequence Detection Sequence (Applied Biosystems, Foster City, CA, USA) implemented in 384-well format for all the African American studies. The single exception was rs2981582, which was not genotyped in CARE-AA. Each 384-well plate included duplicate samples which demonstrated average concordance >98%. Call rates for each assay were over 95%. Assays were not repeated for failed genotype calls. As in the African American studies, for all other studies the average replication rate for assays was >98%, and frequencies of the SNP genotypes did not deviate from those expected under Hardy–Weinberg equilibrium (P > 0.01).
Three SNPs those were not amenable to the TaqMan® assay (rs7895676, rs2912781 and rs2912778) were genotyped by direct sequencing of the SNP and surrounding region. One 96-well plate of CARE-AA samples was sequenced bidirectionally. PCRs were performed in a 10 µl volume containing 5 ng of genomic DNA, 1 mm dNTPs, 1.5 or 2.5 mm MgCl2, 1 µm of both the forward and the reverse primers, and 0.25 U Amplitaq DNA polymerase (Applied Biosystems). PCR products were treated using the ExoSAP-IT method (USB Corporation, Cleveland, OH, USA). DNA-sequencing reactions were carried out using BigDye Terminator v3.1 Cycle Sequencing Kits (Applied Biosystems) and sequence data were obtained on both forward and reverse PCR primers using the ABI 3730xl Genetic Analyzer (Applied Biosystems). Contigs were assembled using PhredPhrap/Consed (40). Variants were detected using Polyphred (41).
Statistical analysis
Each of the five SNPs genotyped by TaqMan® was assessed for association with disease status using the Cochran-Armitage 1 d.f. score test. Per-allele ORs and CIs were estimated by logistic regression stratified by study and CARE-AA site using Intercooled Stata v9. Standard tests of homogeneity of ORs for SNP genotypes across the study strata were computed. Significant SNPs (Ptrend < 0.05) were further analyzed adjusting for age at diagnosis (categorized as: <40, 40–44, 45–49, 50–54, 55–59, 60–64, 65–69, 70–74, >75 years) and family history of BC (categorized as at least one or no first-degree relatives with BC). Family history was adjusted for because the estimated ORs would be expected to be higher in families with a history of BC (1). Age was adjusted for because ORs might vary by age, and different study sets had different selection criteria. To assess potential differences in genetic effects associated with receptor status (ER and PR), separate logistic regression models were used to compare receptor positive cases to controls, receptor-negative cases to controls and receptor-positive cases to receptor-negative cases.
Sampling weights were developed for the CARE-AA data to account for the non-random selection of women from the study population, as described in greater detail elsewhere (30). When these sampling weights were applied to the FGFR2 SNP data, the OR estimates were very similar to those generated using an unweighted analysis (Supplementary Material, Table S1). For simplicity and consistency with the fine-scale mapping analyses, unweighted analyses are presented in the main text.
For the fine-scale mapping analyses, we first estimated haplotype frequencies using the 95 African American individuals for whom all candidate SNPs were typed, using the haplo.stats package in S-plus (42). The haplotype frequencies were used to impute genotype probabilities for each SNP for each individual. Missed calls for TaqMan® genotyped SNPs (rs10736303, rs2981578, rs35054928, rs2981575) were imputed, as were missing data for SNPs genotyped in a subset of the African American subjects (rs7895676, rs2912781, rs2912778, rs2981582). An expectation-maximization algorithm was then used to fit a logistic regression model, allowing uncertainty in the genotypes of the untyped SNPs and assuming that each SNP in turn was the causal variant. By this method, we were able to calculate the likelihood that each SNP was the causal variant (1). Likewise, in Europeans, missed calls were imputed for SNPs genotyped by all studies (rs10736303, rs2981578, rs2981575, rs2981582) as were missing data for SNPs genotyped in a subset of European subjects (rs7895676, rs2912781, rs2912778, rs35054928). In the Asian studies, missed calls were imputed for SNPs (rs10736303, rs11200014, rs2912780, rs2981578, rs4752571, rs2981575, rs1219642, rs2912774, rs2936870, rs2420946, rs2981582, rs3135718) and for SNPs genotyped in a subset of individuals (rs7895676, rs2912781, rs2912778, rs35054928) (Supplementary Material, Table S2b).
