Abstract
The increasing use of nanotechnology in consumer products and medical applications underlies the importance of understanding its potential toxic effects to people and the environment. Although both fullerene and carbon nanotubes have been demonstrated to accumulate to cytotoxic levels within organs of various animal models and cell types and carbon nanomaterials have been exploited for cancer therapies, the molecular and cellular mechanisms for cytotoxicity of this class of nanomaterial are not yet fully apparent. To address this question, we have performed whole genome expression array analysis and high content image analysis based phenotypic measurements on human skin fibroblast cell populations exposed to multiwall carbon nano-onions (MWCNOs) and multiwall carbon nanotubes (MWCNTs). Here we demonstrate that exposing cells to MWCNOs and MWCNTs at cytotoxic doses induces cell cycle arrest and increases apoptosis/necrosis. Expression array analysis indicates that multiple cellular pathways are perturbed after exposure to these nanomaterials at these doses, with material-specific toxigenomic profiles observed. Moreover, there are also distinct qualitative and quantitative differences in gene expression profiles, with each material at different dosage levels (6 and 0.6 μg/mL for MWCNO and 0.6 and 0.06 μg/mL for MWCNT). MWCNO and MWCNT exposure activates genes involved in cellular transport, metabolism, cell cycle regulation, and stress response. MWCNTs induce genes indicative of a strong immune and inflammatory response within skin fibroblasts, while MWCNO changes are concentrated in genes induced in response to external stimuli. Promoter analysis of the microarray results demonstrate that interferon and p38/ERK-MAPK cascades are critical pathway components in the induced signal transduction contributing to the more adverse effects observed upon exposure to MWCNTs as compared to MWCNOs.
Introduction
The emerging field of nanotechnology is part of a new industrial revolution being applied to a diverse array of consumer products and medical applications, ranging from cosmetics to electronics and to drug delivery vehicles. With this revolution, methods to reduce the potential toxic effects of nanoparticles both in the environment and for medical applications should be addressed (for review see Colvin1 and Science Highlights2–4). Defining any potential toxicity will aid the nanotechnology industry to minimize the environmental impact of nanomaterials, leading to reduced concern from the public and policymakers and a more successful industry.
Carbon nanomaterials,5–7 including carbon nanoparticles and nanotubes, have been one of the most extensively used nanoparticles, because of their unique and superior properties, including large surface areas, high electrical conductivity, and excellent strength. Multiwall carbon nanotubes (MWCNTs) and multiwall carbon nano-onions (MWCNOs), which will be the focus of this study, represent a relatively recently discovered allotrope of carbon derived from the more intensively studied fullerene (C60).8,9 Single-walled,10 double-walled,11 and multiwalled12 carbon nanotubes, with their diverse chemical and physical properties, have led them to be used in applications ranging from nanowires, electronic components, catalyst supports, electronic displays, to drug delivery, and may even be used for hydrogen storage.9,10,13–16 Giant, nested fullerenes, generally called nano-onions (MWC-NOs),17,18 comprise the least studied class of carbon nanoparticles. MWCNOs are usually produced by an underwater carbon-arc discharge.19–21 Our arc-produced MWCNOs are typically about 30 nm in diameter.22 Although the applications of MWCNOs have lagged behind those of MWCNTs, they have been used as components of nanocomposites for applications including solar cells, light-emitting devices,23,24 and fuel-cell electrodes.25
The increase in commercial interest of nanomaterials and their subsequent production en masse will lead to greater potential for exposure to individuals. Fortunately, aerosol release of the MWCNOs and MWCNTs during manufacturing is limited.26 However, because of the increase use, the risk associated with exposure and the molecular mechanisms of any cytotoxicity need to be well understood. Some of the primary questions that should be addressed include the following: (i) what are likely routes and location of exposure, (ii) what are the molecular mechanisms of toxicity induced by exposure, (iii) does observed toxicity correlate most to size, shape, or composition, (iv) is there any concentration-dependent toxicity, and (v) are byproducts of production or decomposition toxic. The scientific community is beginning to address these concerns, but information is scant. To date, most toxicity studies have been performed on ultrafine particles, which, interestingly, are more toxic than equivalent micrometer-sized material.27 Other studies, however, have demonstrated that toxicity is more highly correlated with particle composition and surface chemistry rather than size.28
Recently, single-walled carbon nanotubes (SWCNTs) have been demonstrated to be an effective infrared photosensitizer for cancer cells,29 and a C2B10 carborane cage-coated SWCNT has been constructed as the delivery vehicle for boron neutron capture therapy for cancer.30 Fullerene has been suggested to be a promising carcinotoxic chemical.28,31 Therefore, we speculate that multiwalled carbon nanomaterials such as MWCNO and MWCNT will be more effective cancer killing agents than the SWCNT and single-walled fullerene. It is even more important for us to decipher the cytotoxicity and molecular mechanism of the multiwalled carbon nanomaterials.
Early studies have indicated that a repeated subchronic topical dose of fullerenes on mouse skin for up to 24 weeks, after initiation with a polycyclic aromatic hydrocarbon, does not result in either benign or malignant skin tumors, in contrast to development of benign skin tumors when a phorbol ester control is used for promotion.32 More recent studies have begun to indicate some adverse effects from carbon nanomaterial exposure. For example, water-soluble fullerenes demonstrate cytotoxicity as a function of surface derivatization28 and C60 derivative molecules also demonstrate superoxide dismutase mimetic properties in Jurkat cells.33,34 In addition, fullerenes induce oxidative stress in the brains of juvenile largemouth bass,35 possibly through free radical generation. Oberdörster has demonstrated that inhaled nanosized carbon particles accumulate in the nasal cavities, lungs, and brains of rats. The suggested route of migration of these nanomaterials into the brain was from lung to blood to brain where they were speculated to accumulate and cause inflammation, brain damage, or central nervous system disorders.35,36 It has also been reported that there is an increased susceptibility to blood clotting in rabbits inhaling carbon nanospheres.27 Validation of this toxicity comes from studies in vitro, using nanosized diesel exhaust particles (DEP). DEP, which includes carbon nanoparticles, selectively damages dopaminergic neurons through the phagocytic activation of microglial NADPH oxidase and consequential oxidative insult.37
Lam and co-workers have demonstrated that inhaled MWCNTs are more toxic than carbon black and quartz, thus posing a serious occupational health hazard for people who are chronically exposed.38 Monteiro-Riviere and colleagues report that at all examined time points, chemically unmodified MWCNTs were present within cytoplasmic vacuoles of exposed kerotinocytes HEK.39 In addition, MWCNT exposed HEK cells released interleukin-8, a pro-inflammatory cytokine. This release was postulated to result in the skin irritation associated with exposure. In a separate study, epidermal keratinocytes exposed to MWCNTs demonstrated free radical generation, accumulation of peroxidative products, and antioxidant depletion, all indicators of oxidative stress.40 Observations of exposed cells also indicated morphological changes and reduced cell viability.40 These data indicate that dermal exposure to unrefined SWCNTs may lead to dermal toxicity due to accelerated oxidative stress in the exposed skin.40 SWCNT exposures by intratracheal installation in rats also produced transient inflammatory and cell injury. When rat lungs were exposed to SWCNTs, a series of multifocal granulomas were induced, both nonuniform in distribution and in a non-dose-dependent fashion, indicative of a foreign tissue body reaction.41
Evidence thus far suggests that the key factors contributing to nanomaterial-related cytotoxicity are size/mass, shape, surface charge, and surface functionalization. The cytotoxicity with equal mass basis shows an order of: SWNTs > MWNT10 > C60.42 Investigations with 2 nm gold nanoparticles in different cell types, tested by MTT, hemolysis, and bacterial viability assays, showed that surface charge was a key factor in inducing toxicity. This indicates that cationic nanoparticles are moderately toxic and have an immediate toxic effect at the blood/brain barrier, whereas anionic particles are relatively nontoxic.43,44 Different surface coatings also have been shown to change the cytotoxicity profiles of quantum dots (CdSe nanocrystals) dramatically, and modifications may attenuate the toxicity.45
As the exact molecular mechanisms for the damages inflicted are still not fully understood, the above-mentioned analyses have demonstrated the urgency of a more thorough molecular characterization of nanomaterial toxicity. Expression array analysis and phenotypic measurements of exposed cell populations may provide insight into the mechanisms responsible for adverse events observed in these models. For example, a recent preliminary unpublished investigation demonstrated gene expression changes associated with the toxicity of nanoscale materials46 and, thus, the potential benefit for using microarray technology to perform high throughput characterization of nanomaterial toxigenomics.
