Abstract
Chromosome rearrangement, a hallmark of cancer, has profound effects on carcinogenesis and tumor phenotype. We used a panel of 60 human cancer cell lines (the NCI-60) as a model system to identify relationships among DNA copy number, mRNA expression level, and drug sensitivity. For each of 64 cancer-relevant genes, we calculated all 4,096 possible Pearson's correlation coefficients relating DNA copy number (assessed by comparative genomic hybridization using bacterial artificial chromosome microarrays) and mRNA expression level (determined using both cDNA and Affymetrix oligonucleotide microarrays). The analysis identified an association of ERBB2 overexpression with 3p copy number, a finding supported by data from human tumors and a mouse model of ERBB2-induced carcinogenesis. When we examined the correlation between DNA copy number for all 353 unique loci on the bacterial artificial chromosome microarray and drug sensitivity for 118 drugs with putatively known mechanisms of action, we found a striking negative correlation ( −0.983; 95% bootstrap confidence interval, −0.999 to −0.899) between activity of the enzyme drug L-asparaginase and DNA copy number of genes near asparagine synthetase in the ovarian cancer cells. Previous analysis of drug sensitivity and mRNA expression had suggested an inverse relationship between mRNA levels of asparagine synthetase and L-asparaginase sensitivity in the NCI-60. The concordance of pharmacogenomic findings at the DNA and mRNA levels strongly suggests further study of L-asparaginase for possible treatment of a low-synthetase subset of clinical ovarian cancers. The DNA copy number database presented here will enable other investigators to explore DNA transcript-drug relationships in their own domains of research focus.
Introduction
The phenotype of cancer is a dynamic interplay of changes at the DNA, mRNA, and protein levels that results in altered responses to extracellular stimuli and in unregulated growth. Further improvements in cancer treatment will depend, in considerable part, on knowledge of how those disparate factors interact with one another. The complexity of DNA-RNA-protein relationships provides ample opportunity for several levels of regulation leading to a phenotype. There is no reason to expect a priori that such relationships would be linear, consistent across genes, or even consistent for a particular gene under different cellular conditions. For example, there are several cases in which gene copy number has been reported to correlate with expression, others in which gene copy number does not seem to correlate with expression, and instances in which both cases exist for the same gene (1 - 5). However, a strong correlation between DNA copy number and gene expression or drug response increases the likelihood that the gene is subject to selective pressure.
Acquisition of both gene copy number and expression data for the same set of samples presents an opportunity to ask how gene copy number influences mRNA levels for the same gene or different genes. However, we wanted to take another step with respect to pharmacogenomic issues (6). We wanted an experimental system in which we could assess the relationship of DNA copy number and mRNA level to sensitivity of the cells to a variety of drugs and potential drugs. The natural choice was the set of 60 human cancer cell lines (the NCI-60) used by the Developmental Therapeutics Program of the National Cancer Institute for screening and secondary testing of potential anticancer agents. The NCI-60 panel is diverse. It includes leukemias, melanomas, and carcinomas of breast, ovary, kidney, colon, prostate, lung, and central nervous system origin. Since 1990 when the NCI-60 assay went into full operation, >100,000 chemical compounds (plus a large number of natural product extracts) have been tested in a 48-hour growth inhibition assay (7, 8). In addition to the resulting pharmacologic profiles of the NCI-60, the cells have been more fully characterized at the molecular level than any other set of cells in existence. We and our collaborators have profiled them for mRNA expression using cDNA microarrays (9, 10) and oligonucleotide chips (11), for protein expression using two-dimensional protein gel electrophoresis (12, 13) and “reverse-phase” lysate arrays (14, 15), and for chromosomal aberrations (16).
Here, we present the first array comparative genomic hybridization (CGH) characterization of the NCI-60 and relate the resulting profiles to mRNA expression and drug sensitivity. Genes represented on the arrays used in the study included cancer-related genes involved in carcinogenesis, proliferation, apoptosis, cell cycle regulation, signal transduction, and drug resistance. One particular aim was to follow up on our earlier observation in the NCI-60 of a relationship between sensitivity to the enzyme drug L-asparaginase and expression of asparagine synthetase mRNA (9). L-asparaginase has been used since the early 1970s to treat acute lymphoblastic leukemia; thus, we were intrigued to find a very strong negative correlation [−0.98; two-tailed 95% bootstrap confidence interval (95% CI), −1.00 to −0.928], between activity of L-asparaginase and expression of asparagine synthetase for the NCI-60 leukemias. We were not surprised to see an inverse relationship, but the near-perfect correlation across a diverse set of cell lines (and in the face of experimental error) was unexpected. Still more interesting, there was also a strong negative correlation (−0.88; 95% CI, −0.231 to −0.987) for the ovarian cell types. As indicated by the latter CI, the correlation for ovarian lines was statistically significant when considered in relation to a single hypothesis but not after appropriate correction for multiple comparisons testing. Hence, we thought of it as a clue to formulate the hypothesis that a subset of ovarian cancers low in asparagine synthetase would be sensitive to the drug. In the present study, we therefore sought evidence at the DNA copy number level that would either support or contradict the hypothesis and also perhaps cast light on the role of copy number in determining asparagine synthetase expression levels. The results, as will be described in detail later, were surprisingly definite.
Materials and Methods
Gene Expression Data
Gene expression data on the NCI-60 cell lines were obtained by hybridization to cDNA microarrays (9, 10) and Affymetrix (Santa Clara, CA) oligonucleotide chips (11) as described previously. In the cDNA array studies, Cy5-labeled cDNA from the test cells was cohybridized with Cy3-labeled cDNA from an index standard pool (9) consisting of 12 representative cell lines. Measured Cy5/Cy3 ratios were normalized using Gaussian-windowed moving-average fits (without background subtraction; ref. 17) to correct for curvature in the red channel versus green channel scatter plots. For the oligonucleotide data (11), average difference values were floored at 30 (i.e., all values <30 were set to 30) based on empirical determination of the minimum level for which measurements reliably reflected signal rather than noise (18).10
Array CGH Using the Oncobacterial Artificial Chromosome DNA Microarray
DNA harvested from the 60 cell lines was purified using the QIAmp DNA Blood Maxi kit (Qiagen, Inc., Valencia, CA) and quantitated fluorimetrically. Normal female genomic DNA, obtained from Promega (Madison, WI), was used as a reference. Probes for the cell lines and reference sample were prepared by digesting 1 μg of DNA with DpnII (NEB, Boston, MA) and then treating it with a QIAquick PCR purification kit (Qiagen). Cell line and reference DNA samples were labeled with Cy3-dUTP and Cy5-dUTP (Amersham Pharmacia Biotech, Piscataway, NJ), respectively, as described elsewhere (19). Labeled probes were purified using MicroSpin G-50 columns (Amersham Pharmacia Biotech). Approximately 250 ng of each labeled probe was ethanol precipitated with 50 μg of human cot-1 DNA (Invitrogen, Carlsbad, CA) and resuspended in 20 μL of hybridization buffer (50% formamide, 10% dextran sulfate, 2× SSC, 4% SDS, and 1% yeast tRNA). Probes were denatured at 73°C for 5 minutes and reannealed at 37°C for 60 to 90 minutes before applying them to the slides.
The OncoBAC DNA microarrays used for CGH comprised 450 bacterial artificial chromosome, P1-derived artificial chromosome, and P1 clones printed in quadruplicate. The clone set was selected to include well-known and/or previously reported genes and loci associated with carcinogenesis, cell cycle regulation, cell proliferation, or apoptosis, as well as loci located in known amplicons. These clones and their genomic locations are listed at (20).11
The arrays were prepared as described previously (20). Briefly, the DNA was amplified by PCR with degenerate oligonucleotide primers containing a 5′ amine group. The resulting PCR products were printed in quadruplicate onto 3D-Link activated slides (Motorola Life Sciences, Northbrook, IL) using a custom array-printing robot (19). The slides were incubated overnight in a box with saturated NaCl vapor for post-print coupling and then stored in a desiccator.
Array hybridizations were done as described elsewhere (19-22). Briefly, the array slides were pretreated in blocking solution [50 mmol/L ethanolamine, 0.1 mol/L Tris (pH 9), 0.1% SDS] for 15 minutes at 50°C and then immersed in boiling water to denature the DNA. A rubber cement dam was placed around the array, and a probe mixture (described above) denatured at 73°C was applied. The slide was then placed without cover glass in a humidified chamber (50% formamide, 2× SSC) on a rocker at 37°C for 48 to 72 hours. Control hybridizations comparing normal male and female reference DNA were run to check assay quality. After hybridization, the slides were washed for 15 minutes at 50°C in 50% formamide/2× SSC (pH 7), for 30 minutes at 50°C in 2× SSC/0.1% SDS, and finally for 15 minutes at room temperature in PN buffer [0.1 mol/L sodium phosphate buffer, 0.1% NP40 (pH 8)]. The slides were then stained with 4′,6-diamidino-2-phenylindole (1 μmol/L DAPI, 1× PBS, 90% glycerol) for imaging.
TIFF format and 16-bit gray scale images were collected using a custom CCD camera imager fitted with CY3, CY5, and DAPI filters. The images were then analyzed as described elsewhere (19, 23). Spots with low DAPI intensity, low correlation between CY3 and CY5 pixel intensities, or low pixel numbers for the DAPI base segmented spot were eliminated from further analyses. Data were normalized to the median raw CY3/CY5 ratio and converted to log 2 to weight gains and losses equally. The mean and SD of the normalized log 2 ratio were calculated for each of the quadruplicate spots. Clones were eliminated from further analyses if the log 2 SD of the four replicate values exceeded 0.332 or if the ratio measurement was based on a single spot.
Spectral Karyotyping
Spectral karyotyping studies were carried out for 59 of the 60 cell lines (16). The 60th cell line, MDA-N, is no longer being made available for analysis. NCI UNK/ADR RES was not included in the breakpoint analysis presented here because we found that it is almost certainly a derivative of OVCAR8; hence, including it would have introduced bias. The evidence has been reported in detail elsewhere (16).