To give an estimate of the accuracy of the imputation, we calculated the estimated r2 between the imputed and actual SNP (43), which we estimated by:
where var(X) is the sample variance of the imputed genotypes and var(Y) is the variance of the true genotypes, estimated from the overall genotype frequencies.
DNase I hypersensitivity mapping
The BC cell lines MCF-7 and PMC42 [Cambridge Research Institute (CRI) culture collection] were maintained in RPMI with 10% fetal calf serum. RWPE-1 (ATCC) was cultured in Keratinocyte-Serum Free Medium (Gibco) supplemented with 5 ng/ml human recombinant EGF and 0.05 mg/ml bovine pituitary extract from Sigma Aldrich, UK. Cells were harvested while in the exponential growth phase. DNase I hypersensitivity experiments were carried out as described in Follows et al. (44), except that 1 × 106 nuclei were harvested per experiment using a microcentrifuge at full speed for 20 s and RNase treatment was at 55°C for 1 h. Three picomole of annealed primer were ligated to 3 µg of genomic DNA, and the subsequent amplification reaction with biotin-labeled primers was carried out as described except that dNTPs were at 200 µm. A library of enriched fragments was generated from a single PCR and purified on streptavidin coated magnetic beads. Enrichment of open chromatin was ascertained by RT-PCR using probes in the MYC and HMBS locus, before the library was hybridized to an Agilent 4 x44K custom array, where the FGFR2 gene was tiled using the Agilent High Density probe set (May 2007). The libraries were labeled with the Bioprime Array CGH (Invitrogen) labeling kits, using Cy3-dUTP (Enzo Life Sciences, Inc.) for the library and Cy5-dUTP (Enzo Life Sciences, Inc.) for the input genomic DNA in each experiment. A minimum of 1.5 µg of probe were hybridized to each array by the CRI Genomics Core facility according to manufacturer's instructions. The array data were analyzed by ACME (45), using a 95% cut-off and a sliding window size of 500 bp by the CRI Computational Biology group. The data were visualized using the Affymetrix Integrated Genome Browser.
NOTE ADDED IN PROOF
Rebbeck et al. (46) recently tested SNP rs2981582 in a case-control study including 584 African American and 1225 European American Women. They found no evidence of association with breast cancer risk in African American women, however, none of the candidate causal variants were tested in this study. In European Americans, they reported significant associations in the subgroups of ER+, PR+ and HER− breast tumors.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
The genotyping and analysis of this study, and the conduct of the SEARCH study, was funded by Cancer Research UK. DFE is a Principal Research Fellow of CR-UK and PDPP is a CR-UK Senior Clinical Research Fellow. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute and National Human Genome Research Institute, US Department of Health and Human Services. M.U. is supported by the NIH-Oxford/Cambridge PhD program. The Multiethnic Cohort Study was supported by the US National Cancer Institute [CA 54281, CA 63464]. The CARE study was supported by the National Institute of Child Health and Human Development, with additional support from the National Cancer Institute, through contracts with Emory University [N01 HD 3-3168], Fred Hutchinson Cancer Research Center [N01 HD 2-3166], Karmanos Cancer Institute at Wayne State University [N01 HD 3-3174], University of Pennsylvania [N01 HD 3-3176], University of Southern California [N01 HD 3-3175], and through an intra-agency agreement with the Centers for Disease Control and Prevention [Y01 HD 7022]. General support through SEER contracts [N01-PC-67006 (Atlanta), N01-CN-65064 (Detroit), N01-CN-67010 (Los Angeles), and N01-CN-0532 (Seattle)] are also acknowledged. BAJP is Li Ka Shing Professor of Oncology and we acknowledge Hutchison Whampoa Limited.
