Abstract
In humans and non-obese diabetic (NOD) mice, defects in immune tolerance result in the spontaneous development of type-1-diabetes. Recent studies have ascribed a breakdown in tolerance to dysfunction in regulatory T-cells (Tregs) that is secondary to reduced IL-2 production by T-cells having the NOD diabetes susceptibility region insulin-dependent diabetes 3 (Idd3). Here we demonstrate a peripheral tolerance defect in the dendritic cells (DCs) of NOD mice that is independent of Tregs. NOD CD8 T-cells specific for islet antigens fail to undergo deletion in the pancreatic lymph nodes. Deletion was promoted by expression of the protective alleles of both Idd3 (Il2) and Idd5 in DCs. We further identify a second tolerance defect that involves endogenous CD4 T-cell expression of the disease promoting NOD alleles of these genetic regions. Pervasive insulitis can be reduced by expression of the Idd3 and Idd5 protective alleles by either the antigen-presenting cell or lymphocytes.
Keywords: Autoimmunity; dendritic cells; tolerance; T cells, diabetes
Introduction
Type-1-diabetes (T1D) is an autoimmune disease caused by T-cell mediated destruction of the insulin producing beta cells in the pancreatic islets. Over 20 insulin-dependent diabetes (Idd) regions have been identified in the NOD mouse model of spontaneous diabetes, and similar genetic complexity exists in humans (1). Several genes have been implicated, including those that encode MHC class II, insulin, CTLA-4 and PTPN22; and significant overlap exists between the protective gene pathways identified in mice and humans, including CTLA-4 and IL-2/CD25 (1, 2). This genetic complexity highlights that multiple cell types acting at different checkpoints contribute to disease etiology and progression.
NOD mice exhibit defects in thymic and peripheral tolerance of CD4 and CD8 T-cells (3, 4). In the periphery, the first opportunity for T-cell tolerance to islet antigens to be established occurs when T-cells are activated by cross-presented islet antigens in the pancreatic lymph nodes (PcLN) and are then deleted (5, 6). In contrast to such abortive activation, islet-antigen specific T-cells survive following activation in the PcLN of NOD mice and migrate to the islets (7). Congenic NOD mice expressing resistance alleles at both Idd3 and Idd5 (Idd3/5), are highly protected from disease (8), and were found to restore CD8 tolerance to islet antigens in the endogenous repertoire (4), by promoting abortive activation of islet antigen specific CD8 T-cells in the PcLN without infiltrating the islets (7). Importantly, expression of protective genes by the host, rather than the transferred antigen specific CD8 T-cells, determined whether tolerance occurred.
Recent studies have identified a dysfunction in Tregs in NOD mice that is secondary to genetic defects in IL-2 production encoded by Idd3 (9, 10). These studies support the hypothesis that defective Treg homeostasis is a primary factor in disease progression and that provision of IL-2, either therapeutically or by genetic modification, restores protection from diabetes. Controversy remains however as to whether Treg dysfunction is a general consequence of an inflammatory environment or an underlying causative factor in diabetes progression (11). The fact that many protective alleles avert disease suggests that other defects in tolerance exist that may precede the requirement for functional Tregs.
Candidate genes encoded in the Idd3 and Idd5 regions include Il2 (Idd3), Ctla4 (Idd5.1), Slc11a1 (Idd5.2) and Acadl (Idd5.3) (8, 10, 12-14). As these genes are expressed in multiple cell types it is likely that they function pleiotropically to affect disease. Here, we identify cells in the periphery that are primarily responsible for defective deletion of activated islet-specific CD8 T-cells in the PcLNs.
Materials and Methods
Mice
Experimental procedures were performed according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals. NOD/MrkTac and NOD-SCID mice were purchased from Taconic. NOD-InsHA mice were previously described (4). The NOD Idd3/5, Idd3 and Idd5 congenic strains have been described previously (8, 10). Idd3/5, Idd3 and Idd5 congenic mice were backcrossed to NOD-InsHA transgenic mice and are referred to as Idd3/5-InsHA, Idd3-InsHA and Idd5-InsHA. Idd3/5-SCID mice were generated by repeated intercrossing of Idd3/5 and NOD-SCID mice and selecting mice homozygous for the appropriate alleles. Idd3/10/18-SCID mice were generated by the same procedure using Line 1538 (NOD.B6 Idd3/10/18) from the Emerging Models Program at Taconic. NOD.129P2(B6)-B2mtm1Unc/J mice were purchased from The Jackson Laboratory and are referred to as NOD-β2M−/−. NOD mice expressing the human CD20 transgene were generated by backcrossing HuCD20Tg mice (15) to NOD/MrkTac mice for >10 generation and then intercrossed with NOD-InsHA mice to obtain InsHA expressing HuCD20Tg mice. Clone-4 TCR NOD mice were previously described (7). NOD-8.3 TCR transgenic mice were purchased from Jackson Laboratories. NOD CD11c-DTR mice were previously described (16). The NOD.B6 Ptprc (Ptprc encodes CD45; NOD mice have the CD45.1 allotype whereas B6 mice have the CD45.2 allotype; and the strain is referred to as NOD-CD45.2 in the manuscript) strain was initiated by gene-selective backcrossing of the B6 Ptprc allele to NOD/JRC for 11 generations. The strain was rederived at Taconic and subsequently backcrossed to the NOD/MrkTac substrain 5 more generations with selection for NOD/B6 recombination events near the proximal and distal ends of Ptprc. The NOD-CD45.2 congenic segment is defined at its proximal boundary by the SNPs rs31730929 and ss49506637 and at its distal boundary by the SNPs rs33594855 and rs32501384. The maximum size of the congenic interval is 1.067 Mb (Mouse Ensembl release 52).
In vivo cell depletion, antibody treatment and virus
CD4 T-cells were depleted in vivo by injecting 300 μg of GK 1.5 on days −10 and −3. Three doses of 500 μg of MR1 mAb (anti-CD40L) kindly provided by Dr. S. Schoenberger (La Jolla Institute for Allergy and Immunology, La Jolla, CA), were injected at day −10, −4 and 0. B7 interactions were blocked with 100 μg anti B7.1 (16-10A1) and 100 μg of anti B7.2 (GL-1) mAbs on days 0, 1, 2 and 3. Rabbit anti-Asialo GM1 polyclonal Ab (300 μg, Cederlane Laboratories) that transiently depletes NK cells was injected on days −10, −5 and 0. Control animals received the equivalent amount of the corresponding rat or rabbit or hamster normal gamma globulin (Jackson ImmunoResearch Laboratories). B cells were depleted in NOD-InsHA HuCD20Tg mice by injecting either 3 mg of anti-CD20 monoclonal antibody, rituximab (Rituxan, MabThera) on days −10 and day−3 or 2H7 on days −10 (2 mg), −3 (0.5 mg) and 0 (0.5 mg). The degree of depletion was determined by flow cytometry in the PcLN resulting in greater than 95% CD4+ T-cell-depletion in GK 1.5 treated mice, 87% of DX5+ depletion in Asialo GM1 treated mice and 85-95% B220+ cell depletion in Rituxan or 2H7 treated mice. Dendritic cells were depleted by treatment with 120 ng/mouse i.p. of DT (List Biological Laboratories, Campbell CA) on days −4, −1 and +2. Depletion of DCs was assessed in spleen cell preparations enriched for DCs by centrifugation over Nycoprep 1.077 (Axis-Shield, Oslo, Norway). Interface cells were harvested before staining with anti-CD11c-APC (BD-Pharmingen San Diego, CA). Recombinant vaccinia virus expressing the H-2Kd restricted epitope IYSTVASSL, amino acid residue 518-526 (VacKdHA) was provided by J.R. Bennink and J. Yewdell (National Institutes of Health, Bethesda, MD). Mice were infected with 1×107 pfu, by i.p. injection.
Purification, adoptive transfer of T-cells and FACS analysis
Naive CD8+Thy1.1+ clone-4 TCR cells or CD8+Thy1.1+ 8.3 TCR cells were isolated from lymph nodes and spleen of NOD clone-4 Thy1.1+ TCR mice or NOD-scid 8.3 Thy1.1+ TCR mice as described (7). CD8+ T-cells were purified using the CD8+ T-cell isolation kit (Miltenyi Biotec) and CFSE labeling was performed as described (17). Recipient mice were injected with 3-4×106 purified CD8+ cells i.v. on day 0. Four days after the transfer PcLN and non-draining LNs from individual mice or pooled from 2-3 mice were prepared. NOD clone-4 cells and 8.3 cells were stained with for Thy1.1 and CD8. All mAbs were obtained from BD Pharmingen (San Diego, CA). Cells were analyzed with a FACS Calibur (Becton Dickinson, Mountain View, CA.) and FlowJo software (Tree Star, Inc.).
