Summary
We used DNA microarrays to identify panels of transcriptional markers of aging that are differentially expressed in young (5-month) and old (25-month) mice of multiple inbred strains (129sv, BALB/c, CBA, DBA, B6, C3H, and B6C3F1). In the heart, age-related changes of five genes were studied throughout the mouse lifespan: complement component 4, chemokine ligand 14, component of Sp100-rs, phenylalanine hydroxylase, and src family associated phosphoprotein 2. A similar analysis in the brain (cerebellum) involved complement component 1q (alpha polypeptide), complement component 4, P lysozyme structural, glial fibrillary acidic protein, and cathepsin S. Caloric restriction inhibited age-related expression of these genes in both tissues. Parametric analysis of gene set enrichment (PAGE) identified several biological processes that are induced with aging in multiple mouse strains. We also tested the ability of dietary antioxidants to oppose these transcriptional markers of aging. Lycopene, resveratrol, acetyl-L-carnitine, and Tempol were as effective as caloric restriction in the heart, and α-lipoic acid and coenzyme Q10 were as effective as caloric restriction in the cerebellum. These findings suggest that transcriptional biomarkers of aging in mice can be used to estimate the efficacy of aging interventions on a tissue-specific basis.
Keywords: aging, mouse, biomarkers, microarray, antioxidants, caloric restriction
Introduction
Gene expression profiling with DNA microarrays can measure the transcriptional changes of thousands of genes simultaneously providing a useful tool for the study of complex biological processes, such as aging. In order to understand the molecular basis of aging and to identify biomarkers of aging, we have used DNA microarrays to characterize tissue-specific gene expression profiles associated with aging in mice (Higami et al., 2004; Lee et al., 2002; Lee et al., 1999; Lee et al., 2000). Heart aging is characterized by a transcriptional profile suggestive of induction of cellular structural proteins involved in cardiomyocyte hypertrophy, a metabolic shift from fatty acid toward carbohydrate metabolism, and reduced protein biosynthesis (Lee et al., 2002; Park et al., 2008). In contrast, brain aging in mice is associated with a heightened cellular immunity, inflammation and a concerted induction of genes involved in stress response (Lee et al., 2000; Park et al., 2008). In addition, the gene expression pattern is suggestive of reduced protein turnover and a decreased expression of genes encoding growth and trophic factors. Because these studies were performed in a very limited set of mouse inbred strains, it is unclear if the main observations represent general aging features, or if they are secondary to strain-specific pathology.
Transcriptional profiles of tissues from animals on caloric restriction (CR) suggest that CR reduces endogenous damage and induces metabolic shifts and thus opposes the aging process (Lee et al., 2002; Lee et al., 1999; Lee et al., 2000; Park & Prolla, 2005a; Park & Prolla, 2005b). Genome-wide microarray analysis of hepatic RNA show that shifting from control diet to CR induces a rapid shift toward the gene expression profile of long-term CR and shifting from long-term CR to control diet reverses 90% of the CR effect on gene expression within 8 weeks, suggesting a cause-and-effect relationship between the rate of aging and the CR-associated alterations in gene expression (Dhahbi et al., 2004).
Several natural and synthetic compounds with antioxidant activity also have the potential to slow specific aspects of the aging process. In a previous study in the mouse heart, α-lipoic acid (LA) and coenzyme Q10 (CQ) inhibited age-related alterations in the expression of genes involved in the extracellular matrix, cellular structure, and protein turnover, but had no impact on longevity or tumor patterns compared with control mice (Lee et al., 2004). Resveratrol (RE), a polyphenol compound found in red wine, retards cardiac aging in mice (Barger et al., 2008) and increases survival of mice fed a high fat diet (Baur et al., 2006). The yellow curry spice curcumin (CU) is more potent than vitamin E in scavenging free radicals (Zhao et al., 1989) and reduces oxidative damage in an Alzheimer transgenic mouse model (Lim et al., 2001), suggesting that it may also retard brain aging. Lycopene (LY), a major carotenoid present in tomato, reduces lipid peroxidation induced by oxidative stress (Parfitt et al., 1994). Astaxantin (AS) is a carotenoid responsible for the pink color of the flesh of salmon and also exhibits potent antioxidant properties in membranes (Palozza & Krinsky, 1992). Dietary supplementation with acetyl-L-carnitine (AC) in rats reverses the age-associated decline of mitochondrial functions (Hagen et al., 1998a; Hagen et al., 1998b). Chronic treatment of superoxide dismutase mimetic Tempol (TP) shows protective effects on age-related vascular dysfunction in rats (Tatchum-Talom & Martin, 2004). Despite compelling evidence that these agents may retard specific aspects of aging, they have not been systematically or comparatively evaluated in their ability to modify aging parameters. Because lifespan studies are time consuming and costly, the ability to screen compounds for their ability to impact aging or mimic CR in specific tissues would be useful in deciding what compounds to pursue in further study, and also in deciding the most effective combinations of compounds.
In this study, age-related differential expression of genes from several strains of mice, 129sv, BALB/c, CBA, DBA, B6, C3H, and B6C3F1, was screened using Affymetrix high-density oligonucleotide arrays to establish a panel of transcriptional markers of aging and to identify pathways significantly altered with aging in multiple mouse strains. We identified tissue-specific panels of biomarkers of aging that are common in multiple strains of mice in the heart and brain (cerebellum). Using these panels, we tested the effect of middle-age (15-month) onset CR and dietary supplementation of eight antioxidants (LA, CQ, RE, CU, LY, AC, AS, and TP) on the expression of each transcriptional biomarker of aging.
Results
Identification of transcriptional biomarkers of heart aging
Comparison between 5-month-old (C5) and 25-month-old (C25) heart tissues with Affymetrix Mouse Genome 430A arrays representing 22,626 transcripts resulted in age-related changes (P < 0.05) of 3,383 (15%) transcripts in the 129sv strain, 2,552 (11%) in BALB/c, 1,363 (6%) in CBA, 2,449 (11%) in DBA, 2,845 (13%) in B6, 1,452 (6%) in C3H, and 1,718 (8%) in B6C3F1. Among these, only 20 genes were common in at least six of the seven strains of mice tested (Table S1). From the binomial distribution, the probability of at least 6 of 7 tests being significant at the .05 level by chance alone is 1.05e-7. Interestingly, 19 of the genes meeting these criteria were up-regulated with aging. There was only one down-regulated gene, enoyl coenzyme A hydratase 1, and its fold change (FC) with aging is relatively small in all strains (~0.85). Table 1 contains a panel of biomarkers of heart aging that were common in all seven strains of mice tested. Three genes are known to be involved in cellular immune and inflammatory responses, which is suggestive of heightened immunity in the aged heart. Complement component 4 (C4) is involved in the classical complement activation and chemokine (C-X-C motif) ligand 14 (Cxcl14) is a cytokine involved in immune responses (Shurin et al., 2005). Src family associated phosphoprotein 2 (Scap2) is a specific substrate for the Src family protein tyrosine kinase Fyn (Marie-Cardine et al., 1998) and is also known to negatively regulate cell proliferation. A recent study in Scap2−/− mice suggests that Scap2 is required for proper activation of the immune system (Togni et al., 2005). Phenylalanine hydroxylase (Pah) metabolizes aromatic amino acids and is involved in clearing circulating phenylalanine in blood and body fluids, increased levels of which can cause phenylketonuria (Christensen et al., 2005). The biggest FC with aging was observed in component of Sp100-rs (Csprs) which encodes a putative G-protein coupled receptor (Weichenhan et al., 2001). The biological function of F-box only protein 23, also known as tetraspanin 17 (Kawai et al., 2001), is unknown. We note that the method used for identification of aging transcriptional markers is based on analysis of two time points, and therefore does not identify genes that are significantly altered only after 25-months of age, or genes that reach significance at earlier ages but at 25-moths return to an expression level more similar to that of young animals.