In this study, the goal was to assess if changes in gene expression in cells exposed to carbon-based nanomaterials shows a correlation to phenotypic observations. Reported here are two approaches to evaluate toxicity in nanomaterial exposed cells: (i) the measurement of phenotypic changes in large populations of cells by high content analysis and (ii) gene expression array analysis in exposed cells. Phenotypically, cells exposed to high concentrations of nanomaterials were observed to undergo apoptosis/necrosis with a concomitant reduction in proliferation indicative of an inflammation response. We found that carbon nanomaterials generated mRNA level changes in exposed skin fibroblasts, including changes in mRNA levels from genes involved in metabolism, apoptosis, cell cycle, stress response, cellular transport, and inflammatory response. Of interest was our observation that many of the genes that increased in expression in nanomaterial-exposed cells are often associated with a type I interferon response, which is known to be activated during viral infection and lead to antiviral and antiproliferative responses. Promoter analysis, derived from gene expression data, indicates that the primary mechanism for cell effects from MWCNO and MWCNT treatment is through the p38/ERK MAPK kinase and interferon response pathways. Of interest is the observation that MWCNTs appear to induce a greater amount of stress upon the cells than MWCNOs, even though the dosage is 1/10 by weight/volume concentration. This may have far reaching ramifications for the deployment of specific types of nanomaterials in the future. Clearly this study underscores the importance of the potential toxic side effects in this burgeoning field.
Materials and Methods
Details for materials and methods can be found in the Supporting Information. The carbon MWCNOs used in this study were produced by using a modified direct-current electric-arc discharge method19 (Figure 1). The multiwalled carbon nanotubes (MWCNTs) were synthesized by using a chemical vapor deposition (CVD) method47 (Figure 1). Cellomics-based high content image analysis (HCA) has been used for phenotypical measurement of cell apoptosis, necrosis, cell numbers, proliferation, and cell cycle distribution. Apoptotic cells and necrotic cells were detected using DNA dyes that only traverse membranes of necrotic or apoptotic cells.48 The DNA stain, YO-PRO-1 can transverse the slightly permeable membranes of apoptotic cells while propidium iodide requires the greater membrane permeability of necrotic cells. An Affymetrix high-throughput analysis (HTA) automated GeneChip system was used for acquisition of the microarray data for the gene expression profiling. Target preparation, washing, and staining have been performed on an Affymetrix/Caliper robotic system, and scanning was performed on a CCD-based Affymetrix high throughput (HT) scanner, which is a fully automated epiflourescent imaging system. More details for the HTA protocols can be found in the Supporting Information. Data analysis has been performed using GeneSpring, Bioconductor, GeneTraffic, Cluster 3.0, PAINT, GoMiner, and PathwayAssist, with more details in Supporting Information.
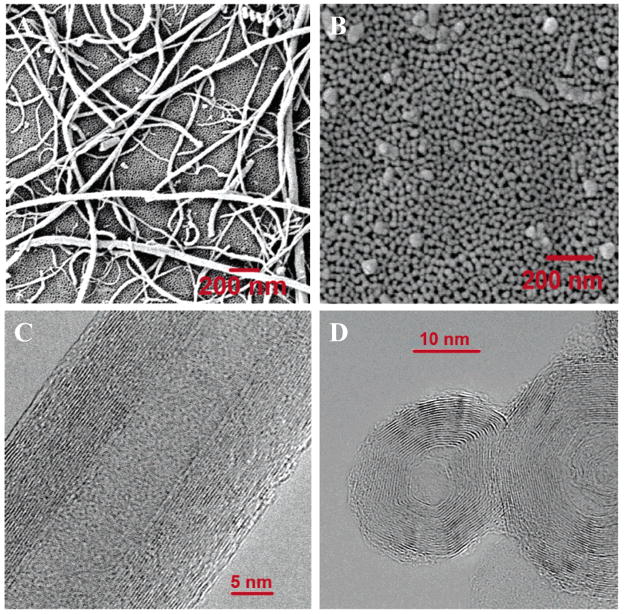
Figure 1.
Scanning electron microscopy (SEM) images and high-resolution transmission electron microscopy (HRTEM) images of carbon nanomaterials used in this study. (A) SEM image of multiwalled carbon nanotubes (scale bar = 200 nm). (B) SEM image of carbon nano-onions (scale bar = 200 nm). (C) HRTEM images of multiwalled carbon nanotubes (MWCNTs) (scale bar = 5 nm). (D) HRTEM images of multiwalled carbon nano-onions (MWCNO) (scale bar = 10 nm).
Results
In Vitro Toxicity
Human skin fibroblasts (HSF42) (Figures 2 and 3) and human embryonic lung fibroblasts (IMR-90) (Supporting Information Figure S1), both untransformed cells, were used to evaluate the cytotoxic and proliferative effects of carbon nanomaterials. Lung and skin cells were selected because entry through the skin or respiratory tract is the most likely route of exposure to nanomaterials. Cells were added to 96-well plates (BD Biosciences), grown to approximately 70% confluency in a CO2 incubator, and then exposed to several concentrations of MWCNOs and MWCNTs (Figure 1). To determine the cytotoxic dose to be used for this study, cells were treated with serial dilutions of MWCNO and MWCNT (data not shown), and we chose doses of 0.6 and 6 mg/L for MWCNO and doses of 0.06 and 0.6 mg/L for MWCNT, so that the cells show approximately 2-fold increase in apoptosis/necrosis from the untreated baseline cells and a ~50% reduction in proliferation (measured by end point cell numbers) after a treatment of 48 h at the low dose. The 2-fold increase of apoptosis/necrosis from the baseline is an artificially defined point, an approach previously used.49 The high doses are chosen as 10 times that of the low dose, so that pronounced gene expression changes can be observed to mimic the acute exposure to carbon nanomaterials. Cells were exposed for 24 or 48 h and counted, and various measurements were made to evaluate cytotoxicity and proliferation. The MWCNTs seem to be 10 times more toxic than the MWCNOs, which is the reason that the amount of MWCNTs used in our studies is only one-tenth that of the amount of MWCNOs used, at both the low dose and high dose levels.
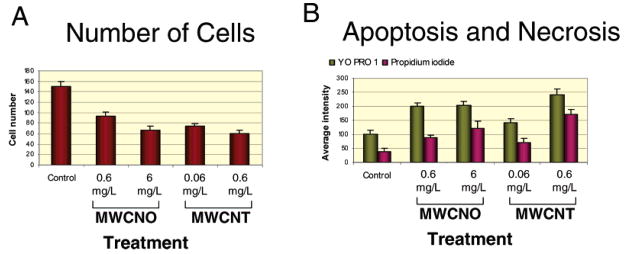
Figure 2.
Cell viability measurements after treatment with carbon nanomaterials at cytotoxic doses. Cells were plated on 96-well plates, treated for 48 h, and then stained with Hoechst (nucleus stain for cell number indicator), YO-PRO 1 (apoptosis indicator) and PI (necrosis indicator). Plates were transported to KineticScan (KSR, Cellomics, Pittsburgh, PA) for image collection, then automated analysis was performed on the collected images. (A) The number of skin fibroblast cells per well 48 h after mock treatment with ethanol or treatment with either MWCNOs (NO) or nanotubes (NT). The numbers of low doses (0.6 μg/mL for MWCNO and 0.06 μg/mL for MWCNT) and high doses (6 μg/mL for MWCNO and 0.6 μg/mL for MWCNT) represent the nanomaterial concentration used for treatment. Bars represent the mean of cell numbers from 10 imaged view fields in 10 treated wells, and error bars represent a 95% confidence interval. Each nuclei imaged by the KSR was identified with the Cell Health Profiling software in the blue channel by Hoechst staining. (B) YO-PRO 1 is visualized in the green channel and PI is visualized in the red channel, where measurement such as dye intensity and area can be made using the Cell Health Profiling algorithm. Average intensity of YO-PRO 1 intensity and PI intensity of mock treated and treated skin fibroblasts at 48 h. The YO-PRO 1 intensity is proportional to apoptosis and the PI intensity correlates to necrosis. Bars represent the mean of cell numbers from eight treated wells and the error bars represent a 95% confidence interval. Data for lung fibroblast treated under the same condition are presented in Supporting Information Figure S1.