Bone marrow chimeras, reconstituted SCID mice and histology
BM cells were harvested and mature T-cells depleted by incubation with anti-Thy 1.2 (J1j.20) supernatant followed by complement-mediated lysis. Recipient mice were lethally irradiated with a single 1100 or 1200 rad dose and provided with antibiotic (neomycin sulfate, 2mg/ml) supplemented water for 10 days. At day 0, 1×107 T-cell depleted donor BM cells were administered i.v. Recipient mice were treated with depleting doses of either anti-CD4 (GK1.5) and anti-CD8 (53-6.2) or anti-Thy1.2 (30H12) mAbs at day + 1 to deplete residual T-cells. In some experiments, recipients were depleted of NK cells by two doses of 300 μg of rabbit anti-Asialo GM1 polyclonal Ab (days −3 and−1) to prevent rejection of non-MHC I expressing cells. Mice were allowed to reconstitute for a period of at least seven weeks. Donor T-cell chimerism as assessed by Thy1.1/1.2 staining was at least 80% and CD11c chimerism as assessed by CD45.1/2 staining was >98% donor origin. MHC-I expression on lymphocytes was determined by staining with anti-H-2Kd(SF1-1.1). SCID mice were reconstituted with total spleen and lymph node cells prepared from 3-week-old donor mice. DCs were depleted with Pan-DC microbeads (Miltenyi Biotec) according to the manufacturer's instructions before transfer of 2-3 × 107 cells i.v. Mice were rested for 6-8 weeks before adoptive transfer of CD8+Thy1.1+8.3 TCR cells. Pancreata were fixed in 10% neutral buffered formalin and paraffin embedded. Hematoxylin and eosin stained sections were scored for insulitis, with at least 30 islets scored per mouse.
BM-derived dendritic cells and macrophages: growth and stimulation conditions
DCs were derived by culturing 2 × 106 BM cells in RPMI-1640 + 10% FCS + 15 ng/ml GM-CSF (PeproTech). Macrophages were derived from BM using a similar protocol, but with 10 ng/ml M-CSF (PeproTech) replacing the GM-CSF. After 8 days the DCs or macrophages were stimulated with 75 U/ml γIFN (Pharmingen) and 0.1 μg/ml LPS (E. coli, Sigma) for 24 or 48 h. DCs were stimulated with zymosan (InvivoGen) 10 μg/ml for 5 h. All cells were harvested and prepared for FACS and RNA or protein analyses. The DCs were 81 to 91 % CD11c+ and the macrophages were 85 to 96% CD11b+ as analyzed by flow cytometry.
Western blots, quantitative PCR and Elisa.
For the analysis of SLC11a1 (NRAMP1) protein expression, BM-derived DCs or macrophages were harvested and then washed in medium containing 2.7 Kunitz Units/μl DNase (Qiagen DNase Set #79254), then lysed with protein lysis buffer containing 1% Nonidet P-40 and 1% Sigma protease inhibitor cocktail (Sigma-Aldrich #P8340). The samples were run on a 10% acrylamide gel, transferred to a Protran nitrocellulose membrane and stained with rabbit anti-mouse Nramp1 primary antibody at 3 μg/ml (Alpha Diagnostic International, #NRAMP12-A) and goat anti-rabbit IgG HRP secondary antibody (Santa Cruz Biotechnology, #sc-2004) at 1.6 ng/ml. Bands were detected using the ECL Advance Western Blotting Detection kit (GE Healthcare, Amersham. #RPN2135). RNA was extracted from DCs or macrophages in TRIZOL® (Invitrogen) and 1000 ng of total RNA were used to make cDNA with Superscript II reverse transcriptase (Invitrogen). TaqMan-based QPCR assays including primers and probes have been described previously for detecting ACADL (12), CTLA-4, and β2M (normalization control) (14) mRNAs. IL-2 was measured in the supernatants by an antibody-capture assay using purified anti-mouse IL-2 (clone JES6-1A12, Pharmingen) bound to 96-well plates to immobilize the secreted IL-2 and biotinylated anti-mouse IL-2 (clone JES6-5H4, Pharmingen) and europium-streptavidin (DELFIA) to detect the captured IL-2. A standard curve of recombinant IL-2 was used to calculate IL-2 protein levels in the supernatants.
Results
CD4 T-cells are not required for survival of islet-specific CD8 T-cells in NOD PcLN
NOD clone-4 TCR transgenic mice express a Kd-restricted TCR with specificity for influenza hemagglutinin (HA) that is expressed under the control of the insulin promoter in NOD-InsHA mice (4). In comparing the fate of CFSE-labeled clone-4 T-cells in NOD-InsHA mice, diabetes resistant Idd3/5-InsHA mice, and conventional mice, we found that deletion of islet-antigen specific T-cells in the PcLNs was defective in NOD mice, yet restored to normal levels in Idd3/5-InsHA recipients (7). Survival was evident as a higher percentage of activated clone-4 cells accumulating in the PcLN (approximately 70%) as compared with mice in which deletion occurred, such as BALB/c, B10.D2 and Idd3/5 (25-35%). The surviving cells then infiltrated the islets of NOD-InsHA mice but not Idd3/5-InsHA mice (7). In order to confirm that clone-4 T-cells transferred into Idd3/5-InsHA recipients are indeed deleted, a small number of Thy1.1+ clone 4 cells (1×104) were transferred into NOD, Idd3/5, NOD-InsHA and Idd3/5-InsHA mice. The recipients were rested for 5 weeks to allow time for activation and possible deletion of the transferred cells in the PcLNs. After 5 weeks any remaining responsive clone-4 cells were expanded by infection with recombinant vaccinia virus expressing the H-2Kd restricted epitope of 518-526 of HA to facilitate detection. One week after infection the frequency of Thy1.1+ NOD clone-4 cells in the spleen was determined (Figure 1A). As expected in the absence of HA antigen, a high frequency of clone-4 cells were recovered from both NOD and Idd3/5 mice. Clone-4 cells were also present at high frequency in NOD-InsHA mice, however, in Idd3/5-InsHA mice they were absent. A somewhat reduced frequency of clone-4 cells was observed in NOD-InsHA mice compared with non-transgenic NOD mice, similar to what we have previously observed in the endogenous HA-specific repertoire (4). This reduction suggests that partial tolerance does result from InsHA expression in NOD mice, however a large population of HA-specific CD8 T-cells still remained when compared with Idd3/5-InsHA mice. InsHA specific CD8 T-cells do not expand following infection with wild type vaccinia (data not shown). We conclude that activation of clone-4 cells in the PcLN of Idd3/5-InsHA mice results in their deletion, a process that is defective in NOD-InsHA mice.
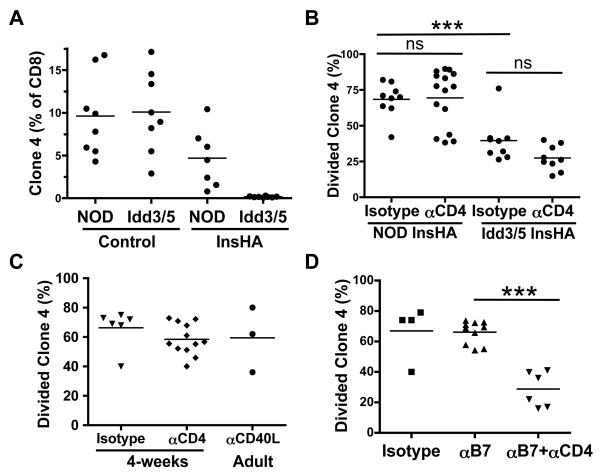
Figure 1. A role for CD4 T-cells in accumulation of NOD clone-4 cells in the PcLN of NOD-InsHA is revealed by co-stimulation blockade.
(A) Thy1.1+ NOD clone-4 cells (1×104) were transferred into NOD, Idd3/5, NOD-InsHA and Idd3/5-InsHA mice. After 5 weeks the mice were infected with Vac-KdHA and 7 days later the frequency of Thy1.1+ clone-4 cells was determined in the spleen by FACS analysis (two pooled experiments). (B) On days −10 and −3, anti-CD4 or isotype control Abs were injected into NOD-InsHA or Idd3/5-InsHA mice. On day 0 CSFE-labeled Thy1.1+ NOD clone-4 cells (3×106), were injected and PcLN analyzed by FACS four days later (three to four pooled experiments). ***p<0.0001, ns: not significant. (C) Adult or 4 week old NOD-InsHA mice were injected with anti-CD4 or anti-CD40L mAb or isotype control on days −10, −3 and 0 before transfer of CFSE labeled clone-4 cells and analysis of PcLN on day 4 (two pooled experiments). (D) NOD-InsHA mice were injected with anti B7.1and anti B7.2 mAbs was on days 0, 1, 2 and 3 with or without CD4 depletion before transfer of CFSE labeled clone-4 cells and analysis of PcLN on day 4 (two pooled experiments). ***p<0.0001.