Table 1.
Age-related fold change of selected biomarkers of heart aging
| Strain |
|||||||
|---|---|---|---|---|---|---|---|
| Gene | 129 | BalbC | CBA | DBA | B6 | C3H | B6/C3H |
| C4 | 2.1 | 2.1 | 2.1 | 3.7 | 1.9 | 2.0 | 1.8 |
| Csprs | 13.4 | 7.1 | 1.8 | 4.8 | 10.0 | 3.8 | 11.1 |
| Pah | 2.3 | 4.5 | 2.1 | 2.0 | 3.7 | 2.9 | 4.2 |
| Cxcl14 | 2.0 | 2.8 | 1.6 | 2.0 | 1.5 | 1.7 | 2.4 |
| Scap2 | 1.9 | 1.5 | 1.3 | 1.5 | 1.4 | 1.9 | 1.4 |
Values are the fold change (FC) of genes obtained by comparing 25-month-old control group (C25) with 5-month-old control group (C5). All genes are significantly (P < 0.05) changed in expression in all strains tested.
Identification of transcriptional biomarkers of cerebellum aging
Of the 45,037 transcripts screened using Affymetrix Mouse Genome 430 2.0 arrays, 3,752 (8%) transcripts were significantly changed by aging in the cerebellum of the 129sv strain, 4,317 (10%) in BALB/c, 7,273 (16%) in CBA, 4,635 (10%) in B6, 9,020 (20%) in C3H, and 5,948 (13%) in B6C3F1. In the cerebellum, 99 genes were common in all strains tested: 82 of these were increased in expression and 12 were decreased in all strains tested (Table S2). Among them, we selected five genes having a higher fold changes with aging for further study (Table 2). Many genes involved in cellular immune and inflammatory responses were induced with aging. Four initiators of the classical complement cascade, C4 and three polypeptides of complement component 1q (C1q), were increased in expression by aging in all strains of mice, which supports a chronic inflammatory state of the aged cerebellum. Several cathepsins were also up-regulated in the aged cerebellum, including cathepsin D, S, and Z. Cathepsin S (Ctss) plays a key role in major histocompatability complex (MHC) class II-mediated antigen presentation (Boes et al., 2005; Hsieh et al., 2002). Lysozyme and P lysozyme structural (Lzp-s, also known as Lzp-1) are bacteriolytic lysosomal hydrolases, and involved in human hereditary amyloydosis (Rocken et al., 2006). In all six strains of mice, Lzp-s showed the largest FC with aging. Glial fibrillary acidic protein (Gfap) was first discovered as an astrocyte-specific intermediate filament (Eng et al., 1971) and is widely used as a marker of neurodegeneration (Nawashiro et al., 2002; Rozovsky et al., 2005; Wei et al., 2002). In addition, two lipid molecule transporters, apolipoprotein D (ApoD) and E (ApoE), were induced with aging in the cerebellum. ApoD is also known to be involved in the response to oxidative stress in the brain (Navarro-Incio & Tolivia-Fernandez, 2004), and we have previously shown that its expression is increased with aging in the brain of mice, rhesus monkeys, and humans (Loerch et al., 2008).
Table 2.
Age-related fold change of selected biomarkers of cerebellum aging
| Strain |
||||||
|---|---|---|---|---|---|---|
| Gene | 129 | BalbC | CBA | B6 | C3H | B6/C3H |
| C4 | 1.9 | 1.8 | 3.9 | 3.3 | 3.5 | 2.3 |
| C1qa | 4.3 | 2.7 | 2.8 | 3.0 | 4.0 | 3.3 |
| Ctss | 2.6 | 1.8 | 2.7 | 2.3 | 2.2 | 2.7 |
| Lzp-s | 13.5 | 5.3 | 8.1 | 4.8 | 5.1 | 5.4 |
| Gfap | 3.6 | 2.6 | 3.2 | 2.3 | 4.0 | 2.6 |
Values are the fold change (FC) of genes obtained by comparing 25-month-old control group (C25) with 5-month-old control group (C5). All genes are significantly (P < 0.05) changed in expression in all strains tested.
The kinetics of expression of transcriptional biomarkers of aging and the effect of caloric restriction
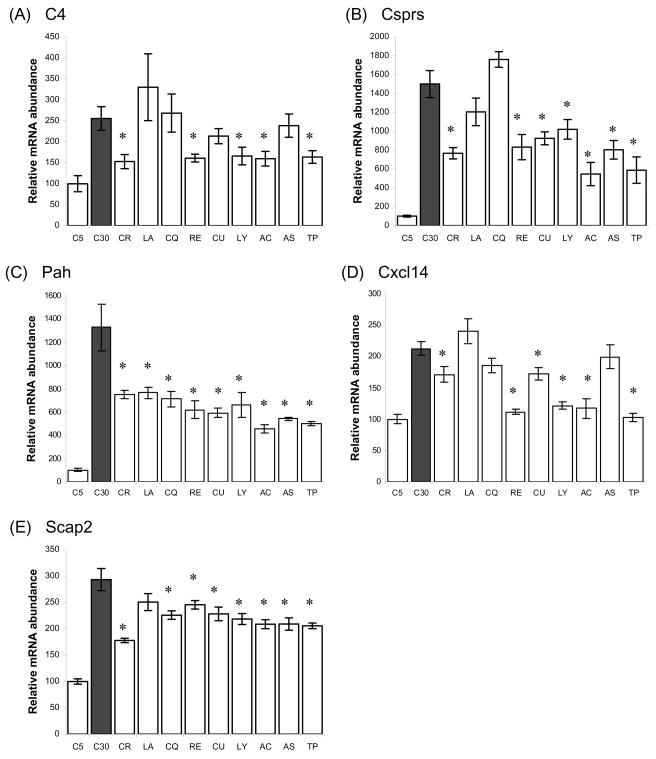
We measured changes in mRNA levels for selected biomarkers of aging at various points of the mouse lifespan using real-time quantitative RT-PCR, and also tested the ability of CR to inhibit these changes. mRNA was extracted from the tissues of B6 mice (n ≥ 5) sacrificed at 5, 10, 15, 20, 25, 30 months of age and used as a template for real-time quantitative RT-PCR. In the heart, there was an age-associated, approximate linear induction of expression in Csprs and Pah (Fig. 1). The expression of C4 was only slightly increased until 25 months of age and then greatly increased in 30-month-old animals. Both Cxcl14 and Scap2 showed a rising and falling expression pattern between 15 and 25 months and a significant induction at the age of 30 months. In the heart, CR appeared to prevent the late (25–30 months) age-related induction of these markers, with minimal effects earlier in life. Measurement of heart function using echocardiogram analysis showed that there was no decline in heart function with aging until late age (25 months) in these strains (data not shown).
Fig. 1.
Time-course quantitative RT-PCR of biomarkers of heart aging in B6 mice. The effect of aging and CR on the expression of selected biomarkers of heart aging is measued. Relative amount of mRNA compared to young control is shown for each sample. (A) C4, (B) Csprs, (C) Pah, (D) Cxcl14; (E) Scap2. C5, 5-month-old control grouop. Lines above/below bars indicate standard error.
In the cerebellum, the expression of C1q alpha polypeptide (C1qa), Ctss, and Gfap displayed an age-related linear increase throughout the life span of B6 mice (Fig. 2). The age-associated induction of C4 and Lzp-s was observed largely after 20 months of age. Interestingly, C4, a biomarker of aging common in both heart and cerebellum, followed the same expression pattern throughout the life span in both tissues, suggesting that this gene may be a very good biomarker of aging in postmitotic tissues. CR significantly opposed age-related transcriptional changes in the cerebellum (Fig. 2). CR showed a consistent effect on aging markers throughout the lifespan, including C1qa, Ctss, and Gfap. CR reduced expression of Gfap and C1qa as early as 5 months of age (Fig. 2).