Drug Activity Profiles
For this analysis, we focused on a set of 118 compounds (24) with known or experimentally supported mechanisms of action. The data used were 50% growth inhibitory concentrations of the compounds in the NCI Developmental Therapeutics Program's NCI-60 screen (7).12 Each compound had been profiled multiple, independent times against the cell lines in a 48-hour sulforhodamine B assay. The data were filtered and analyzed as described previously (9, 24).10
Included are antimitotic agents, DNA antimetabolites, RNA antimetabolites, topoisomerase 1 inhibitors, topoisomerase 2 inhibitors, and several subtypes of alkylating agents. A listing of characteristics and mechanistic subclassifications can be found elsewhere (Table 1 in ref. 12).
Table 1.
Pearson's correlations of array CGH and expression for each gene in the analysis (i.e., self-self comparisons)
| Cytogenetic location |
Symbol | Gene name | cDNA expression array |
Oligonucleotide expression array |
||||
|---|---|---|---|---|---|---|---|---|
| R* | P† | Q‡ | R | P | Q | |||
| 1p36.13 | EPHA2 | Ephrin receptor EphA2 | −0.01 | 0.99 | 1.00 | −0.07 | 0.99 | 1.00 |
| 1p31.2 | GADD45A | Growth arrest- and DNA damage–inducible gene GADD45, α | 0.56 | 0.04 | 0.02 | 0.46 | 0.03 | 0.01 |
| 1p13.3 | CSF1 | Colony-stimulating factor 1 (macrophage) | 0.18 | 0.84 | 0.66 | 0.09 | 0.98 | 1.00 |
| 1q23.3 | DDR2 | Discoidin domain receptor family, member 2 | 0.15 | 0.90 | 0.89 | 0.11 | 0.98 | 1.00 |
| 1q32.1 | ELF3 | E74-like factor 3 (ets domain transcription factor, epithelial specific) | 0.33 | 0.46 | 0.14 | 0.27 | 0.53 | 0.20 |
| 2p21 | MSH2 | mutS homologue 2, colon cancer, nonpolyposis type 1 (Escherichia coli) | 0.19 | 0.84 | 0.56 | 0.17 | 0.90 | 0.64 |
| 2q23.3 | ARHE | ras homologue gene family, member E | 0.21 | 0.84 | 0.49 | 0.17 | 0.90 | 0.64 |
| 2q24.1 | ACVR1 | activin A receptor, type I | 0.22 | 0.81 | 0.43 | 0.09 | 0.98 | 1.00 |
| 3p25.3 | VHL | von Hippel-Lindau syndrome | 0.32 | 0.46 | 0.14 | 0.29 | 0.46 | 0.17 |
| 3p25.2 | RAF1 | v-raf-1 murine leukemia viral oncogene homologue 1 | 0.61 | 0.02 | 0.02 | 0.41 | 0.07 | 0.03 |
| 3p21.31 | CDC25A | Cell division cycle 25A | 0.59 | 0.03 | 0.02 | 0.34 | 0.25 | 0.08 |
| 3p21.31 | ARHA | ras homologue gene family, member A | 0.61 | 0.02 | 0.02 | 0.34 | 0.25 | 0.08 |
| 3p21.1 | TKT | Transketolase (Wernicke-Korsakoff syndrome) | 0.32 | 0.47 | 0.15 | 0.24 | 0.65 | 0.28 |
| 3q12.1 | DOC1 | Down-regulated in ovarian cancer 1 | −0.28 | 0.99 | 1.00 | −0.14 | 0.99 | 1.00 |
| 3q23 | PLS1 | Plastin 1 (I isoform) | −0.25 | 0.99 | 1.00 | −0.06 | 0.99 | 1.00 |
| 3q24 | AGTR1 | Angiotensin II receptor, type 1 | 0.25 | 0.72 | 0.32 | −0.02 | 0.99 | 1.00 |
| 3q27.3 | BCL6 | B-cell CLL/lymphoma 6 (zinc finger protein 51) | 0.28 | 0.61 | 0.23 | 0.16 | 0.91 | 0.73 |
| 3q29 | TFRC | Transferrin receptor (p90, CD71) | 0.35 | 0.40 | 0.11 | 0.48 | 0.03 | 0.01 |
| 4p16.3 | FGFR3 | Fibroblast growth factor receptor 3 (achondroplasia, thanatophoric dwarfism) | 0.33 | 0.46 | 0.14 | 0.38 | 0.12 | 0.04 |
| 4q12 | PDGFRA | Platelet-derived growth factor receptor, α polypeptide | 0.49 | 0.10 | 0.03 | 0.35 | 0.22 | 0.07 |
| 4q27 | CCNA2 | Cyclin A2 | 0.63 | 0.02 | 0.02 | 0.51 | 0.02 | 0.01 |
| 5q13.2 | CCNB1 | Cyclin B1 | 0.37 | 0.34 | 0.09 | 0.34 | 0.26 | 0.08 |
| 5q31.2 | CDC25C | Cell division cycle 25C | 0.29 | 0.57 | 0.20 | 0.31 | 0.33 | 0.11 |
| 6p21.31 | CDKN1A | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) | −0.03 | 0.99 | 1.00 | 0.1 | 0.98 | 1.00 |
| 6p21.2 | PIM1 | Proto-oncogene serine/threonine-protein kinase pim-1 (ec 2.7.1.37) | 0.36 | 0.39 | 0.11 | 0.27 | 0.48 | 0.18 |
| 6q21 | FYN | FYN tyrosine kinase proto-oncogene | 0.11 | 0.95 | 1.00 | 0.12 | 0.98 | 1.00 |
| 8p12 | FGFR1 | Fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome) | 0.33 | 0.46 | 0.14 | 0.31 | 0.33 | 0.11 |
| 8q12.1 | LYN | v-yes-1 Yamaguchi sarcoma viral related oncogene homologue | 0.26 | 0.69 | 0.28 | 0.38 | 0.11 | 0.04 |
| 8q12.1 | RAB2 | RAB2, member RAS oncogene family | 0.26 | 0.69 | 0.28 | 0.23 | 0.73 | 0.34 |
| 9q31.3 | GNG10 | Guanine nucleotide binding protein (G protein), γ 10 | 0.2 | 0.84 | 0.51 | 0.2 | 0.86 | 0.51 |
| 10p12.2 | BMI | Murine leukemia viral (bmi-1) oncogene homologue | 0.26 | 0.69 | 0.29 | 0.07 | 0.98 | 1.00 |
| 11p15.4 | ARHG | ras homologue gene family, member G (ρ G) | 0.33 | 0.46 | 0.14 | 0.34 | 0.23 | 0.08 |
| 11q13.1 | MEN1 | Multiple endocrine neoplasia I | 0.44 | 0.18 | 0.05 | −0.05 | 0.99 | 1.00 |
| 11q13.3 | CCND1 | Cyclin D1 (PRAD1: parathyroid adenomatosis 1) | 0.43 | 0.19 | 0.05 | 0.45 | 0.04 | 0.01 |
| 11q13.3 | EMS1 | Src substrate cortactin (amplaxin; oncogene ems1) | 0.57 | 0.04 | 0.02 | 0.4 | 0.08 | 0.03 |
| 11q13.5 | PAK1 | p21/Cdc42/Rac1-activated kinase 1 (STE20 homologue, yeast) | 0.01 | 0.99 | 1.00 | 0.04 | 0.98 | 1.00 |
| 11q14.2 | CTSC | Cathepsin C | 0.23 | 0.79 | 0.40 | 0.22 | 0.77 | 0.39 |
| 11q21 | MRE11A | MRE11 meiotic recombination 11 homologue A (Saccharomyces cerevisiae) | 0.63 | 0.02 | 0.02 | 0.34 | 0.25 | 0.08 |
| 12q14.1 | SAS | Sarcoma-amplified sequence | 0.18 | 0.84 | 0.64 | 0.0019 | 0.99 | 1.00 |
| 12q23.3 | TRA1 | Tumor rejection antigen (gp96) 1 | 0.28 | 0.62 | 0.23 | 0.15 | 0.91 | 0.76 |
| 12q24.12 | ALDH2 | Aldehyde dehydrogenase 2 family (mitochondrial) | 0.08 | 0.97 | 1.00 | 0.11 | 0.98 | 1.00 |
| 12q23.3 | TDG | Thymine-DNA glycosylase | 0.33 | 0.46 | 0.14 | 0.24 | 0.63 | 0.26 |
| 14q11.2 | APEX | APEX nuclease (multifunctional DNA repair enzyme) 1 | 0.58 | 0.04 | 0.02 | 0.3 | 0.37 | 0.13 |
| 14q11.2 | PSME1 | Proteasome (prosome, macropain) activator subunit 1 (PA28 α) | 0.06 | 0.98 | 1.00 | 0.27 | 0.50 | 0.19 |
| 14q32.33 | AKT1 | v-akt murine thymoma viral oncogene homologue 1 | 0.42 | 0.20 | 0.05 | 0.36 | 0.17 | 0.06 |
| 15q24.1 | PML | acute promyelocytic leukemia, inducer | −0.04 | 0.99 | 1.00 | 0.17 | 0.90 | 0.64 |
| 15q24.1 | CSK | c-src tyrosine kinase | 0.51 | 0.08 | 0.02 | 0.28 | 0.46 | 0.17 |
| 16q22.1 | CDH1 | Cadherin 1, type 1, E-cadherin (epithelial) | −0.01 | 0.99 | 1.00 | −0.03 | 0.99 | 1.00 |
| 17q12 | ERBB2 | v-erb-b2 avian erythroblastic leukemia viral oncogene homologue 2 | 0.46 | 0.13 | 0.04 | 0.5 | 0.02 | 0.01 |
| 17q21.2 | TOP2A | Topoisomerase (DNA) II α, 170 kDa | 0.44 | 0.17 | 0.05 | 0.31 | 0.33 | 0.11 |
| 18p11.23 | PTPRM | Protein tyrosine phosphatase, receptor type, M | 0.22 | 0.81 | 0.43 | 0.11 | 0.98 | 1.00 |
| 19p13.2 | JUNB | Jun B proto-oncogene | 0.19 | 0.84 | 0.56 | 0.2 | 0.87 | 0.54 |
| 19q13.33 | LIG1 | Ligase I, DNA, ATP dependent | 0.32 | 0.46 | 0.14 | 0.1 | 0.98 | 1.00 |
| 20p13 | FKBP1A | FK506 binding protein 1A, 12 kDa | 0.49 | 0.09 | 0.03 | 0.53 | 0.01 | 0.01 |
| 20p13 | CDC25B | Cell division cycle 25B | 0.31 | 0.51 | 0.17 | 0.28 | 0.48 | 0.17 |
| 20p11.23 | SEC23B | Sec23 homologue B (S. cerevisiae) | 0.53 | 0.06 | 0.02 | 0.49 | 0.02 | 0.01 |
| 20q13.12 | MYBL2 | v-myb myeloblastosis viral oncogene homologue (avian)-like 2 | 0.22 | 0.81 | 0.43 | 0.19 | 0.88 | 0.59 |
| 20q13.12 | PRKCBP1 | protein kinase C binding protein 1 | 0.2 | 0.84 | 0.54 | 0.24 | 0.69 | 0.31 |
| 20q13.13 | CSE1L | CSE1 chromosome segregation 1-like (yeast) | 0.58 | 0.04 | 0.02 | 0.44 | 0.05 | 0.02 |
| 20q13.13 | PTPN1 | Protein tyrosine phosphatase, non-receptor type 1 | −0.03 | 0.99 | 1.00 | 0.18 | 0.90 | 0.64 |
| 21q22.3 | MX2 | Myxovirus (influenza virus) resistance 2 (mouse) | 0.38 | 0.32 | 0.09 | 0.05 | 0.98 | 1.00 |
| 21q22.3 | SMT3H1 | SMT3 suppressor of mif two 3 homologue 3 (yeast) | 0.43 | 0.19 | 0.05 | 0.41 | 0.07 | 0.03 |
| 22q13.1 | ATF4 | Activating transcription factor 4 (tax-responsive enhancer element B67) | 0.05 | 0.98 | 1.00 | 0.28 | 0.48 | 0.17 |
| Xq23§ | PRKCI | Protein kinase C, iota | −0.01 | 0.99 | 1.00 | −0.11 | 0.99 | 1.00 |
Pearson's correlation coefficient.