Cognate CD4 T-cells activate DCs through CD40L-CD40 interaction, enhancing DC expression of B7 costimulatory molecules that promote survival of clone-4 cells (17-20). We hypothesized that effector CD4 T-cells undermine CD8 T-cell tolerance and that removal of CD4 T-cells would restore deletion. To test this, CD4 T-cells were depleted by at least 95% in vivo with anti-CD4 mAb for 10-14 days prior to injection of CFSE-labeled clone-4 cells, to allow time for replacement of DCs previously activated by CD4 T-cells (21). Surprisingly, the frequency of divided clone-4 cells was similarly high in CD4 depleted and non-depleted animals (Figure 1B). Furthermore when CD4 T-cells were depleted in Idd3/5–InsHA mice, clone-4 cells still underwent abortive activation, indicating that CD4 T-cells are not required for deletion mediated by protective alleles at Idd3/5 (Figure 1B, Idd3/5 anti-CD4 vs. isotype control p>0.05). To confirm further that autoreactive CD4 T-cells are not required to prevent deletion of CD8 T-cells, CD4 depletion was begun in 2-week old neonates, before the onset of insulitis and generation of autoreactive CD4 T-cells (22). Again, a high level of accumulation of clone-4 cells was observed in CD4 depleted NOD neonates (p>0.05, Figure 1C). Finally, blocking CD40-CD40L interactions with anti-CD40L mAb did not reduce accumulation of clone-4 cells in the PcLN of NOD-InsHA mice (p>0.05, Figure. 1C). Thus, survival of clone-4 CD8 T-cells in the PcLN of NOD-Ins-HA mice, and abortive activation in Idd3/5 mice occurs independently of CD4 T-cells.
Both removal of CD4 T-cells and costimulation blockade is required to restore deletion of clone-4 cells in NOD PcLNs.
An important survival signal for newly activated CD8 T-cells is provided by B7.1/2 expression on activated DCs (23). Surprisingly, blocking CD28-B7 interaction with a mixture of anti-B7.1 and B7.2 mAbs did not reduce the accumulation of activated clone-4 cells in the PcLN (Figure 1D). However, activated clone-4 cells were significantly reduced by such treatment if the mice were first depleted of CD4 T-cells (p<0.0001 Figure 1D). These results suggest the NOD DCs provide B7 costimulatory signals to clone-4 cells, but when NOD CD4 T-cells are present, such costimulation is either not required or another CD4 T-cell-mediated costimulatory pathway becomes available. Since clone-4 cells are deleted in Idd3/5 PcLNs, we conclude that protective alleles at Idd3 and Idd5 restore the ability of both CD4 T-cells and DCs to promote deletion.
It remained possible that expression of B7 by another cell type (eg. B-cells) is involved and/or that activation of the CD8 T-cell is downstream of a B7 dependent effect on another cell type in which endogenous expression of differential alleles of Idd3/5 genes primarily acts. Consequently, we continued to investigate possible effects from other cell types present in the PcLN.
Neither B cells, nor NK cells are needed to prevent CD8 T-cell tolerance in the PcLN.
To determine whether B cells contribute to CD8 T-cell tolerance in the PcLNs, NOD InsHA mice were mated with huCD20TgNOD mice expressing the human CD20 molecule on all B cells (15). Following treatment with anti-huCD20mAb, which depleted 85-95% of B cells, no reduction was observed in the accumulation of transferred clone-4 cells in NOD-InsHA huCD20Tg mice (p>0.05, Figure 2A). Simultaneous depletion of both B cells and CD4 T-cells also did not reduce the frequency of divided clone-4 cells (p>0.05, Figure 2A).
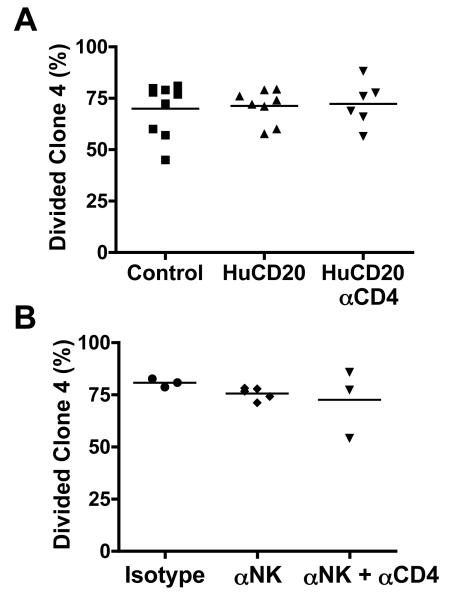
Figure 2. Accumulation of clone-4 CD8 T-cells in NOD-InsHA hosts is not dependent on B cells or NK cells.
(A) CFSE-labeled Thy1.1+ clone-4 T-cells (3×106) were injected into either NOD-InsHA mice (controls) or Human CD20 Tg NOD-InsHA mice that had been injected with Rituxan or 2H7 mAb with or without anti-CD4 on days −10 and −3. Four days after clone-4 cell transfer PcLN were analyzed by FACS (three pooled experiments). (B) NOD-InsHA recipients were injected with anti-Asialo GM1 Ab with or without anti-CD4 on days −10 and −3 before transfer of CFSE labeled clone-4 cells and analysis of PcLN on day 4 (two pooled experiments).
Depletion of NK cells in BDC2.5/NOD mice prevents the occurrence of diabetes and may play a role in the loss of T-cell tolerance (24). To assess the role of NK cells in promoting accumulation of clone-4 cells in the PcLN, NK cells were depleted from NOD-InsHA mice with anti-asialo ganglio-N-tetraosylceramide for 10 days prior to clone-4 transfer. This resulted in greater than 87% depletion of NK cells, but accumulation of clone-4 cells in the PcLN was not decreased as compared to controls either with or without simultaneous depletion of CD4 T-cells (p>0.05, Figure 2B). Collectively, these results suggest that neither B cells nor NK cells contribute to the accumulation of islet-specific CD8 T-cells in the PcLN of NOD mice.
Expression of protective Idd3/5 genes by DCs is necessary to promote deletion of autoreactive CD8 T-cells
We next sought to test directly whether expression of protective Idd3/5 alleles by APCs is able to promote deletion of CD8 T-cells in the PcLN. Mixed bone marrow (BM) chimeric mice were constructed in which NOD APCs, but not Idd3/5 APCs, were rendered dysfunctional. Idd3/5-InsHA mice were lethally irradiated and reconstituted with a mixture of BM from Idd3/5 mice and β-microglobulin (β2M) deficient NOD mice. In such chimeras, only MHC class-I-expressing Idd3/5 APCs can present islet-derived antigens to CD8 T-cells. To prevent NK cell mediated cytotoxicity towards the β2M−/− BM, NK cells were depleted with asialo GM1 for two days before BM transfer. After seven weeks, between 35% and 75% (mean 48%) of the lymphocytes in the chimeras were derived from the β2M deficient NOD BM cells. CD4 T-cells were depleted for 10 days before adoptive transfer of CFSE-labeled clone-4 cells since NOD CD4 T-cells can prevent deletion of islet-specific CD8 T-cells (Figure 1D). In control chimeras, a mixture of class I sufficient NOD BM and Idd3/5 BM resulted in high accumulation of divided clone-4 cells relative to the abortive activation observed when only Idd3/5 bone marrow was present (Figure 3A). This occurred even when the proportion of Idd3/5 lymphocytes to NOD lymphocytes ranged between 26-68% (mean 52%), indicating that when both NOD and Idd3/5 APCs are present, NOD APCs are dominant and cause accumulation of clone-4 cells. When a mixture of NOD β2M−/− and Idd3/5 BM was used for reconstitution, clone-4 cells underwent abortive activation (Figure 3A). The presence of β2M−/− BM per se did not reduce accumulation as when a mixture of NOD β2M−/− BM and NOD BM was used for reconstitution, a high percentage of activated clone-4 cells survived (Figure 3A).
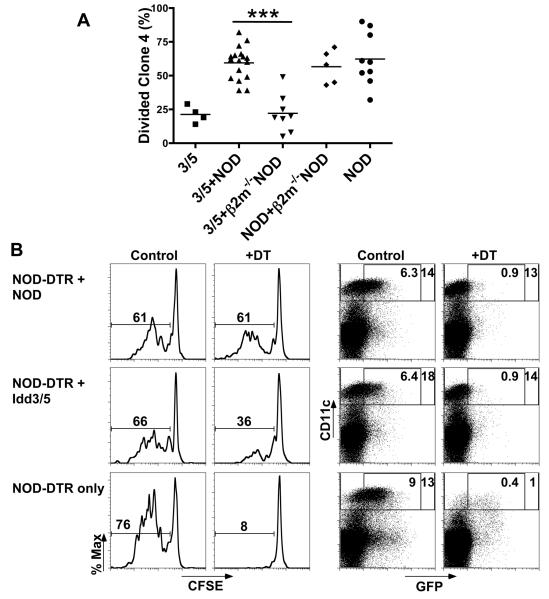
Figure 3. Expression of protective Idd3 and Idd5 alleles by DCs is sufficient to promote deletion of autoreactive CD8 T-cells.