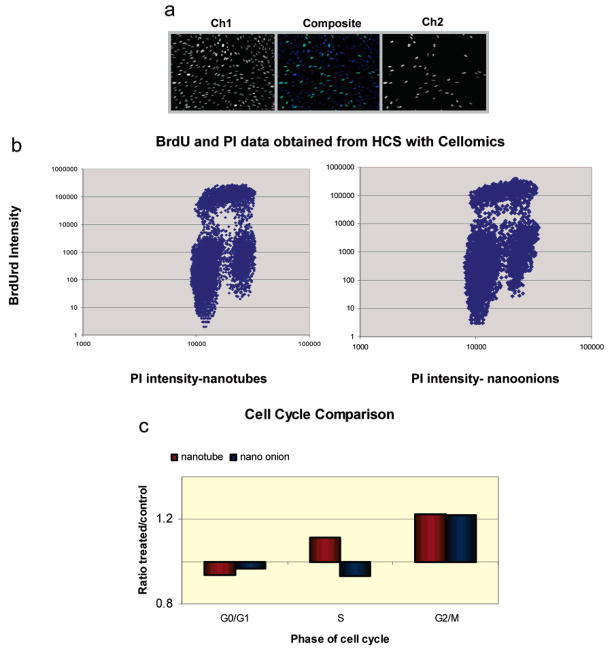
Figure 3.
Measurement of cell proliferation after treatment with carbon nanomaterials at cytotoxic doses. Cells were plated on 96-well plates, treated, pulsed with BrdU, fixed, and then stained with anti-BrdU and PI. Plates were transported to the KSR for image collection and then automated analysis was performed on the collected images. (A) Images generated by the KSR. Channel 1 is images of PI stained nuclei, and this is used for cell identification, counting, and DNA content. Channel 2 represents BrdU staining, and this shows cells that have passed through the S-phase during the pulse with BrdU. The composite image is also shown. (B) Typical scatter plot of BrdU staining intensity versus PI intensity. This is used for calculating the number of cells in G0/G1, S, and G2/M phases. (C) Summary of cell cycle data for nanomaterial-treated cells as compared to controls. An average of 20 000 cells were measured for each treatment condition.
Cell counts were obtained by staining live cells with Hoechst 33342 (Sigma), 48 h post-treatment, and then using high content imaging in the KineticScan (KSR, Cellomics, Pittsburgh, PA) to visualize the cells. Hoechst will stain DNA in both live and dead cells; however the intensity of staining is higher in apoptotic cells because of the condensed chromosomes. The image analysis software, Cell Health Profiling (Cellomics), was then used with the images obtained with the KSR to identify and count cells. The bars in the graphs in Figure 2A and Supporting Information Figure S1 show cell numbers. This graph demonstrates that treatment with either the MWCNOs or MWCNTs reduces cell number in a dose-dependent fashion, with the higher concentrations of MWCNTs creating the greatest effect (Figure 2). This reduction in cell number could result from apoptosis/necrosis and/or reduced proliferation.
Apoptosis and Necrosis
Cytotoxicity was evaluated by staining live cells for 30 min with YO-PRO 1 (Invitrogen, Molecular Probes), propidium iodide (PI, Sigma), and Hoechst. Live cells are impermeable to YO-PRO 1 and PI, both of which are intercalating DNA dyes. Apoptotic cells are permeable to YO-PRO 1, while PI only stains necrotic cells.48 Stained culture plates were analyzed using the KSR and images were acquired at each appropriate fluorescence channel for Hoechst, YO-PRO 1, and PI. The image analysis software establishes average and total intensity for each nucleus in all channels. In the experiments pictured in Figure 2, wells were exposed to either the indicated concentration of nanomaterials or the same volume of ethanol solvent as a control, both kept at less than 1% of the total volume. Ten replicates were done for each condition with intensities for both YO-PRO 1 and PI averaged by well, to obtain the bars in Figure 2B. Student t-tests indicated that all treatment groups demonstrated significant differences from the control group, with p < 0.01.
The following observations were made: (1) Apoptosis and necrosis were observed for both MWCNO and MWCNT treated cells with MWCNTs having the most detrimental effect on both types of cells at the highest concentration (Figure 2). (2) MWCNO treatment to lung fibroblasts, however, demonstrated less of an effect as compared to treatment of skin fibroblasts (Figure 2 and Supporting Information Figure S1). (3) Skin fibroblasts treated with the higher concentration of MWCNOs demonstrated a cell count less than half of that observed in the control, more closely reflecting the results obtained with MWCNT treatment (Figure 2). (4) The average intensity of YO-PRO 1 and propidium iodide staining in the cell types treated with both MWCNOs and MWCNTs went up in a dose-dependent manner (Figure 2) with one exception. The exception is average YO-PRO 1 staining in MWCNO-treated skin fibroblasts, and it remains similar at both concentrations. The PI staining, however, gains intensity at the higher concentration of MWCNOs, indicating a greater number of necrotic cells. These observations indicate the induction of apoptosis and necrosis in nanomaterial-treated cells that is dose and material dependent. It cannot be ruled out that some of the reduction in cell number was a result of reduced proliferation, so this was also tested.
Cell Proliferation
Proliferation was measured in skin fibroblasts by incorporating BrdU for 30 min, fixing cells, staining for BrdU with an antibody, and then counterstaining the DNA with PI.50 Figure 3A shows images from one field, generated by the KSR for image analysis, with PI staining pictured in channel 1, BrdU antibody staining in channel 2, and the composite pictured in the middle. After images from stained culture plates were obtained using the KSR, intensity measurements for both BrdU and DNA staining were made for each identified cell to generate a scatter plot with the intensity of BrdU antibody staining on the Y-axis and PI intensity on the X-axis. Analysis of these scatter plots allowed us to obtain percentages of cells in G0/G1, S, and G2/M phases during cell cycle (Figure 3B). Data from these scatter plots are summarized in Figure 3C as a ratio of the percentages of cells in each phase of the cell cycle in treated cells as compared to control cells. The ratio of treated to control cells in G0/G1 is very close to 1.00, suggesting that nanomaterial treatment does not induce a block in G1. Ratios of cells in S-phase of treated to control were also similar, with a Student t-test demonstrating borderline significance. The p-values for both nanomaterial treatments were both slightly over 0.05. The largest difference in ratio, approximately 1:1.2 for both treatments, was in G2/M (Figure 3C), indicating a possible G2/M block and S phase delay during cell cycle.