Westfall-Young P for finding a correlation for any gene as big as the observed correlation under the null hypothesis of zero correlation (27).
Benjamini-Hochberg procedure Q representing the smallest false discovery rate for declaring the gene positively correlated (28).
Reassigned in July 2003 to 3q26.2.
Gene Matching between Data Sets
Before analysis could begin, it was necessary to identify the genes represented in the array CGH and in the two expression data sets and to determine the intersection of those three sets. The data set intersections (merge lists) were generated using a Python script, parseUniGene,13 an early developmental version of our publicly available, web-based MatchMiner program (25).14 The program matched IMAGE clone IDs for the cDNA arrays with Genbank accession numbers for both the oligonucleotide chips and the CGH arrays through their UniGene cluster assignments (build 132). After this matching procedure, the resulting data were screened for missing and floored values. For each of the three data sets, only those sequences or clones with ≤45 missing or floored values were carried forward.
Because some genes were represented by more than one sequence on the array, the next step was to ensure that each entry in a merge list represented a one-to-one relationship between array CGH and expression. Therefore, we derived, on a gene-by-gene basis, a single array CGH or expression value per cell line for those genes represented by more than one sequence on an array. For the array CGH data, the log-mean of the copy number ratio was computed after confirming a similar pattern of hybridization between clones representing the same locus across the 60 cell lines by computing the Pearson's correlation coefficient. That procedure yielded a total of 353 unique genes from the array CGH data. To maximize the reliability of the expression data, we restricted our attention in the present analyses to a 64-gene subset that satisfied two conditions: (a) each gene was present in the array CGH, cDNA, and oligonucleotide data sets, and (b) each gene showed an expression pattern reasonably concordant between the cDNA and oligonucleotide data sets (i.e., correlation coefficient > 0.3; ref. 18). For genes with multiple representations in the expression data sets, we used the sequence that gave the highest correlation between cDNA and oligonucleotide array expression data (18). That selection process would not be expected to bias statistical calculations relating expression to the array CGH data.
Statistical Analysis
Except when specified, all analyses were done using SAS (SAS Software, Inc., v8.2). Once the gene lists were finalized, array CGH-cDNA array Pearson's correlation coefficients were calculated for all 64 × 64 = 4,096 possible pairs of genes (with all data log 2 transformed) after a detailed examination of the data for outliers (see Supplementary Materials and Supplementary Tables S3 and S4 for a description).15 The same was done for all 4,096 array CGH-oligonucleotide array gene pairs. Histograms showing the distributions of correlation coefficients for those comparisons can be found in Supplementary Figs. S1 and S2.15 Confidence limits for the correlations were estimated by bootstrap resampling (26) using the empirical percentile method with balanced resampling of 10,000 iterations. By using bootstrap resampling, we avoided parametric assumptions about the distributions of the variables.
The reliability of the correlation between each gene's copy number and its expression was assessed in two ways. First the Westfall-Young (27) Ps for each gene expresses the probability of finding a correlation for any gene as big as that gene's correlation but under the null hypothesis of zero correlation (assuming no systematic relationship between gene expression and array CGH). The null distribution was estimated by recalculating the correlations many times after permuting the correspondence between expression values and array CGH. Such resampling preserves the correlations between different genes. The second estimate of reliability used the notion of false discovery rate, the expected fraction of false positives among all positives. The expected false discovery rate was computed using the Benjamini-Hochberg (28) procedure. The q (29) for each gene is the smallest false discovery rate at which that gene would be declared as positively correlated. We calculated qs for each of the 64 correlations. Those calculations were done using the R statistical language.16
The 64 × 64 matrix of array CGH-cDNA array Pearson's correlation coefficients was row- and column- ordered by chromosomal location (i.e., from chromosome 1 to X) to create what we here term a “genomic image map” using our CIMminer program package with the “no-clustering” option.17 Analogous genomic image maps were also generated for the 64 × 64 array CGH-array CGH and array CGH-oligonucleotide array correlation matrices. To begin analysis of the relationship between DNA copy number and drug sensitivity, similar pairwise correlations were calculated between the array CGH 353-gene data set and the 118 tested compounds. Genomic image maps and clustered image maps (6) of the resulting matrices of drug-array CGH correlation coefficients were ordered by hierarchical agglomerative clustering with a correlation distance metric and average linkage using CIMminer.
Results
Array CGH Expression Comparisons
We explored the relationships between DNA copy number and mRNA expression. A total of 165 genes (represented by 204 sequences on the array CGH array and 206 sequences on the cDNA array) were found in both the array CGH and cDNA sets. 212 genes (represented by 263 sequences and 252 sequences, respectively) were found in both the array CGH and oligonucleotide sets. To ensure a robust analysis, we focused our attention on 64 genes that were represented in all three data sets, had no more than 45 missing or floored values in any of the data sets, and had a Pearson's correlation coefficient of >0.30 when comparing the cDNA and oligonucleotide expression data (Table 1). The latter criterion defines a set of genes whose expression patterns have been validated in silico. This validation process lent confidence that we were measuring expression levels and DNA copy numbers for the same genes (18).
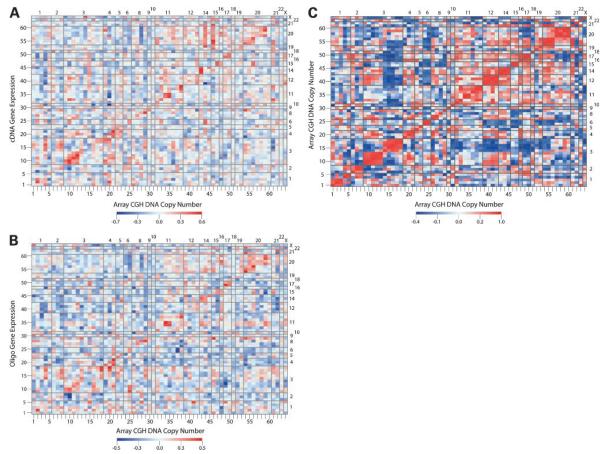
Using CIMminer, we created genomic image maps (Fig. 1A and B) containing the Pearson correlation coefficients for all possible combinations of array CGH-cDNA array and array CGH-oligonucleotide array data, respectively. “Self-self” (i.e., same gene) correlations seem on the major diagonal in each figure. The average ± SE of the array CGH expression self-self correlation (over the 64 genes) was 0.29 ± 0.03 (range, −0.28 to 0.63) for the array CGH-cDNA and 0.23 ± 0.0.02 (range, −0.29 to 0.51) for the array CGH-oligonucleotide comparisons. There were no statistically significant negative self-self correlations (Table 1). Those findings across disparate cell types and across the genome support the generalization that DNA copy number is one factor (among others) that can influence gene expression.
Figure 1.
Genomic image maps showing the Pearson's correlation of DNA copy number with gene expression level across the NCI-60 cell lines. Genes are listed on the axes in chromosomal order (corresponding to the order in Table 1). A, correlation of DNA copy number with expression levels measured using cDNA arrays. B, correlation of DNA copy number with expression levels measured using Affymetrix oligonucleotide arrays. C, correlation of DNA copy number with itself. Red and blue indicate high and low correlations, respectively. Top right, chromosome numbers from 1 to X.
Figure 1A and B shows several interesting patterns off diagonal, potentially representing associations between the DNA copy number of one gene and the expression of another gene (on the same chromosome or a different one). Two such patterns of positive correlation lie close to the diagonal and correspond to genes at 3p21.31-p25.3 (i.e., #9–12, Fig. 1, bottom) and 11q13.3 (i.e., #34–35). Those regions contain several genes (VHL, von Hippel-Lindau tumor suppressor; RAF1, v-raf-1 murine leukemia viral oncogene homologue 1; CDC25A, cell division cycle 25A; and ARHA, ras homologue gene family member A on 3p21.31-p25.3; CCND1, cyclin D1, and EMS1, cortactin on 11q13.3) with positive correlations between DNA copy number and gene expression that were reciprocal (i.e., the copy number of VHL was positively correlated with expression of RAF1 and vice versa). The 3p pattern was most striking in the array CGH-cDNA comparison (Fig. 1A). Several of the 3p relationships were statistically significant (Table 2; Supplementary Table S1).15 The same pattern was seen in the array CGH-oligonucleotide comparison (Fig. 1B; Table 2; Supplementary Table S2).15 The relationship between CCND1 and EMS1 on 11q13.3 was significant and reciprocal in both comparisons (Table 2).
Table 2.