(A) Idd3/5-InsHA mice were irradiated and reconstituted with BM from Idd3/5, mixed Idd3/5+NOD, mixed Idd3/5+ NOD β2M−/−, mixed NOD+ NOD β2M−/− or NOD. After 7 weeks anti-CD4 was injected on days −10 and −3 and 3×106 purified CSFE-labeled CD8+Thy1.1+ clone-4 NOD T-cells were transferred on day 0. PcLN were analyzed on day 4, data pooled from 2 experiments. ***P<0.0001. (B) NOD-CD45.2 congenic mice were irradiated and reconstituted with BM from the following mice: mixed NOD-CD11c-DTR and NOD, mixed NOD-CD11c-DTR and Idd3/5 or NOD-CD11c-DTR alone. After 6 weeks anti-CD4 was injected on days −10 and −3, DT or saline was injected on days −4, −1 and +2 and 3×106 purified CSFE-labeled CD8+Thy1.1+ 8.3 NOD T-cells were transferred on day 0. PcLN were analyzed on day 4 for divided 8.3 cells (left panels) and spleen cells enriched for DCs and analyzed for CD11c and GFP expression (right panels).
We next sought to determine whether the relevant APC population was a CD11c+ DC. We made use of BM cells from mice that express the diphtheria toxin receptor (DTR) on CD11c+ DCs (16) to construct mixed BM radiation chimeras. Treatment of such mice with diphtheria toxin (DT) causes selective depletion of CD11c+ DCs, lasting about 3 days. Additionally, NOD-CD11c-DTR derived DCs and non-DTR DCs could be distinguished by the expression of GFP that is included in the CD11c-DTR transgene. We also made use of NOD CD45.2 hosts, which allowed confirmation that all CD11c+ DCs in the resultant chimeras expressed CD45.1 and were of donor origin. NOD CD45.2 congenic mice were lethally irradiated and reconstituted with a mixture of BM from Idd3/5 or NOD mice and NOD-CD11c-DTR mice or NOD-CD11c-DTR BM only. After 6 weeks reconstitution, CD4 T-cells were depleted for 10 days before adoptive transfer of CFSE-labeled 8.3 cells. Transgenic 8.3 cells are CD8 T-cells that recognize an endogenous islet antigen IGRP (25) and like clone-4 cells, also undergo high-level accumulation in NOD PcLN but low-level accumulation in Idd3/5 PcLN (26). The genotype of the remaining DC was determined in the spleen based on GFP expression and CD45 allotype (Figure 3B). When mice were reconstituted with NOD-CD11c-DTR BM only, transferred 8.3 cells accumulated following activation (Figure 3B). When these mice were treated with DT, CD11cHI DC were >95% depleted and proliferation of 8.3 cells was completely abolished (Figure 3B). Therefore CD11c+ DC are absolutely required to activate islet-specific CD8 T-cells in the PcLN. Mice reconstituted with a mixture of either NOD-CD11c-DTR + NOD BM or NOD-CD11c-DTR + Idd3/5 BM resulted in high proliferation of transferred 8.3 cells (Figure 3B). Treatment with DT specifically depleted GFP+ CD11c+ cells, but did not reduce accumulation in chimeras reconstituted with NOD-CD11c-DTR and NOD BM, however treatment of NOD-CD11c-DTR + Idd3/5 BM chimeras with DT reduced proliferation (Figure 3B). Thus, when naïve CD8 T-cells are first exposed to islet antigens in the periphery, the allelic form of Idd3/5 genes expressed by the cross-presenting DCs determines deletion versus survival of the activated cells.
Both Idd3 and Idd5 expression in DCs is required for optimal deletion of islet-antigen specific CD8 T-cells in the PcLN.
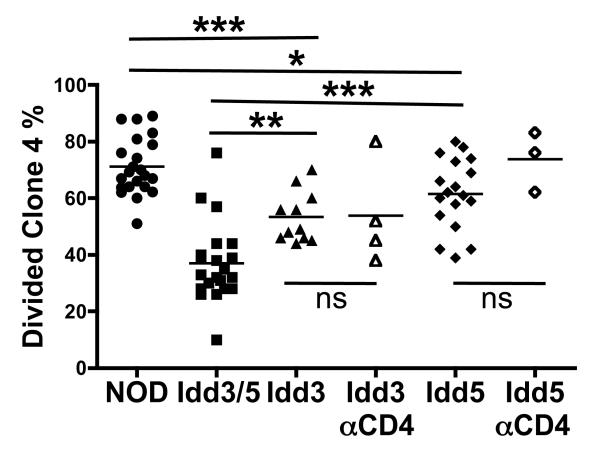
We next asked whether deletion in the PcLNs could be restored if DCs expressed only one protective region, Idd3 or Idd5. CFSE labeled clone-4 cells were transferred into NOD, Idd3/5, Idd5 or Idd3 Ins-HA mice, and the percent of divided clone-4 cells in the PcLN were assessed. The proportion of activated clone-4 cells in Idd5-InsHA was significantly higher than that seen in Idd3/5-InsHA recipients (p<0.0001, Figure 4) and slightly lower than that in NOD-InsHA mice (p<0.05, Figure 4). In Idd3-InsHA mice, the frequency of divided clone-4 cells was intermediate between NOD-InsHA mice (p<0.0001, Figure 4) and Idd3/5-InsHA mice (p<0.01, Figure 4). Thus neither the Idd3 nor the Idd5 region alone could recapitulate the degree of deletion achieved by expression of protective alleles at both regions. We next depleted CD4 T-cells from Idd3 and Idd5 InsHA mice in order to assess the contributions of these genes on the DCs alone. Removal of CD4 T-cells did not reduce clone-4 accumulation in either Idd3 or Idd5 InsHA mice (Figure 4), which would be expected if only one of the regions was required for correcting DC function. These data support the hypothesis that the DCs must express protective alleles of both Idd3 and Idd5 genes in order to fully restore deletion of autoreactive CD8 T-cells in the PcLN.
Figure 4. DC expression of both Idd3 and Idd5 is required for optimal deletion of islet-specific CD8 T-cells in the PcLN.
Purified CSFE-labeled CD8+Thy1.1+ clone-4 NOD T-cells (3×106) were injected into NOD-InsHA, Idd3/5-InsHA, Idd3-InsHA and Idd5-InsHA recipients with or without anti-CD4 treatment on days −10 and −3. Four days after clone-4 cell transfer PcLN were analyzed by FACS for divided clone-4 cells (pooled data from three to seven experiments). ***p<0.0001, **p<0.01,*p<0.05, ns: not significant.
Differential expression of candidate genes in NOD and Idd3/5 DCs
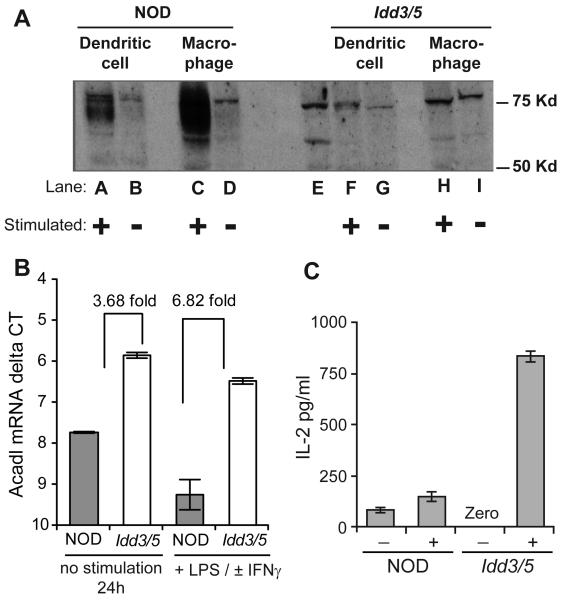
We next sought to determine which of the candidate genes in the Idd3/5 regions exhibit allelic differences in expression in DCs. Differential expression of the Idd3/5 candidate genes Il2, Ctla4 and Acadl have previously been described between NOD and Idd3 or Idd5 T-cells and therefore we did not perform additional expression studies in CD4 T-cells (10, 12, 14). BM derived DCs from NOD and Idd3/5 mice were assessed for their expression levels of CTLA-4 (Idd5.1), SLC11a1 (Idd5.2), ACADL (Idd5.3) and IL-2 (Idd3) either directly, or after stimulation with either γIFN and LPS or LPS alone or zymosan. CTLA-4 mRNA was not detected in BM derived DCs under any condition. DCs and control BM derived macrophages from NOD and Idd3/5 mice were tested for the induction of SLC11a1 protein following 48 h of stimulation with γIFN and LPS (Figure 5A). Low expression of SlC11a1 protein in Idd3/5 cells is predicted by the presence of the G169D mutation which prevents the proper folding and/or insertion of SLC11a1 into the membrane thereby subjecting the misfolded protein to degradation, especially upon its upregulation (27). NOD cells demonstrated an induction of protein consistent with the wild type Slc11a1 allele (Lanes A to D) (13). Without stimulation, NOD and Idd3/5 DC and macrophages had a similar level of SLC11a1 protein (Lanes B vs G and D vs I), but with stimulation, Idd3/5 DCs and macrophages failed to detectably increase SLC11a1 protein (Lanes F to I).
Figure 5. Differential expression of SLC11a1 and IL-2 protein and ACADL mRNA by NOD and Idd3/5 DCs.
