Abstract
The ability of the host to distinguish between self and foreign nucleic acids is one of the critical factors contributing to the recognition of pathogens by Toll-like receptors (TLRs). Under certain circumstances, eukaryotic self-RNA may reach TLR-containing compartments allowing for self-recognition. Specific modifications were previously demonstrated to suppress immune activation when placed at several positions in an immune stimulatory RNA or silencing RNA (siRNA). However, we show that even a simple natural modification such as a single 2′-O-methylation at different nucleotide positions throughout a sequence derived from a self-RNA strongly interferes with TLR-mediated effects. Such a single modification can even have an inhibitory effect in vitro and in vivo when placed in a different than the immune stimulatory RNA strand acting as suppressive RNA. Several safeguard mechanisms appear to have evolved to avoid cellular TLR-mediated activation by self-RNAs that may under other circumstances result in inflammatory or autoimmune responses. This knowledge can be used to include as few as a single 2′-O-methyl modification at a specific position in a siRNA sense or anti-sense strand to avoid TLR immune effects.
Keywords: methylation, oligoribonucleotide, RNA, siRNA, TLR7, TLR8
Introduction
The recognition of molecules expressed by specific pathogens is mediated by several families of pattern recognition receptors (PRRs). Toll-like receptors (TLRs) are one of the best characterized class of PRRs and belong to the IL-1-receptor (IL-1R) superfamily that recognizes pathogen-associated molecular patterns (1). Ligation of TLRs triggers innate immune responses to bacterial, viral or fungal threats. Natural pathogen-derived ligands have been identified for most of the 11 reported mammalian TLRs. Pathogen nucleic acids such as single-stranded, non-methylated, CpG-containing DNA of bacterial or viral origin have been found to activate TLR9 (2). Other TLRs sensing nucleic acids include TLR7 and TLR8 recognizing single-stranded viral RNA (3, 4), whereas double-stranded viral RNA was demonstrated to target TLR3 (5). In addition to the TLRs, cytoplasmic receptors such as retinoic acid-inducible gene-I (RIG-I) or the cellular RNA helicase melanoma differentiation-associated gene-5 (mda-5) participate in the recognition of pathogen double-stranded RNA (6–8). All these receptors have a cell type- and compartmental-specific distributions (9–11). Human TLR7 and TLR9 are expressed in the same sub-populations of immune cells, B cells and plasmacytoid dendritic cells (pDCs), whereas strongest expression of human TLR8 is found within monocytes. The cell type-specific expression usually results in specific cytokine profiles upon TLR7 (strong IFN-α production from pDC) or TLR8 [strong tumor necrosis factor (TNF)-α production from monocytes] activation. The combined responses of endolysosomal (TLR3, 7, 8 and 9) and cytoplasmic (RIG-1, mda-5) PRRs result in an efficient cell type and pathogen molecule-dependent defense mechanism of the host.
Eukaryotic RNAs are heavily modified, and ∼100 different post-transcriptionally modified nucleotides have been reported (12, 13). These encompass complex nucleotide modifications such as N6-methyladenosine or N7-methylguanosine as well as chemically more simple modifications such as 2′-O-methyl modifications. The most ancient 2′-O-methyl or pseudouridine modifications are widespread and can be found in many types of RNAs, in all kingdoms and probably in all species (13). Modifications are usually found in clusters in RNA that are functionally important (12–14), suggesting that the modifications are essential for the function of the RNA type. RNA modifications appear to be involved mainly in not only stabilizing RNA structures but also in processing of RNA as well as providing alternative hydrogen bonding capabilities (12–14). Although modifications can be found throughout different RNA types and species, differences in the number of modified nucleotides can be observed, either dependent on the presence of proteins that help to maintain specific structures as is the case for small nuclear RNA (snRNA) containing relatively few modifications or dependent on the organism with prokaryotic tRNAs bearing less modifications than eukaryotic tRNAs (15–17).
Several nucleotide modifications found in natural RNAs appear to interfere with their reported TLR-dependent immune modulatory effects (15). Long natural-modified RNAs are significantly less stimulatory than their unmodified counterparts. In addition, double-stranded silencing RNAs (siRNAs) that trigger immune responses via signaling through TLR7 and TLR8 lose their activity if they contain several 2′ modifications (18–20). Specific modifications can also affect TLR3 signaling responding to viral double-stranded RNA molecules (21). However, nucleotide modifications are usually found in clusters, leaving a significant number of RNA nucleotides in natural RNAs such as rRNAs unmodified (12). If specific modifications would as predicted help eukaryotic cells to differentiate between self and foreign RNA (15, 22), how may this be achieved for the non-modified self-RNA regions?
In this study, the impact of natural occurring modifications in eukaryotic U1 snRNA on its TLR-dependent immune effects was investigated. By introducing a single 2′-O-methyl modification in a given sequence contained in the U1 snRNA, we surprisingly found that immune modulation is decreased independent of the position of the modified nucleotide. Using a simplified oligoribonucleotide (ORN) sequence with a defined stimulatory GU-rich region, we further demonstrate that the 2′-O-methyl modification imposes its negative effect also when placed upstream or downstream of the immune modulatory region itself. Moreover, a simple 2′-O-methyl modification can transform a stimulatory into a suppressive RNA sequence, suggesting that modified RNA can act itself as an inhibitor of TLR7 and TLR8 immune responses. In summary, our data extend the understanding of how a eukaryotic organism discriminates between self and pathogen RNA to avoid inflammatory responses induced by endogenous self-RNA.
Methods
Reagents
ORN were obtained from BioSpring (Frankfurt, Germany) and were controlled for identity and purity by Coley Pharmaceutical GmbH and had undetectable endotoxin levels (<0.1 EU ml−1) measured by the Limulus assay (BioWhittaker, Verviers, Belgium). ORN were suspended in sterile endotoxin-free DNAse- and RNAse-free water (Life Technologies, Eggenstein, Germany) and were stored and handled under aseptic conditions to prevent contamination. Sequences are listed in Table 1. Chloroquine was obtained from Sigma (Deisenhofen, Germany) and 1,3-dioleoyloxy-3-(trimethylammonium)propane (DOTAP) from Roche (Mannheim, Germany).
Table 1.
ORN sequences used for the study
| ORN | Sequence 5′–3′ |
| HIV R-1075 | CCGUCUGUUGUGUGACUC |
| HIV 2′OMe R-1238 | CCGUCUGUUGUGUGACUC |
| HIV PO R-1688 | ccgucuguugugugacuc |
| snRNA Loop II R-1171 | GGCUUAUCCAUUGCACUCCGGA |
| Loop II 2′OMe A R-1336 | GGCUUAUCCAUUGCACUCCGG |
| snRNA 5′ R-1300 | GAUACUUACCUG |
| snRNA 5′ 2′OMe 5′ AU R-1301 | GAUACUUACCUG |
| snRNA 5′ 2′OMe 5′ U R-1334 | GAUACUUACCUG |
| snRNA 5′ 2′OMe 5′ A R-1335 | GAUACUUACCUG |
| snRNA 5′ 2′OMe centre U R-1634 | GAUACUUACCUG |
| snRNA 5′ 2′OMe 3′ A R-1635 | GAUACUUACCUG |
| snRNA 5′ 2′OMe 3′ U R-1636 | GAUACUUACCUG |
| snRNA PO 2′OMe 5′ U R-1683 | gauacuuaccug |
| R-1505 (parent) | CCGAGCCGAUUGUACC |
| 2′OMe first C R-1628 | CCGAGCCGAUUGUACC |
| 2′OMe first G R-1768 | CCGAGCCGAUUGUACC |
| 2′OMe second G R-1629 | CCGAGCCGAUUGUACC |
| 2′OMe second C R-1766 | CCGAGCCGAUUGUACC |
| 2′OMe third G R-1765 | CCGAGCCGAUUGUACC |
| 2′OMe first A R-1630 | CCGAGCCGAUUGUACC |
| 2′OMe first U R-1863 | CCGAGCCGAUUGUACC |
| 2′OMe second U R-1685 | CCGAGCCGAUUGUACC |
| 2′OMe fourth G R-1686 | CCGAGCCGAUUGUACC |
| 2′OMe third U R-1687 | CCGAGCCGAUUGUACC |
| 2′OMe second A R-1631 | CCGAGCCGAUUGUACC |
| 2′OMe third C R-1769 | CCGAGCCGAUUGUACC |
| 2′OMe fourth C R-1632 | CCGAGCCGAUUGUACC |
| R-1505 2′OMe UUGU R-1993 | CCGAGCCGAUUGUACC |
| R-1505 PO R-1874 | ccgagccgauuguacc |
| PO 2′OMe first U R-1864 | ccgagccgauuguacc |
| PO 2′OMe second U R-1865 | ccgagccgauuguacc |
| PO 2′OMe fourth G R-1866 | ccgagccgauuguacc |
| PO 2′OMe third U R-1867 | ccgagccgauuguacc |
| R-1507 (control) | CCGAGCCGAAGGCACC |
| R-1862 (R-1507 2′OMe 1st A) | CCGAGCCGAAGGCACC |
| R-1507 2′OMe first G R-1991 | CCGAGCCGAAGGCACC |
| R-1507 2′OMe first C R-1992 | CCGAGCCGAAGGCACC |
| R-1507 PO R-1873 | ccgagccgaaggcacc |
| Sendai R-1159 | UGUUUUUUCUCUUGUUUGGU |
| Sendai 2′OMe U R-2538 | UGUUUUUUCUCUUGUUUGGU |
| Sendai 2′OMe G R-2539 | UGUUUUUUCUCUUGUUUGGU |
| Influenza R-1160 | AUAAUUGACCUGCUUUCGCU |
| Influenza 2′OMe A R-2540 | AUAAUUGACCUGCUUUCGCU |
| Influenza 2′OMe U R-2541 | AUAAUUGACCUGCUUUCGCU |
| R-0006 | UGUUGUUGUUGUUGUUGUU |
ORN in upper case are completely phosphorothioate (PS), ORN in lower case have phosphodiester (PO) linkages. Underlined nucleotides are 2′-O-methylated.