Top 50 positive and negative array CGH expression correlations statistically significant (two-tailed 95% bootstrap) for both cDNA and oligonucleotide expression array comparisons
| Array CGH gene |
Expression level gene |
R* (cDNA array) | Lower 95% CI bound (cDNA array) |
Upper 95% CI bound (cDNA array) |
R (Oligo array) | Lower 95% CI bound (oligo array) |
Upper 95% CI bound (oligo array) |
|---|---|---|---|---|---|---|---|
| CCND1 | EMS1 | 0.58 | 0.32 | 0.75 | 0.45 | 0.13 | 0.67 |
| TFRC | PDGFRA | 0.51 | 0.21 | 0.81 | 0.52 | 0.28 | 0.75 |
| LYN | RAB2 | 0.57 | 0.38 | 0.71 | 0.45 | 0.23 | 0.62 |
| PSME1 | APEX | 0.62 | 0.45 | 0.76 | 0.39 | 0.13 | 0.62 |
| CTSC | MRE11A | 0.54 | 0.34 | 0.70 | 0.46 | 0.17 | 0.67 |
| EMS1 | CCND1 | 0.48 | 0.21 | 0.67 | 0.51 | 0.29 | 0.68 |
| RAB2 | FKBP1A | 0.56 | 0.36 | 0.73 | 0.41 | 0.23 | 0.58 |
| FGFR3 | CCNA2 | 0.48 | 0.29 | 0.67 | 0.46 | 0.27 | 0.64 |
| VHL | RAF1 | 0.56 | 0.40 | 0.71 | 0.35 | 0.08 | 0.59 |
| CDC25B | PML | 0.61 | 0.12 | 0.85 | 0.28 | 0.05 | 0.48 |
| ATF4 | FKBP1A | 0.49 | 0.22 | 0.71 | 0.41 | 0.13 | 0.63 |
| PTPN1 | CSE1L | 0.48 | 0.18 | 0.74 | 0.39 | 0.06 | 0.65 |
| MYBL2 | CSE1L | 0.48 | 0.19 | 0.74 | 0.39 | 0.07 | 0.64 |
| DOC1 | DDR2 | 0.43 | 0.14 | 0.65 | 0.41 | 0.18 | 0.60 |
| AKT1 | APEX | 0.53 | 0.30 | 0.70 | 0.30 | 0.06 | 0.51 |
| AGTR1 | ACVR1 | 0.48 | 0.21 | 0.68 | 0.35 | 0.05 | 0.59 |
| PRKCBP1 | CDH1 | 0.42 | 0.10 | 0.62 | 0.41 | 0.02 | 0.63 |
| MX2 | FKBP1A | 0.36 | 0.10 | 0.58 | 0.47 | 0.23 | 0.67 |
| PDGFRA | CCNA2 | 0.47 | 0.26 | 0.63 | 0.36 | 0.16 | 0.51 |
| RAF1 | VHL | 0.46 | 0.22 | 0.63 | 0.35 | 0.09 | 0.56 |
| PRKCI | DOC1 | 0.52 | 0.25 | 0.71 | 0.28 | 0.05 | 0.48 |
| BCL6 | TFRC | 0.31 | 0.01 | 0.57 | 0.48 | 0.35 | 0.61 |
| ERBB2 | TOP2A | 0.44 | 0.22 | 0.61 | 0.34 | 0.05 | 0.55 |
| PLS1 | RAB2 | 0.43 | 0.21 | 0.60 | 0.34 | 0.14 | 0.54 |
| CDC25A | CCNA2 | 0.42 | 0.20 | 0.60 | 0.35 | 0.10 | 0.57 |
| BMI | FYN | −0.32 | −0.54 | −0.07 | −0.40 | −0.56 | −0.22 |
| BMI | DDR2 | −0.46 | −0.64 | −0.25 | −0.27 | −0.45 | −0.07 |
| MRE11A | CSF1 | −0.35 | −0.59 | −0.06 | −0.39 | −0.59 | −0.15 |
| CDH1 | FYN | −0.31 | −0.55 | −0.05 | −0.42 | −0.63 | −0.17 |
| MEN1 | RAB2 | −0.41 | −0.60 | −0.12 | −0.33 | −0.54 | −0.19 |
| EMS1 | PDGFRA | −0.50 | −0.71 | −0.19 | −0.24 | −0.38 | −0.07 |
| CCNA2 | ACVR1 | −0.44 | −0.62 | −0.22 | −0.30 | −0.52 | −0.07 |
| MYBL2 | LYN | −0.45 | −0.66 | −0.12 | −0.29 | −0.49 | −0.06 |
| CSE1L | LYN | −0.41 | −0.62 | −0.12 | −0.33 | −0.52 | −0.08 |
| ALDH2 | ACVR1 | −0.39 | −0.59 | −0.19 | −0.36 | −0.55 | −0.15 |
| PTPN1 | LYN | −0.46 | −0.67 | −0.14 | −0.29 | −0.48 | −0.05 |
| PIM1 | CTSC | −0.32 | −0.51 | −0.09 | −0.44 | −0.61 | −0.24 |
| ALDH2 | RAB2 | −0.40 | −0.62 | −0.12 | −0.36 | −0.54 | −0.20 |
| SAS | FYN | −0.36 | −0.62 | −0.04 | −0.41 | −0.63 | −0.15 |
| CCNA2 | DDR2 | −0.43 | −0.63 | −0.18 | −0.36 | −0.53 | −0.16 |
| CDKN1A | CTSC | −0.34 | −0.50 | −0.15 | −0.46 | −0.60 | −0.28 |
| CSF1 | CDH1 | −0.43 | −0.65 | −0.13 | −0.38 | −0.61 | −0.10 |
| ACVR1 | RAF1 | −0.34 | −0.61 | −0.04 | −0.48 | −0.65 | −0.28 |
| CCND1 | PDGFRA | −0.57 | −0.77 | −0.27 | −0.27 | −0.42 | −0.10 |
| SAS | DDR2 | −0.53 | −0.73 | −0.25 | −0.33 | −0.53 | −0.11 |
| CDH1 | DDR2 | −0.47 | −0.68 | −0.25 | −0.42 | −0.61 | −0.22 |
| ARHA | ERBB2 | −0.42 | −0.61 | −0.15 | −0.48 | −0.68 | −0.19 |
| FGFR3 | FKBP1A | −0.54 | −0.72 | −0.30 | −0.41 | −0.61 | −0.18 |
| ARHE | RAF1 | −0.51 | −0.68 | −0.30 | −0.50 | −0.65 | −0.34 |
| BMI | PDGFRA | −0.66 | −0.87 | −0.42 | −0.38 | −0.60 | −0.15 |
Pearson's Correlation Coefficient.
The observation of reciprocity between neighboring genes could be interpreted as evidence for an underlying structural mechanism that links gene expression to DNA copy number. Array CGH-array CGH correlations showed strong relationships between neighboring genes in the same region (Fig. 1C). Interestingly, spectral karyotyping data (16) for 58 of the same 60 cell lines indicate that those regions are bounded by zones with significantly more breaks than expected by chance. In addition, there is a lack of breaks within the region, suggesting that the regions have remained relatively intact during genomic rearrangements in the NCI-60 cell lines. Analysis of the Mitelman Database of Chromosome Aberrations in Cancer (30) indicates this pattern of breakpoints on 3p (increased breakage at 3p21 with limited breakage telomeric to that) to be a feature of clinical tumor samples as well.
Off-diagonal relationships between gene expression and gene copy number may reflect any number of different phenomena: direct interaction between the gene products, “linkage effects” of neighboring genes that interact directly, indirect interactions involving other genes in a pathway, or selective pressure for underlying chromosomally mediated mechanisms. In the last case, we might expect to find that array CGH-expression correlations are not a result of the copy number of one gene influencing the expression of another but rather reflect a correlation at the DNA level. To address this, we compared the instances of high positive and negative array CGH-expression correlation with those of high positive and negative array CGH self-correlation. There was little overlap between the two types of correlations, indicating a lack of selection for coordinate rearrangement at the chromosomal level (Fig. 1). Those associations that did show overlap between the array CGH-expression correlations and the array CGH self-correlation included chromosome 3p25-p21 (i.e., #9–12, Fig. 1) and chromosome 11q13 (i.e., #33–34, Fig. 1). Additionally, a positive correlation between 20p11-13 (encompassing FKBP1A, FK506 binding protein 1A, 12 kDa; CDC25B, cell division cycle 25B; and SEC23B, Sec23 homologue B; i.e., #54–56, Fig. 1) and MX2, myxovirus (influenza virus) resistance 2 mouse (i.e., #61, Fig. 1) at 21q22.3 in the array CGH expression data was also present in the array CGH self-comparison.
Most of the statistically significant off-diagonal relationships corresponding to genes on different chromosomes could not be explained on the basis of the literature. However, four such relationships were identified as interesting based on known biology:
(a) There was a positive correlation between DNA copy number for CDC25A and expression of CCNA2 (cyclin A2) in both the array CGH-cDNA (0.42; 95% CI, 0.2–0.59) and array CGH-oligonucleotide (0.35; 95% CI, 0.10–0.57) comparisons. Cdc25A is a phosphatase that directly interacts with cyclin-dependent kinase 2/cyclin A complexes, dephosphorylating cyclin A and activating the complex during the G1-S transition. Both CDC25A and CCNA2 showed significant self-self correlations in the array CGH-cDNA array comparisons (Table 1). CCNA2 also showed a significant self-self correlation in the array CGH-oligonucleotide array comparison (Table 1). Those observations suggest that DNA copy number plays a role in determining the expression levels of those genes. To check whether the correlation between CDC25A copy number and CCNA2 expression reflected selection at the DNA level, we examined the correlation between CDC25A and CCNA2 DNA copy number. The correlation was 0.34 (95% bootstrap CI, 0.07–0.58), suggesting that there may be selective pressure to keep the DNA copy numbers of those two genes in some degree of balance.
(b) CCNA2 copy number was significantly positively correlated with cell division cycle 25C (CDC25C) expression in the cDNA array comparison (0.44; 95% CI, 0.25–0.61). As mentioned above, CCNA2 showed a significant self-self correlation, whereas CDC25C did not. However, the transcription of CDC25C and CCNA2 is mediated through the CDE-CHR complex (31). That shared regulation, combined with the fact that CCNA2 expression was significantly correlated with its copy number, suggests that the association between CCNA2 copy number and CDC25C reflects the shared transcriptional control of those two genes.