(A) NOD and Idd3/5 BM derived DCs and macrophages were unstimulated or stimulated with γIFN and LPS for 48 hours and analyzed by western blot for SLC11a1 protein expression. Lane E shows the positive control, a lysate from the Slc11a1-expressing NAMALWA cell line (Santa Cruz Biotech). This is one of two experiments yielding similar results. (B) BM derived DC were analyzed by qRT-PCR for ACADL mRNA levels. Representative data from one experiment of three performed is shown. Comparisons of the delta Ct values using β2M as the normalization control of ACADL mRNA levels obtained from NOD vs Idd3/5 DCs stimulated with γIFN and LPS (or LPS alone) demonstrated significant differences in both unstimulated and stimulated samples (P = 0.031 using the nonparametric Wilcoxon matched pairs test). Data are represented as mean +/− SEM. (C) NOD and Idd3/5 BM derived DCs were unstimulated or stimulated with zymosan for 5 hours before analysis of supernatants for IL-2 protein. Representative data from one experiments of 7 performed using various NOD strains having the B6 versus NOD allele at Idd3. DCs were stimulated with zymosan (10 μg/ml) or LPS (1 μg/ml) for 5 hours in these seven experiments. DCs from strains having the B6 allele at Idd3 consistently produced more IL-2 than DCs having the NOD allele (P = 0.0156) using the nonparametric Wilcoxon matched pairs test).
Acyl-coenzyme A dehydrogenase long chain (ACADL) is an ubiquitously expressed enzyme whose mRNA is differentially expressed in CD4 T-cells from NOD mice having B10-derived alleles at Idd5.3 as compared with the NOD parental strain (8, 12). The levels of ACADL mRNA in BM-derived DCs from NOD and Idd3/5 mice were tested (Figure 5B) and found to be 2 to 6-fold higher in DCs from Idd3/5 mice, which have B10-derived ACADL alleles, with and without stimulation as compared to DCs from NOD mice. Therefore, differential expression by genotype of ACADL mRNA is observed in DCs as well as in T-cells.
Stimulation of BM derived DCs with zymosan or LPS has been shown to induce IL-2 by this cell population (28). Five hours after zymosan was added, 83 and 832 pg/ml IL-2 had been produced by the NOD and NOD Idd3/5 DCs respectively (Figure 5C). To avoid any potential influences from T-cells present in the BM derived DCs, we also used NOD-SCID BM and BM from a NOD-SCID congenic strain expressing a protective allele at Idd3, NOD.B6 Idd3/10/18-SCID. Zymosan stimulated NOD-SCID derived DCs produced 400 pg/mL and Idd3/10/18-SCID DCs produced 800 pg/mL IL-2 (data not shown). Higher production of IL-2 by DCs having the B6-derived Il2 allele as compared with DCs having the NOD allele at Il2 mirrors the results obtained with stimulated T-cells (10).
Both lymphocytes and non-lymphocytes can provide protection from insulitis
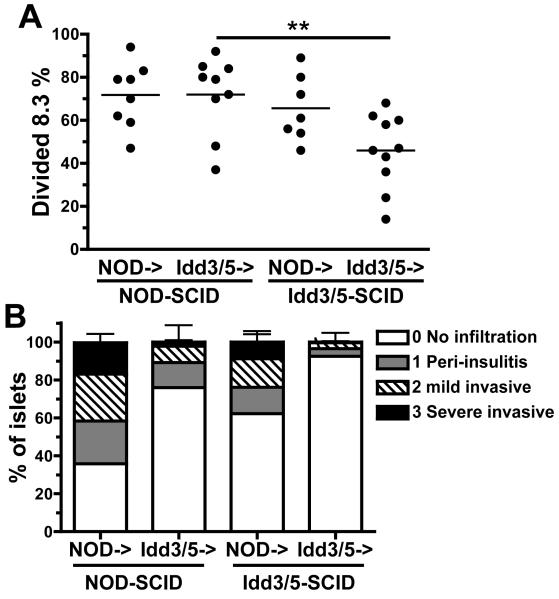
To determine whether DC expression of Idd3/5 genes could potentially protect islet beta cells from autoimmune attack we reconstituted either NOD-SCID or Idd3/5-SCID mice with spleen and lymph node cells depleted of CD11c+ cells, derived from 3-week old NOD or Idd3/5 female donors. In these animals the SCID hosts provide the DCs and the lymphocytes are of donor origin. Six to 8 weeks following reconstitution the mice received 3 × 106 CFSE labeled NOD 8.3 CD8 T-cells. CD4 T-cells were not depleted, as they are required to promote insulitis. On day 4 after transfer a high proportion of activated 8.3 cells survived in PcLNs of both NOD->NOD-SCID and NOD->Idd3/5-SCID mice (Figure 6A). This confirms that NOD CD4 T-cells prevent deletion of 8.3 cells even when DCs express a protective genotype. However, when lymphocytes expressed protective Idd3/5 genes, the genotype of the host determined the survival of activated 8.3 cells, with significantly greater levels of accumulation of activated 8.3 CD8 T-cells in Idd3/5->NOD-SCID as compared with Idd3/5->Idd3/5-SCID mice (p<0.005, Figure 6A). Thus NOD-SCID host DCs were sufficient to drive high-level accumulation of islet-specific CD8 T-cells in the PcLN. We next assessed the degree of insulitis in these same mice and mice that did not receive 8.3 cells (Figure 6B). NOD->NOD-SCID mice had extensive insulitis which was reduced in both NOD->Idd3/5-SCID mice and Idd3/5->NOD-SCID mice. Idd3/5->Idd3/5-SCID mice were highly protected with very few infiltrated islets. Thus, expression of protective Idd3/5 alleles by either lymphocytes or a host cell was able to provide significant protection from the development of insulitis. The greatest protection from insulitis, as well as maximal CD8 T-cell tolerance induction was achieved when both lymphocytes and host cells expressed protective Idd3/5 alleles.
Figure 6. Idd3/5-SCID lymphocytes and host-derived cells protect from insulitis development.
(A) NOD-SCID or Idd3/5-SCID mice were reconstituted with spleen and lymph node cells depleted of CD11c+ DCs isolated from 3-week-old donor NOD or Idd3/5 mice. After 6-8 weeks CFSE labeled 8.3 Thy1.1+ CD8 T-cells were transferred (3×106) and the PcLN analyzed by FACS on day 4 for divided 8.3 cells (three pooled experiments). **p<0.005. (B) H&E stained pancreas sections from the following mice were scored for insulitis: NOD->NOD-SCID n=6, NOD->Idd3/5-SCID n=8, Idd3/5->NOD-SCID n=14, Idd3/5->Idd3/5-SCID n=15. Some reconstituted SCID mice did not receive CFSE labeled 8.3 cells. Mice receiving 8.3 cells and those not injected with the cells had similar degrees of insulitis and pooled data are shown. Data represent mean +/− SEM.
Discussion
NOD mice exhibit a defective ability to tolerize islet-specific CD8 T-cells as compared with non-diabetes prone strains such as Balb/c, B10.D2 and Idd3/5. We have observed that although transferred islet-specific CD8 T-cells undergo a similar number of divisions in NOD and Idd3/5 mice (7), they accumulate to a lower level in Idd3/5 PcLN and are deleted (Figure 1). Of interest, other than the difference in levels of accumulation, we have found no evidence for a difference in function between CD8 T-cells activated in the PcLNs of Idd3/5 and NOD. Transferred clone-4 cells produce very little IFNγ in either NOD or Idd3/5 InsHA mice (unpublished observations, EHW and XM). The functional capacity of the transferred cells was also assessed by the highly sensitive in vivo CTL assay and we found low levels of specific killing by clone-4 cells in the PcLN of NOD and Idd3/5 InsHA mice, proportional to the frequency of divided cells (unpublished observations XM). Consequently, the primary difference between clone-4 cells activated in NOD or Idd3/5 PcLN is survival.
The goal of the current study was to determine which cell(s) are responsible for defective CD8 T-cell deletion in the PcLN of NOD mice. Genes within Idd3 and Idd5 regulate this tolerance checkpoint (7). Our initial candidate for a cell type capable of preventing deletion was host CD4 T-cells. The presence of islet-antigen specific CD4 T-cells prevents deletion of CD8 T-cells (17), and NOD mice harbor high levels of islet-antigen specific CD4 T-cells (29, 30). Surprisingly, we found that removal of CD4 T-cells was insufficient to restore deletion of islet specific CD8 T-cells in the PcLN. In previous experiments, we found that B7 costimulatory signals were critical to achieve high-level accumulation of clone-4 cells in the PcLN of BALB/c InsHA recipients (31). Thus we hypothesized that NOD DCs may deliver enhanced costimulatory signals to CD8 T-cells in the steady state. Indeed we found that in the absence of CD4 T-cells, CD8 T-cell accumulation that occurred in the PcLN in NOD-InsHA recipients was inhibited by anti-B7.1/2. As inhibition did not occur in the presence of CD4 T-cells, it is possible that additional costimulatory pathways are activated on DCs by CD4 T-cells. Alternatively, activated CD4 T-cells may provide an inflammatory milieu that either directly, or indirectly, enhances survival of activated CD8 T-cells.