PBMC isolation and purification of cell subsets
PBMC preparations from healthy male and female human donors were obtained from the Institute for Hemostaseology and Transfusion Medicine of the University of Düsseldorf (Germany). PBMCs were purified by centrifugation over Ficoll-Hypaque (Sigma). Purified PBMCs were washed twice with 1× PBS and re-suspended in RPMI 1640 culture medium supplemented with 5% (v/v) heat inactivated human AB serum (BioWhittaker) or 10% (v/v) heat inactivated FCS, 1.5 mM L-glutamine, 100 U ml−1 penicillin and 100 mg ml−1 streptomycin (all from Sigma).
Measurement of cytokine and chemokine production
Freshly isolated PBMCs were re-suspended at a concentration of 3 ×1 06 ml−1 to 5 × 106 ml−1 and added to 96-well round-bottomed plates (200 μl per well), which had previously received nothing or ORN complexed to DOTAP or DOTAP alone. Cells were cultured in a humidified incubator at 37°C for the indicated time points. Culture supernatants (SNs) were collected and, if not used immediately, were frozen at −20°C until required. Amounts of cytokines in the SNs were assessed using commercially available ELISA kits (IFN-γ or TNF-α, Diaclone; or IL-12p40, BD Pharmingen), an in-house ELISA (IFN-α) developed using commercially available antibodies (from BD Pharmingen or PBL, New Brunswick, NJ, USA, respectively), or the Luminex technology using a cytokine 25-Plex (BioSource, Camarillo, CA, USA).
Reporter assays
Human embryonic kidney cells (HEK293) containing a NFκB-luciferase reporter construct and expressing human TLR7 or TLR8 or without TLR expression were used as described before (23). Cells were plated on 96-well plates at 1.5 × 104 per well and allowed to attach overnight. Cells were subsequently incubated for 16 h with the indicated amount of ORN complexed to DOTAP (Roche) and then tested for luciferase expression. Each data point was done in duplicate.
In vitro murine assays
Naive sv129 mouse splenocytes were used for all in vitro assays. Animals were anesthetized with isofluorane and euthanized by cervical dislocation. Spleens were removed under aseptic conditions and placed in PBS + 0.2% BSA (Sigma, St Louis, MO, USA). Spleens were then homogenized and splenocytes were re-suspended in RPMI 1640 (Life Technologies) medium supplemented with 2% normal mouse serum (Cedarlane Laboratories, Ontario, Canada), 2 mM L-glutamine, penicillin–streptomycin solution (final concentration of 1000 U ml−1 and 1 mg ml−1, respectively) and 5 × 10−5 M β-mercaptoethanol (all from Sigma). Splenocytes pooled from the spleens of five mice were plated in 96-well round-bottomed plates (5 × 105 cells per well). Each splenocyte sample was plated in quadruplicate and the cells were incubated in a humidified 5% CO2 incubator at 37°C for 20 h. SNs were harvested and a commercially available assay kit for IL-6 or IL-12p40 (mouse OptEIA kit; PharMingen, Mississauga, Ontario, Canada) was used according to the manufacturer’s instructions to assay cytokine levels.
In vivo murine assays
Female Balb/c or sv129 mice (6–8 weeks of age) were used for the in vivo experiments; sv129 mice were purchased from Charles River Canada (Quebec, Canada), and mice were housed in micro isolators at the animal care facility of Pfizer Vaccine Ottawa (Kanata, Canada). All studies were conducted under approval of the institutional animal care committees and in accordance with the guidelines set forth by the Canadian Council on Animal Care. For post-injection experiments, phosphorothioate-modified suppressive S-Class ODN 2088 [5′-TCCTGGCGGGGAAGT-3′ (24)] was administered intra-peritoneally to Balb/c mice (n = 5 per group). After 1 h, animals were injected subcutaneously with ORN HIV formulated with DOTAP. Plasma was collected at 3 h post subcutaneous injections for ELISA assays. Suppressive ORN R-1862 or control ORN R-1507 were formulated with DOTAP and, for post-injection experiments, were administered intravenously to sv129 mice (n = 5 per group). After 1 h, animals were injected intravenously with ORN HIV formulated with DOTAP. Plasma was collected at 3 h post intravenous injections with ORN HIV for ELISA assays. For co-injection experiments, suppressive ORN R-1862 or control ORN R-1507 were formulated with DOTAP in the presence of ORN HIV. ORN were administered intravenously to sv129 mice (n = 5 per group) and plasma was collected at 3 h post intravenous injections for ELISA assays.
Results
Single 2′ methylation in self-RNA suppresses TLR-dependent effects
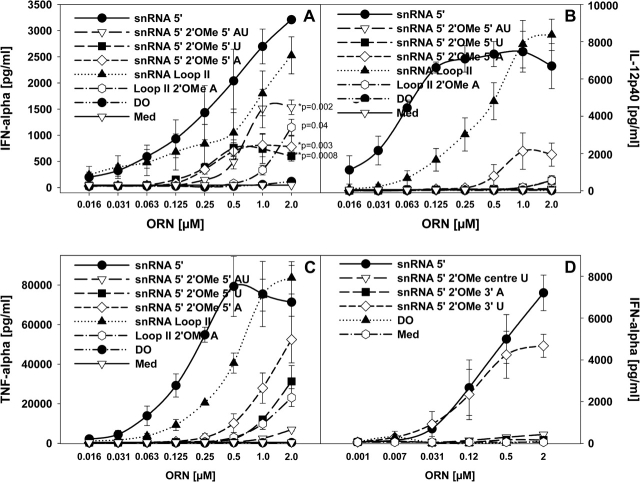
Self-RNAs such as U1 snRNA in immune complexes or upon enforced uptake activate human immune cells via TLR7 and TLR8 (25, 26). The cytokine response by the U1 snRNA is essentially dependent on the RNA itself as incubation with RNAse decreases IFN-α induction to the background levels (25). Three out of only five modified nucleotides in the U1 snRNA are 2′-O-methylated, two at the very 5′ end and one in stem loop II (Table 2) (27). To investigate a potential impact of these 2′ modifications on immune stimulation, we used short ORN with sequences of different lengths derived from the 5′ end or stem loop II of the U1 snRNA covering modified U1 snRNA regions and that were previously examined for their immune stimulatory effects (25). These sequences were either unmodified or modified at the ‘natural’ modified sites (see Tables 1 and 2). Compared with the unmodified ORN, all 2′-O-methylated ORN stimulated significantly lower IFN-α levels, and there was no considerable difference between ORN R-1301 (the 5′ U1 snRNA sequence with both A and U 2′-O-methylated) and ORN R-1334 (2′-O-methylated at the U) or R-1335 (2′-O-methylated at the A) each with only one modification (Fig. 1A). In contrast, the IL-12 and TNF-α responses showed more pronounced differences between the modified and unmodified ORN (Fig. 1B and C). These data cannot be explained by an impact of the 2′-O-methyl modification on the potential formation of higher ordered RNA structures known to be stabilized by 2′-O-methyl modifications, since none of the ORN formed secondary structures (data not shown).