(c) Thymine-DNA glycosylase (TDG) copy number was inversely correlated with SMT3 suppressor of mif two 3 homologue 3 (SMT3H1) expression in the array CGH-oligonucleotide comparison (−0.37; 95% CI, −0.54 to −0.16) although not in the array CGH-cDNA comparison (−0.11; 95% CI, −0.32 to 0.11). TDG encodes thymidine-DNA glycosylase, and SMT3H1 encodes SUMO-3. SUMO-3 sumoylates TDG, a modification proposed as the link between TDG and AP glycosylases in the base excision repair pathway (32).
(d) ERBB2 (c-erb B2/neu) expression was inversely correlated with ARHA and CDC25A DNA copy number. For the cDNA array expression data, the values were −0.42 (95% CI, −0.61 to −0.15) and −0.30 (95% CI, −0.52 to −0.08), respectively. For the oligonucleotide array expression data, the corresponding figures were −0.48 (95% CI, −0.68 to −0.19) and −0.31 (95% CI, −0.53 to −0.12), respectively. This inverse relationship has been documented in the literature as the observation of ERBB2 amplification and/or over-expression and loss of 3p, as determined by CGH or loss of heterozygosity (33, 34). Neither study was able to show a statistical relationship between ERBB2 amplification and 3p loss and did not address whether ERBB2 overexpression was associated with 3p loss. Similarly, the analysis of array CGH-array CGH correlations did not show a negative correlation between 3p copy number and ERBB2 copy number. That observation makes it clear that the previously established association is a result of ERBB2 overexpression, and that the association is not dependent on overexpression as a result of ERBB2 amplification.
Array CGH and Drug Response
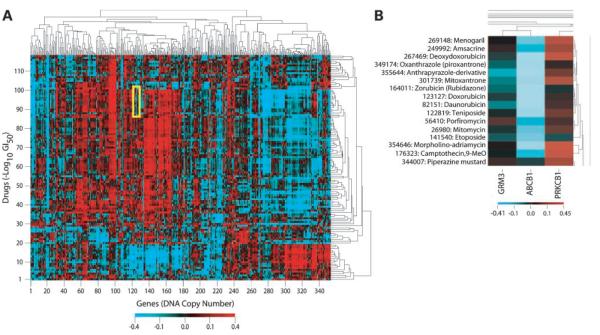
In addition to examining the relationship between DNA copy number and mRNA expression levels, we analyzed the correlation between DNA copy number and drug activity against the NCI-60 for a set of 118 compounds whose mechanisms of action are putatively known. Computing the Pearson's correlation coefficients for these “mechanism of action” drugs (24) and clustering (6) yielded the clustered image map in Fig. 2A. The correlations between drugs and DNA copy number ranged from −0.57 to 0.54. Drugs with similar mechanisms of action, such as the taxol analogues and camptothecin derivatives, clustered together (Supplementary Fig. S3).15 Known relationships, such as the inverse correlation between ATP-binding cassette, subfamily B (MDR/TAP), member 1 (ABCB1) DNA copy number and activity of ABCB1/MDR1 substrates, were present (Fig. 2B), as were several novel relationships. Some chromosome regions (e.g., 3q) clustered tightly together, whereas others (e.g., chromosome 7) scattered widely (see Supplementary Fig. S4).15 Table 3 and Supplementary Table S515 highlight some of those DNA copy number-drug relationships with bootstrap-estimated 95% CIs that do not include zero.
Figure 2.
Clustered image map showing the Pearson's correlation of DNA copy number with drug sensitivity across the NCI-60 cell lines. A, clustered image map showing the correlation for 353 genes and the −log(GI50) of 118 “mechanism of action” drugs. The negative correlation of DNA copy number for ABCB1 (MDR1) and substrates of P-glycoprotein is highlighted in yellow and enlarged in B. Red and blue indicate high and low correlations, respectively. Cluster trees of both axes with labels can be viewed in Supplementary Figs. S3 and S4 (available at http://mct.aacrjournals.org).
Table 3.
Top 50 positive and negative correlations between array CGH and drug sensitivity for 118 “mechanism of action” compounds (with two-tailed 95% boostrap CIs)
| Locus | Gene name | NSC No. | Drug | R* | Lower 95% CI bound |
Upper 95% CI bound |
|---|---|---|---|---|---|---|
| BTK | Bruton agammaglobulinemia tyrosine kinase | 176323 | Camptothecin,9-MeO | 0.54 | 0.27 | 0.72 |
| ZNF70 | Zinc finger protein 70 (Cos17) | 658831 | Taxol analogue | 0.53 | 0.24 | 0.74 |
| CTSB | Cathepsin B | 153858 | Maytansine | 0.51 | 0.13 | 0.75 |
| AFM267xd9 | 658831 | Taxol analogue | 0.51 | 0.31 | 0.67 | |
| BRCA1 | Breast cancer 1, early onset | 109229 | l-Asparaginase | 0.50 | 0.32 | 0.66 |
| FGFR3 | Fibroblast growth factor receptor 3 (achondroplasia, thanatophoric dwarfism) | 143095 | Pyrazofurin | 0.50 | 0.32 | 0.66 |
| GPRK2L | G protein–coupled receptor kinase 4 | 143095 | Pyrazofurin | 0.50 | 0.32 | 0.66 |
| 276n12rev | 658831 | Taxol analogue | 0.49 | 0.28 | 0.66 | |
| TEC | Tec protein tyrosine kinase | 143095 | Pyrazofurin | 0.49 | 0.26 | 0.68 |
| EST_185259 | 658831 | Taxol analogue | 0.49 | 0.23 | 0.68 | |
| ETV6 | ets variant gene 6 (TEL oncogene) | 167780 | Asaley | 0.48 | 0.28 | 0.73 |
| LPL | Lipoprotein lipase | 49842 | Vinblastine-sulfate | 0.47 | 0.15 | 0.67 |
| ETV6 | ets variant gene 6 (TEL oncogene) | 172112 | Spiromustine | 0.47 | 0.12 | 0.74 |
| 139k18rev | 658831 | Taxol analogue | 0.46 | 0.23 | 0.65 | |
| TCL1A | T-cell leukemia/lymphoma 1A | 249992 | Amsacrine | 0.46 | 0.25 | 0.64 |
| AFMa082zd9 | 658831 | Taxol analogue | 0.46 | 0.24 | 0.63 | |
| 191e05rev | 658831 | Taxol analogue | 0.46 | 0.22 | 0.65 | |
| ETV6 | ets variant gene 6 (TEL oncogene) | 34462 | Uracil mustard | 0.46 | 0.06 | 0.76 |
| CDH1 | Cadherin 1, type 1, E-cadherin (epithelial) | 606985 | Camptothecin,20-ester (S) | 0.46 | 0.26 | 0.64 |
| SHGC_6945 | 658831 | Taxol analogue | 0.46 | 0.24 | 0.64 | |
| CDH1 | Cadherin 1, type 1, E-cadherin (epithelial) | 249992 | Amsacrine | 0.46 | 0.25 | 0.62 |
| PRKCI | Protein kinase C, ι | 176323 | Camptothecin,9-MeO | 0.46 | 0.12 | 0.67 |
| S1B11_PRLL | 658831 | Taxol analogue | 0.46 | 0.20 | 0.65 | |
| PPP2R1B | Protein phosphatase 2 (formerly 2A), regulatory subunit A (PR 65), β isoform | 143095 | Pyrazofurin | 0.45 | 0.19 | 0.64 |
| CDH1 | Cadherin 1, type 1, E-cadherin (epithelial) | 94600 | Camptothecin | 0.45 | 0.24 | 0.63 |
| ETV6 | ets variant gene 6 (TEL oncogene) | 664402 | Taxol analogue | −0.45 | −0.77 | −0.01 |
| ABR | Active BCR-related gene | 118994 | Inosine-glycodialdehyde | −0.45 | −0.63 | −0.21 |
| AFM289zh5 | 656178 | Taxol analogue | −0.45 | −0.62 | −0.24 | |
| PTEN | Phosphatase and tensin homologue (mutated in multiple advanced cancers 1) | 330500 | Geldanamycin | −0.45 | −0.64 | −0.21 |
| PTEN | Phosphatase and tensin homologue (mutated in multiple advanced cancers 1) | 673187 | Taxol analogue | −0.45 | −0.66 | −0.21 |
| CCNC | Cyclin C | 671870 | Taxol analogue | −0.46 | −0.71 | −0.15 |
| CEBPB | CCAAT/enhancer binding protein (C/EBP), β | 338947 | Clomesone | −0.46 | −0.67 | −0.15 |
| GADD45A | Growth arrest and DNA damage–inducible, α | 118994 | Inosine-glycodialdehyde | −0.46 | −0.63 | −0.26 |
| RET | ret proto-oncogene (multiple endocrine neoplasia and medullary thyroid carcinoma 1, Hirschsprung disease) | 656178 | Taxol analogue | −0.46 | −0.65 | −0.21 |
| CH1 | Chromosome 1 open reading frame 9 | 118994 | Inosine-glycodialdehyde | −0.46 | −0.63 | −0.26 |
| ABCB1 | ATP-binding cassette, subfamily B (MDR/TAP), member 1 | 666608 | Taxol analogue | −0.46 | −0.70 | −0.03 |
| AFM289zh5 | 666608 | Taxol analogue | −0.47 | −0.64 | −0.24 | |
| GRM3 | Glutamate receptor, metabotropic 3 | 671867 | Taxol analogue | −0.47 | −0.64 | −0.25 |
| RARB | Retinoic acid receptor, β | 600222 | Taxol analogue | −0.47 | −0.66 | −0.20 |
| PTEN | Phosphatase and tensin homologue (mutated in multiple advanced cancers 1) | 664404 | Taxol analogue | −0.47 | −0.66 | −0.25 |
| PTEN | Phosphatase and tensin homologue (mutated in multiple advanced cancers 1) | 664402 | Taxol analogue | −0.47 | −0.68 | −0.22 |
| SAS | Sarcoma-amplified sequence | 656178 | Taxol analogue | −0.47 | −0.64 | −0.23 |
| RET | ret proto-oncogene (multiple endocrine neoplasia and medullary thyroid carcinoma 1, Hirschsprung disease) | 666608 | Taxol analogue | −0.47 | −0.69 | −0.19 |
| CCNC | Cyclin C | 666608 | Taxol analogue | −0.48 | −0.76 | −0.12 |
| ZNF70 | Zinc finger protein 70 (Cos17) | 107124 | Camptothecin,10-OH | −0.48 | −0.69 | −0.19 |
| GLI | Glioma-associated oncogene homologue (zinc finger protein) | 656178 | Taxol analogue | −0.49 | −0.65 | −0.24 |
| SRC | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homologue (avian) | 153858 | Maytansine | −0.50 | −0.65 | −0.31 |
| AFMa303yb1 | 600222 | Taxol analogue | −0.51 | −0.69 | −0.26 | |
| ETV6 | ets variant gene 6 (TEL oncogene) | 671867 | Taxol analogue | −0.56 | −0.76 | −0.18 |
| ARHI | ras homologue gene family, member I | 118994 | Inosine-glycodialdehyde | −0.57 | −0.71 | −0.42 |
Pearson's correlation coefficient.