Other types of cells that have been reported to play a role in progression of disease in NOD mice included NKT-cells, B cells, and NK cells. We did not consider further the role of NKT-cells, as the variation in numbers of NKT-cells in NOD and C57BL/6 mice does not map to the Idd3 or Idd5 regions (32). Furthermore, NKT-cells do not affect initial activation of T-cells but rather, their survival at later time points (33). Idd5 genes have been reported to reduce diabetes incidence in part through effects on B-cells (34). Depletion of B cells or NK cells, either alone or in combination with CD4 T-cells, did not reduce accumulation of activated CD8 T-cells. As depletion of the candidate cells was not complete (87% depletion of NK and 85-95% depletion of B cells), we can not exclude the possibility that a small number of B cells or NK cells present in the PcLN may be sufficient to prevent deletion. However, the results obtained using radiation bone marrow chimeras (discussed below) makes this possibility unlikely.
A number of laboratories have reported abnormalities in NOD DC populations (35-39). We next sought direct evidence that NOD DCs prevent deletion of activated CD8 T-cells in the PcLN observed when CD4 T-cells are removed. To this end we produced mice in which bone marrow derived cells of both NOD and Idd3/5 origin coexisted, yet only DCs of Idd3/5 origin were functional for antigen-presentation to CD8 T-cells. Irradiated Idd3/5 mice were reconstituted with a mixture of bone marrow from Idd3/5 and NOD-β2M−/− mice. As we could not completely exclude the possibility that loss of γ2M expression by antigen presenting cells other than DCs contributed to the observed decrease in proliferation, we also took a second approach. We utilized the newly available NOD CD11c-DTR strain (16) to construct mixed bone marrow chimeras. This experiment identified the genotype of the DC as liable for the loss of tolerance within islet-antigen specific CD8 T-cells in the PcLN. This is a particularly striking result since in this model bone marrow cells of NOD and Idd3/5 origin develop together and the NOD DCs are dominant with regard to activating CD8 T-cells when both genotypic sources of DCs are present. Under these conditions, removal of the NOD DCs is sufficient to promote CD8 T-cell deletion in the PcLN. Unfortunately in this model depletion of CD11c-DTR expressing cells is only temporary and it is not possible to use this same system to assess the role of DCs in the development of insulitis or diabetes. Another limitation of the CD11c-DTR transgenic model is a small amount of ectopic expression of the transgene on other cell types including a small subset of macrophages (40). We think it is unlikely that a macrophage contributes to CD8 tolerance in the PcLN as the Idd3/5 + NOD-β2M−/− mixed chimeras indicated that a cell presenting antigen to the CD8 T-cell is critical and evidence in the literature supports a role for DCs, not macrophages, in presentation of islet antigens to CD8 T-cells in the PcLN (41, 42).
Although CD11c+ DCs are generally believed to be the only antigen-presenting cell capable of cross-presenting exogenously derived antigen to activate naïve CD8 T-cells, it has recently been reported that lymph node stromal cells endogenously express a range of peripheral antigens and can activate CD8 T-cells (43). Depletion of DCs in NOD CD11c-DTR->NOD chimeric mice completely abolished proliferation of transferred 8.3 cells. Thus we found no evidence that cells other than DCs could activate IGRP-specific CD8 T-cells in the PcLN.
To determine whether NOD DCs may potentially support insulitis development in the presence of Idd3/5 T and B-lymphocytes, we reconstituted SCID mice. We found a reduction in insulitis when either donor lymphocytes or the host SCID cells expressed protective Idd3/5 genes and maximal protection when both compartments were Idd3/5. This was consistent with our findings that both CD4 T-cells and DC are involved in CD8 T-cell tolerance induction. However expression of Idd3/5 genes on either CD4 T-cells or DC alone did not have a measurable effect on accumulation of CD8 T-cells in the PcLN. Collectively, these data suggest there may be protective effects from Idd3/5 genes that prevent insulitis that are distinct from reduced accumulation of islet specific CD8 T-cells in the PcLN. For example Idd3/5 genes may have protective effects in the islet as was previously observed for Idd9 genes (7).
Our results suggest that Idd3 (Il2) and one or more of the Idd5 region genes (Ctla4, Acadl, and Slc11a1), exert an effect on tolerance through expression in both CD4 T-cells and DCs. It is the subject of ongoing investigations to identify exactly which combination of protective genes must be expressed in each cell for maximal tolerogenic effect. Both protective regions were required for optimal tolerance induction and depletion of CD4 T-cells in either Idd3 or Idd5 mice did not significantly alter the degree of accumulation observed in each strain. The literature provides evidence for expression of Il2, Ctla4 and Acadl by NOD CD4 T-cells (12, 44, 45) but there is no evidence of expression of Slc11a1 outside of macrophages, polymorphonuclear leukocytes and DCs (46). DC expression of IL-2 has been reported to be of importance in immune responses to pathogens (47). Our results suggest it also plays a role in T-cell tolerance. As our expression studies were performed with BM derived DC using strong stimulatory conditions, it is unclear which conditions and locations in vivo such differences would be observed. Ongoing investigations are attempting to address this technically challenging issue. It is possible that DC-produced IL-2 has autocrine effects on DC development or function rather than being required by the T-cells at the time of tolerance induction. As such restoration of DC function by increased IL-2 may require precise timing during DC maturation.
SLC11a1 protein is involved in resistance to intracellular pathogens and influences phagosomal acidification which may alter antigen processing and thereby quantitatively affect cross-presented peptide antigen (48). This could contribute to the amount of CD8 T-cell proliferation. However we do not believe that the difference in proliferation of islet-specific CD8 T-cells attributable to CD8-DC interaction in the PcLNs is principally due to differences in amount of antigen. Differences in the amount of co-stimulatory molecules and cytokines can also affect the level of T cell activation. In an attempt to remove these additional signals, we eliminated CD4 cells and provided co-stimulation blockade. This lowered the level of clone-4 accumulation in PcLNs of NOD mice to a level similar to Idd3/5. This suggests that the amount of antigen presented to CD8 T-cells is similar between NOD and Idd3/5 DCs, and that they differ in delivery of costimulation.
Differential expression of the enzyme ACADL by DCs may confer altered fatty acid metabolism and it remains to be investigated how this could affect DC function. The DCs were tested for expression of all CTLA-4 mRNA isoforms (14), but none were detected in either stimulated or unstimulated cells, however, this locus could influence the function of NOD vs Idd3/5 CD4 T-cells. As maximal tolerance was only achieved when both Idd3 and Idd5 genes were expressed, it is possible that epistatic gene interactions occur to enhance protection.
It has recently been reported that the protective allele of Idd3 (Il2) correlates with enhanced production of IL-2 (10). In the 8.3 CD8 T-cell transgenic model of diabetes, enhanced levels of IL-2 production correlated with protection from diabetes by a mechanism involving enhanced homeostasis of Tregs, which are dependent on IL-2 for survival. In fact, although we have found the level of accumulation of clone-4 cells does increase somewhat following removal of CD25+ Tregs in Idd3/5 mice (7), the level of accumulation is never as high as that seen in NOD mice and, importantly, the CD8 T-cells do not migrate to the islets following removal of Tregs (XM, unpublished results) suggesting only brief accumulation. Anderson et al have presented evidence supporting the hypothesis that the reduction of Treg function in the NOD mouse as compared with mice expressing protective Idd3 genes is in part due to the effects of Idd3 on CD11b+CD11c- APC (49). We think it is likely that the Idd3/Il2 gene acts indirectly on the development or function of CD11b+CD11c- cells, as we have been unable to detect expression of IL-2 by macrophages (LW unpublished results) and it has not been reported elsewhere in the literature. Thus, expression of IL-2 by CD4 T-cells may in part affect CD8 T-cell tolerance through mechanisms involving Tregs. Taken together, these results demonstrate a participation of IL-2 in multiple layers of tolerance.
Due to the complex nature of the Idd3/5 loci it is not surprising that we have identified two cell types that contribute to restoration of CD8 T-cell tolerance to islet antigens. It also remains possible that Idd3/5 genes expressed in other cell types may impact CD8 T-cell independent aspects of diabetes progression. For example, the diabetes-protective effect of Idd5 expression in B cells could act by increasing regulatory T cell function (34). In conclusion our data demonstrate that Idd3 and Idd5 genes establish multiple layers of tolerance. We have identified the islet-antigen presenting DC as a key modulator of autoreactive CD8 T-cells. The DC has not previously been identified as a cell type in which Idd-mediated gene variation influences the disease state. Dissection of complex, multi-genetic disease susceptibility pathways could lead to identification of novel targets for potentially modulating tolerance.
Acknowledgements
We thank Dr. Judith A. Shizuru (Stanford University) for her contribution to developing the NOD-CD45.2 congenic strain. The authors thank all members of the Sherman laboratory for helpful discussion, Kristi Marquardt, Rebecca Trenney and Judith Biggs for excellent technical assistance and Shelly Gassert for excellent secretarial assistance.