Fig. 2.
Time-course quantitative RT-PCR of biomarkers of cerebellum aging in B6 mice. The effect of aging and CR on the expression of selected biomarkers of cerebellum aging is measued. Relative amount of mRNA compared to young control is shown for each sample. (A) C4, (B) C1qa, (C) Ctss, (D) Lzp-s, (E) Gfap. C5, 5-month-old control grouop. Lines above/below bars indicate standard error.
Identification of common pathways in heart and cerebellum aging of multiple mouse strains
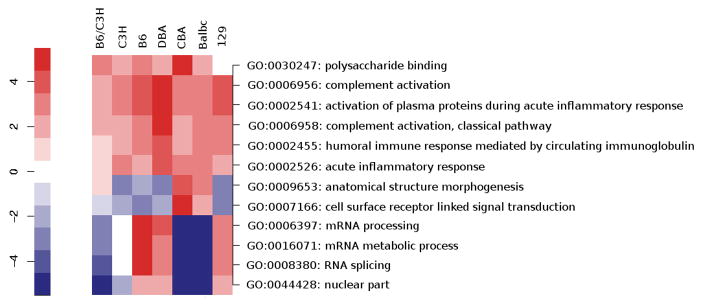
To determine to what extent aging is associated with the alteration of shared pathways in multiple inbred mouse strains, we performed parametric analysis of gene set enrichment (PAGE), a computational method that allows determination of significant changes in defined gene sets (Kim & Volsky, 2005). For our analysis we used GO Biological Pathway, Cellular component, and Molecular function gene sets. We also calculated z ratios for each gene set, which serve as a normalization factor (Cheadle et al., 2003). An initial comparative analysis between the 7 mouse strains revealed no gene sets significantly enriched (P < 0.05) among all strains in the heart. Relaxing the criteria to at least 6 strains significantly changed out of 7 tested identified several enriched GO gene sets. A robust observation was the induction of genes involved in the complement and innate immune response, including complement activation (GO:0006956), acute inflammatory response (GO:0002526), and activation of plasma proteins during inflammation (GO:0002541) (Fig 3). We had previously reported age-related induction of the complement system in the heart of both B6C3H F1 hybrid mice, and outbred mice (Leinhase et al., 2006; Gold et al., 2006). Our study extends this finding to an additional six strains of inbred mice, and shows that complement activation is a universal feature of aging in the mouse heart. Gene sets involved in mRNA processing and splicing were also altered in expression, but the direction of change as determined by the z ratio varied between strains (Fig. 3). Interestingly, the GO term Polyssacharide Binding (GO:0030247) was significantly and consistently up-regulated in most strains. A gene significantly up-regulated in this gene set in some strains is Chitinase-3-Like 1(CHI3L1). CHI3L1 is a secreted 40 kDa glycoprotein that is up-regulated in a number of human cancers and in non-neoplastic disease states characterized by chronic inflammation and tissue remodeling (Coffman, 2008). Stabilin 1, a receptor expressed on both macrophages and different subtypes of endothelial cells that is induced during chronic inflammation and tumorigenesis (Kzhyshkowska et al., 2006), was also consistently induced. Several other members of this gene set are consistent with alterations in the extracellular matrix (ECM) with aging in the heart.
Fig. 3.
Common pathways of heart aging in multiple mouse strains.
Parametric analysis of gene set enrichment (PAGE) identified gene sets significantly enriched (P < 0.05) in heart aging among at least 6 strains out of 7 tested. Each row corresponds to the transcriptional alteration of each gene set with aging. Gene sets up-regulated with aging are shown in red, while gene sets down-regulated with aging are shown in blue. Labels indicate the GO numbers and GO term of each pathway.
A similar analysis in cerebellum revealed common gene sets altered in multiple mouse strains with aging. Inflammatory gene sets such as positive regulation of immune system process (GO:0002684) and regulation of immune response (GO:0050776) were induced with aging. A key feature of induction of these gene sets is also the activation of complement genes, including C4B (Rostagno et al., 2002). Gene sets related to lysosomomal activity, such as lysosome (GO:0005764) and lytic vacuole (GO:0000323) were also consistently induced across multiple mouse strains (Fig. 4). Gene set members changed in expression included several lysosomal proteases, such as Cathepsins S, D, A and H. Cathepsins are involved in extracellular matrix proteolytic degradation, and the induction of cathepsins in the brain is a marker of astrogliosis and microglial activation (Akahoshi et al., 2007). Interestingly, the expression of Cathepsin D appears to be a hallmark of aging in dogs and the human Alzheimer brain as well (Bi et al., 2003). An imbalance of cathepsins, and defective lysosomes has been postulated to play an important role in human age-related neuronal dysfunction (Nakanishi, 2003). Many other GO terms were significantly changed in expression in at least 5 out of 6 strains analyzed, but the direction of change as evidenced by the z score, was not uniform. Surprisingly, gene sets related to mitochondria were strongly and significantly induced in some strains, but reduced in others (Fig. 4). Our overall analysis suggests induction of transcription of innate immunity genes in both tissues examined, as well as multiple tissue-specific patterns of aging.
Fig. 4.

Common pathways of cerebellum aging in multiple mouse strains.
Parametric analysis of gene set enrichment (PAGE) identified gene sets significantly enriched (P < 0.05) in cerebellum aging among at least 5 strains out of 6 tested. Each row corresponds to the transcriptional alteration of each gene set with aging. Gene sets up-regulated with aging are shown in red, while gene sets down-regulated with aging are shown in blue. Labels indicate the GO numbers and GO term of each pathway.
The effect of antioxidant supplementation on the expression of transcriptional biomarkers of aging
To determine the influences of selected antioxidants on the expression of biomarkers of aging in each tissue, 30-month-old mice fed various antioxidants from 15 months of age were compared with 30-month-old control fed mice (C30). As a positive control for aging retardation, we also analyzed 30-month-old mice on CR from 15 months of age to 30 months of age. We determined the effects of each intervention on the panel of aging markers, generating a tissue-specific “aging prevention index” (API), representing the average effect of an intervention on all biomarkers. In the heart, middle age-onset CR significantly prevented age-related up-regulation of all six biomarkers of aging tested with an API value of 51 (Fig. 5). Among biomarkers of heart aging, only Pah was affected by all antioxidants tested. LA and CQ were not strongly effective in inhibiting age-related increases in expression of biomarkers of heart aging: only one gene, Pah, was affected by LA supplementation and two genes, Pah and Scap2, were affected by CQ (Table 3). There was a moderate anti-aging effect in CU- and AS-supplemented groups (API of CU and AS was 41% and 45%, respectively). Remarkably, AC and TP were even more effective than CR in terms of the API, and also significantly reduced the age-related expression of all biomarkers tested. RE, which we have previously shown to retard cardiac aging (Barger et al., 2008), and LY also showed strong efficacy in inhibiting the cardiac aging biomarkers.
Fig. 5.
The effect of middle age-onset CR and antioxidant supplementation on the expression of biomarkers of heart aging. Relative amount of mRNA compared to young control is shown for each group. (A) C4, (B) Csprs, (C) Pah, (D) Cxcl14, (E) Scap2. C5, 5-month-old control; C30, 30-month-old control; CR, caloric restriction; LA, α-lipoic acid; CQ, coenzyme Q10; RE, resveratrol; CU, curcumin; LY, lycopene; AC, acetyl-L-carnitine; AS, astaxanthin; TP, tempol. Lines above/below bars indicate standard error. The one-way ANOVA test showed significant p-value (P < 0.01) in all genes tested. * Significantly different from C30 (P < 0.05).
Table 3.