The results outlined above indicate that the reduction in cell number is due to both apoptosis/necrosis and a possible G2/M block. As measured by Student t-test and percentage change from control, apoptosis/necrosis appears to be a more significant mechanism for the reduced cell number after nanomaterial treatment. These results are consistent with other studies done with water-soluble fullerenes28,38,41,51 and toxicity studies using MWCNTs.40
Gene Expression
Treating human skin fibroblast with carbon nanomaterials induced profound gene expression changes. Gene expression profiling was performed with the new generation Affymetrix HTA GeneChip system. Figure 4A lists numbers of genes whose expression levels changed after treatment with different particles and doses. We compared gene expression changes using different doses of the same particle structure (Figure 4B,C). These data indicate that, although higher doses induced a greater number of genes expression changes than low doses, there are no global dose-dependent responses to both particles. This is demonstrated by the small portion of genes that were changed commonly at both low and high doses (Figure 4B,C, Supporting Information Tables S2 and S5). The data indicated that distinct pathways were activated in cells treated with low dose or high dose nanomaterials. This is a phenomenon reported before for other cellular stress factors; we have observed similar qualitative differences between carefully chosen low and high doses of radiation.49
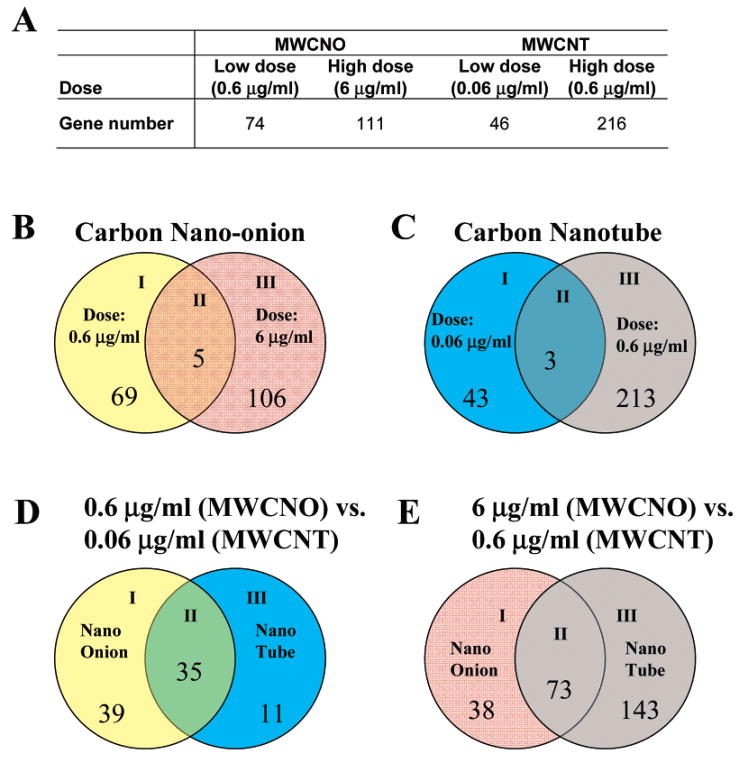
Figure 4.
(A) Numbers of genes whose expression levels changed after treatment with carbon nanomaterials at cytotoxic doses. (B–E) Venn diagrams comparing numbers of genes that showed expression changes. Each Venn diagram is divided into three areas and labeled as I, II, and III. Area II is the overlapping area of two circles, representing commonly changed genes in both conditions. Area I and III represent genes that changed only in the condition specified in the circle. Bioconductor software was used to perform significance analysis to determine the difference between expression levels in treated sample, and the control sample possesses statistical significance. The empirical Bayesian model was used with Bonferroni’s multitest correction. The cutoff of p-values produced through the analysis was determined by at least 10 times less than the p-values of the smallest p-value of control probe sets on the chip. (B) Comparing different doses for the nano-onions. (C) Comparing different doses for the nanotubes. (D) Comparing different particles at low doses (0.6 μg/mL for MWCNO and 0.06 μg/mL for MWCNT). (E) Comparing different particles at high doses (6 μg/mL for MWCNO and 0.6 μg/mL for MWCNT).
We also compared genes that demonstrate altered expression after treatment with different types of carbon nanomaterials (Figure 4D,E). The number of genes in the area of intersection in the Venn diagram in parts D and E of Figure 4 indicates a large percentage of genes show a common expression changes after treatment with both types of particles (Supporting Inormation Tables S8 and S11). However, unique genes were also induced in response to MWCNO or MWCNT and more genes demonstrated changes in levels of expression at the lower concentration of MWCNOs than that with lower concentration MWCNT treatment (Figure 4). Interestingly, it is the dosage of carbon nanomaterials that appears to have the greatest influence on gene expression changes in common between MWCNOs and MWCNTs, not the specific nanomaterial. This could be similar to the threshold effect that is observed after cells are treated with other insults, such as radiation.49
Specific Transcriptional Changes
Genes that demonstrated expression level changes after nanoparticle treatment were placed into functional categories, evaluated for statistical significance, and then sorted by significance (Table 1). The top 10 categories are listed in Table 1 with the percentages of genes over- and underexpressed calculated. At the low dose, MWCNO and MWCNT treatment caused expression changes in similar groups of genes, including Golgi vesicle transport, secretory pathway, fatty acid biosynthesis, protein metabolism, and G1/S transition of mitotic cell cycle (Table 1), with down-regulated genes dominating in all of these categories. An additional group of genes, involved in protein ubiquitination, were up-regulated (Supporting information Table S13). These data suggest that when cells are treated with a low dose of carbon nanomaterials there is decreased cell growth and metabolism, but increased protein degradation. Conversely, treatment with both MWCNOs and MWCNTs at high dosages induced up-regulated genes in tRNA aminoacylation and amino acid metabolism pathways, indicating positive regulation of amino acid and protein biosynthesis.
Table 1.
Most Significantly Changed Gene Categories after Treating HSF42 Cells with Carbon Nanomaterials at Cytotoxic Dosesa
| gene category | p-value | % underexpressed | % overexpressed | |
|---|---|---|---|---|
| carbon nano-onion 0.6 μg/mL | Golgi vesicle transport | 0.0000 | 8.51 | 2.13 |
| membrane fusion | 0.0002 | 15.79 | 0.00 | |
| secretory pathway | 0.0003 | 4.35 | 1.09 | |
| protein ubiquitination | 0.0139 | 0.68 | 2.05 | |
| intracellular transport | 0.0166 | 1.23 | 0.62 | |
| cell growth and/or maintenance | 0.0201 | 0.66 | 0.27 | |
| fatty acid biosynthesis | 0.0208 | 5.71 | 0.00 | |
| protein metabolism | 0.0323 | 0.71 | 0.29 | |
| ubiquitin cycle | 0.0342 | 0.70 | 1.06 | |
| G1/S transition of mitotic cell cycle | 0.0361 | 4.26 | 0.00 | |
| carbon nanotube 0.06 μg/mL | Golgi vesicle transport | 0.0007 | 4.26 | 2.13 |
| protein metabolism | 0.0020 | 0.65 | 0.18 | |
| secretory pathway | 0.0049 | 2.17 | 1.09 | |
| fatty acid biosynthesis | 0.0076 | 5.71 | 0.00 | |
| G1/S transition of mitotic cell cycle | 0.0135 | 4.26 | 0.00 | |
| protein ubiquitination | 0.0174 | 0.68 | 1.37 | |
| mitotic cell cycle | 0.0200 | 1.95 | 0.00 | |
| ubiquitin cycle | 0.0214 | 0.70 | 0.70 | |
| cell homeostasis | 0.0228 | 3.23 | 0.00 | |
| protein prenylation | 0.0262 | 14.29 | 0.00 | |
| carbon nano-onion 6 μg/mL | L-serine metabolism | 0.0000 | 0.00 | 40.00 |
| tRNA aminoacylation | 0.0000 | 0.00 | 23.81 | |
| amine metabolism | 0.0000 | 0.00 | 5.42 | |
| amine transport | 0.0000 | 0.00 | 12.20 | |
| dicarboxylic acid transport | 0.0020 | 0.00 | 25.00 | |
| response to extracellular stimulus | 0.0063 | 0.00 | 14.29 | |
| heterocycle metabolism | 0.0076 | 0.00 | 6.38 | |
| porphyrin metabolism | 0.0139 | 0.00 | 9.52 | |
| TGF beta receptor signaling pathway | 0.0139 | 4.76 | 4.76 | |
| pigment metabolism | 0.0194 | 0.00 | 8.00 | |
| carbon nanotube 0.6 μg/mL | tRNA aminoacylation | 0.0000 | 0.00 | 33.33 |
| L-serine metabolism | 0.0000 | 0.00 | 50.00 | |
| amine metabolism | 0.0000 | 0.00 | 6.90 | |
| amine transport | 0.0000 | 0.00 | 14.63 | |
| response to stimulus | 0.0000 | 0.16 | 2.86 | |
| immune response | 0.0000 | 0.18 | 4.50 | |
| water-soluble vitamin biosynthesis | 0.0024 | 0.00 | 40.00 | |
| inflammatory response | 0.0034 | 0.00 | 5.06 | |
| heterocycle metabolism | 0.0062 | 2.13 | 6.38 | |
| dicarboxylic acid transport | 0.0065 | 0.00 | 25.00 |
The categories are generated by the GoMiner program (Materials and Methods, Supporting Information), using p-values as the evaluation criteria of statistically significant changes. for each category. The p-value was calculated by conducting a two-sided Fisher’s exact test, which reflects the statistical significance for that category being enriched in changed genes. The p-values were used to sort categories to identify those gene functional groups that have responded the most after treatments.