Abbreviations
- T1D
type-1-diabetes
- Idd
insulin-dependent diabetes
- β2M
beta-2 microglobulin
- DT
diphtheria toxin
- NOD
non-obese diabetic
- PcLN
pancreatic lymph node
- Treg
regulatory T cell
- HA
hemagglutinin
- DTR
diphtheria toxin receptor
- ACADL
acyl-coenzyme A dehydrogenase long chain
Footnotes
This work was supported by National Institute of Allergy and Infectious Disease (NIAID) grant AI 070351. EHW and XM supported by postdoctoral fellowships from the Juvenile Diabetes Research Foundation (JDRF). LSW is supported by grants from the JDRF and the Wellcome Trust and LSW is a Wellcome Trust Principal Research Fellow. The Cambridge Institute for Medical Research is the recipient of a Wellcome Trust Strategic Award (079895). The availability of NOD congenic mice through the Taconic Farms Emerging Models Program has been supported by grants from the Merck Genome Research Institute, NIAID, and the JDRF.
References
- 1.Wicker LS, Clark J, Fraser HI, Garner VE, Gonzalez-Munoz A, Healy B, Howlett S, Hunter K, Rainbow D, Rosa RL, Smink LJ, Todd JA, Peterson LB. Type 1 diabetes genes and pathways shared by humans and NOD mice. J Autoimmun. 2005;25(Suppl):29–33. doi: 10.1016/j.jaut.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Dendrou CA, Wicker LS. The IL-2/CD25 Pathway Determines Susceptibility to T1D in Humans and NOD Mice. J Clin Immunol. 2008;28:685–696. doi: 10.1007/s10875-008-9237-9. [DOI] [PubMed] [Google Scholar]
- 3.Liston A, Lesage S, Gray DH, O'Reilly LA, Strasser A, Fahrer AM, Boyd RL, Wilson J, Baxter AG, Gallo EM, Crabtree GR, Peng K, Wilson SR, Goodnow CC. Generalized resistance to thymic deletion in the NOD mouse; a polygenic trait characterized by defective induction of Bim. Immunity. 2004;21:817–830. doi: 10.1016/j.immuni.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 4.Kreuwel HT, Biggs JA, Pilip IM, Pamer EG, Lo D, Sherman LA. Defective CD8+ T cell peripheral tolerance in nonobese diabetic mice. J Immunol. 2001;167:1112–1117. doi: 10.4049/jimmunol.167.2.1112. [DOI] [PubMed] [Google Scholar]
- 5.Morgan DJ, Kreuwel HT, Sherman LA. Antigen concentration and precursor frequency determine the rate of CD8+ T cell tolerance to peripherally expressed antigens. J Immunol. 1999;163:723–727. [PubMed] [Google Scholar]
- 6.Redmond WL, Wei CH, Kreuwel HT, Sherman LA. The apoptotic pathway contributing to the deletion of naive CD8 T cells during the induction of peripheral tolerance to a cross-presented self-antigen. J Immunol. 2008;180:5275–5282. doi: 10.4049/jimmunol.180.8.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez X, Kreuwel HT, Redmond WL, Trenney R, Hunter K, Rosen H, Sarvetnick N, Wicker LS, Sherman LA. CD8+ T cell tolerance in nonobese diabetic mice is restored by insulin-dependent diabetes resistance alleles. J Immunol. 2005;175:1677–1685. doi: 10.4049/jimmunol.175.3.1677. [DOI] [PubMed] [Google Scholar]
- 8.Hunter K, Rainbow D, Plagnol V, Todd JA, Peterson LB, Wicker LS. Interactions between Idd5.1/Ctla4 and other type 1 diabetes genes. J Immunol. 2007;179:8341–8349. doi: 10.4049/jimmunol.179.12.8341. [DOI] [PubMed] [Google Scholar]
- 9.Tang Q, Adams JY, Penaranda C, Melli K, Piaggio E, Sgouroudis E, Piccirillo CA, Salomon BL, Bluestone JA. Central role of defective interleukin-2 production in the triggering of islet autoimmune destruction. Immunity. 2008;28:687–697. doi: 10.1016/j.immuni.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamanouchi J, Rainbow D, Serra P, Howlett S, Hunter K, Garner VE, Gonzalez-Munoz A, Clark J, Veijola R, Cubbon R, Chen SL, Rosa R, Cumiskey AM, Serreze DV, Gregory S, Rogers J, Lyons PA, Healy B, Smink LJ, Todd JA, Peterson LB, Wicker LS, Santamaria P. Interleukin-2 gene variation impairs regulatory T cell function and causes autoimmunity. Nature genetics. 2007;39:329–337. doi: 10.1038/ng1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lazarski C, Hughson A, Sojka D, Fowell D. Regulating Treg Cells at Sites of Inflammation. Immunity. 2008;29:511. doi: 10.1016/j.immuni.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 12.Irie J, Reck B, Wu Y, Wicker LS, Howlett S, Rainbow D, Feingold E, Ridgway WM. Genome-wide microarray expression analysis of CD4+ T Cells from nonobese diabetic congenic mice identifies Cd55 (Daf1) and Acadl as candidate genes for type 1 diabetes. J Immunol. 2008;180:1071–1079. doi: 10.4049/jimmunol.180.2.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kissler S, Stern P, Takahashi K, Hunter K, Peterson LB, Wicker LS. In vivo RNA interference demonstrates a role for Nramp1 in modifying susceptibility to type 1 diabetes. Nature genetics. 2006;38:479–483. doi: 10.1038/ng1766. [DOI] [PubMed] [Google Scholar]
- 14.Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, Rainbow DB, Hunter KM, Smith AN, Di Genova G, Herr MH, Dahlman I, Payne F, Smyth D, Lowe C, Twells RC, Howlett S, Healy B, Nutland S, Rance HE, Everett V, Smink LJ, Lam AC, Cordell HJ, Walker NM, Bordin C, Hulme J, Motzo C, Cucca F, Hess JF, Metzker ML, Rogers J, Gregory S, Allahabadia A, Nithiyananthan R, Tuomilehto-Wolf E, Tuomilehto J, Bingley P, Gillespie KM, Undlien DE, Ronningen KS, Guja C, Ionescu-Tirgoviste C, Savage DA, Maxwell AP, Carson DJ, Patterson CC, Franklyn JA, Clayton DG, Peterson LB, Wicker LS, Todd JA, Gough SC. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423:506–511. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- 15.Ahuja A, Shupe J, Dunn R, Kashgarian M, Kehry MR, Shlomchik MJ. Depletion of B cells in murine lupus: efficacy and resistance. J Immunol. 2007;179:3351–3361. doi: 10.4049/jimmunol.179.5.3351. [DOI] [PubMed] [Google Scholar]
- 16.Saxena V, Ondr JK, Magnusen AF, Munn DH, Katz JD. The countervailing actions of myeloid and plasmacytoid dendritic cells control autoimmune diabetes in the nonobese diabetic mouse. J Immunol. 2007;179:5041–5053. doi: 10.4049/jimmunol.179.8.5041. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez J, Aung S, Marquardt K, Sherman LA. Uncoupling of proliferative potential and gain of effector function by CD8(+) T cells responding to self-antigens. J Exp Med. 2002;196:323–333. doi: 10.1084/jem.20011612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett SR, Carbone FR, Karamalis F, Flavell RA, Miller JF, Heath WR. Help for cytotoxic-T-cell responses is mediated by CD40 signalling. Nature. 1998;393:478–480. doi: 10.1038/30996. [DOI] [PubMed] [Google Scholar]
- 19.Ridge JP, Di Rosa F, Matzinger P. A conditioned dendritic cell can be a temporal bridge between a CD4+ T-helper and T-killer cell. Nature. 1998;393:474–478. doi: 10.1038/30989. [DOI] [PubMed] [Google Scholar]
- 20.Schoenberger SP, Toes RE, van der Voort EI, Offringa R, Melief CJ. T-cell help for cytotoxic T lymphocytes is mediated by CD40-CD40L interactions. Nature. 1998;393:480–483. doi: 10.1038/31002. [DOI] [PubMed] [Google Scholar]
- 21.Liu K, Waskow C, Liu X, Yao K, Hoh J, Nussenzweig M. Origin of dendritic cells in peripheral lymphoid organs of mice. ature immunology. 2007;8:578–583. doi: 10.1038/ni1462. [DOI] [PubMed] [Google Scholar]
- 22.Delovitch TL, Singh B. The nonobese diabetic mouse as a model of autoimmune diabetes: immune dysregulation gets the NOD. Immunity. 1997;7:727–738. doi: 10.1016/s1074-7613(00)80392-1. [DOI] [PubMed] [Google Scholar]
- 23.Prilliman KR, Lemmens EE, Palioungas G, Wolfe TG, Allison JP, Sharpe AH, Schoenberger SP. Cutting edge: a crucial role for B7-CD28 in transmitting T help from APC to CTL. J Immunol. 2002;169:4094–4097. doi: 10.4049/jimmunol.169.8.4094. [DOI] [PubMed] [Google Scholar]