Effect of CR or antioxidant supplementation on the expression of biomarkers of heart aging
| Antioxidant |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | CR | LA | CQ | RE | CU | LY | AC | AS | TP |
| C4 | *66 | −48 | −8 | *61 | 27 | *58 | *62 | 11 | *59 |
| Csprs | *52 | 21 | −19 | *48 | *41 | *34 | *68 | *50 | *65 |
| Pah | *47 | *46 | *50 | *58 | *60 | *55 | *71 | *64 | *67 |
| Cxcl14 | *37 | −24 | 24 | *90 | *36 | *81 | *85 | 12 | *98 |
| Scap2 | *60 | 22 | *35 | *25 | *34 | *39 | *44 | *44 | *45 |
| API | 51 | 16 | 19 | 52 | 41 | 51 | 66 | 45 | 66 |
The % inhibition effect of CR or antioxidants was computed as ((O−S)/(O−Y)) × 100, where O, S, and Y are the average signal intensities of the 30-month-old control, 30-month-old CR or antioxidant-supplemented group, and 5-month-old control, respectively. API represents the average effect of the intervention on all markers tested. CR, caloric restriction; LA, α-lipoic acid; CQ, coenzyme Q10; RE, resveratrol; CU, curcumin; LY, lycopene; AC, acetyl-L-carnitine; AS, astaxanthin; TP, tempol.
Significantly different between S and O groups (P < 0.05).
The impact of middle age-onset CR and antioxidant supplementation on the expression of biomarkers of cerebellum aging is shown in Figure 6 and Table 4. Similar to biomarkers of heart aging, all six biomarkers of cerebellum aging were markedly inhibited by CR (a 59% API). The age-associated up-regulation of C1qa was decreased significantly by all eight antioxidant interventions (Fig. 6). As opposed to the effect on heart aging, LA and CQ were the two most effective antioxidants in suppressing age-related induction of biomarkers of aging in the cerebellum, nearly as effective as CR (API of LA and CQ were 58% and 50%, respectively). Supplementation of CU, LY, and AC showed significant effects on the expression of five biomarkers of aging in the cerebellum. However, these antioxidants did not prevent the age-related up-regulation of C4 (Table 4). RE and TP, which were among the most effective antioxidants in heart aging, displayed only marginal efficacy in the cerebellum. These observations suggest that unlike the effect CR, the effects of dietary antioxidant supplementation are tissue-specific.
Fig. 6.
The effect of middle age-onset CR and antioxidant supplementation on the expression of biomarkers of cerebellum aging. Relative amount of mRNA compared to young control is shown for each group. (A) C4, (B) C1qa, (C) Ctss, (D) Lzp-s, (E) Gfap. C5, 5-month-old control; C30, 30-month-old control; CR, caloric restriction; LA, α-lipoic acid; CQ, coenzyme Q10; RE, resveratrol; CU, curcumin; LY, lycopene; AC, acetyl-L-carnitine; AS, astaxanthin; TP, tempol. Lines above/below bars indicate standard error. The one-way ANOVA test showed significant p-value (P < 0.01) in all genes tested. * Significantly different from C30 (P < 0.05).
Table 4.
Effect of CR or antioxidant supplementation on the expression of biomarkers of cerebellum aging
| Antioxidant |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | CR | LA | CQ | RE | CU | LY | AC | AS | TP |
| C4 | *34 | *48 | *43 | 8 | −2 | −5 | 0 | −8 | −21 |
| C1qa | *61 | *54 | *45 | *83 | *54 | *90 | *93 | *76 | *87 |
| Ctss | *62 | *63 | *58 | 18 | *45 | *42 | *56 | *41 | *37 |
| Lzp-s | *87 | *77 | 59 | 53 | *76 | *70 | *68 | 60 | 45 |
| Gfap | *54 | *57 | *48 | 17 | *53 | *35 | *30 | *36 | 23 |
| API | 59 | 58 | 50 | 37 | 46 | 46 | 49 | 42 | 36 |
The % inhibition effect of CR or antioxidants was computed as ((O−S)/(O−Y)) × 100, where O, S, and Y are the average signal intensities of the 30-month-old control, 30-month-old CR or antioxidant-supplemented group, and 5-month-old control, respectively. API represents the average effect of the intervention on all markers tested. CR, caloric restriction; LA, α-lipoic acid; CQ, coenzyme Q10; RE, resveratrol; CU, curcumin; LY, lycopene; AC, acetyl-L-carnitine; AS, astaxanthin; TP, tempol.
Significantly different between S and O groups (P < 0.05).
Discussion
In short-lived organisms, the examination of survival curves is practical, and therefore useful in assaying aging rates. However, for most mammalian species, survival curves are less practical due to their relatively long lifespan and complexity. Given the long lifespan of mammals, it would be useful to establish organ-specific biomarkers of aging for evaluating the efficacy of interventions. Our comparison of gene expression patterns in heart and cerebellum of multiple strains of mice revealed tissue-specific biomarkers of aging that can be used to measure the aging process. Although thousands of genes were changed in expression with aging in each individual strain, there was a relatively small number of common genes changed in all strains tested: 20 genes in the heart and 99 genes in the cerebellum. One possible explanation for this finding is that many biological processes associated with aging are strain-specific. Aging resulted in up-regulation of several genes that are involved in immune and inflammatory responses related to innate immunity. The expression of C4 was induced with aging in both heart and cerebellum. C4 prevents early stage autoimmune disease (Paul et al., 2002) and mice with a disrupted C4 locus showed an impaired immune response (Gadjeva et al., 2002). In the heart, two more genes involved in the immune response were identified as biomarkers of aging. Cxcl14 is a potent chemoattractant that is ubiquitously expressed in normal tissues, but absent in many tumor cell lines (Frederick et al., 2000) and Scap2 is required for the activation of immune system (Togni et al., 2005).
In addition to C4, several genes involved in the immune and inflammatory response were increased in expression with aging in the cerebellum, including the lysosomal proteases cathepsin D, cathepsin S, cathepsin Z, and three components of C1q (alpha, beta, and gamma polypeptide) involved in innate imunity. The lysosomal protease cathepsin S is involved in degradation of protein antigens and controls intracellular trafficking of class II MHC molecules (Hsieh et al., 2002). The activation of the classical complement system was reported in the nondemented aged human brain and also in early-stage Alzheimer’s disease (Zanjani et al., 2005), and an increase of C1q beta polypeptide mRNA was found in aging rats (Pasinetti et al., 1999). Taken together, our observations suggest that normal aging in the heart and brain is associated with a transcriptional pattern indicative of heightened immune and inflammatory responses. Interestingly, the expression of complement activation genes has been shown in skeletal muscle, kidney, and brain in humans (Zahn et al., 2006). The expression of innate immunity genes may be due to the activation of an ancient NF-κB signaling pathway of host defense in multicellular organisms (Salminen et al., 2008). This signaling system may connect genotoxic stress, inflammation, and apoptosis, and therefore play an important role in the origin of aging phenotypes and age-related diseases (Salminen et al., 2008). Previous studies have shown that the expression of Gfap, the first validated brain aging transcriptional marker, increases progressively during aging in humans and rodent models (Nichols et al., 1993). It is reassuring that our screen identified Gfap, and also that the expression of this gene is reduced by CR at all ages examined. Some of the inflammatory markers reported in this study were also identified in our original DNA microarray analysis of heart (Lee et al., 2002) and brain (Lee et al., 2000). Interestingly, gene sets related to the immune system, such as complement activation (GO:0006958) and regulation of the immune system (GO:0050776), were induced in both heart and cerebellum. Examination of these gene sets suggests that genes involved in innate immunity account for the majority of genes induced. Possibly, induction of these and other genes related to the immune system is a consequence of either increased levels of monocytes/macrophages in tissues, or increased levels of cytokines, as demonstrated in adipose (Wu et al., 2007) and brain (Ye & Johnson, 1999) tissues of aged mice. In contrast, GO categories related to mitochondria were induced in some strains, but suppressed in others. This observation suggests that a reduction in the expression of genes related to mitochondria and energy metabolism is not a universal feature of aging in mice.