Table 2.
U1 snRNA sequence and positions of nucleotide modifications
| 5′-m32,2,7GpppAmUmACΨΨACCUGGCAGGGGAGAUACCAUGAUCACGAAGGUGGUUUUCCCAGGGCGAGGCUUAUCCAUUGCAmCUCCGGAUGUGCUGACCCCUGCGAUUUCCCCAAAUGUGGGAAACUCGACUGCAUAAUUUGUGGUAGUGGGGGACUGCGUUCGCGCUUUCCCCUG-3′ |
Underlined are sequences used for oligonucleotide synthesis. Highlighted are U1 snRNA modifications: Am, Um: 2′OMe modification; Ψ: Pseudouridine.
Fig. 1.
Single 2′-O-methyl modifications in U1 snRNA sequences affect cytokine induction. (A–C) Human PBMCs were stimulated for 24 h with the indicated ORN complexed to DOTAP (DO) or DOTAP alone and cytokines measured. Significance of differences (in A) at 2 μM to the parent compounds snRNA 5′ and snRNA Loop II were determined by Student's t-test (*P < 0.01, **P < 0.001). (D) PBMCs were cultured with the indicated ORN as in (A–C) and IFN-α measured. Med, medium control. Mean ± SEM of one representative out of at least two independent experiments (n = 3 donors).
From these data, although demonstrating that a single 2′-O-methyl modification is sufficient to reduce the cytokine induction, it was unclear if a modification at any position in the ORN would have a suppressive effect. Therefore, additional ORNs with 2′-O-methylation at various other positions of the 5′ U1 snRNA sequence were tested for their IFN-α-inducing activity (Fig. 1D and Table 1). Although most modifications again interfered with cytokine induction, an ORN with a 3′ U modification (R-1636) showed only a minimal reduction of IFN-α stimulation. The 5′ snRNA ORN R-1300 contains Us at various positions throughout its sequence. The U and G content of an ORN determines its immune stimulatory activity (4, 25, 28), but from these data it appears as if either 2′-O-methylation affects immune stimulation in a position-dependent way or not all Us are equally important for mediating a cytokine response.
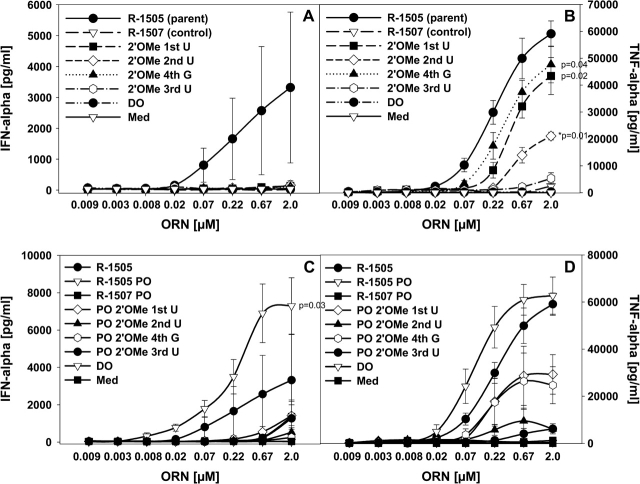
A single 2′-O-methyl modification inside or outside of an immune modulatory GU-rich RNA sequence suppresses RNA-mediated immune effects
To better understand a potential position dependency of the 2′ modification on immune activation, we used a non-stimulatory control ORN sequence (R-1507) and introduced a GU-rich 4mer sequence 5′-UUGU-3′ (in R-1505) that is sufficient to stimulate cytokine production from human PBMC (Table 1 and Fig. 2) (29). Whereas ORN R-1507 was inactive in all assays, ORN R-1505 showed immune stimulatory effects. Single 2′-O-methyl modifications at each position of the GU-rich 4mer sequence in the parent ORN R-1505 resulted in substantial loss of IFN-α stimulation (Fig. 2A), highlighting the importance of all positions of this 4mer sequence for IFN-α induction. In contrast, the effect on TNF-α production was less pronounced, but a clear difference in the importance of the nucleotide positions was observed (Fig. 2B). Whereas ORN with a 2′ modification at the first U and fourth G (positions 1 and 3 of the 4mer) of the parent ORN R-1505 still stimulated substantial although somewhat reduced TNF-α secretion, ORN modified at the second and third U (positions 2 and 4 of the 4mer) induced significantly lower cytokine levels. Similar results for IFN-α and TNF-α were obtained with PO ORN (Fig. 2C and D), with the PO form of R-1505 stimulating only somewhat higher cytokine levels compared with the PS form. These data suggest that at least with these defined RNA sequences, putative TLR8-mediated immune effects (e.g. TNF-α production) may be more forgiving to RNA 2′ modifications than TLR7-mediated effects (e.g. IFN-α production).
Fig. 2.
Effect of 2′ modifications in GU-rich regions on cytokine production. (A and B) Human PBMCs were stimulated for 24 h with the indicated ORN complexed to DOTAP (DO) or DOTAP alone and cytokines measured. (C and D) PBMCs were cultured with the indicated ORN as in (A and B) and cytokines measured. Med, medium control. Mean ± SEM of at least one experiment (n = 3 donors). Significance of differences (in A–C) at 2 μM to the parent compound R-1505 were determined for data points as indicated by Student’s t-test (*P < 0.01, **P < 0.001) (in A: P = 0.1).
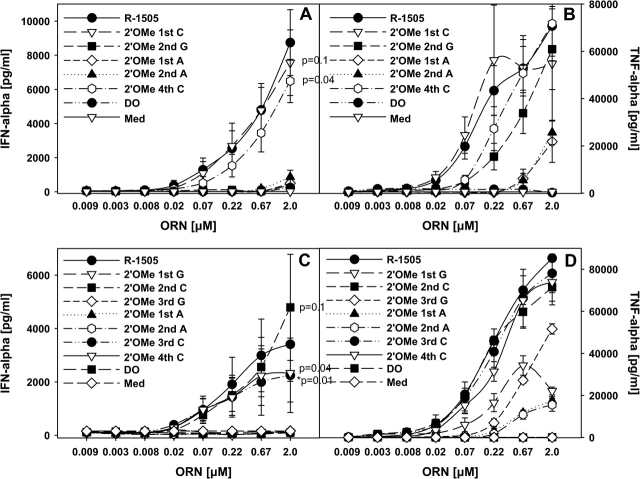
In addition to the modifications in the 4mer GU sequence, we tested ORN with single 2′ modifications outside of this sequence (Fig. 3A and B). Surprisingly, all but the ORN with C modifications induced background IFN-α stimulation. ORN with a 2′-O-methyl-C (R-1628 and R-1632) at different positions had similar not statistically significant IFN-α activity as the parent ORN. The same result was in principal observed for TNF-α induction (Fig. 3B), although the G modification still mediated substantial TNF-α secretion, whereas the two ORN with an A modification adjacent to the GU-rich 4mer sequence had strongly decreased effects.
Fig. 3.
Effect of 2′ modifications outside of GU-rich regions on cytokine production. (A and B) Human PBMCs were stimulated for 24 h with the indicated ORN complexed to DOTAP (DO) or DOTAP alone and cytokines measured. Significance of differences at 2 μM to the parent compound R-1505 were determined for data points as indicated by Student’s t-test (*P < 0.01, **P < 0.001). (C and D) PBMCs were cultured with the indicated ORN as in (A and B) and cytokines measured. Med, medium control. Mean ± SEM of at least one experiment (n = 3 donors).
To further investigate the obvious lack of a negative effect of 2′-O-methyl-C modifications on immune stimulation, we generated additional C-modified ORN derived from R-1505 (Table 1 and data not shown). As above, most C-modified ORN had activities not significantly different to the parent ORN, whereas modifications at any other position completely (IFN-α) or strongly (TNF-α) inhibited cytokine secretion (Fig. 3C and D).