Changes in the expression of functionally related genes were found at high doses of CMWNT treatment. These included genes involved in the inflammatory and immune response (Table 2). Most of the genes in this category can be ascribed to the innate immune system and generally are induced in response to interferon (IFN) and the defense against virus. STAT1 (for signal transduction and activator of transcription-1) (Table 5) is activated by a number of different ligands, including interferon-alpha (IFNA), interferon-gamma (IFNG), and IL6 52 and in turn regulates IFN7 production. Treatment with MWCNTs up-regulates STAT1 leading to an observed IRF7 induction in these cells. IFN7 was recently demonstrated to regulate all elements of IFN responses, including the systemic production of IFN in innate immunity.53 IRF1, also up-regulated, has been demonstrated to play an important role in transcription activation of type I IFN genes.54 Additionally, most of the genes in Table 2 are IFN inducible including ADAR,55 CXCL10,56,57 G1P2,58 G1P3,59 IFI44, IFIT1,60 IFIT2,61 IFIT3,62 and IFIT563 among others (Table 2). Several induced genes are also specifically associated with an antiviral response including MX1, MX2, OAS1, OAS2, and OAS3. The MX proteins are related to an interferon-regulated mouse protein induced by influenza virus,64,65 and the OAS proteins have been observed to be induced as a response to the yellow fever vaccine.66,67 These data indicate that MWCNTs may interact with cells differently than MWCNOs, and this type of interaction influences the cellular response. On the basis of the large number of genes associated with cellular response to viral infection and an IFN type I response, MWCNT treatment may mimic viral infection in some respects.
Table 2.
Immune-Response and Inflammatory Genes that Over- or Underexpressed after Treating HSF42 Cells with Cytotoxic Dose (0.6 μg/mL) of Multiwall Carbon Nanotubes
| gene symbol | gene name | fold changea |
|---|---|---|
| ADAR | adenosine deaminase, RNA-specific | 1.44 |
| BDKRB1 | bradykinin receptor B1 | 1.59 |
| CEBPB | CCAAT/enhancer binding protein (C/EBP), beta | 1.53 |
| CXCL10 | chemokine (C–X–C motif) ligand 10 | 4.82 |
| CXCL3 | chemokine (C–X–C motif) ligand 3 | 2.71 |
| G1P2 | interferon, alpha-inducible protein (clone IFI-15K) | 2.51 |
| G1P3 | interferon, alpha-inducible protein (clone IFI-6–16) | 2.03 |
| IFI44 | interferon-induced protein 44 | 3.50 |
| IFIT1 | interferon-induced protein with tetratricopeptide repeats 1 | 6.99 |
| IFIT2 | interferon-induced protein with tetratricopeptide repeats 2 | 5.99 |
| IFIT3 | interferon-induced protein with tetratricopeptide repeats 3 | 5.85 |
| IFIT5 | interferon-induced protein with tetratricopeptide repeats 5 | 1.76 |
| IRF1 | interferon regulatory factor 1 | 2.02 |
| IRF7 | interferon regulatory factor 7 | 2.47 |
| ISGF3G | interferon-stimulated transcription factor 3, gamma 48kDa | 1.55 |
| LIF | leukemia inhibitory factor (cholinergic differentiation factor) | 2.67 |
| MGST2 | microsomal glutathione S-transferase 2 | 0.67 |
| MX1 | Homo sapiens myxovirus (influenza) resistance 1 | 11.18 |
| MX2 | myxovirus (influenza virus) resistance 2 (mouse) | 6.88 |
| NFE2L1 | nuclear factor (erythroid-derived 2)-like 1 | 1.70 |
| NR4A2 | nuclear receptor subfamily 4, group A, member 2 | 3.26 |
| OAS1 | 2′,5′-oligoadenylate synthetase 1, 40/46kDa | 2.82 |
| OAS2 | 2′,5′-oligoadenylate synthetase 2, 69/71kDa | 2.79 |
| OAS3 | 2′,5′-oligoadenylate synthetase 3, 100kDa | 2.21 |
| RIPK2 | receptor-interacting serine-threonine kinase 2 | 1.45 |
| TNFAIP6 | tumor necrosis factor, alpha-induced protein 6 | 1.82 |
Fold changes represent the ratio of mRNA amount of treated samples divided by those of control samples.
Table 5.
Genes Changed by Nano-Onion and Carbon Nanotubes but Fall in the Category of Apoptosisa
| gene symbol | gene name | fold change of gene expression |
|---|---|---|
| Fold Change of Gene Expression for 0.6 μg/mL MWCNO | ||
| EGFR | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 0.17 |
| MCL1 | myeloid cell leukemia sequence 1 (BCL2-related) | 0.19 |
| BCL2L1 | BCL2-like 1 | 0.24 |
| PPM1F | protein phosphatase 1F (PP2C domain containing) | 1.63 |
| TGM2 | transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) | 0.35 |
| FGFR1 | fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome) | 1.72 |
| CRKL | v-crk sarcoma virus CT10 oncogene homologue (avian)-like | 0.20 |
| EXTL3 | exostoses (multiple)-like 3 | 0.44 |
| MAPK14 | mitogen-activated protein kinase 14 | 0.31 |
| MACF1 | microtubule-actin cross-linking factor 1 | 1.81 |
| Fold Change of Gene Expression for 0.06 μg/mL MWCNT | ||
| TGM2 | transglutaminase 2 (C polypeptide, protein-glutamine-γ -glutamyltransferase) | 0.40 |
| MCL1 | myeloid cell leukemia sequence 1 (BCL2-related) | 0.18 |
| FGFR1 | fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome) | 1.72 |
| CRK | v-crk sarcoma virus CT10 oncogene homologue (avian) | 0.48 |
| MAPK14 | mitogen-activated protein kinase 14 | 0.30 |
| Fold Change of Gene Expression for 6 μg/mL MWCNO | ||
| YARS | tyrosyl-tRNA synthetase | 1.62 |
| Fold Change of Gene Expression for 0.6 μg/mL MWCNT | ||
| YARS | tyrosyl-tRNA synthetase | 1.75 |
| MX1 | myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) | 11.55 |
| BIRC3 | baculoviral IAP repeat-containing 3 | 2.16 |
| RIPK2 | receptor-interacting serine-threonine kinase 2 | 1.38 |
| STAT1 | signal transducer and activator of transcription 1, 91kDa | 2.22 |
| TNFAIP3 | tumor necrosis factor, alpha-induced protein 3 | 1.95 |
| AHR | aryl hydrocarbon receptor | 1.68 |
| TNFRSF10B | tumor necrosis factor receptor superfamily, member 10b | 1.62 |
Fold change of gene expression is given for the low dose (0.6 μg/mL for MWCNO and 0.06 μg/mL for MWCNT). Fold changes represent the ratio of MRNA amount of treated samples divided by those of control samples.
Many of the genes altered in expression after treatment with the lower concentration of nanomaterials are those involved in transport, membrane fusion, and secretion (Table 3). These genes did not show discernible changes in expression with higher concentrations of MWCNOs and MWCNTs. Many of the genes in this category, SNAP23, NAPG, NAPA and GBF1, are involved in the process of docking and fusion of vesicles to their target membranes.68–71 Most of the genes in this category are underexpressed indicating that the cells may be slowing secretion of proteins. Treatment of cells with the lower concentrations of nanomaterials also has an impact on the expression of cell cycle genes (Table 4) and genes involved in ubiquitination (Supporting information Table S13). Again, many of these genes are down-regulated, indicating a slowing of cell proliferation and protein degradation.
Table 3.