The array CGH-drug genomic image map (not shown) yielded interesting observations. Certain regions of the genome showed correlations for entire classes of drugs, such as the tubulin-interacting agents and those that act through incorporation into DNA. For example, we saw a block of three genes from chromosome 4q13-21 (PDGFRA, platelet-derived growth factor receptor, α polypeptide; EPHA5, EPH receptor A5; and ALB, albumin) that were inversely correlated with almost all of the drugs thought to act through incorporation into DNA. None of the genes has any known association with DNA metabolism, suggesting that we had observed a linkage effect and that the gene responsible for the correlations was in the same region of 4q. We used MatchMiner (25) to retrieve a list of the HUGO symbols of genes in the region of 4q13-21. That list was then used as input to GoMiner (35) to identify genes that might be involved in DNA metabolism, particularly in DNA repair. Electronic annotation of one of those genes, ankyrin repeat domain 17 (ANKRD17), indicate that it binds damaged DNA and that it is involved in mismatch repair.
Two regions (3q26-qter and 12p12-13) were correlated with the tubulin-interacting agents. Genes from 3q26 [SLC2A, solute carrier family 2 (facilitated glucose transporter), member 2; PIK3CA, phosphoinositide-3-kinase, catalytic, α polypeptide; and TERC, telomerase RNA component, as well as several sequence-tagged sites] were negatively correlated with the activity patterns of maytansine and vincristine sulfate, both microtubule destabilizers, and positively correlated with those of taxol and its derivatives, which are microtubule stabilizers. Those observations suggest that the correlations related in some fashion to microtubule chemistry or function, probably focused on β-tubulin (36-38). However, there was no statistically significant correlation with some of the other tubulin-destabilizing agents, such as the colchicines, dolastatin-10, halichondrin B, trityl-cysteine, or vinblastine sulfate. None of the genes on the array that map to 3q26 or to 3q have any known interaction with tubulins in general or β-tubulin specifically.
Following a similar approach to that described above, we retrieved the gene symbols for genes on 3q26-3qter and used GoMiner to map them into the Gene Ontology. There were several genes known to be involved in transport (including some ABC transporters), genes involved in cytoskeleton organization and maintenance, and LOC389192, a gene for a hypothetical protein similar in sequence to the tubulin β-4q chain. However, not one gene stood out as the likely driver of the correlations with tubulin-interacting agents.
On chromosome 12, ets variant gene 6 (ETV6/TEL oncogene, 12p12-p13) was negatively correlated with all of the tubulin-interacting agents. Several tubulin or tubulin-associated genes are on chromosome 12. However, all of them are on the q arm, and there were no significant negative correlations between 12q genes and the activity patterns of the tubulin-interacting agents.
From DNA to RNA to Drug Sensitivity
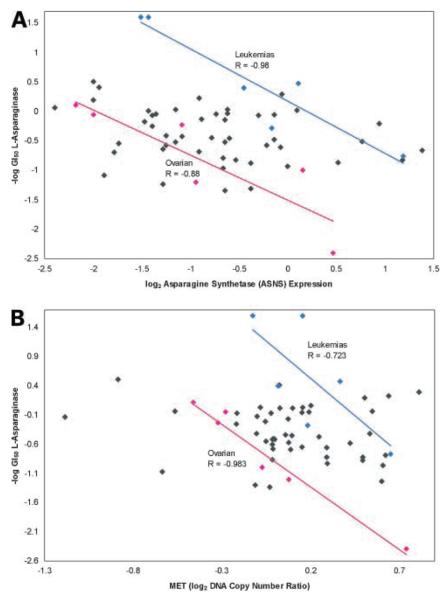
One particularly interesting relationship we showed in a previous analysis was that between asparagine synthetase expression and sensitivity to L-asparaginase (9). Although the correlation across all 60 cell lines was only moderately high (−0.44), an analysis of the data by tissue of origin revealed strong negative correlations for leukemias (−0.98; 95% bootstrap CI, −1.00 to −0.93) ovarian cancers (−0.88; 95% bootstrap CI, −0.99 to −0.23). When we asked whether a similar correlation would be seen at the DNA level, loci on 7q showed a negative correlation in both the ovarian and leukemic cell lines. In particular, MET (met proto-oncogene), which is 18.9 Mb telomeric of asparagine synthetase (asparagine synthetase itself was not present on the array), showed a striking correlation of −0.983 (95% bootstrap CI, −0.999 to −0.899) for the ovarian cell lines and −0.723 (95% bootstrap CI, −0.991 to 0.0388) for the leukemic lines (Fig. 3A), similar to what was found previously with the expression data (Fig. 3B).
Figure 3.
Relationship of L-asparaginase activity to asparagine synthetase (ASNS) expression level and DNA copy number in the NCI-60 cell lines. A, L-asparaginase activity [from data in Scherf et al. (12)] versus asparagine synthetase expression level. These data generated the hypothesis that a subset of ovarian cancers might respond to L-asparaginase. B, L-asparaginase activity versus DNA copy number for a clone (MET) near asparagine synthetase on chromosome 7. Blue, pink, and gray data points are from leukemic, ovarian, and other cancer cell types, respectively. The blue and pink lines represent linear least-squares fits. CIs for the correlation coefficient estimates are given in Results.
Discussion
The new technologies for high-resolution, genome-wide profiling at the DNA and mRNA levels make it possible to study the effect of genomic rearrangements and DNA copy number changes on gene expression. By performing such studies for the NCI-60 cell line panel, we have taken advantage of the rich characterization of those cells in terms of drug sensitivities (to >100,000 chemically defined compounds) to map DNA and mRNA profiles into the domains of molecular pharmacologic and drug discovery (6, 39). The methods of analysis used here enable us to correlate the DNA copy number of a gene, not just with its own expression but also with the expression of other genes (on the same or a different chromosome).
The data show a generally positive correlation between a given gene's copy number and its expression at the mRNA level. The association of gene copy number with gene expression has also been found in breast cell lines and primary breast tumors (40, 41), as well as in a model of prostate cancer (42) and in HL60 (43). Hyman et al. (40) showed that in 14 breast cancer cell lines, 44% of highly amplified genes were highly expressed and found that ∼10.5% of highly expressed genes were amplified. They also saw a positive trend in expression level with copy number when the DNA copy number and mRNA expression were classified as “gain,” “loss,” or “no change” without regard to magnitude. The results were similar for a panel of 44 primary breast tumors and 10 cell lines (41). Sixty-two percent of highly amplified genes showed increased gene expression of at least a moderate level, and 42% of the highly amplified genes showed comparable overexpression of mRNA. Similarly, average RNA expression tracked with DNA copy number across all classes of DNA copy number alteration (deletion, no change, and low-level, medium-level, and high-level amplification). Twelve percent of the variation in gene expression was directly attributable to gene copy number. In a prostate carcinogenesis model, Phillips et al. (42) found that 51% of genes up-regulated during the transition to malignancy mapped to DNA copy number gains, and that 42% of down-regulated genes were found in regions of DNA copy number loss. When there was gain of a whole chromosome arm or whole chromosome, the average gene expression of all of the involved genes increased, although the degree of increase varied on a gene-by-gene basis.
Very few of the apparent relationships in our data between DNA copy number of one gene and expression of another are reflected in the literature. Most of them probably reflect statistical coincidence or else the large chromosomal rearrangements that characterize most solid tumors and result in “linkage effects.” We would expect such “linkage effects” when the true relationship driving the correlation actually involves genes close to those identified in our data, but our resolution along the chromosome is not sufficient to determine the true source of the correlation. Beyond the question of resolution, it should be recognized that the present analysis has the following limitations: (a) Pearson's correlation is a fundamentally linear relationship; nonlinear relationships might well be missed; (b) Single cell lines may sometimes be unduly influential in determining the correlations of particular gene-gene pairs. The data are shown in Supplementary Tables S3 and S4,15 and the calculations (based on the Cook's D statistic) are described in Supplementary Information.15 Removal of outlier cell lines did, in fact, alter the significance of the correlation in some cases. (c) The data presented here give an incomplete biological picture of the relationship between DNA and RNA. Cancer cells are rarely diploid, and they almost always show large-scale genomic rearrangements. The NCI-60 cell lines are no exception (16). The DNA copy numbers as measured in this study are relative to the ploidy of the cells and cannot convey information about the genomic context of gains and losses or information about rearrangements that do not lead to a net gain or loss.
We observed three cases in which there were strong correlations between genes whose products are known to interact with one another directly: CDC25A and CCNA2, CCNA2, and CDC25C, and TDG and SMT3H1. In the cases of the positive correlation of CDC25A copy number-CCNA2 expression and CCNA2 copy number-CDC25C expression, we questioned whether the correlation observed reflected a strong correlation between the genes at the DNA level or at the transcriptional level, rather than an interaction between the DNA copy number of one gene and the transcript of the other. In the case of CDC25A and CCNA2, the correlation reflected a relationship at the DNA level between the two genes. As described more fully in Results, we determined this because several types of correlation were seen: CDC25A copy number correlated with CCNA2 expression and both CDC25A and CCNA2 showed significant self-self correlations. The latter suggests that DNA copy number plays a large role in determining the expression levels of these genes. Therefore, the expression level of CCNA2 likely reflects to a large degree its DNA copy number. If true, then a correlation at the DNA level between the two genes would yield the observed correlation between CDC25A copy number and CCNA2 expression. This is indeed the case as CDC25A copy number correlated significantly with CCNA2 copy number.