ACKNOWLEDGEMENTS
The authors thank the women who participated in this research.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Easton D.F., Pooley K.A., Dunning A.M., Pharoah P.D., Thompson D., Ballinger D.G., Struewing J.P., Morrison J., Field H., Luben R., et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunter D.J., Kraft P., Jacobs K.B., Cox D.G., Yeager M., Hankinson S.E., Wacholder S., Wang Z., Welch R., Hutchinson A., et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adnane J., Gaudray P., Dionne C.A., Crumley G., Jaye M., Schlessinger J., Jeanteur P., Birnbaum D., Theillet C. BEK and FLG, two receptors to members of the FGF family, are amplified in subsets of human breast cancers. Oncogene. 1991;6:659–663. [PubMed] [Google Scholar]
- 4.Penault-Llorca F., Bertucci F., Adelaide J., Parc P., Coulier F., Jacquemier J., Birnbaum D., deLapeyriere O. Expression of FGF and FGF receptor genes in human breast cancer. Int. J. Cancer. 1995;61:170–176. doi: 10.1002/ijc.2910610205. [DOI] [PubMed] [Google Scholar]
- 5.Adelaide J., Finetti P., Bekhouche I., Repellini L., Geneix J., Sircoulomb F., Charafe-Jauffret E., Cervera N., Desplans J., Parzy D., et al. Integrated profiling of basal and luminal breast cancers. Cancer Res. 2007;67:11565–11575. doi: 10.1158/0008-5472.CAN-07-2536. [DOI] [PubMed] [Google Scholar]
- 6.Moffa A.B., Tannheimer S.L., Ethier S.P. Transforming potential of alternatively spliced variants of fibroblast growth factor receptor 2 in human mammary epithelial cells. Mol. Cancer Res. 2004;2:643–652. [PubMed] [Google Scholar]
- 7.Theodorou V., Kimm M.A., Boer M., Wessels L., Theelen W., Jonkers J., Hilkens J. MMTV insertional mutagenesis identifies genes, gene families and pathways involved in mammary cancer. Nat. Genet. 2007;39:759–769. doi: 10.1038/ng2034. [DOI] [PubMed] [Google Scholar]
- 8.Lu P., Ewald A.J., Martin G.R., Werb Z. Genetic mosaic analysis reveals FGF receptor 2 function in terminal end buds during mammary gland branching morphogenesis. Dev. Biol. 2008;321:77–87. doi: 10.1016/j.ydbio.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jang J.H., Shin K.H., Park J.G. Mutations in fibroblast growth factor receptor 2 and fibroblast growth factor receptor 3 genes associated with human gastric and colorectal cancers. Cancer Res. 2001;61:3541–3543. [PubMed] [Google Scholar]
- 10.Pollock P.M., Gartside M.G., Dejeza L.C., Powell M.A., Mallon M.A., Davies H., Mohammadi M., Futreal P.A., Stratton M.R., Trent J.M., et al. Frequent activating FGFR2 mutations in endometrial carcinomas parallel germline mutations associated with craniosynostosis and skeletal dysplasia syndromes. Oncogene. 2007;26:7158–7162. doi: 10.1038/sj.onc.1210529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katoh M. Cancer genomics and genetics of FGFR2 (Review) Int. J. Oncol. 2008;33:233–237. [PubMed] [Google Scholar]
- 12.Shifman S., Kuypers J., Kokoris M., Yakir B., Darvasi A. Linkage disequilibrium patterns of the human genome across populations. Hum. Mol. Genet. 2003;12:771–776. doi: 10.1093/hmg/ddg088. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez-Neira A., Calafell F., Navarro A., Lao O., Cann H., Comas D., Bertranpetit J. Geographic stratification of linkage disequilibrium: a worldwide population study in a region of chromosome 22. Hum. Genomics. 2004;1:399–409. doi: 10.1186/1479-7364-1-6-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans D.M., Cardon L.R. A comparison of linkage disequilibrium patterns and estimated population recombination rates across multiple populations. Am. J. Hum. Genet. 2005;76:681–687. doi: 10.1086/429274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sawyer S.L., Mukherjee N., Pakstis A.J., Feuk L., Kidd J.R., Brookes A.J., Kidd K.K. Linkage disequilibrium patterns vary substantially among populations. Eur. J. Hum. Genet. 2005;13:677–686. doi: 10.1038/sj.ejhg.5201368. [DOI] [PubMed] [Google Scholar]