Genes Changed by Nano-Onion and Nanotube but Fall in the Category of “Transport” (Golgi Vesicle Transport, Membrane Fusion, Secretory Pathway, Intracellular Transport)a
| gene category | gene symbol | gene name | fold change of gene expression |
|---|---|---|---|
| Fold Change of Gene Expression for Onion, 0.6 μg/mL | |||
| Golgi vesicle transport | COPA | coatomer protein complex, subunit alpha | 0.57 |
| SNAP23 | synaptosomal-associated protein | 0.30 | |
| GBF1 | golgi-specific brefeldin A resistance factor 1 | 2.45 | |
| NAPG | N-ethylmaleimide-sensitive factor attachment protein, gamma | 0.48 | |
| NAPA | N-ethylmaleimide sensitive fusion protein attachment protein alpha | 0.60 | |
| membrane fusion | NAPA | N-ethylmaleimide sensitive fusion protein attachment protein alpha | 0.60 |
| NAPG | N-ethylmaleimide-sensitive factor attachment protein, gamma | 0.48 | |
| SNAP23 | synaptosomal-associated protein | 0.30 | |
| secretory pathway | NAPA | N-ethylmaleimide sensitive fusion protein attachment protein alpha | 0.60 |
| COPA | coatomer protein complex, subunit alpha | 0.57 | |
| GBF1 | golgi-specific brefeldin A resistance factor 1 | 2.45 | |
| NAPG | N-ethylmaleimide-sensitive factor attachment protein, gamma | 0.48 | |
| SCD | stearoyl-CoA desaturase | 0.19 | |
| SNAP23 | synaptosomal-associated protein | 0.30 | |
| intracellular transport | GBF1 | golgi-specific brefeldin A resistance factor 1 | 2.45 |
| DST | dystonin | 0.40 | |
| NAB2 | NGFI-A binding protein 2 | 0.43 | |
| SNAP23 | synaptosomal-associated protein | 0.30 | |
| KDELR3 | KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 | 0.76 | |
| NAPG | N-ethylmaleimide-sensitive factor attachment protein, gamma | 0.48 | |
| COPA | coatomer protein complex, subunit alpha | 0.57 | |
| HNRPA1 | heterogeneous nuclear ribonucleoprotein A 1 | 2.49 | |
| NAPA | N-ethylmaleimide sensitive fusion protein attachment protein alpha | 0.60 | |
| nucleocytoplasmic transport | NAB2 | NGFI-A binding protein 2 | 0.43 |
| HNRPA1 | heterogeneous nuclear ribonucleoprotein A 1 | 2.49 | |
| Fold Change of Gene Expression for Tube, 0.06 μg/mL | |||
| Golgi vesicle transport | COPA | coatomer protein complex, subunit alpha | 0.57 |
| NAPA | N-ethylmaleimide sensitive fusion protein attachment protein alpha | 0.60 | |
| GBF1 | Golgi-specific brefeldin A resistance factor 1 | 2.45 | |
| membrane fusion | NAPA | N-ethylmaleimide sensitive fusion protein attachment protein alpha | 0.60 |
| secretory pathway | COPA | coatomer protein complex, subunit alpha | 0.57 |
| NAPA | N-ethylmaleimide sensitive fusion protein attachment protein alpha | 0.60 | |
| SCD | stearoyl-CoA desaturase | 0.19 | |
| GBF1 | Golgi-specific brefeldin A resistance factor 1 | 2.45 | |
| intracellular transport | GBF1 | Golgi-specific brefeldin A resistance factor 1 | 2.45 |
| NAPA | N-ethylmaleimide sensitive fusion protein attachment protein alpha | 0.60 | |
| COPA | coatomer protein complex, subunit alpha | 0.57 | |
Fold change of gene expression is given for the low dose (0.6 μg/mL for MWCNO and 0.06 μg/mL for MWCNT). Fold changes represent the ratio of MRNA amount of treated samples divided by those of control samples.
Table 4.
Genes Changed by Nano-Onion and Carbon Nanotubes but Fall in the Category of Cell Cycle Regulatory Genes (G1/S Transition of Mitotic Cell Cycle, Mitotic Cell Cycle, and Cell Growth of Maintenance)a
| gene category | gene symbol | gene name | fold change of gene expression |
|---|---|---|---|
| Fold Change of Gene Expression for Nano-Onion | |||
| cell proliferation | EXTL3 | exostoses (multiple)-like 3 | 0.44 |
| FGFR1 | fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome) | 1.72 | |
| NAB2 | NGFI-A binding protein 2 (EGR1 binding protein 2) | 0.43 | |
| cell cycle | DUSP1 | dual specificity phosphatase 1 | 0.17 |
| TRIM33 | tripartite motif-containing 33 | 1.60 | |
| HSF1 | heat shock transcription factor 1 | 0.52 | |
| BCAT1 | branched chain aminotransferase 1, cytosolic | 0.17 | |
| regulation of cell cycle | SKP2 | S-phase kinase-associated protein 2 (p45) | 0.21 |
| MCL1 | myeloid cell leukemia sequence 1 (BCL2-related) | 0.19 | |
| EGFR | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 0.24 | |
| MAPK14 | mitogen-activated protein kinase 14 | 0.25 | |
| CRKL | v-crk sarcoma virus CT10 oncogene homologue (avian)-like | 0.20 | |
| cell cycle arrest | MACF1 | microtubule-actin cross-linking factor 1 | 1.81 |
| DST | dystonin | 0.40 | |
| cell differentiation | PDLIM7 | PDZ and LIM domain 7 (enigma) | 0.60 |
| BSG | basigin (OK blood group) | 0.44 | |
| NAPA | N-ethylmaleimide sensitive fusion protein attachment protein alpha | 0.60 | |
| EGR1 | early growth response 1 | 0.39 | |
| Fold Change of Gene Expression for Nanotube | |||
| cell proliferation | FGFR1 | fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome) | 1.72 |
| cell cycle | DUSP1 | dual specificity phosphatase 1 | 0.23 |
| BCAT1 | branched chain aminotransferase 1, cytosolic | 0.24 | |
| CDK2 | cyclin-dependent kinase 2 | 0.58 | |
| regulation of cell cycle | SKP2 | S-phase kinase-associated protein 2 (p45) | 0.18 |
| MCL1 | myeloid cell leukemia sequence 1 (BCL2-related) | 0.18 | |
| MAPK14 | mitogen-activated protein kinase 14 | 0.30 | |
| CRK | v-crk sarcoma virus CT10 oncogene homologue (avian) | 0.53 | |
| SLC12A4 | solute carrier family 12 (potassium/chloride transporters), member 4 | 0.23 | |
| cell differentiation | PDLIM7 | PDZ and LIM domain 7 (enigma) | 0.62 |
| NAPA | N-ethylmaleimide sensitive fusion protein attachment protein alpha | 0.62 | |
Fold change of gene expression is given for the low dose (0.6 μg/mL for MWCNO and 0.06 μg/mL for MWCNT). Fold changes represent the ratio of MRNA amount of treated samples divided by those of control samples.
Table 5 lists genes involved in apoptosis that were induced or repressed with nanomaterial treatment. A greater number of genes involved in apoptosis were observed to be up-regulated with MWCNT treatment at the higher dose, possibly explaining the greater number of apoptotic and dead cells observed with high content screening (Figure 2). Of interest was the up-regulation of the cytokine and TNF family member, TNFRSF10B (TRAILR2) in cells treated with the highest concentration of MWCNTs, which is known to induce apoptosis.72 Also, the RIPK273 gene contributes to the induction of apoptosis and was observed to be up-regulated in these treated cells. At lower doses, many of the genes related to apoptosis listed in Table 5 are down-regulated and are anti-apoptotic; examples include EGFR,74 MCL1,75 BCL2L1,76 and CRKL.77 Up-regulation of YARS was observed with both nanomaterial treatments, especially with the higher concentrations. YARS is believed to contribute to apoptosis by arresting translation and producing cytokines.78
Large numbers of stimuli response genes were observed to be up-regulated with the higher concentration of nano-materials (Table 6). These include the immune response genes pictured in Table 2. A few were down-regulated in this category, including FOS,79 which is related to an increase in cell proliferation. Again, most of the observed transcriptional changes were observed with treatment with MWCNTs at high concentration, although stimulus response genes were also induced with MWCNO treatment. These results point to a concerted cellular reaction to offset a cellular insult from the addition of nanomaterials, with the greatest response being observed with MWCNT treatment at the higher concentration. This is consistent with the greatest phenotypic response with respect to apoptosis, cell death, and proliferation also being observed at the higher concentration of nanomaterials.