Our data revealed two biomarkers of aging that are common in both heart and cerebellum: C4 and tissue inhibitor of metalloproteinase 2 (TIMP2). Aging is a major risk factor for the development of arterial stiffness and vascular disease such as hypertension and atherosclerosis (Lakatta, 2002), and it is associated with the imbalance between matrix metalloproteinases and their endogenous inhibitors, tissue inhibitors of metalloproteinases (Dollery et al., 1995; Zervoudaki et al., 2003). In mice, new fibrovascular tissue from old animals expressed more TIMP2 than did corresponding tissue from young mice (Koike et al., 2003). Comparison between young and old human microvascular endothelial cell lines revealed that TIMP2 is expressed at higher levels in cell lines from old humans (McNulty et al., 2005). Interestingly, a transcriptional profiling of human tissues with aging identified TIMP1 as the gene displaying the highest change in gene expression in multiple tissues in humans (Zahn et al., 2006). The transcriptional alterations of TIMP2 in multiple strains of aged mice are consistent with these previous observations, suggesting that elevated levels of TIMP2 may modulate impaired angiogenesis and fibrosis in aged tissues.
Previous studies reported the impact of antioxidants on age-related gene expression patterns in mice. Dietary supplementation with LA or CQ results in transcriptional changes associated with reduced oxidative stress in heart, but these antioxidants did not extend maximum life span and reduce tumor incidence (Lee et al., 2004). Middle age-onset dietary supplementation of vitamin E also showed a partial inhibitory effect on age-related transcriptional alteration in heart and brain, but was not as effective as CR (Park et al., 2008; Lee et al., 2002). We have previously shown that RE can prevent age-related cardiac dysfunction and transcriptional alterations associated with cardiac aging (Barger et al., 2008), and these findings are in agreement with the strong effect of RE in inhibiting transcriptional markers of aging reported in this study. In the heart, AC was the natural compound that displayed the largest inhibition in the expression of the transcriptional markers. In rats, short-term supplementation with AC reduced age-related alterations in lipid metabolism in multiple tissues including normalization of the cholesterol/phospholipid ratio (Tanaka et al., 2004) and reduced DNA damage in the brain (Haripriya et al., 2005). We have previously identified transcriptional evidence for alterations in lipid metabolism as a major feature of aging in the heart, and showed that CR, but not dietary antioxidants, can prevent these alterations (Park et al., 2008; Lee et al., 2004). Thus, we postulate that similar to RE, AC may be acting to mimic the metabolic effects of CR in the heart.
We note that a major finding of this study is the remarkable difference in efficacy of the tested compounds. In the heart, RE, LY, AC, and TP were at least as effective as CR, whereas LA and CQ were the most effective antioxidants tested in the cerebellum. Our studies provide support for an important role of oxidative stress in aging, but suggest that the effects of individual antioxidants are tissue-specific. Robust transcriptional biomarkers of aging will be useful for designing combinations of dietary antioxidants that will be effective in inhibiting the aging process in individual tissues in mammals.
Experimental procedures
Animals and dietary manipulations
Different strains of male mice were purchased from Harlan Sprague-Dawley at 6–7 weeks of age. Mice were housed singly in a pathogen-free facility and provided acidified water ad libitum. Each mouse was fed 84 kcal per week of AIN-76A diet. For the study of CR, B6 mice were used: the control group was fed 84 kcal per week of AIN-76A and the CR group was fed 63 kcal per week (a 25% CR) from 5 months of age. In order to avoid malnutrition, the restricted diet was enriched in protein, vitamins, and minerals. The effect of dietary supplementation of antioxidant was performed in B6C3F1 mice. The each group was fed 84 kcal per week of AIN-93M diet mixed with each antioxidant: LA (600 mg kg−1 of diet), CQ (100 mg kg−1 of diet), RE (50 mg kg−1 of diet), CU (500 mg kg−1 of diet), LY (250 mg kg−1 of diet), AC (1 g kg−1 of diet), AS (1 g kg−1 of diet), and TP (5 mg kg−1 of diet). The antioxidant groups were supplemented with each antioxidant since middle age, since we intended to test the effect of middle-age onset dietary supplementation. As a control, we also included mice under CR since 15 months of age. At the age of 30 months, mice were euthanized by rapid cervical dislocation and tissues were immediately frozen in liquid nitrogen and stored at −80 °C. All aspects of animal care were approved by the appropriate university committees and conformed to institutional guidelines.
RNA sample preparation and hybridization
Total RNA was extracted from frozen tissue and converted to double-stranded cDNA after purifying mRNA. Biotin-labeled cRNA was made from double-stranded cDNA and then hybridized to the gene chip as previously described (Lee et al., 1999). Following hybridization, gene chip was installed in a fluidics system for washes and staining. The signals on gene chip were read using a Hewlett Packard GeneArray Scanner (Affymetrix, Santa Clara, CA, USA). The averaged images collected from two scanned images were used as raw data for statistical analysis. We used five animals per group, and hybridized each sample to independent DNA chips, because previous work from our laboratory suggests that variability between individuals is higher than variability observed in replicate hybridizations of the same samples (Weindruch et al., 2002).
Microarray data analysis
Preliminary data analysis was done using Affymetrix algorithms for microarray data analysis, GCOS (GeneChip Operating Software). Detailed protocols for data analysis and extensive documentation of sensitivity and quantitative aspects of the method have been described previously (Lee et al., 2002). Gene expression change was called significant when the p-value (P) was < 0.05. To obtain posterior true positive probabilities (pp) for each gene, we used a mixture modeling approach (Allison, 2002) that uses the frequentist Ps and incorporates them into a mixture model. The pp is the Bayesian probability that a gene is truly different between groups in mean expression level. The raw data of each DNA chip are provided as supporting information (Table S3 and S4).
Pathway analysis of microarray data
To identify common pathways with aging in multiple inbred mouse strains, parametric analysis of gene set enrichment (PAGE) was employed (Kim & Volsky, 2005). We input a list of probe set IDs and their t-test statistic values to PAGE. For multiple test correction, we used Benjamini-Hochberg FDR estimate. Only GO terms that have at least 10 and at most 1,000 genes and have level 3 and below were analyzed. Probe set IDs filtered from the original series were used. The filtering process deletes the following probe set ids: probe sets ending with x_at and s_at, probe sets annotated with more than one gene, probe sets not mapped to a gene, and probe sets not with the highest average SI for corresponding gene. z ratios was also determined for each gene set (Cheadle et al., 2003). All GO terms and pathways significantly altered with aging in each strain were shown in Table S5 and S6.
Real-time quantitative RT-PCR
mRNA quantification was performed using real-time quantitative RT-PCR with ABI prism 7000 Sequence Detection System (TaqMan) (Applied Biosystems, Foster City, CA, USA. Template mRNA was first converted to double-stranded cDNA and then amplified by Taq DNA polymerase. Gene-specific TaqMan probes contain a fluorescent reporter at the 5′ end of the probe. As the PCR cycle progresses, the degradation and release of the fluorescent reporter by Taq DNA polymerase results in fluorescence at 518 nm. The accumulation of PCR products, therefore, is detected directly by monitoring the increase in fluorescence during the amplification process. Gene-specific primers and probe sets were purchased from Assays-on-Demand Gene Expression probes (Applied Biosystems, Foster City, CA, USA): Pah (Mm00500918_m1), C4 (Mm00437890_m1), Cxcl14 (Mm00444699_m1), Scap2 (Mm00490022_m1), C1qa (Mm00432142_m1), Lzp-s (Mm00657323_m1), Gfap (Mm00546086_m1), Ctss (Mm00457902_m1), and TATA binding protein (Tbp) (Mm00446973_m1). For Csprs and copine 2 (Cpne2), we designed our own primers and probe sets: for Csprs, 5′-GTT ATC CAT TGA ACT CTC CAT CCT TT-3′ (forward primer), 5′-TGG TCC AAG TCC CAG CTA GAA-3′ (reverse primer), and 5′-FAM-TGG ATT CTG CTA AGT ACA G-TAMRA-3′ (fluorescent probe); for Cpne2, 5′-TCT CAG TGC TGT GTG TGC AAA G-3′ (forward primer), 5′-TCC CGG TCC AGC AGG TT-3′ (reverse primer), and 5′-FAM-CTG TCA GTG AGT GGC CA-TAMRA-3′ (fluorescent probe). Tbp and Cpne2 were used as control genes for normalization.