- 16.Meyer K.B., Maia A.T., O'Reilly M., Teschendorff A.E., Chin S.F., Caldas C., Ponder B.A. Allele-specific up-regulation of FGFR2 increases susceptibility to breast cancer. PLoS Biol. 2008;6:e108. doi: 10.1371/journal.pbio.0060108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Birney E., Stamatoyannopoulos J.A., Dutta A., Guigo R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyle A.P., Davis S., Shulha H.P., Meltzer P., Margulies E.H., Weng Z., Furey T.S., Crawford G.E. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang J., Chen P., Hu Z., Zhou X., Chen L., Li M., Wang Y., Tang J., Wang H., Shen H. Genetic variants in fibroblast growth factor receptor 2 (FGFR2) contribute to susceptibility of breast cancer in Chinese women. Carcinogenesis. 2008;29:2341–2346. doi: 10.1093/carcin/bgn235. [DOI] [PubMed] [Google Scholar]
- 20.Gold B., Kirchhoff T., Stefanov S., Lautenberger J., Viale A., Garber J., Friedman E., Narod S., Olshen A.B., Gregersen P., et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc. Natl Acad. Sci. U S A. 2008;105:4340–4345. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raskin L., Pinchev M., Arad C., Lejbkowicz F., Tamir A., Rennert H.S., Rennert G., Gruber S.B. FGFR2 is a breast cancer susceptibility gene in Jewish and Arab Israeli populations. Cancer Epidemiol. Biomarkers Prev. 2008;17:1060–1065. doi: 10.1158/1055-9965.EPI-08-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Closas M., Hall P., Nevanlinna H., Pooley K., Morrison J., Richesson D.A., Bojesen S.E., Nordestgaard B.G., Axelsson C.K., Arias J.I., et al. Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genet. 2008;4:e1000054. doi: 10.1371/journal.pgen.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Curado M.P., Edwards B., Shin H.R., Storm H., Ferlay J., Heanue M., Boyle P. Cancer Incidence in Five Continents. IX. Lyon: IARC; 2007. IARC Scientific Publications No. 160. [Google Scholar]
- 24.Parkin D.M., Bray F., Ferlay J., Pisani P. Global cancer statistics, 2002. CA Cancer J. Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 25.Pike M.C., Kolonel L.N., Henderson B.E., Wilkens L.R., Hankin J.H., Feigelson H.S., Wan P.C., Stram D.O., Nomura A.M. Breast cancer in a multiethnic cohort in Hawaii and Los Angeles: risk factor-adjusted incidence in Japanese equals and in Hawaiians exceeds that in whites. Cancer Epidemiol. Biomarkers Prev. 2002;11:795–800. [PubMed] [Google Scholar]
- 26.Breast cancer and breastfeeding: collaborative reanalysis of individual data from 47 epidemiological studies in 30 countries, including 50302 women with breast cancer and 96973 women without the disease. Lancet. 2002;360:187–195. doi: 10.1016/S0140-6736(02)09454-0. [DOI] [PubMed] [Google Scholar]
- 27.Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van C.S., Qu C., Ching K.A., et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 28.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchbanks P.A., McDonald J.A., Wilson H.G., Burnett N.M., Daling J.R., Bernstein L., Malone K.E., Strom B.L., Norman S.A., Weiss L.K., et al. The NICHD Women's Contraceptive and Reproductive Experiences Study: methods and operational results. Ann. Epidemiol. 2002;12:213–221. doi: 10.1016/s1047-2797(01)00274-5. [DOI] [PubMed] [Google Scholar]
- 30.Malone K.E., Daling J.R., Doody D.R., Hsu L., Bernstein L., Coates R.J., Marchbanks P.A., Simon M.S., McDonald J.A., Norman S.A., et al. Prevalence and predictors of BRCA1 and BRCA2 mutations in a population-based study of breast cancer in white and black American women ages 35 to 64 years. Cancer Res. 2006;66:8297–8308. doi: 10.1158/0008-5472.CAN-06-0503. [DOI] [PubMed] [Google Scholar]
- 31.Ma H., Bernstein L., Ross R.K., Ursin G. Hormone-related risk factors for breast cancer in women under age 50 years by estrogen and progesterone receptor status: results from a case–control and a case–case comparison. Breast Cancer Res. 2006;8:R39. doi: 10.1186/bcr1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haiman C.A., Setiawan V.W., Xia L.Y., Le M.L., Ingles S.A., Ursin G., Press M.F., Bernstein L., John E.M., Henderson B.E. A variant in the cytochrome p450 oxidoreductase gene is associated with breast cancer risk in African Americans. Cancer Res. 2007;67:3565–3568. doi: 10.1158/0008-5472.CAN-06-4801. [DOI] [PubMed] [Google Scholar]