Removal of the 2′-OH at the As adjacent to the 4mer GU-rich sequence by introducing deoxynucleotides in contrast to the introduction of 2′-O-methyl residues did not alter the immune modulatory effects, indicating that the 2′ residue itself is involved in the suppression (data not shown).
2′ methylation suppresses RNA-dependent immune responses even when not in the same immune stimulatory RNA sequence
Our data demonstrate that 2′ methylation interferes with RNA-mediated immune stimulation when placed in an immune stimulatory ORN. The possibility exists that 2′ modified RNA suppresses RNA immune activation even when co-incubated with a stimulatory unmodified ORN. Therefore, human PBMCs were stimulated with ORN R-1300 derived from the U1 snRNA and the same sequence with a 2′-O-methyl modification at the 5′ U (R-1334) was added. The modified ORN indeed suppressed IFN-α secretion stimulated by ORN R-1300 in a dose-dependent manner (Fig. 4A and data not shown) and the suppression was similar as observed with chloroquine. Similar results were obtained with the same modified sequence with a PO backbone, although the PS ORN was significantly more efficient most probably due to its enhanced nuclease stability (Fig. 4B).
Fig. 4.
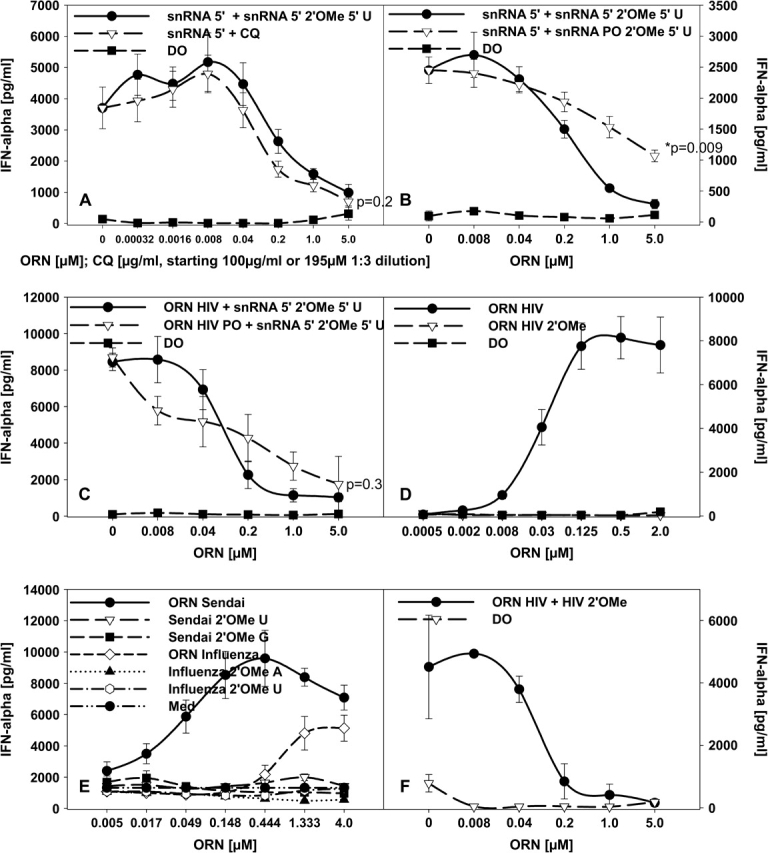
ORN with 2′-O-methyl modifications suppress cytokine induction stimulated by unmodified ORN. (A) Human PBMCs were incubated for 24 h with 1.0 μM of the stimulatory snRNA 5′ ORN (R-1300) complexed to DOTAP (DO) or in the presence of the indicated concentrations of chloroquine (CQ) or the same sequence with a 2′-O-methyl modification or DO alone and IFN-α measured. (B) Human PBMCs were cultured as in (A) with 1.0 μM of the snRNA ORN, and the indicated concentrations of the 2′-O-methyl modified PS snRNA ORN or the same sequence with a PO backbone were added. (C) The stimulatory ORN HIV, derived from the 5′ untranslated region of HIV, was used with a PS (ORN HIV, R-1075) or PO (ORN HIV PO) backbone at 1.0 μM complexed to DOTAP to stimulate human PBMC. The indicated concentrations of the 2′-O-methyl-modified snRNA sequence (snRNA 5′ 2′OMe 5′ U) was added and after 24 h IFN-α was measured. (D) Human PBMCs were cultured for 24 h with the indicated concentrations of the PS HIV ORN or the same sequence containing 2′-O-methyl modifications complexed to DOTAP and IFN-α measured. (E) Human PBMCs were cultured as in (D) with increasing concentrations of Sendai-derived or influenza-derived ORN and the same sequences each with a single 2′-O-methyl modification were added. (F) ORN HIV at 1.0 μM was used to stimulate human PBMC as in (A) and the indicated concentrations of the same sequence with 2′-O-methyl modifications were added. IFN-α was measured after 24 h. Mean ± SEM of one representative out of at least one to two independent experiments (n = 3 donors). Significance of differences at 2 μM (A–C) to the addition of R-1334 were determined for data points as indicated by Student’s t-test (*P < 0.01, **P < 0.001).
We further wanted to explore if a 2′-O-methylated ORN is also capable to suppress immune stimulatory effects induced by ORN derived from virus RNA. Indeed, when using a stimulatory PO or PS sequence derived from the 5′ untranslated region of the HIV genome (ORN HIV, R-1075) [Table 1 and (4)], the addition of the 2′-O-methylated U1 snRNA ORN (modified at the 5′ U, R-1334, compare Fig. 4A and B) completely suppressed IFN-α secretion (Fig. 4C). Moreover, the same phosphorothioate HIV sequence containing 2′-O-methylated bases lost its immune stimulatory effects (Table 1 and Fig. 4D). The same result was observed for two other ORN derived from the 3′ untranslated regions of the Sendai and influenza virus [Table 1 and (30)] with a single modification at U, A or G (Fig. 4E). Although pathogen RNA is hypomethylated, we wanted to investigate if similar to the U1 snRNA 2′-O-methylation of the HIV sequence would result in a suppressive ORN. Therefore, the HIV ORN was cultured with increasing concentrations of the same sequence with 2′-O-methylated bases (Table 1, R-1238). As observed for the methylated U1 snRNA sequence, the 2′-O-methylated HIV ORN did decrease the IFN-α production induced by the HIV ORN to the background levels (Fig. 4F).
Characterization of the 2′-O-methyl-mediated suppression
We generated an ORN with the sequence of the non-stimulatory ORN R-1507 (see Fig. 2), but with a single 2′-O-methyl modification at a central A (R-1862, see Table 1). The non-modified control ORN R-1507 did have a suppressive effect, although only at the highest concentrations tested (Fig. 5A). In contrast, a snRNA-derived modified ORN (R-1334) was a significantly stronger inhibitor and the same sequence as ORN R-1507 but with the 2′-O-methyl modification (R-1862) was strongest to significantly suppress IFN-α induction even at low concentrations (Fig. 5A). The difference became even more apparent in the suppression of TNF-α secretion where the unmodified ORN was non-inhibitory at the concentrations tested, and the 2′ modified ORN did suppress TNF-α secretion, although at higher concentrations compared with the IFN-α inhibition (Fig. 5B).
Fig. 5.