Supplementary Material
Table S1 Age-related fold change of biomarkers of heart aging
Table S2 Age-related fold change of biomarkers of cerebellum aging
Table S3 Normalized signal intensities and absent/present calls of microarray data of heart
Table S4 Normalized signal intensities and absent/present calls of microarray data of cerebellum
Table S5 Pathways significantly altered with heart aging
Table S6 Pathway significantly altered with cerebellum aging
Acknowledgments
We thank Roger Klopp for maintaining mouse strains and help with tissue collection. We also wish to thank members of the Prolla and Weindruch labs for helpful advice and critical discussion. This work was supported by NIH grant RO1AG020681.
References
- Akahoshi N, Murashima YL, Himi T, Ishizaki Y, Ishii I. Increased expression of the lysosomal protease cathepsin S in hippocampal microglia following kainate-induced seizures. Neurosci Lett. 2007;429:136–141. doi: 10.1016/j.neulet.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Allison DB, Gadbury GL, Heo M, Fernandez JR, Lee CK, Prolla TA, Weindruch R. A mixture model approach for the analysis of microarray gene expression data. Comput Statist Data Anal. 2002;39:1–20. [Google Scholar]
- Barger JL, Kayo T, Pugh TD, Prolla TA, Weindruch R. Short-term consumption of a resveratrol-containing nutraceutical mixture mimics gene expression of long-term caloric restriction in mouse heart. Exp Gerontol. 2008;43:859–866. doi: 10.1016/j.exger.2008.06.013. [DOI] [PubMed] [Google Scholar]
- Barger JL, Kayo T, Vann JM, Arias EB, Wang J, Hacker TA, Wang Y, Raederstorff D, Morrow JD, Leeuwenburgh C, Allison DB, Saupe KW, Cartee GD, Weindruch R, Prolla TA. A low dose of dietary resveratrol partially mimics caloric restriction and retards aging parameters in mice. PLoS One. 2008;3:e2264. doi: 10.1371/journal.pone.0002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barger JL, Walford RL, Weindruch R. The retardation of aging by caloric restriction: its significance in the transgenic era. Exp Gerontol. 2003;38:1343–1351. doi: 10.1016/j.exger.2003.10.017. [DOI] [PubMed] [Google Scholar]
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Head E, Cotman CW, Lynch G. Spatial patterns of mammalian brain aging: distribution of cathepsin D-immunoreactive cell bodies and dystrophic dendrites in aging dogs resembles that in Alzheimer’s disease. J Comp Neurol. 2003;464:371–381. doi: 10.1002/cne.10795. [DOI] [PubMed] [Google Scholar]
- Boes M, van der Wel N, Peperzak V, Kim YM, Peters PJ, Ploegh H. In vivo control of endosomal architecture by class II-associated invariant chain and cathepsin S. Eur J Immunol. 2005;35:2552–2562. doi: 10.1002/eji.200526323. [DOI] [PubMed] [Google Scholar]
- Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z score transformation. J Mol Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen R, Alhonen L, Wahlfors J, Jakobsen M, Jensen TG. Characterization of transgenic mice with the expression of phenylalanine hydroxylase and GTP cyclohydrolase I in the skin. Exp Dermatol. 2005;14:535–542. doi: 10.1111/j.0906-6705.2005.00326.x. [DOI] [PubMed] [Google Scholar]
- Coffman FD. Chitinase 3-like-1 (CHI3L1): a putative disease marker at the interface of proteomics and glycomics. Crit Rev Clin Lab Sci. 2008;45:531–562. doi: 10.1080/10408360802334743. [DOI] [PubMed] [Google Scholar]
- Dhahbi JM, Kim HJ, Mote PL, Beaver RJ, Spindler SR. Temporal linkage between the phenotypic and genomic responses to caloric restriction. Proc Natl Acad Sci USA. 2004;101:5524–5529. doi: 10.1073/pnas.0305300101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollery CM, McEwan JR, Henney AM. Matrix metalloproteinases and cardiovascular disease. Circ Res. 1995;77:863–868. doi: 10.1161/01.res.77.5.863. [DOI] [PubMed] [Google Scholar]
- Eng LF, Vanderhaeghen JJ, Bignami A, Gerstl B. An acidic protein isolated from fibrous astrocytes. Brain Res. 1971;28:351–354. doi: 10.1016/0006-8993(71)90668-8. [DOI] [PubMed] [Google Scholar]
- Frederick MJ, Henderson Y, Xu X, Deavers MT, Sahin AA, Wu H, Lewis DE, El-Naggar AK, Clayman GL. In vivo expression of the novel CXC chemokine BRAK in normal and cancerous human tissue. Am J Pathol. 2000;156:1937–1950. doi: 10.1016/S0002-9440(10)65067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadjeva M, Verschoor A, Brockman MA, Jezak H, Shen LM, Knipe DM, Carroll MC. Macrophage-derived complement component C4 can restore humoral immunity in C4-deficient mice. J Immunol. 2002;169:5489–5495. doi: 10.4049/jimmunol.169.10.5489. [DOI] [PubMed] [Google Scholar]
- Gold B, Merriam JE, Zernant J, Hancox LS, Taiber AJ, Gehrs K, Cramer K, Neel J, Bergeron J, Barile GR, Smith RT, Hageman GS, Dean M, Allikmets R AMD Genetics Clinical Study Group. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet. 2006;38:458–462. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber J, Tang SY, Halliwell B. Evidence for a trade-off between survival and fitness caused by resveratrol treatment of Caenorhabditis elegans. Ann N Y Acad Sci. 2007;1100:530–542. doi: 10.1196/annals.1395.059. [DOI] [PubMed] [Google Scholar]
- Hagen TM, Ingersoll RT, Wehr CM, Lykkesfeldt J, Vinarsky V, Bartholomew JC, Song MH, Ames BN. Acetyl-L-carnitine fed to old rats partially restores mitochondrial function and ambulatory activity. Proc Natl Acad Sci USA. 1998a;95:9562–9566. doi: 10.1073/pnas.95.16.9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen TM, Wehr CM, Ames BN. Mitochondrial decay in aging. Reversal through supplementation of acetyl-L-carnitine and N-tert-butyl-alpha-phenyl-nitrone. Ann N Y Acad Sci. 1998b;854:214–223. doi: 10.1111/j.1749-6632.1998.tb09904.x. [DOI] [PubMed] [Google Scholar]
- Haripriya D, Sangeetha P, Kanchana A, Balu M, Panneerselvam C. Modulation of age-associated oxidative DNA damage in rat brain cerebral cortex, striatum and hippocampus by L-carnitine. Exp Gerontol. 2005;40:129–135. doi: 10.1016/j.exger.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Higami Y, Pugh TD, Page GP, Allison DB, Prolla TA, Weindruch R. Adipose tissue energy metabolism: altered gene expression profile of mice subjected to long-term caloric restriction. FASEB J. 2004;18:415–417. doi: 10.1096/fj.03-0678fje. [DOI] [PubMed] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. [see comment] Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- Hsieh CS, deRoos P, Honey K, Beers C, Rudensky AY. A role for cathepsin L and cathepsin S in peptide generation for MHC class II presentation. J Immunol. 2002;168:2618–2625. doi: 10.4049/jimmunol.168.6.2618. [DOI] [PubMed] [Google Scholar]