Table 6.
Genes Changed by Nano-Onion and Carbon Nanotubes but Fall in the Category of External Stimuli Response Genesa
| gene category | gene symbol | gene name | fold change of gene expression |
|---|---|---|---|
| Fold Change of Gene Expression for MWCNO 6 mg/L | |||
| immune response | EGR1 | early growth response 1 | 0.37 |
| FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | 0.14 | |
| stress response | DDIT3 | DNA-damage-inducible transcript 3 | 2.39 |
| SLC3A2 | solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 | 2.46 | |
| STC2 | stanniocalcin 2 | 2.38 | |
| VEGF | vascular endothelial growth factor | 2.18 | |
| DDIT3 | DNA-damage-inducible transcript 3 | 2.39 | |
| FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | 0.14 | |
| SQSTM1 | sequestosome 1 | 2.00 | |
| VEGF | vascular endothelial growth factor | 2.18 | |
| Fold Change of Gene Expression for MWCNT 0.6 mg/L | |||
| immune response | CXCL10 | chemokine (C–X–C motif) ligand 10 | 4.82 |
| IFIT2 | interferon-induced protein with tetratricopeptide repeats 2 | 5.99 | |
| IFIT3 | interferon-induced protein with tetratricopeptide repeats 3 | 5.85 | |
| IRF1 | interferon regulatory factor 1 | 2.02 | |
| IRF7 | interferon regulatory factor 7 | 2.47 | |
| CXCL3 | chemokine (C–X–C motif) ligand 3 | 2.71 | |
| MX2 | myxovirus (influenza virus) resistance 2 (mouse) | 6.88 | |
| NR4A2 | nuclear receptor subfamily 4, group A, member 2 | 2.62 | |
| PLSCR1 | phospholipid scramblase 1 | 2.38 | |
| response to DNA damage stimulus | DDIT3 | DNA-damage-inducible transcript 3 | 2.70 |
| IRF7 | interferon regulatory factor 7 | 2.47 | |
| stress response | CXCL10 | chemokine (C–X–C motif) ligand 10 | 4.82 |
| CXCL3 | chemokine (C–X–C motif) ligand 3 | 2.71 | |
| DDIT3 | DNA-damage-inducible transcript 3 | 2.70 | |
| IRF7 | interferon regulatory factor 7 | 2.47 | |
| MKNK2 | MAP kinase interacting serine/threonine kinase 2 | 2.11 | |
| MX2 | myxovirus (influenza virus) resistance 2 (mouse) | 6.88 | |
| NR4A2 | nuclear receptor subfamily 4, group A, member 2 | 2.62 | |
| OAS1 | 2′,5′-oligoadenylate synthetase 1, 40/46 kDa | 2.82 | |
| OAS2 | 2′,5′-oligoadenylate synthetase 2, 69/71 kDa | 2.79 | |
| OAS3 | 2′,5′-oligoadenylate synthetase 3, 100 kDa | 2.21 | |
| PLSCR1 | phospholipid scramblase 1 | 2.38 | |
| SLC3A2 | solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 | 2.58 | |
| STAT1 | signal transducer and activator of transcription 1, 91 kDa | 2.22 | |
| VEGF | vascular endothelial growth factor | 2.24 | |
Fold change of gene expression is given for the low dose (0.6 μg/mL for MWCNO and 0.06 μg/mL for MWCNT). Fold changes represent the ratio of MRNA amount of treated samples divided by those of control samples.
Promoter Analysis
According to our analysis of regulatory elements (cis elements) within the promoters of genes altered in expression upon carbon nanomaterial treatment, different pathways appear to be activated depending upon the nano-material dosage. As gene expression patterns observed in microarray experiments reflect the activity of transcription factors (TFs) in trans, we can trace back the regulatory cascades upstream of the physiological effect. This is performed by identifying the enriched transcription regulatory elements (TRE) on the promoters of genes demonstrating altered expression profiles. These analyses were performed using the microarray data from MWCNT- and MWCNO-treated HSF cells at low and high dosages.
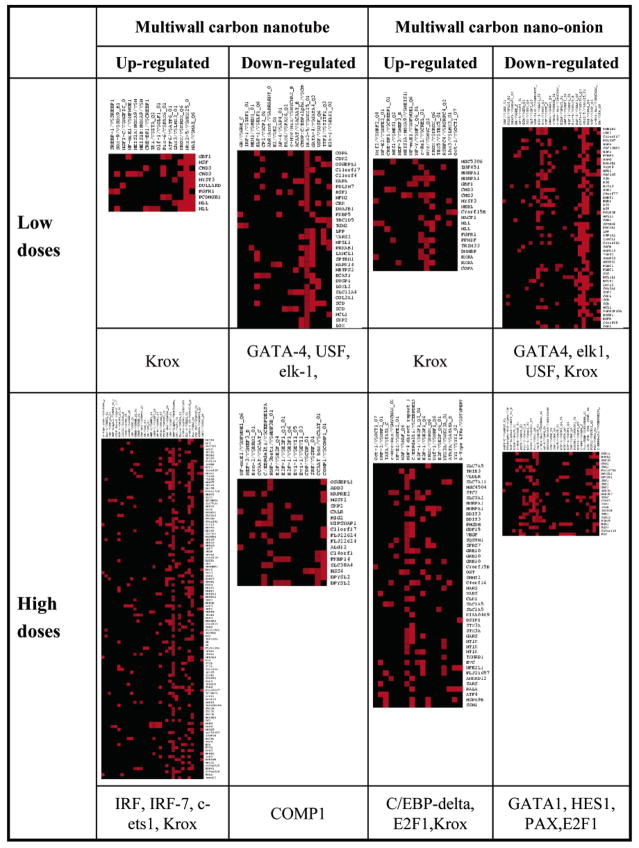
Promoter analysis of the predominantly down-regulated genes at the lower dosages points to the enrichment of EGR1-(KROX1), GATA4, ELK1, and USF regulatory elements in cells treated with MWCNO versus GATA4, ELK1, and USF regulatory elements in cells treated with MWCNTs (Figure 5). Promoters in genes of up-regulated transcripts demonstrate the enrichment of EGR1 binding elements. However, the transcription of EGR1 is down-regulated after MWCNO treatment indicating that up-regulation of some transcripts may be a consequence of relieved repression as opposed to activation. GATA4, EGR1, USF, and ELK1 TFs have all been shown to be phosphorylated and activated by ERK and p38 MAPK cascades.80–86 The down-regulation of these TFs may reflect the down-regulation of the MAPK cascades. This hypothesis is partially validated by the observation that p38 (MAPK14) expression is down-regulated in both experiments with lower dosages of MWCNOs and MWCNTs.
Figure 5.
Promoter analysis. The interaction matrix for the differentially expressed genes (horizontal) and transcription regulatory elements (vertical) in the up- and down-regulated gene sets at different dosage using different carbon nanoparticles. The PAINT software (Supporting Information) then computes p-values to look for the overrepresented TREs in the set of promoters analyzed in reference to all the genes in the PAINT database to generate filtered (p-value < 0.1) interaction matrixes. Individual elements of the matrix are colored by the significance p-values: over-representation in the matrix is colored in red. The brightest red represents low p-value (most significantly over-represented). Enriched transcription regulatory elements for the nanoparticle dataset.