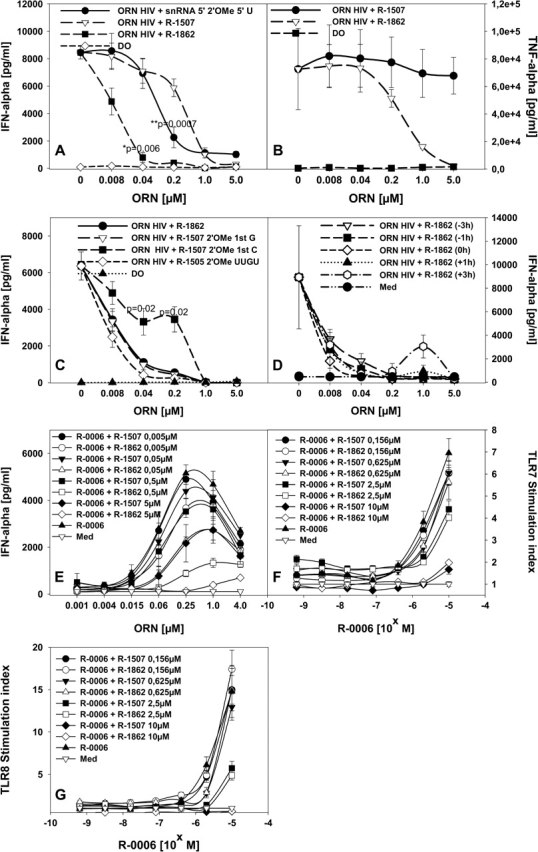
Strong suppressive activity of ORN depends on the presence of 2′-O-methyl modification. (A and B) Human PBMCs were incubated for 24 h with 0.25 μM HIV ORN complexed to DOTAP (DO) or in the presence of the indicated concentrations of the unmodified non-stimulatory ORN R-1507, the same sequence with a single 2′-O-methyl modification (R-1862), or the snRNA-derived 2′ modified ORN (snRNA 5′ 2′OMe 5′ U), or DOTAP alone and cytokines measured. (C) Human PBMCs were cultured as in (A and B) with 0.25 μM HIV ORN, and the indicated concentrations of R-1507-derived ORN with single 2′-O-methyl modifications at different positions or a R-1505-derived ORN with all nucleotides in the GU-rich 4mer motif 2′-O-methylated. (D) Human PBMCs were cultured as in (C) with or without (medium, Med) 0.25 μM HIV ORN, and the 2′-O-methylated ORN R-1862 complexed to DOTAP was added at different time points before or after or at the same time as the stimulatory ORN. (E) Cross-titration studies were performed using the GU-rich ORN R-0006 at the concentrations indicated, and increasing concentrations (0.005, 0.05, 0.5 and 5.0 μM) of the non-stimulatory unmodified ORN R-1507 or the same sequence with a 2′-O-methyl modification (R-1862) were added. PBMCs were cultured for 24 h and IFN-α measured. Mean ± SEM of at least one experiment (n = 3 donors). (F and G) HEK293 cells stably transfected with human TLR7 or TLR8 were incubated with the indicated concentrations of ORN R-0006 complexed to DOTAP for 16 h in the presence of increasing concentrations of unmodified ORN R-1507 or 2′-O-methyl modified ORN R-1862. NFκB activation was measured by assaying luciferase activity. Results are given as fold induction above background (medium). Significance of differences at 0.4 and 2 μM (A and C) to the addition of R-1334 (A) or R-1862 (C) were determined for data points as indicated by Student’s t-test (*P < 0.01, **P < 0.001). No statistical significant difference was observed for (F and G) between the addition of R-1862 and R-1507.
ORN R-1862 contains a single 2′-O-methyl modification at an A, so that we also wanted to explore if 2′ modifications at other positions are inhibitory. Indeed, ORN with single 2′-O-methyl modifications at a G (R-1991) or even 2′ methylated in the 4mer GU-rich sequence (R-1993) suppressed IFN-α induction by the HIV-derived ORN (Fig. 5C). A modified ORN with a 2′-O-methyl-C (Fig. 5C, R-1992) appeared not as potent as the other ORN, although a significant inhibition of the IFN-α response could not be observed. Similar results were observed for other cytokines such as IL-12p40 and IFN-γ (data not shown). In principal, the lower inhibitory activity of C-modified ORN may have similarities to the finding of a lack of a negative effect of 2′ C methylation in an immune stimulatory ORN (Fig. 3).
To further characterize the suppressive effects mediated by 2′-O-methyl-modified ORN, we wanted to investigate the time dependency of the addition of the suppressive ORN (R-1862) to confer the inhibitory effect (Fig. 5D). Similar to the formerly described suppressive ODN (S-Class ODN) (31) delayed addition of suppressive ORN R-1862 to the stimulatory HIV ORN still inhibited IFN-α induction. There was no substantial difference between the addition of inhibitory ORN R-1862 up to 3 h before or after the stimulatory HIV-derived ORN.
We performed cross-titration experiments to further investigate the suppression of IFN-α secretion by the unmodified or modified ORN R-1507 and R-1862, respectively. In these experiments, we used the GU-rich immune stimulatory TLR7, 8 agonist ORN R-0006 (29) and measured IFN-α production from PBMC upon addition of increasing concentrations of R-1507 and R-1862 (Fig. 5E). As observed for the U1 snRNA and HIV ORN, the suppression of IFN-α secretion was strongest for the 2′-O-methylated compared with the unmodified ORN and was concentration dependent, although the unmodified ORN also resulted in some degree of IFN-α inhibition. However, in contrast to the results obtained in the cytokine secretion experiments, there was no statistical significant difference between the modified and unmodified ORN in suppressing NFκB signaling in TLR7 and TLR8 recombinant cells (Fig. 5F and G).
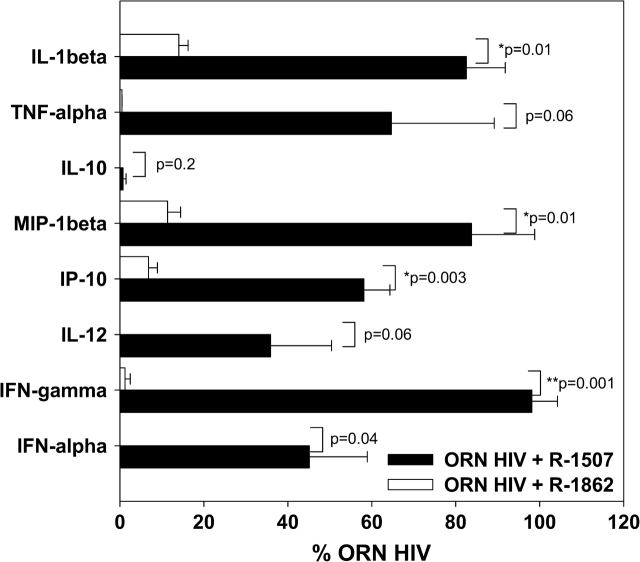
To further investigate the suppressive effect of the 2′-O-methylated ORN R-1862 versus the unmodified control ORN R-1507, we investigated their effect on a panel of different cytokines and chemokines (Fig. 6). The unmodified non-stimulatory ORN at the concentration tested did partially suppress the response induced by ORN HIV, although the 2′-O-methylated ORN was partially significantly stronger and inhibited cytokine secretion to ∼100%. The differences observed between the strength of suppression by unmodified ORN R-1507 versus R-1862 are also reflected in Fig. 5(B) where ORN R-1507 only partially affected TNF-α secretion, but had a stronger effect on IFN-α secretion, probably indicating a higher sensitivity of TLR7 to 2′ modification as suggested above.
Fig. 6.
Profiling of suppressive effects of a 2′-O-methyl-modified sequence. Human PBMCs (n = 3 donors) were incubated for 24 h with 0.25 μM ORN R-1075 complexed to DOTAP (DO), or in the presence of 1.0 μM of the unmodified non-stimulatory ORN R-1507, or in the presence of the same sequence with a single 2′-O-methyl modification, and cytokines measured by Luminex. Given is the mean percentage ± SD of remaining cytokine-inducing activity of ORN R-1075 after co-culture. Significance of differences were determined by Student’s t-test (*P < 0.01, **P < 0.001).
2′-O-methyl inhibition is also observed on murine immune cells
The above data demonstrate that 2′-O-methyl modifications negatively affect responses induced by immune stimulatory ORN in vitro on human immune cells. To further evaluate the immune suppressive effect, the mouse macrophage cell line RAW264.7 or mouse splenocytes were used (Fig. 7 for splenocytes and data not shown). In contrast to human immune cells, murine cells appear to express only one receptor (TLR7) that is stimulated by ligands identified to induce human TLR7 responses (29, 32). It is possible that murine TLR8 needs additional co-factors or signal transduction molecules allowing for TLR8-dependent signaling stimulated by RNA (29). Similar to the data on human PBMC, the 2′-O-methylated ORN R-1862 was significantly more potent in inhibiting cytokine secretion stimulated by the HIV-derived ORN (Fig. 7A and B). In contrast, the non-modified ORN R-1507 suppressed only at substantially higher concentrations.
Fig. 7.