- Kawai J, Shinagawa A, Shibata K, Yoshino M, Itoh M, Ishii Y, Arakawa T, Hara A, Fukunishi Y, Konno H, Adachi J, Fukuda S, Aizawa K, Izawa M, Nishi K, Kiyosawa H, Kondo S, Yamanaka I, Saito T, Okazaki Y, Gojobori T, Bono H, Kasukawa T, Saito R, Kadota K, Matsuda H, Ashburner M, Batalov S, Casavant T, Fleischmann W, Gaasterland T, Gissi C, King B, Kochiwa H, Kuehl P, Lewis S, Matsuo Y, Nikaido I, Pesole G, Quackenbush J, Schriml LM, Staubli F, Suzuki R, Tomita M, Wagner L, Washio T, Sakai K, Okido T, Furuno M, Aono H, Baldarelli R, Barsh G, Blake J, Boffelli D, Bojunga N, Carninci P, de Bonaldo MF, Brownstein MJ, Bult C, Fletcher C, Fujita M, Gariboldi M, Gustincich S, Hill D, Hofmann M, Hume DA, Kamiya M, Lee NH, Lyons P, Marchionni L, Mashima J, Mazzarelli J, Mombaerts P, Nordone P, Ring B, Ringwald M, Rodriguez I, Sakamoto N, Sasaki H, Sato K, Schönbach C, Seya T, Shibata Y, Storch KF, Suzuki H, Toyo-oka K, Wang KH, Weitz C, Whittaker C, Wilming L, Wynshaw-Boris A, Yoshida K, Hasegawa Y, Kawaji H, Kohtsuki S, Hayashizaki Y RIKEN Genome Exploration Research Group Phase II Team and the FANTOM Consortium. Functional annotation of a full-length mouse cDNA collection. Nature. 2001;409:685–690. doi: 10.1038/35055500. [DOI] [PubMed] [Google Scholar]
- Kim SY, Volsky DJ. PAGE: parametric analysis of gene set enrichment. BMC Bioinformatics. 2005;6:144. doi: 10.1186/1471-2105-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike T, Vernon RB, Gooden MD, Sadoun E, Reed MJ. Inhibited angiogenesis in aging: a role for TIMP-2. J Gerontol A Biol Sci Med Sci. 2003;58:B798–805. doi: 10.1093/gerona/58.9.b798. [DOI] [PubMed] [Google Scholar]
- Kzhyshkowska J, Gratchev A, Goerdt S. Stabilin-1, a homeostatic scavenger receptor with multiple functions. J Cell Mol Med. 2006;10:635–649. doi: 10.1111/j.1582-4934.2006.tb00425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakatta EG. Age-associated cardiovascular changes in health: impact on cardiovascular disease in older persons. Heart Fail Rev. 2002;7:29–49. doi: 10.1023/a:1013797722156. [DOI] [PubMed] [Google Scholar]
- Lee CK, Allison DB, Brand J, Weindruch R, Prolla TA. Transcriptional profiles associated with aging and middle age-onset caloric restriction in mouse hearts. Proc Natl Acad Sci USA. 2002;99:14988–14993. doi: 10.1073/pnas.232308999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CK, Klopp RG, Weindruch R, Prolla TA. Gene expression profile of aging and its retardation by caloric restriction. Science. 1999;285:1390–1393. doi: 10.1126/science.285.5432.1390. [DOI] [PubMed] [Google Scholar]
- Lee CK, Pugh TD, Klopp RG, Edwards J, Allison DB, Weindruch R, Prolla TA. The impact of alpha-lipoic acid, coenzyme Q10 and caloric restriction on life span and gene expression patterns in mice. Free Radic Biol Med. 2004;36:1043–1057. doi: 10.1016/j.freeradbiomed.2004.01.015. [DOI] [PubMed] [Google Scholar]
- Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nat Genet. 2000;25:294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- Leinhase I, Holers VM, Thurman JM, Harhausen D, Schmidt OI, Pietzcker M, Taha ME, Rittirsch D, Huber-Lang M, Smith WR, Ward PA, Stahel PF. Reduced neuronal cell death after experimental brain injury in mice lacking a functional alternative pathway of complement activation. BMC Neurosci. 2006;7:55. doi: 10.1186/1471-2202-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim GP, Chu T, Yang F, Beech W, Frautschy SA, Cole GM. The curry spice curcumin reduces oxidative damage and amyloid pathology in an Alzheimer transgenic mouse. J Neurosci. 2001;21:8370–8377. doi: 10.1523/JNEUROSCI.21-21-08370.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loerch PM, Lu T, Dakin KA, Vann JM, Isaacs A, Geula C, Wang J, Pan Y, Gabuzda DH, Li C, Prolla TA, Yankner BA. Evolution of the aging brain transcriptome and synaptic regulation. PLoS One. 2008;3:e3329. doi: 10.1371/journal.pone.0003329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie-Cardine A, Verhagen AM, Eckerskorn C, Schraven B. SKAP-HOM, a novel adaptor protein homologous to the FYN-associated protein SKAP55. FEBS Lett. 1998;435:55–60. doi: 10.1016/s0014-5793(98)01040-0. [DOI] [PubMed] [Google Scholar]
- McNulty M, Spiers P, McGovern E, Feely J. Aging is associated with increased matrix metalloproteinase-2 activity in the human aorta. Am J Hypertens. 2005;18:504–509. doi: 10.1016/j.amjhyper.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Nakanishi H. Neuronal and microglial cathepsins in aging and age-related diseases. Ageing Res Rev. 2003;2:367–381. doi: 10.1016/s1568-1637(03)00027-8. [DOI] [PubMed] [Google Scholar]
- Navarro-Incio AM, Tolivia-Fernandez J. The involvement of apolipoprotein D in pathologies affecting the nervous system. Rev Neurol. 2004;38:1166–1175. [PubMed] [Google Scholar]
- Nawashiro H, Huang S, Brenner M, Shima K, Hallenbeck JM. ICP monitoring following bilateral carotid occlusion in GFAP-null mice. Acta Neurochir Suppl. 2002;81:269–270. doi: 10.1007/978-3-7091-6738-0_69. [DOI] [PubMed] [Google Scholar]
- Nichols NR, Day JR, Laping NJ, Johnson SA, Finch CE. GFAP mRNA increases with age in rats and human brain. Neurobiol Aging. 1993;14:421–429. doi: 10.1016/0197-4580(93)90100-p. [DOI] [PubMed] [Google Scholar]
- Palozza P, Krinsky NI. Astaxanthin and canthaxanthin are potent antioxidants in a membrane model. Arch Biochem Biophysic. 1992;297:291–295. doi: 10.1016/0003-9861(92)90675-m. [DOI] [PubMed] [Google Scholar]
- Parfitt VJ, Rubba P, Bolton C, Marotta G, Hartog M, Mancini M. A comparison of antioxidant status and free radical peroxidation of plasma lipoproteins in healthy young persons from Naples and Bristol. Eur Heart J. 1994;15:871–876. doi: 10.1093/oxfordjournals.eurheartj.a060603. [DOI] [PubMed] [Google Scholar]