- 33.Setiawan V.W., Chu L.H., John E.M., Ding Y.C., Ingles S.A., Bernstein L., Press M.F., Ursin G., Haiman C.A., Neuhausen S.L. Mitochondrial DNA G10398A variant is not associated with breast cancer in African American women. Cancer Genet. Cytogenet. 2008;181:16–19. doi: 10.1016/j.cancergencyto.2007.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolonel L.N., Henderson B.E., Hankin J.H., Nomura A.M., Wilkens L.R., Pike M.C., Stram D.O., Monroe K.R., Earle M.E., Nagamine F.S. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am. J. Epidemiol. 2000;151:346–357. doi: 10.1093/oxfordjournals.aje.a010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lesueur F., Pharoah P.D., Laing S., Ahmed S., Jordan C., Smith P.L., Luben R., Wareham N.J., Easton D.F., Dunning A.M., et al. Allelic association of the human homologue of the mouse modifier Ptprj with breast cancer. Hum. Mol. Genet. 2005;14:2349–2356. doi: 10.1093/hmg/ddi237. [DOI] [PubMed] [Google Scholar]
- 36.Lee K.M., Choi J.Y., Park S.K., Chung H.W., Ahn B., Yoo K.Y., Han W., Noh D.Y., Ahn S.H., Kim H., et al. Genetic polymorphisms of ataxia telangiectasia mutated and breast cancer risk. Cancer Epidemiol. Biomarkers Prev. 2005;14:821–825. doi: 10.1158/1055-9965.EPI-04-0330. [DOI] [PubMed] [Google Scholar]
- 37.Lo Y.L., Yu J.C., Chen S.T., Yang H.C., Fann C.S., Mau Y.C., Shen C.Y. Breast cancer risk associated with genotypic polymorphism of the mitosis-regulating gene Aurora-A/STK15/BTAK. Int. J. Cancer. 2005;115:276–283. doi: 10.1002/ijc.20855. [DOI] [PubMed] [Google Scholar]
- 38.Cheng T.C., Chen S.T., Huang C.S., Fu Y.P., Yu J.C., Cheng C.W., Wu P.E., Shen C.Y. Breast cancer risk associated with genotype polymorphism of the catechol estrogen-metabolizing genes: a multigenic study on cancer susceptibility. Int. J. Cancer. 2005;113:345–353. doi: 10.1002/ijc.20630. [DOI] [PubMed] [Google Scholar]
- 39.Sangrajrang S., Schmezer P., Burkholder I., Boffetta P., Brennan P., Woelfelschneider A., Bartsch H., Wiangnon S., Cheisilpa A., Popanda O. The XRCC3 Thr241Met polymorphism and breast cancer risk: a case–control study in a Thai population. Biomarkers. 2007;12:523–532. doi: 10.1080/13547500701395602. [DOI] [PubMed] [Google Scholar]
- 40.Gordon D., Abajian C., Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 41.Nickerson D.A., Tobe V.O., Taylor S.L. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 1997;25:2745–2751. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schaid D.J., Rowland C.M., Tines D.E., Jacobson R.M., Poland G.A. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am. J. Hum. Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Bakker P.I., Ferreira M.A., Jia X., Neale B.M., Raychaudhuri S., Voight B.F. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Follows G.A., Janes M.E., Vallier L., Green A.R., Gottgens B. Real-time PCR mapping of DNaseI-hypersensitive sites using a novel ligation-mediated amplification technique. Nucleic Acids Res. 2007;35:e56. doi: 10.1093/nar/gkm108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scacheri P.C., Crawford G.E., Davis S. Statistics for ChIP-chip and DNase hypersensitivity experiments on NimbleGen arrays. Methods Enzymol. 2006;411:270–282. doi: 10.1016/S0076-6879(06)11014-9. [DOI] [PubMed] [Google Scholar]
- 46.Rebbeck T.R., DeMichele A., Tran T.V., Panossian S., Bunin G.R., Troxel A.B., Strom B.L. Hormone-dependent effects of FGFR2 and MAP3K1 in breast cancer susceptibility in a population-based sample of post-menopausal African-American and European-American women. Carcinogenesis. 2009;30:269–274. doi: 10.1093/carcin/bgn247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.