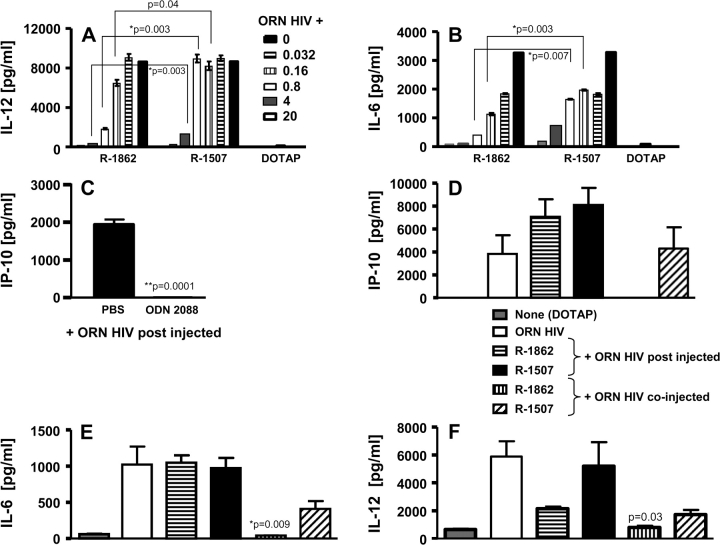
Effect of 2′-O-methylation on murine TLR7-dependent immune responses. (A and B) Murine splenocytes were stimulated for 20 h with 1.0 μM HIV ORN complexed to DOTAP, in the presence of 0.032, 0.16, 0.8, 4.0 or 20.0 μM of the unmodified non-stimulatory ORN R-1507 or the same sequence with a single 2′-O-methyl modification and cytokines were measured. (C) BALB/c mice (n = 5) were pre-treated with suppressive ODN 2088 (240 μg) by intra-peritoneal injection. After 1 h, animals were injected subcutaneously with ORN HIV (240 μg) complexed to DOTAP. Plasma was collected at 3 h post subcutaneous injections for IP-10 assay by ELISA. Given is the mean ± SD. (D–F) sv129 mice (n = 5) were pre-treated intravenously with 200 μg of non-stimulatory unmodified (R-1507) or 2′-O-methylated (R-1862) ORN complexed to DOTAP (400 μg). After 1 h, animals were injected intravenously with ORN HIV (100 μg) complexed to DOTAP (200 μg) (post-injection). In addition, mice (n = 5) were injected intravenously with 100 μg of ORN HIV complexed together with 100 μg ORN R-1507 or R-1862 to DOTAP (400 μg) (co-injection). Plasma was collected at 3 h post intravenous injections for cytokine assays by ELISA. Significance of differences between R-1862 and R-1507 in (C) ODN 2088 were determined by Student’s t-test (*P < 0.01, **P < 0.001).
We also wanted to investigate the suppressive effect of 2′-O-methyl-modified ORN on in vivo activation of the TLR7 pathway. As a first step, we used the well-defined inhibitory S-Class ODN 2088 (24) to significantly suppress in vivo cytokine secretion induced by the HIV ORN (Fig. 7C). Animals were pre-treated intra-peritoneally with ODN 2088 and 1 h after were injected subcutaneously with the HIV ORN complexed to DOTAP. The IP-10 plasma levels found 3 h after injection of the HIV ORN were decreased to the background levels when the S-Class ODN was pre-injected. However, the same protocol used for the 2′-O-methyl-modified ORN did not result in an inhibitory effect (data not shown). Therefore, we choose to inject the stimulatory and inhibitory ORN via the same intravenous route of injection (Fig. 7D–F). We tested two different protocols, either as above injected ORN R-1862 or R-1507 complexed to DOTAP 1 h before the HIV ORN (HIV ORN post injected) or complexed the HIV ORN and ORN R-1862 or R-1507 to DOTAP and injected the two ORN together (HIV ORN co-injected). When the HIV ORN was administered after the modified or unmodified ORN, no significant effect on IP-10, IL-6 or IL-12 induction could be observed (Fig. 7D–F). However, co-injection of the 2′-O-methyl-modified ORN with the stimulatory HIV ORN resulted in partially significant and complete inhibition of the immune modulatory effects otherwise induced by the HIV ORN. Similar to the human PBMC data, the non-modified ORN also resulted in partial inhibition, although not to the background levels.
Discussion
The immune system appears to be enabled to discriminate between self and invading DNAs because they differ markedly in their CpG dinucleotide content and methylation. Bacterial and many viral DNAs compared with vertebrate DNAs not only contain a higher frequency of CpG dinucleotides that are recognized by TLR9 but also contain unmodified CpG dinucleotides, and methylation at the C5 position in the CpG dinucleotide greatly decreases immune stimulatory effects (33, 34). In contrast, enforced uptake and distribution of self-DNA to TLR9-expressing endosomal compartments lead to TLR9-dependent immune activation, demonstrating that the segregation of self-DNA and TLR9 plays a critical role (35, 36).
TLR7 and TLR8 respond to pathogen RNA and are expressed in endosomal compartments making it difficult for self-RNAs to reach these receptors (37–39). In contrast, self-RNAs coated with cationic liposomes or in immune complexes are taken up in TLR7, 8-containing compartments resulting in immune activation (25, 40, 41). The host must have evolved an additional mechanism to be enabled to discriminate between self and foreign RNAs reaching endosomal compartments. Indeed, certain naturally occurring modifications in RNAs can limit their detection by TLRs (15). In contrast to other eukaryotic RNAs, U1 snRNA contains only few modifications concentrated at the 5′ end that appear to have a role in pre-mRNA splicing (42). Comparison of the immune stimulatory effects of isolated 2′-O-methyl-modified snRNA sequences and the unmodified same sequences demonstrate that simple modifications such as 2′-O-methylation have a strong suppressive effect and that even only one such a 2′ modification is sufficient to strongly affect immune recognition.
Recognition of RNA by TLR7 and TLR8 depends on the U content, and the modification of Us is detrimental to its immune effects (4, 28, 43). However, RNA modifications not only occur at U nucleotides but can also be found at other nucleotides and may not be localized to U-containing and stimulatory regions of self-RNAs (12). In addition, modifications in eukaryotic RNAs are usually clustered in conserved areas (17). From our data, it appears possible that modifications such as 2′-O-methylation are enabled to impose a suppressive effect even on unmodified sites further up or downstream of stimulatory RNA regions. This effect cannot be easily explained by the modification imposing a certain structure on the RNA as suggested before (15, 44), as 2′-O-methylated ORN used in our experiments do not form secondary structures that may prevent or alter the interaction with the receptor. Placing a single 2′-O-methyl modification at any G or A 5′ or 3′ of a defined stimulatory GU-rich region has a strong negative impact on most immune effects. In contrast, 2′-O-methylation of Cs appears to only slightly affect TLR7 or TLR8 activation. One explanation may be that the number of 2′-O-methyl-modified Cs in a given endogenous RNA is under-represented compared with modified Us, As or Gs as can be found in rRNA from the small ribosomal subunit or in snRNA (12, 27). An inhibition of the TLR response due to these 2′-modified bases would then be sufficient to avoid inflammatory processes potentially induced by certain self-RNAs.
Activation of the IFN-α pathway and increased expression of type I IFN in the pDCs of systemic lupus erythematosus patients are associated with disease severity and may contribute to disease development and perpetuation of inflammatory responses (45). Whereas IFN-α stimulation appears in most experiments to be completely suppressed by 2′-O-methylation, the effect on other cytokines such as TNF-α is varying. This may indicate that especially pDCs secreting IFN-α upon RNA-mediated TLR7 stimulation are more sensitive to RNA modifications and utilize such modifications to discriminate between microbial RNAs and abundant endogenous RNAs. However, although there was suppression of RNA-mediated TLR7- as well as TLR8-dependent NFκB signaling in recombinant cells expressing these receptors observed for 2′-O-methyl-modified RNA, the same RNA sequence without such a modification had a similar negative effect on NFκB activation. It may be possible that the TLR7 and TLR8 recombinant cells used for these experiments are less sensitive to suppressive ORN as described previously for inhibition of responses stimulated by small molecule TLR7, 8 agonists (23) or they do not primarily affect NFκB but other signaling pathways that cannot be tested with these cells such as IRF-7 signaling in pDCs leading to IFN-α production.