Exploration of the correlation of CCNA2 copy number and CDC25C expression did not support a correlation at the DNA level but rather pointed to a correlation at the transcriptional level. Our data showed a significant, moderate CCNA2 self-self correlation, but CDC25C showed no significant self-self correlation. Therefore, it was unlikely that CDC25C expression would approximate its copy number. Indeed, there was no correlation between CCNA2 copy number and CDC25C copy number. However, because CCNA2 had a significant self-self correlation, DNA copy number might approximate expression level. Therefore, we looked at the transcript level for possible explanations for the observed correlation. CCNA2 and CDC25C share a common repressor binding site, CDE-CHR. Binding to that site results in coordinate transcription of the two genes during S phase in preparation for the G2-M transition (31). The correlation of CCNA2 copy number with CDC25C expression is probably a reflection of this common transcriptional control.
It is unclear what to make of the inverse correlation of TDG copy number with SMT3H1 expression. TDG encodes thymidine-DNA glycosylase, a mismatch-specific uracil/thymine-DNA glycosylase involved in base excision repair. SMT3H1 encodes SUMO-3, a small ubiquitin-like protein. In in vitro base release assays, TDG binds the mismatched G*T or G*U and hydrolyzes the thymidine/uracil but does not release the abasic site (44, 45). Hardeland et al. (32) have shown that sumoylation results in an ∼3-fold increase in the turnover of TDG. Introduction of APEX nuclease (multifunctional DNA repair enzyme) 1 (APE1) increased the turnover rate by an additional 3-fold (32). The hypothesis is that unmodified TDG binds with high affinity to mismatched G*T or G*U, catalyzes removal of the T or U, and remains bound to protect the reactive abasic site. Modification of TDG by SUMO-3 in the DNA-bound state then causes a conformational change leading to dissociation, and that reaction may be coordinated with APE1, the AP endonuclease downstream of TDG in the repair process (32). If SUMO-3 speeds up the turnover of TDG, then the inverse correlation seen in our data might reflect a compensatory measure that keeps base excision repair functioning when the amount of TDG has been reduced through copy number loss.
Our data indicate a generally inverse correlation between the copy numbers of genes on chromosome 3p and expression of ERBB2. That observation is consistent with a previously observed ERBB2 overexpression in association with deletion or loss of heterozygosity of 3p in a subset of breast and ovarian carcinomas (33, 34). Similar relationships have been seen in lung cancers (46). It is unclear from the literature whether overexpression alone or overexpression as a result of amplification is responsible for those relationships. Our data did not indicate a relationship between ERBB2 copy number and the copy numbers or expression levels of ARHA or CDC25A, which are located on 3p. Those observations suggest that overexpression of ERBB2 is the important factor. An investigation of loss of heterozygosity at various loci in the primary mammary adenocarcinomas of a transgenic mouse model with ERBB2 under the control of the mouse mammary tumor virus promoter showed the frequency of loss of heterozygosity on mouse chromosomes 3 and 4 to be significantly higher than background (47). The findings in that study suggest that the regions of loss of heterozygosity on chromosomes 3 and 4 may harbor tumor suppressor genes involved in ERBB2-induced carcinogenesis. One marker from the study, D3Mit127, lies in a region of mouse chromosome 3 that is syntenic with human chromosome 3p in the region surrounding CDC25A and ARHA, the two genes whose copy numbers were negatively correlated with ERBB2 expression in our study. The loss of genes on 3p may be a result of ERBB2 overexpression.
In addition to looking for relationships between gene copy number and mRNA level, we examined the relationship between DNA copy number and drug sensitivity for a set of 118 drugs with putatively known mechanisms of action (9, 24). We first looked at the correlations between ABCB1/MDR1 gene copy number and drugs involved in the multidrug resistance phenotype. Consistent with previous literature, we were able to identify significant negative correlations between ABCB1 copy number and the activity of known substrates of the MDR1 efflux pump.
To identify regions of potential interest for follow up, we used the genomic image map analysis and graphical visualization. There were instances in which particular genes or regions of the genome were correlated with entire classes of agents. We found positive correlations between the tubulin interaction agents and genes at 3q26 (SLC2A2, PIK3CA, TERC, and several sequence-tagged sites) and 12p (ETV6). There were negative correlations between the activity of agents that incorporate into DNA and genes on 4q. In each case, the genes involved in the correlations were not related in any obvious way to the mechanisms of action of the drugs. Because no convincing candidate targets or modifiers of action were identified, the correlations involving those loci are probably statistical coincidence or “linkage effects” that might be resolved with higher-resolution studies. ANKRD17 emerged as a possible explanation for the negative correlation between genes on 4q and agents that incorporate into DNA. ANKRD17 has no experimentally known function, but it is hypothesized through electronic annotation to have a role in DNA binding and mismatch repair, functions that are consistent with the negative association with agents that act through DNA incorporation.
The most immediately interesting drug-copy number relationship thus far identified in the present study provides a new dimension to our earlier report of a relationship in the NCI-60 between the expression of asparagine synthetase and sensitivity to the bacterial enzyme L-asparaginase (9). L-asparaginase has been used since the early 1970s to treat acute lymphoblastic leukemia and some other hematogenous malignancies. Those cells are often low or lacking in endogenous asparagine synthetase and must therefore scavenge asparagine from the bloodstream. L-asparaginase depletes circulating asparagine and selectively starves the cancer cells (48, 49). Occasional responses in various types of solid tumors were reported in early clinical trials (48), but the data were apparently insufficient to justify phase II trials for clinical efficacy. As described in more quantitative detail in Results, we found a strong inverse correlation (—0.98; ref. 9) between expression of asparagine synthetase and sensitivity to L-asparaginase for the leukemia cells among the NCI-60. There was a similar correlation (—0.88) for the ovarian cell types (9), but that value was not high enough to survive statistical “multiple comparisons” testing. Hence, we considered it a clue to formulate the null hypothesis that a subset of ovarian cancers would be susceptible to treatment with L-asparaginase. In analyzing data from the present study, we asked whether the same sort of negative correlation would be found between L-asparaginase activity and DNA copy number of loci in the vicinity of asparagine synthetase on 7q. If so, the finding would lend credence to the initial hypothesis and also suggest a possible mechanism for the differences in expression of the enzyme. We did, indeed, find the correlation to be very strong (—0.98; 95% bootstrap CI, —0.999 to —0.899) in the ovarian carcinomas. Furthermore, loss of 7q has been observed in ovarian carcinomas (50-54). Those observations strengthen the rationale for studying L-asparaginase activity in clinical ovarian tumors and suggest a mechanism on which sensitivity or resistance may be based. An additional, potentially important attribute of L-asparaginase's mechanism of action (i.e., depletion of circulating asparagine) is that the enzyme need not penetrate into a tumor to be effective.
In conclusion, we have presented here a new database of array CGH results focused on cancer-related genes in the NCI-60 cells and have analyzed correlations of those data with mRNA expression and drug sensitivities of those same cells. The L-asparaginase/asparagine synthetase story, which will have clinical implications if the relationships hold for clinical tumors, was particularly apparent and interesting to us. However, the DNA copy number database will also serve as a publicly available “time capsule” to be mined by investigators with domain expertise and research focus on their own particular genes or drugs. Integration of the DNA copy number data with the other rich data resources on the NCI-60 can then be used for further exploration and incisive analysis.
Acknowledgments
We thank Daniel Von Hoff for his expertise and advice with respect to the L-asparaginase/asparagine synthetase work.
Grant support: U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research Contract DE-AC03-76SF00098; NIH, National Cancer Institute grants P01 CA 64602 and P50 CA 58207 (J.W. Gray); and Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Array CGH data have been deposited in ArrayExpress (accession no. E-TABM-65).
Data from these studies can be found online (http://discover.nci.nih.gov).
Ajay et al., unpublished.
Supplementary material for this article is available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org) or at the Genomics and Bioinformatics Group web site (http://discover.nci.nih.gov/host/2005_ bussey_supplement/Bussey_et_al_Supplementary_information.jsp).