- 24.Poirot L, Benoist C, Mathis D. Natural killer cells distinguish innocuous and destructive forms of pancreatic islet autoimmunity. Proc Natl Acad Sci U S A. 2004;101:8102–8107. doi: 10.1073/pnas.0402065101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lieberman SM, Evans AM, Han B, Takaki T, Vinnitskaya Y, Caldwell JA, Serreze DV, Shabanowitz J, Hunt DF, Nathenson SG, Santamaria P, DiLorenzo TP. Identification of the beta cell antigen targeted by a prevalent population of pathogenic CD8+ T cells in autoimmune diabetes. Proc Natl Acad Sci U S A. 2003;100:8384–8388. doi: 10.1073/pnas.0932778100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamilton-Williams EE, Martinez X, Lyman M, Hunter K, Wicker LS, Sherman LA. The use of idd congenic mice to identify checkpoints of peripheral tolerance to islet antigen. Annals of the New York Academy of Sciences. 2007;1103:118–127. doi: 10.1196/annals.1394.003. [DOI] [PubMed] [Google Scholar]
- 27.Vidal SM, Malo D, Vogan K, Skamene E, Gros P. Natural resistance to infection with intracellular parasites: isolation of a candidate for Bcg. Cell. 1993;73:469–485. doi: 10.1016/0092-8674(93)90135-d. [DOI] [PubMed] [Google Scholar]
- 28.Rogers NC, Slack EC, Edwards AD, Nolte MA, Schulz O, Schweighoffer E, Williams DL, Gordon S, Tybulewicz VL, Brown GD, Reis e Sousa C. Syk-dependent cytokine induction by Dectin-1 reveals a novel pattern recognition pathway for C type lectins. Immunity. 2005;22:507–517. doi: 10.1016/j.immuni.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 29.Kaufman DL, Clare-Salzler M, Tian J, Forsthuber T, Ting GS, Robinson P, Atkinson MA, Sercarz EE, Tobin AJ, Lehmann PV. Spontaneous loss of T-cell tolerance to glutamic acid decarboxylase in murine insulin-dependent diabetes. Nature. 1993;366:69–72. doi: 10.1038/366069a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tisch R, Yang XD, Singer SM, Liblau RS, Fugger L, McDevitt HO. Immune response to glutamic acid decarboxylase correlates with insulitis in non-obese diabetic mice. Nature. 1993;366:72–75. doi: 10.1038/366072a0. [DOI] [PubMed] [Google Scholar]
- 31.Hernandez J, Aung S, Redmond WL, Sherman LA. Phenotypic and functional analysis of CD8(+) T cells undergoing peripheral deletion in response to cross-presentation of self-antigen. J Exp Med. 2001;194:707–717. doi: 10.1084/jem.194.6.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esteban LM, Tsoutsman T, Jordan MA, Roach D, Poulton LD, Brooks A, Naidenko OV, Sidobre S, Godfrey DI, Baxter AG. Genetic control of NKT cell numbers maps to major diabetes and lupus loci. J Immunol. 2003;171:2873–2878. doi: 10.4049/jimmunol.171.6.2873. [DOI] [PubMed] [Google Scholar]
- 33.Beaudoin L, Laloux V, Novak J, Lucas B, Lehuen A. NKT cells inhibit the onset of diabetes by impairing the development of pathogenic T cells specific for pancreatic beta cells. Immunity. 2002;17:725–736. doi: 10.1016/s1074-7613(02)00473-9. [DOI] [PubMed] [Google Scholar]
- 34.Silveira PA, Chapman HD, Stolp J, Johnson E, Cox SL, Hunter K, Wicker LS, Serreze DV. Genes within the Idd5 and Idd9/11 diabetes susceptibility loci affect the pathogenic activity of B cells in nonobese diabetic mice. J Immunol. 2006;177:7033–7041. doi: 10.4049/jimmunol.177.10.7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fallarino F, Bianchi R, Orabona C, Vacca C, Belladonna ML, Fioretti MC, Serreze DV, Grohmann U, Puccetti P. CTLA-4-Ig activates forkhead transcription factors and protects dendritic cells from oxidative stress in nonobese diabetic mice. J Exp Med. 2004;200:1051–1062. doi: 10.1084/jem.20040942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poligone B, Weaver DJ, Jr., Sen P, Baldwin AS, Jr., Tisch R. Elevated NF-kappaB activation in nonobese diabetic mouse dendritic cells results in enhanced APC function. J Immunol. 2002;168:188–196. doi: 10.4049/jimmunol.168.1.188. [DOI] [PubMed] [Google Scholar]
- 37.Vasquez AC, Feili-Hariri M, Tan RJ, Morel PA. Qualitative and quantitative abnormalities in splenic dendritic cell populations in NOD mice. Clinical and experimental immunology. 2004;135:209–218. doi: 10.1111/j.1365-2249.2003.02359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wheat W, Kupfer R, Gutches DG, Rayat GR, Beilke J, Scheinman RI, Wegmann DR. Increased NF-kappa B activity in B cells and bone marrow-derived dendritic cells from NOD mice. Eur J Immunol. 2004;34:1395–1404. doi: 10.1002/eji.200324490. [DOI] [PubMed] [Google Scholar]
- 39.Marleau AM, Summers KL, Singh B. Differential contributions of APC subsets to T cell activation in nonobese diabetic mice. J Immunol. 2008;180:5235–5249. doi: 10.4049/jimmunol.180.8.5235. [DOI] [PubMed] [Google Scholar]
- 40.Probst HC, Tschannen K, Odermatt B, Schwendener R, Zinkernagel RM, Van Den Broek M. Histological analysis of CD11c-DTR/GFP mice after in vivo depletion of dendritic cells. Clinical and experimental immunology. 2005;141:398–404. doi: 10.1111/j.1365-2249.2005.02868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Belz GT, Behrens GM, Smith CM, Miller JF, Jones C, Lejon K, Fathman CG, Mueller SN, Shortman K, Carbone FR, Heath WR. The CD8alpha(+) dendritic cell is responsible for inducing peripheral self-tolerance to tissue-associated antigens. J Exp Med. 2002;196:1099–1104. doi: 10.1084/jem.20020861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turley S, Poirot L, Hattori M, Benoist C, Mathis D. Physiological beta cell death triggers priming of self-reactive T cells by dendritic cells in a type-1 diabetes model. J Exp Med. 2003;198:1527–1537. doi: 10.1084/jem.20030966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee JW, Epardaud M, Sun J, Becker JE, Cheng AC, Yonekura AR, Heath JK, Turley SJ. Peripheral antigen display by lymph node stroma promotes T cell tolerance to intestinal self. Nature immunology. 2007;8:181–190. doi: 10.1038/ni1427. [DOI] [PubMed] [Google Scholar]
- 44.Sgouroudis E, Albanese A, Piccirillo CA. Impact of protective IL-2 allelic variants on CD4+ Foxp3+ regulatory T cell function in situ and resistance to autoimmune diabetes in NOD mice. J Immunol. 2008;181:6283–6292. doi: 10.4049/jimmunol.181.9.6283. [DOI] [PubMed] [Google Scholar]
- 45.Wicker LS, Chamberlain G, Hunter K, Rainbow D, Howlett S, Tiffen P, Clark J, Gonzalez-Munoz A, Cumiskey AM, Rosa RL, Howson JM, Smink LJ, Kingsnorth A, Lyons PA, Gregory S, Rogers J, Todd JA, Peterson LB. Fine mapping, gene content, comparative sequencing, and expression analyses support Ctla4 and Nramp1 as candidates for Idd5.1 and Idd5.2 in the nonobese diabetic mouse. J Immunol. 2004;173:164–173. doi: 10.4049/jimmunol.173.1.164. [DOI] [PubMed] [Google Scholar]
- 46.Stober CB, Brode S, White JK, Popoff JF, Blackwell JM. Slc11a1, formerly Nramp1, is expressed in dendritic cells and influences major histocompatibility complex class II expression and antigen-presenting cell function. Infection and immunity. 2007;75:5059–5067. doi: 10.1128/IAI.00153-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Granucci F, Zanoni I, Feau S, Ricciardi-Castagnoli P. Dendritic cell regulation of immune responses: a new role for interleukin 2 at the intersection of innate and adaptive immunity. Embo J. 2003;22:2546–2551. doi: 10.1093/emboj/cdg261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dai YD, Marrero IG, Gros P, Zaghouani H, Wicker LS, Sercarz EE. Slc11a1 Enhances the Autoimmune Diabetogenic T-cell Response by Altering Processing and Presentation of Pancreatic Islet Antigens. Diabetes. 2008;58:156–164. doi: 10.2337/db07-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anderson AC, Chandwaskar R, Lee DH, Kuchroo VK. Cutting edge: the Idd3 genetic interval determines regulatory T cell function through CD11b+CD11c- APC. J Immunol. 2008;181:7449–7452. doi: 10.4049/jimmunol.181.11.7449. [DOI] [PMC free article] [PubMed] [Google Scholar]