Viral genomic nucleic acids contain few modifications (7, 13) and can reach TLR-expressing endosomes during virus replication by autophagy (46). However, processed pathogen mRNAs also contain post-translational 2′-O-methyl modifications most probably interfering with an efficient recognition. In contrast to most eukaryotic mRNAs, viral RNA intermediates in the cytoplasm have an uncapped 5′-triphosphate end and can be recognized by another eukaryotic pathogen sensor, RIG-I (6, 7). Although there is not much known about the influence of RNA modifications on the activation of cytoplasmic receptors such as RIG-I or mda-5 (7), it is highly unlikely that the single-stranded modified and unmodified ORN used in our experiments and delivered by DOTAP would have interacted with these receptors. The use of specific delivery techniques such as liposomal formulations (e.g. DOTAP) for nucleic acids can strongly influence organ, tissue, cellular and intracellular distribution (47). Formulations like DOTAP release their cargo mainly in the endosome, whereas other or modified such formulations rather release the payload into the cytoplasm where RIG-I and mda-5 can be found (3, 4).
Modifications in eukaryotic RNAs are clustered in specific sites. Although modifications such as 2′-O-methylation can interfere with TLR activation even when placed outside of GU-rich immune stimulatory RNA regions, in some endogenous RNAs such as rRNA only ∼3% of the nucleotides are modified (12), although this is still significantly more compared with bacteria (15). The relative depression of the ratio of unmodified to modified nucleotides and their clustering leaves relatively long eukaryotic RNA regions unmodified that may stimulate a TLR response when reaching the right endosomal compartments. Self-RNA appears to locate to TLR-expressing compartments only in rare cases such as during defects in apoptotic cell clearance (45). Eukaryotic RNAs are degraded in apoptotic cells and, for example, U1 snRNA is cleaved near the 5′ end resulting in two truncated apoptotic RNA molecules (42). In our experiments, the modified 5′ end of the U1 snRNA not only lacked immune modulatory effects but also suppressed the stimulation induced by another RNA sequence. This observation could be even extended to other RNA sequences with only one 2′-O-methyl modification. The inhibition appeared to be stronger for IFN-α and related responses than for other cytokines similar to the effect imposed by a 2′ modification in the same RNA sequence. In contrast, an unmodified non-stimulatory ORN was much weaker in interfering with the TLR-mediated immune response when co-cultured with a stimulatory RNA sequence. Again, this suggests that pDCs evolved a tight control mechanism not to allow for IFN-α secretion stimulated by self-RNAs that may result in inflammatory responses. The inhibition of RNA-mediated TLR7 activation was also reported for specific DNA and chemically modified siRNA sequences (48–51). Although incompletely understood, inhibitory effects imposed by suppressive DNA sequences appear not to be due to uptake competition, and it was speculated that TLR suppression is mediated through an allosteric regulatory site or competition for the binding site. The same mechanism may apply for self-RNA and would allow for the generation of inhibitory modified sequences during apoptosis or similar processes avoiding unwanted inflammation stimulated by unmodified self-RNA potentially leading to autoimmune responses.
Only when stimulatory and modified suppressive RNAs are co-delivered in vivo by complexing them to the same liposomal particles, inhibition of the murine TLR7-mediated immune responses is observed. This observation make the use of 2′-O-methyl modified ORN as potential TLR7, 8 antagonists difficult. But at the same time, it demonstrates that even a single 2′-O-methyl modification placed in a strand of a siRNA will reduce the non-target-mediated pro-inflammatory effects in vivo otherwise stimulated by the unmodified sense or anti-sense strand of the siRNA.
In summary, the detection of pathogen RNAs mainly depends on endosomal and cytoplasmatic pathogen receptors. Although it is not yet completely clear how cytoplasmatic receptors such as RIG-I distinguish between self and viral RNAs, RNA modifications may also have an impact on RIG-I activation (7). In contrast, for endoplasmatic receptors such as TLR7, 8 and 9, several mechanisms to avoid self-stimulation appear to have evolved. On the one hand, the intracellular localization of TLRs usually ensures the recognition of viral but not self-RNAs. However, under certain circumstances such as during defective apoptotic clearance or in immune complexes (45), self-RNAs can reach these compartments in principle allowing for self-recognition. In these cases, another safeguard mechanism appears to involve the inhibition of TLR7 and TLR8 responses by RNA modifications found more frequently in self than pathogen RNAs. Although RNA modifications in eukaryotic RNAs are clustered in conserved regions, modified RNA sequences appear to impose a suppressive effect even when placed on another RNA strand, helping eukaryotic cells to tolerate their own nucleic acids.
Funding
National Institute of Allergy and Infectious Disease (HHSSN266200400044C).
Acknowledgments
We thank Dr Bernhard Noll and Angela Lohner for performing quality control for identity and purity on the synthetic ORN and for measuring potential formation of higher ordered RNA structures as well as Andrea Kritzler, Carmen Montino, Claudia Schmitz, Qin Zhu and Cristina Vizena for expert technical assistance. We also thank Dr Grayson Lipford for helpful discussions. All the authors except SB are employees of Pfizer and may have a financial interest in the studies outcome.
Glossary
Abbreviations
- DOTAP
1,3-dioleoyloxy-3-(trimethylammonium)propane
- mda-5
melanoma differentiation-associated gene-5
- ODN
Oligodeoxyribonucleotide
- ORN
oligoribonucleotide
- PDC
plasmacytoid dendritic cell
- PO
phosphodiester
- PRR
pattern recognition receptor
- RIG-I
retinoic acid-inducible gene-I
- siRNA
silencing RNA
- SN
supernatant
- SnRNA
small nuclear RNA
- TLR
Toll-like receptor
- TNF
tumor necrosis factor
References
- 1.Akira S. Mammalian Toll-like receptors. Curr. Opin. Immunol. 2003;15:5. doi: 10.1016/s0952-7915(02)00013-4. [DOI] [PubMed] [Google Scholar]
- 2.Vollmer J. Editorial: CpG motifs to modulate innate and adaptive immune responses. Int. Rev. Immunol. 2006;25:125. doi: 10.1080/08830180600743115. [DOI] [PubMed] [Google Scholar]
- 3.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis eSousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 4.Heil F, Hemmi H, Hochrein H, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 5.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 6.Pichlmair A, Schulz O, Tan CP, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science Express. 006;314:997. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 7.Hornung V, Ellegast J, Kim S, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 8.Gitlin L, Barchet W, Gilfillan S, et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl Acad. Sci. USA. 2006;103:8459. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 2004;5:987. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 10.Kato H, Sato S, Yoneyama M, et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 11.Hornung V, Rothenfusser S, Britsch S, et al. Quantitative expression of toll-like receptor 1-10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J. Immunol. 2002;168:4531. doi: 10.4049/jimmunol.168.9.4531. [DOI] [PubMed] [Google Scholar]
- 12.Maden BE. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 1990;39:241. doi: 10.1016/s0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- 13.Helm M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006;34:721. doi: 10.1093/nar/gkj471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bachellerie JP, Cavaille J. Guiding ribose methylation of rRNA. Trends Biochem. Sci. 1997;22:257. doi: 10.1016/s0968-0004(97)01057-8. [DOI] [PubMed] [Google Scholar]
- 15.Kariko K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 16.Calagan JL, Pirtle RM, Pirtle IL, Kashdan MA, Vreman HJ, Dudock BS. Homology between chloroplast and prokaryotic initiator tRNA. Nucleotide sequence of spinach chloroplast methionine initiator tRNA. J. Biol. Chem. 1980;255:9981. [PubMed] [Google Scholar]