References
- 1.Bieche I, Lidereau R. Loss of heterozygosity at 13q14 correlates with RB1 gene underexpression in human breast cancer. Mol Carcinog. 2000;29:151–8. [PubMed] [Google Scholar]
- 2.Bieche I, Laurendeau I, Tozlu S, et al. Quantitation of MYC gene expression in sporadic breast tumors with a real-time reverse transcription-PCR assay. Cancer Res. 1999;59:2759–65. [PubMed] [Google Scholar]
- 3.Smith CA, Pollice AA, Gu LP, et al. Correlations among p53, HER-2/neu, and ras overexpression and aneuploidy by multiparameter flow cytometry in human breast cancer: evidence for a common phenotypic evolutionary pattern in infiltrating ductal carcinomas. Clin Cancer Res. 2000;6:112–26. [PubMed] [Google Scholar]
- 4.Kristiansen G, Yu Y, Petersen S, et al. Overexpression of c-erbB2 protein correlates with disease-stage and chromosomal gain at the c-erbB2 locus in non-small cell lung cancer. Eur J Cancer. 2001;37:1089–95. doi: 10.1016/s0959-8049(01)00096-x. [DOI] [PubMed] [Google Scholar]
- 5.Benz CC, Scott GK, Santos GF, Smith HS. Expression of c-myc, c-Haras1, and c-erbB-2 proto-oncogenes in normal and malignant human breast epithelial cells. J Natl Cancer Inst. 1989;81:1704–9. doi: 10.1093/jnci/81.22.1704. [DOI] [PubMed] [Google Scholar]
- 6.Weinstein JN, Myers TG, O'Connor PM, et al. An information-intensive approach to the molecular pharmacology of cancer. Science. 1997;275:343–9. doi: 10.1126/science.275.5298.343. [DOI] [PubMed] [Google Scholar]
- 7.Shoemaker RH, Monks A, Alley MC, et al. Development of human tumor cell line panels for use in disease-oriented drug screening. Prog Clin Biol Res. 1988;276:265–86. [PubMed] [Google Scholar]
- 8.Boyd MR, Paull KD. Some practical considerations and applications of the National Cancer Institute in vitro anticancer drug discovery screen. Drug Dev Res. 1995;34:91–109. [Google Scholar]
- 9.Scherf U, Ross DT, Waltham M, et al. A gene expression database for the molecular pharmacology of cancer. Nat Genet. 2000;24:236–44. doi: 10.1038/73439. [DOI] [PubMed] [Google Scholar]
- 10.Ross DT, Scherf U, Eisen MB, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–35. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 11.Staunton JE, Slonim DK, Coller HA, et al. Chemosensitivity prediction by transcriptional profiling. Proc Natl Acad Sci U S A. 2001;98:10787–92. doi: 10.1073/pnas.191368598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myers TG, Anderson NL, Waltham M, et al. A protein expression database for the molecular pharmacology of cancer. Electrophoresis. 1997;18:647–53. doi: 10.1002/elps.1150180351. [DOI] [PubMed] [Google Scholar]
- 13.Li G, Waltham M, Anderson NL, et al. Rapid mass spectrometric identification of proteins from two-dimensional polyacrylamide gels after in gel proteolytic digestion. Electrophoresis. 1997;18:391–402. doi: 10.1002/elps.1150180313. [DOI] [PubMed] [Google Scholar]
- 14.Nishizuka S, Chen ST, Gwadry FG, et al. Diagnostic markers that distinguish colon and ovarian adenocarcinomas: identification by genomic, proteomic, and tissue array profiling. Cancer Res. 2003;63:5243–50. [PubMed] [Google Scholar]
- 15.Nishizuka S, Charboneau L, Young L, et al. Proteomic profiling of the NCI-60 cancer cell lines using new high-density reverse-phase lysate microarrays. Proc Natl Acad Sci U S A. 2003;100:14229–34. doi: 10.1073/pnas.2331323100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roschke AV, Tonon G, Gehlhaus KS, et al. Karyotypic complexity of the NCI-60 drug-screening panel. Cancer Res. 2003;63:8634–47. [PubMed] [Google Scholar]
- 17.Zhou Y, Gwadry FG, Reinhold WC, et al. Transcriptional regulation of mitotic genes by camptothecin-induced DNA damage: microarray analysis of dose- and time-dependent effects. Cancer Res. 2002;62:1688–95. [PubMed] [Google Scholar]
- 18.Lee JK, Bussey KJ, Gwadry FG, et al. Comparing cDNA and oligonucleotide array data: concordance of gene expression across platforms for the NCI-60 cancer cells. Genome Biol. 2003;4:R82. doi: 10.1186/gb-2003-4-12-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinkel D, Segraves R, Sudar D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–11. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 20.Massion PP, Kuo WL, Stokoe D, et al. Genomic copy number analysis of non-small cell lung cancer using array comparative genomic hybridization: implications of the phosphatidylinositol 3-kinase pathway. Cancer Res. 2002;62:3636–40. [PubMed] [Google Scholar]
- 21.Snijders AM, Nowak N, Segraves R, et al. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet. 2001;29:263–4. doi: 10.1038/ng754. [DOI] [PubMed] [Google Scholar]
- 22.Hackett CS, Hodgson JG, Law ME, et al. Genome-wide array CGH analysis of murine neuroblastoma reveals distinct genomic aberrations which parallel those in human tumors. Cancer Res. 2003;63:5266–73. [PubMed] [Google Scholar]
- 23.Jain AN, Tokuyasu TA, Snijders AM, Segraves R, Albertson DG, Pinkel D. Fully automatic quantification of microarray image data. Genome Res. 2002;12:325–32. doi: 10.1101/gr.210902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinstein JN, Kohn KW, Grever MR, et al. Neural computing in cancer drug development: predicting mechanism of action. Science. 1992;258:447–51. doi: 10.1126/science.1411538. [DOI] [PubMed] [Google Scholar]
- 25.Bussey KJ, Kane D, Sunshine M, et al. MatchMiner: A tool for batch navigation among gene and gene product identifiers. Genome Biol. 2003;4:R27. doi: 10.1186/gb-2003-4-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Efron B, Gong G. A leisurely look at the bootstrap, the jackknife, and cross-validation. Am Stat. 1983;37:36–48. [Google Scholar]
- 27.Westfall P, Young S. P-values adjustment for multiple tests in multivariate binomial models. J Am Stat Assoc. 1989;84:780–6. [Google Scholar]
- 28.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 29.Storey J, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitelman F, Johansson B, Mertens F, editors. Mitelman Database of Chromosome Aberrations in Cancer. 2005 http://cgap.nci.nih.gov/Chromosomes/Mitelman.
- 31.Zwicker J, Lucibello FC, Wolfraim LA, et al. Cell cycle regulation of the cyclin A, cdc25C and cdc2 genes is based on a common mechanism of transcriptional repression. EMBO J. 1995;14:4514–22. doi: 10.1002/j.1460-2075.1995.tb00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hardeland U, Steinacher R, Jiricny J, Schar P. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 2002;21:1456–64. doi: 10.1093/emboj/21.6.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Isola J, Chu L, DeVries S, et al. Genetic alterations in ERBB2-amplified breast carcinomas. Clin Cancer Res. 1999;5:4140–5. [PubMed] [Google Scholar]
- 34.Zheng JP, Robinson WR, Ehlen T, Yu MC, Dubeau L. Distinction of low grade from high grade human ovarian carcinomas on the basis of losses of heterozygosity on chromosomes 3, 6, and 11 and HER-2/neu gene amplification. Cancer Res. 1991;51:4045–51. [PubMed] [Google Scholar]
- 35.Zeeberg BR, Feng W, Wang G, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson L, Panda D, Jordan MA. Modulation of microtubule dynamics by drugs: a paradigm for the actions of cellular regulators. Cell Struct Funct. 1999;24:329–35. doi: 10.1247/csf.24.329. [DOI] [PubMed] [Google Scholar]
- 37.Gupta S, Bhattacharyya B. Antimicrotubular drugs binding to Vinca domain of tubulin. Mol Cell Biochem. 2003;253:41–7. doi: 10.1023/a:1026045100219. [DOI] [PubMed] [Google Scholar]
- 38.Cassady JM, Chan KK, Floss HG, Leistner E. Recent developments in the maytansinoid antitumor agents. Chem Pharm Bull (Tokyo) 2004;52:1–26. doi: 10.1248/cpb.52.1. [DOI] [PubMed] [Google Scholar]
- 39.Blower PE, Yang C, Fligner MA, et al. Pharmacogenomic analysis: correlating molecular substructure classes with microarray gene expression data. Pharmacogenomics J. 2002;2:259–71. doi: 10.1038/sj.tpj.6500116. [DOI] [PubMed] [Google Scholar]
- 40.Hyman E, Kauraniemi P, Hautaniemi S, et al. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 2002;62:6240–5. [PubMed] [Google Scholar]
- 41.Pollack JR, Sorlie T, Perou CM, et al. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci U S A. 2002;99:12963–8. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phillips JL, Hayward SW, Wang Y, et al. The consequences of chromosomal aneuploidy on gene expression profiles in a cell line model for prostate carcinogenesis. Cancer Res. 2001;61:8143–9. [PubMed] [Google Scholar]
- 43.Ulger C, Toruner GA, Alkan M, et al. Comprehensive genome-wide comparison of DNA and RNA level scan using microarray technology for identification of candidate cancer-related genes in the HL-60 cell line. Cancer Genet Cytogenet. 2003;147:28–35. doi: 10.1016/s0165-4608(03)00155-9. [DOI] [PubMed] [Google Scholar]
- 44.Waters TR, Gallinari P, Jiricny J, Swann PF. Human thymine DNA glycosylase binds to apurinic sites in DNA but is displaced by human apurinic endonuclease 1. J Biol Chem. 1999;274:67–74. doi: 10.1074/jbc.274.1.67. [DOI] [PubMed] [Google Scholar]
- 45.Hardeland U, Bentele M, Jiricny J, Schar P. Separating substrate recognition from base hydrolysis in human thymine DNA glycosylase by mutational analysis. J Biol Chem. 2000;275:33449–56. doi: 10.1074/jbc.M005095200. [DOI] [PubMed] [Google Scholar]
- 46.Gazdar AF. The molecular and cellular basis of human lung cancer. Anticancer Res. 1994;14:261–7. [PubMed] [Google Scholar]
- 47.Ritland SR, Rowse GJ, Chang Y, Gendler SJ. Loss of heterozygosity analysis in primary mammary tumors and lung metastases of MMTV-MTAg and MMTV-neu transgenic mice. Cancer Res. 1997;57:3520–5. [PubMed] [Google Scholar]
- 48.Capizzi RL, Bertino JR, Handschumacher RE. l-asparaginase. Annu Rev Med. 1970;21:433–44. doi: 10.1146/annurev.me.21.020170.002245. [DOI] [PubMed] [Google Scholar]
- 49.Cooney DA, Handschumacher RE. l-asparaginase and l-asparagine metabolism. Annu Rev Pharmacol. 1970;10:421–40. doi: 10.1146/annurev.pa.10.040170.002225. [DOI] [PubMed] [Google Scholar]
- 50.Atkin NB, Baker MC. Chromosome 7q deletions: observations on 13 malignant tumors. Cancer Genet Cytogenet. 1993;67:123–5. doi: 10.1016/0165-4608(93)90164-h. [DOI] [PubMed] [Google Scholar]
- 51.Kerr J, Leary JA, Hurst T, et al. Allelic loss on chromosome 7q in ovarian adenocarcinomas: two critical regions and a rearrangement of the PLANH1 locus. Oncogene. 1996;13:1815–8. [PubMed] [Google Scholar]
- 52.Edelson MI, Scherer SW, Tsui LC, et al. Identification of a 1300 kilobase deletion unit on chromosome 7q31.3 in invasive epithelial ovarian carcinomas. Oncogene. 1997;14:2979–84. doi: 10.1038/sj.onc.1201271. [DOI] [PubMed] [Google Scholar]
- 53.Watson RH, Neville PJ, Roy WJ, Jr., Hitchcock A, Campbell IG. Loss of heterozygosity on chromosomes 7p, 7q, 9p and 11q is an early event in ovarian tumorigenesis. Oncogene. 1998;17:207–12. doi: 10.1038/sj.onc.1201945. [DOI] [PubMed] [Google Scholar]
- 54.Hauptmann S, Denkert C, Koch I, et al. Genetic alterations in epithelial ovarian tumors analyzed by comparative genomic hybridization. Hum Pathol. 2002;33:632–41. doi: 10.1053/hupa.2002.124913. [DOI] [PubMed] [Google Scholar]