- Park SK, Page GP, Kim K, Allison DB, Meydani M, Weindruch R, Prolla TA. alpha- and gamma-Tocopherol prevent age-related transcriptional alterations in the heart and brain of mice. J Nutr. 2008;138:1010–1018. doi: 10.1093/jn/138.6.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SK, Prolla TA. Gene expression profiling studies of aging in cardiac and skeletal muscles. Cardiovasc Res. 2005a;66:205–212. doi: 10.1016/j.cardiores.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Park SK, Prolla TA. Lessons learned from gene expression profile studies of aging and caloric restriction. Ageing Res Rev. 2005b;4:55–65. doi: 10.1016/j.arr.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Pasinetti GM, Hassler M, Stone D, Finch CE. Glial gene expression during aging in rat striatum and in long-term responses to 6-OHDA lesions. Synapse. 1999;31:278–284. doi: 10.1002/(SICI)1098-2396(19990315)31:4<278::AID-SYN5>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Paul E, Pozdnyakova OO, Mitchell E, Carroll MC. Anti-DNA autoreactivity in C4-deficient mice. Eur J Immunol. 2002;32:2672–2679. doi: 10.1002/1521-4141(200209)32:9<2672::AID-IMMU2672>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Prolla TA, Mattson MP. Molecular mechanisms of brain aging and neurodegenerative disorders: lessons from dietary restriction. Trends Neurosci. 2001;24:S21–31. doi: 10.1016/s0166-2236(00)01957-3. [DOI] [PubMed] [Google Scholar]
- Röcken C, Becker K, Fändrich M, Schroeckh V, Stix B, Rath T, Kähne T, Dierkes J, Roessner A, Albert FW. ALys amyloidosis caused by compound heterozygosity in exon 2 (Thr70Asn) and exon 4 (Trp112Arg) of the lysozyme gene. Hum Mutat. 2006;27:119–120. doi: 10.1002/humu.9393. [DOI] [PubMed] [Google Scholar]
- Rostagno A, Revesz T, Lashley T, Tomidokoro Y, Magnotti L, Braendgaard H, Plant G, Bojsen-Møller M, Holton J, Frangione B, Ghiso J. Complement activation in chromosome 13 dementias. Similarities with Alzheimer’s disease. J Biol Chem. 2002;277:49782–29790. doi: 10.1074/jbc.M206448200. [DOI] [PubMed] [Google Scholar]
- Rozovsky I, Wei M, Morgan TE, Finch CE. Reversible age impairments in neurite outgrowth by manipulations of astrocytic GFAP. Neurobiol Aging. 2005;26:705–715. doi: 10.1016/j.neurobiolaging.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Salminen A, Huuskonen J, Ojala J, Kauppinen A, Kaarniranta K, Suuronen T. Activation of innate immunity system during aging: NF-kB signaling is the molecular culprit of inflamm-aging. Ageing Res Rev. 2008;7:83–105. doi: 10.1016/j.arr.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Sharpless NE. Ink4a/Arf links senescence and aging. Exp Gerontol. 2004;39:1751–1759. doi: 10.1016/j.exger.2004.06.025. [DOI] [PubMed] [Google Scholar]
- Shurin GV, Ferris R, Tourkova IL, Perez L, Lokshin A, Balkir L, Collins B, Chatta GS, Shurin MR. Loss of new chemokine CXCL14 in tumor tissue is associated with low infiltration by dendritic cells (DC), while restoration of human CXCL14 expression in tumor cells causes attraction of DC both in vitro and in vivo. J Immunol. 2005;174:5490–5498. doi: 10.4049/jimmunol.174.9.5490. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Sasaki R, Fukui F, Waki H, Kawabata T, Okazaki M, Hasegawa K, Ando S. Acetyl-L-carnitine supplementation restores decreased tissue carnitine levels and impaired lipid metabolism in aged rats. J Lipid Res. 2004;45:729–735. doi: 10.1194/jlr.M300425-JLR200. [DOI] [PubMed] [Google Scholar]
- Tatchum-Talom R, Martin MS. Tempol improves vascular function in the mesenteric vascular bed of senescent rats. Can J Physiol Pharmacol. 2004;82:200–207. doi: 10.1139/y04-010. [DOI] [PubMed] [Google Scholar]
- Togni M, Swanson KD, Reimann S, Kliche S, Pearce AC, Simeoni L, Reinhold D, Wienands J, Neel BG, Schraven B, Gerber A. Regulation of in vitro and in vivo immune functions by the cytosolic adaptor protein SKAP-HOM. Mol Cell Biol. 2005;25:8052–8063. doi: 10.1128/MCB.25.18.8052-8063.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Remmen H, Ikeno Y, Hamilton M, Pahlavani M, Wolf N, Thorpe SR, Alderson NL, Baynes JW, Epstein CJ, Huang TT, Nelson J, Strong R, Richardson A. Lifelong reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiol Genomics. 2003;16:29–37. doi: 10.1152/physiolgenomics.00122.2003. [DOI] [PubMed] [Google Scholar]
- Wei LC, Shi M, Chen LW, Cao R, Zhang P, Chan YS. Nestin-containing cells express glial fibrillary acidic protein in the proliferative regions of central nervous system of postnatal developing and adult mice. Brain Res Dev Brain Res. 2002;139:9–17. doi: 10.1016/s0165-3806(02)00509-6. [DOI] [PubMed] [Google Scholar]
- Weichenhan D, Kunze B, Winking H, van Geel M, Osoegawa K, de Jong PJ, Traut W. Source and component genes of a 6–200 Mb gene cluster in the house mouse. Mamm Genome. 2001;12:590–594. doi: 10.1007/s00335-001-3015-9. [DOI] [PubMed] [Google Scholar]
- Weindruch R, Kayo T, Lee CK, Prolla TA. Gene expression profiling of aging using DNA microarrays. Mech Ageing Dev. 2002;123:177–193. doi: 10.1016/s0047-6374(01)00344-x. [DOI] [PubMed] [Google Scholar]
- Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans.[erratum appears in Nature. 2004 Sep 2;431(7004):107] Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- Wu D, Ren Z, Pae M, Guo W, Cui X, Merrill AH, Meydani SN. Aging up-regulates expression of inflammatory mediators in mouse adipose tissue. J Immunol. 2007;179:4829–4839. doi: 10.4049/jimmunol.179.7.4829. [DOI] [PubMed] [Google Scholar]
- Ye SM, Johnson RW. Increased interleukin-6 expression by microglia from brain of aged mice. J Neuroimmunol. 1999;93:139–148. doi: 10.1016/s0165-5728(98)00217-3. [DOI] [PubMed] [Google Scholar]
- Zanjani H, Finch CE, Kemper C, Atkinson J, McKeel D, Morris JC, Price JL. Complement activation in very early Alzheimer disease. Alzheimer Dis Assoc Disord. 2005;19:55–66. doi: 10.1097/01.wad.0000165506.60370.94. [DOI] [PubMed] [Google Scholar]
- Zervoudaki A, Economou E, Stefanadis C, Pitsavos C, Tsioufis K, Aggeli C, Vasiliadou K, Toutouza M, Toutouzas P. Plasma levels of active extracellular matrix metalloproteinases 2 and 9 in patients with essential hypertension before and after antihypertensive treatment. J Hum Hypertens. 2003;17:119–124. doi: 10.1038/sj.jhh.1001518. [DOI] [PubMed] [Google Scholar]
- Zahn JM, Sonu R, Vogel H, Crane E, Mazan-Mamczarz K, Rabkin R, Davis RW, Becker KG, Owen AB, Kim SK. Transcriptional profiling of aging in human muscle reveals a common aging signature. PLoS Genet. 2006;2:e115. doi: 10.1371/journal.pgen.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao BL, Li XJ, He RG, Cheng SJ, Xin WJ. Scavenging effect of extracts of green tea and natural antioxidants on active oxygen radicals. Cell Biophys. 1989;14:175–185. doi: 10.1007/BF02797132. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Age-related fold change of biomarkers of heart aging
Table S2 Age-related fold change of biomarkers of cerebellum aging
Table S3 Normalized signal intensities and absent/present calls of microarray data of heart
Table S4 Normalized signal intensities and absent/present calls of microarray data of cerebellum
Table S5 Pathways significantly altered with heart aging
Table S6 Pathway significantly altered with cerebellum aging