- 17.Hansen MA, Kirpekar F, Ritterbusch W, Vester B. Posttranscriptional modifications in the A-loop of 23S rRNAs from selected archaea and eubacteria. RNA. 2002;8:202. doi: 10.1017/s1355838202013365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eberle F, Giessler K, Deck C, et al. Modifications in small interfering RNA that separate immunostimulation from RNA interference. J. Immunol. 2008;180:3229. doi: 10.4049/jimmunol.180.5.3229. [DOI] [PubMed] [Google Scholar]
- 19.Judge AD, Sood V, Shaw JR, Fang D, McClintock K, Maclachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat. Biotechnol. 2005;23:457. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 20.Sioud M. Single-stranded small interfering RNA are more immunostimulatory than their double-stranded counterparts: a central role for 2′-hydroxyl uridines in immune responses. Eur. J. Immunol. 2006;36:1222. doi: 10.1002/eji.200535708. [DOI] [PubMed] [Google Scholar]
- 21.Wagner H, Bauer S. All is not Toll: new pathways in DNA recognition. J. Exp. Med. 2006;203:265. doi: 10.1084/jem.20052191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sioud M. Innate sensing of self and non-self RNAs by Toll-like receptors. Trends Mol. Med. 2006;12:167. doi: 10.1016/j.molmed.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 23.Jurk M, Kritzler A, Schulte B, et al. Modulating responsiveness of human TLR7 and 8 to small molecule ligands with T-rich phosphorothiate oligodeoxynucleotides. Eur. J. Immunol. 2006;36:1815. doi: 10.1002/eji.200535806. [DOI] [PubMed] [Google Scholar]
- 24.Stunz LL, Lenert P, Peckham D, et al. Inhibitory oligonucleotides specifically block effects of stimulatory CpG oligonucleotides in B cells. Eur. J. Immunol. 2002;32:1212. doi: 10.1002/1521-4141(200205)32:5<1212::AID-IMMU1212>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 25.Vollmer J, Tluk S, Schmitz C, et al. Immune stimulation mediated by autoantigen binding sites within small nuclear RNAs involves Toll-like receptors 7 and 8. J. Exp. Med. 2005;202:1575. doi: 10.1084/jem.20051696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savarese E, Chae OW, Trowitzsch S, et al. U1 small nuclear ribonucleoprotein immune complexes induce type I interferon in plasmacytoid dendritic cells via TLR7. Blood. 2006;107:3229. doi: 10.1182/blood-2005-07-2650. [DOI] [PubMed] [Google Scholar]
- 27.Krol A, Branlant C, Lazar E, Gallinaro H, Jacob M. Primary and secondary structures of chicken, rat and man nuclear U4 RNAs. Homologies with U1 and U5 RNAs. Nucleic Acids Res. 1981;9:2699. doi: 10.1093/nar/9.12.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diebold SS, Massacrier C, Akira S, Paturel C, Morel Y, Sousa CR. Nucleic acid agonists for Toll-like receptor 7 are defined by the presence of uridine ribonucleotides. Eur. J. Immunol. 2006;36:3256. doi: 10.1002/eji.200636617. [DOI] [PubMed] [Google Scholar]
- 29.Forsbach A, Nemorin JG, Montino C, et al. Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. J. Immunol. 2008;180:3729. doi: 10.4049/jimmunol.180.6.3729. [DOI] [PubMed] [Google Scholar]
- 30.Forsbach A, Nemorin JG, Volp K, et al. Characterization of conserved viral leader RNA sequences that stimulate innate immunity through TLRs. Oligonucleotides. 2007;17:405. doi: 10.1089/oli.2007.0098. [DOI] [PubMed] [Google Scholar]
- 31.Lenert P, Rasmussen W, Ashman RF, Ballas ZK. Structural characterization of the inhibitory DNA motif for the type A (D)-CpG-induced cytokine secretion and NK-cell lytic activity in mouse spleen cells. DNA Cell Biol. 2003;22:621. doi: 10.1089/104454903770238094. [DOI] [PubMed] [Google Scholar]
- 32.Jurk M, Heil F, Vollmer J, et al. Human TLR7 or TLR8 independently confer responsiveness to the antiviral compound R-848. Nat. Immunol. 2002;3:499. doi: 10.1038/ni0602-499. [DOI] [PubMed] [Google Scholar]
- 33.Vollmer J. TLR9 in health and disease. Int. Rev. Immunol. 2006;25:155. doi: 10.1080/08830180600743107. [DOI] [PubMed] [Google Scholar]
- 34.Krieg AM. CpG motifs in bacterial DNA and their immune effects. Annu. Rev. Immunol. 2002;20:709. doi: 10.1146/annurev.immunol.20.100301.064842. [DOI] [PubMed] [Google Scholar]
- 35.Yasuda K, Yu P, Kirschning CJ, et al. Endosomal translocation of vertebrate DNA activates dendritic cells via TLR9-dependent and -independent pathways. J. Immunol. 2005;174:6129. doi: 10.4049/jimmunol.174.10.6129. [DOI] [PubMed] [Google Scholar]
- 36.Barton GM, Kagan JC, Medzhitov R. Intracellular localization of Toll-like receptor 9 prevents recognition of self DNA but facilitates access to viral DNA. Nat. Immunol. 2006;7:49. doi: 10.1038/ni1280. [DOI] [PubMed] [Google Scholar]
- 37.Sioud M. Induction of inflammatory cytokines and interferon responses by double-stranded and single-stranded siRNAs is sequence-dependent and requires endosomal localization. J. Mol. Biol. 2005;348:1079. doi: 10.1016/j.jmb.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 38.Nishiya T, DeFranco AL. Ligand-regulated chimeric receptor approach reveals distinctive subcellular localization and signaling properties of the Toll-like receptors. J. Biol. Chem. 2004;279:19008. doi: 10.1074/jbc.M311618200. [DOI] [PubMed] [Google Scholar]
- 39.Nishiya T, Kajita E, Miwa S, DeFranco AL. TLR3 and TLR7 are targeted to the same intracellular compartments by distinct regulatory elements. J. Biol. Chem. 2005;280:37107. doi: 10.1074/jbc.M504951200. [DOI] [PubMed] [Google Scholar]
- 40.Barrat FJ, Meeker T, Gregorio J, et al. Nucleic acids of mammalian origin can act as endogenous ligands for Toll-like receptors and may promote systemic lupus erythematosus. J. Exp. Med. 2005;202:1131. doi: 10.1084/jem.20050914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lau CM, Broughton C, Tabor AS, et al. RNA-associated autoantigens activate B cells by combined B cell antigen receptor/Toll-like receptor 7 engagement. J. Exp. Med. 005;202:1171. doi: 10.1084/jem.20050630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Degen WG, Aarssen Y, Pruijn GJ, Utz PJ, van Venrooij WJ. The fate of U1 snRNP during anti-Fas induced apoptosis: specific cleavage of the U1 snRNA molecule. Cell Death. Differ. 2000;7:70. doi: 10.1038/sj.cdd.4400617. [DOI] [PubMed] [Google Scholar]
- 43.Forsbach A, Nemorin JG, Montino C, et al. Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. J. Immunol. 2008;180:3729. doi: 10.4049/jimmunol.180.6.3729. [DOI] [PubMed] [Google Scholar]
- 44.Wang JP, Liu P, Latz E, Golenbock DT, Finberg RW, Libraty DH. Flavivirus activation of plasmacytoid dendritic cells delineates key elements of TLR7 signaling beyond endosomal recognition. J. Immunol. 2006;177:7114. doi: 10.4049/jimmunol.177.10.7114. [DOI] [PubMed] [Google Scholar]
- 45.Krieg AM, Vollmer J. Toll-like receptors 7, 8, and 9: linking innate immunity to autoimmunity. Immunol. Rev. 2007;220:251. doi: 10.1111/j.1600-065X.2007.00572.x. [DOI] [PubMed] [Google Scholar]
- 46.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cell. Science. 2007;315:1398. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 47.Oliveira S, Storm G, Schiffelers RM. Targeted delivery of siRNA. J. Biomed. Biotechnol. 2006;2006:63675. doi: 10.1155/JBB/2006/63675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sioud M, Furset G, Cekaite L. Suppression of immunostimulatory siRNA-driven innate immune activation by 2′-modified RNAs. Biochem. Biophys. Res. Commun. 2007;361:122. doi: 10.1016/j.bbrc.2007.06.177. [DOI] [PubMed] [Google Scholar]
- 49.Robbins M, Judge A, Liang L, McClintock K, Yaworski E, Maclachlan I. 2′-O-methyl-modified RNAs Act as TLR7 antagonists. Mol. Ther. 2007;15:1663. doi: 10.1038/sj.mt.6300240. [DOI] [PubMed] [Google Scholar]
- 50.Gursel I, Gursel M, Yamada H, Ishii KJ, Takeshita F, Klinman DM. Repetitive elements in mammalian telomeres suppress bacterial DNA-induced immune activation. J. Immunol. 2003;171:1393. doi: 10.4049/jimmunol.171.3.1393. [DOI] [PubMed] [Google Scholar]
- 51.Lenert P, Stunz L, Yi AK, Krieg AM, Ashman RF. CpG stimulation of primary mouse B cells is blocked by inhibitory oligodeoxyribonucleotides at a site proximal to NF-kappaB activation. Antisense Nucleic Acid Drug Dev. 2001;11:247. doi: 10.1089/108729001317022241. [DOI] [PubMed] [Google Scholar]