Treatment of cells with higher dosages of carbon particles caused a more pronounced effect on gene expression than lower dosages. More transcripts are up-regulated as opposed to down-regulated (Figure 5). The promoters of up-regulated genes in MWCNT treated cells are enriched with IRFs, ETS1, PPAR and EGR1 regulatory elements while MWCNO treated cells are enriched with C/EBPdelta, E2F1, and EGR1 regulatory elements (Figure 5). Mechanistically, cells treated with both of the higher doses of carbon nanomaterials appear to trigger responses from the activated p38 and ERK MAPK cascades, based on transcription factor profiling. In fact, CCAAT enhancer binding protein delta (C/EBPdelta), enriched in MWCNO-treated cells, is a target of p38 MAPK87 and is associated with growth arrest in epithelial cells.88 However, the expression pattern of higher dose MWCNT treatment differs significantly from that of MWCNO treatment. For example, a robust IFN response is observed in MWCNT-treated cells, but not in MWCNO-treated cells. The presence of IRF elements contained within the promoters of many of the up-regulated genes may explain this response. In fact, IRF7 is one of the up-regulated genes observed (Table 2) and is believed to be central to an IFN response53 along with STAT1 (Table 5), another up-regulated gene discussed above, and one of the central signal transduction factors needed for an IFN response. Transcriptional regulatory elements present in the down-regulated genes of cells treated with MWCNOs, such as GATA1, may also contribute to the differences in gene transcription observed (Figure 5). FOS gene expression is also reduced, leading to a lowered activity of AP1 (FOS/JUN) transcription factors (Table 6). These differences may be responsible for the difference in the magnitude of response between these particles, observed phenotypically by high content analysis. Additional experiments monitoring the kinase activities should give us better understanding the underlying mechanism.
Discussion
The results presented here show for the first time both a phenotypic response of cells to carbon nanomaterials (apoptosis, necrosis, cell cycle perturbation, and antiproliferation) and a global gene expression response at a cellular level. Phenotypic effects were confirmed for two different fibroblast cell types, human skin fibroblast (HSF, see figures and tables in text) and IMR-90 (Supporting Information Figure S1). This information will be important for elucidating possible mechanisms responsible for the toxicity observed after exposure to these particles. Important to the validation of this experimental approach was to determine if the microarray results were consistent with our phenotypic observations of exposed cells by high content analysis (HCA). The phenotypic responses of apoptosis, cell death, and proliferation changes were predicted by changes in expression levels of many of the genes we observed.
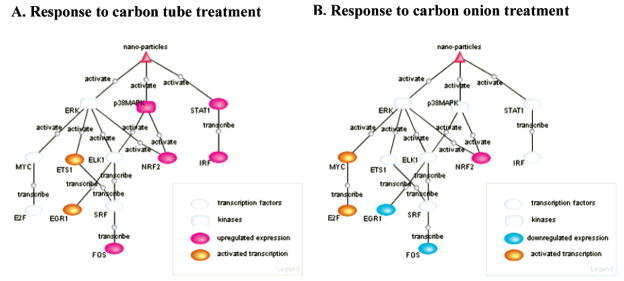
HCA of cells treated with MWCNOs and MWCNTs showed significant changes in cell number that, upon further investigation, was shown to be due to apoptosis, cell death, and proliferation changes. Therefore nanomaterials do demonstrate toxicity, especially at higher concentrations. MWCNTs appear to be more toxic to cells than MWCNOs as demonstrated by the greater number of cells undergoing apoptosis or necrosis after treatment, even at one-tenth the amount of the carbon nano-onions. This response, at least with the MWCNT treatment may be due to a type I INF response, which has been shown to lead to apoptosis and cell death. This type of response also leads to changes in cell proliferation, which were also observed. The phenotypic response is dose-dependent, even though the molecular mechanisms causing the phenotypic changes may be different depending on the dose or particle type. The magnitude of the response could be a reflection of differential pathway activation (Figure 6). One limitation of this study is that it has been performed on cells and not on whole organisms. A living system may have several lines of defense to prevent or minimize some of the toxic effects of exposure to small particles. However, because toxicity has been indicated from this analysis, workers that come in contact with large amounts of nanomaterials should protect their skin and lungs from potential exposure.
Figure 6.
A comparison of activated signal transduction networks for higher dose responses to carbon tubes and carbon onions. PathwayBuilder software (Arkin Group, LBNL) is used to analyze and create pathways differentially activated with the treatment matrix based on published literature.
Gene expression changes in human skin fibroblasts serve as readout for cellular responses to the stimulus of carbon nanomaterials. By applying significance analysis with very conservative Bonferroni multitesting correction, we found a profound number of genes with statistically significant expression level changes (Figure 4, Supporting Information Table S1–S12). Treating cells with a high dose of carbon particles caused more gene expression changes than the low dose treatment (Figure 4). However, it would be misleading to say that the responses were dose-dependent, at least for the two doses in this study. As shown in parts B and C of Figure 4, only a small portion of genes with altered transcription were found in common between the low and high dose profiles, when treating with same type of particle. This indicates that distinct gene expression profiles were induced at low and high dose treatment. In contrast, if we compare two types of particles, they induced similar transcriptional changes in cells at the same high and low doses (Figure 4D,E). The unique genes flanking the overlapping area in Figure 4D,E may indicate cellular responses unique to exposure with MWCNOs or MWCNTs (Supporting Information Tables S7, S9, S10, and S12).
Gene ontology analysis gave further evidence supporting the qualitative differences of cell responses to low and high doses of carbon nanomaterials. The percentages of overexpressed and underexpressed genes in the top 10 most changed gene categories are shown in Table 1. At low dose of both of MWCNO and MWCNT, genes were down-regulated in most of the categories. Many of these genes involve Golgi vesicle transport, secretory pathway, fatty acid biosynthesis, protein metabolism, and G1/S transition of mitotic cell cycle. The only up-regulated category was protein ubiquitination (Table 1, Supporting Information Table 13). Collectively, these data suggest a reduction of cell growth and metabolism but an acceleration of protein degradation at the low dosage. Reduction of cell growth was consistent with our phenotypic data. In contrast, a high dose of MWCNO and MWCNT resulted in the up-regulation of protein and amino acid metabolism; with additional up-regulation of genes involved in a type I IFN response (Table 1). The outcome is an increase in apoptosis and reduction in cell growth. However, the distinct gene expression profiles induced at low and high doses may indicate that different mechanisms are responsible for our phenotypic observations or that the response occurs at a different rate and we are observing two “snapshots” of a temporal progression of a single mechanism. This observation agrees with the our earlier experience with gene expression changes induced by radiation.49
Structure-specific cellular responses were also observed in this experiment. At high dose, only MWCNT caused overexpression of a significant number of immune and inflammatory response genes (Table 1 and Table 2). Totally 25 genes in this category were overexpressed and only one gene was underexpressed, indicating a robust response of this function group (Tables 1 and 2). Most of these genes are involved in innate immune response system and are induced by interferon or interferon-related proteins. Many of these genes are implicated in an interferon type I response, which is potently antiviral and antiproliferative. Some of the genes that are typically induced by an interferon type I response include Irf7, Isgf3g, Stat1 Adar, Cxcl10, Irf1, Isgf3g, IFIT1, and MX2, all found in Table 2. Interestingly the dimension of carbon tubes is similar to that of a virus, and the cellular response may mimic the response observed with viral infection. Certainly the induction of many of the same genes during viral infection is observed. Our observation fits a previous report that kerotinocytes (HEKs) exposed to chemically unmodified MWCNTs released interleukin-8, a pro-inflammatory cytokine, which was postulated to result in the skin irritation associated with exposure.39
In addition to the IFN type I response genes, we also observed up-regulation of intracellular aryl hydrocarbon (AHR) in MWCNT-treated cells (Table 5). This gene is typically expressed in cells or animals exposed to polycyclic aromatic hydrocarbons and is believed to mediate the teratogenesis, immune suppression, epithelial disorders, and tumor production in exposed experimental animals.89 Transcription of Bax, driven by AHR, is part of an evolutionarily conserved cell-death-signaling pathway response, responsible for ovarian failure induced by environmental toxins.90 Overexpression of this gene is consistent with the cell death we observed with the carbon nanomaterials. In addition, the cytokine and TNF family member TNFRSF10B (TRAILR2) is up-regulated in cells treated with the highest concentration of nanotubes, and this protein induces apoptosis in a wide variety of cells.72 Additional apoptosis genes involved include BCL2L2 and MCL1. Finally, RIPK2 and TNFAIP3, genes that contribute to the induction of apoptosis, were also observed to be up-regulated in these treated cells.
Data from Tables 4 and 5 indicate that FGFR1 and EGFR are involved in the response. We suspect that the cells are using strategies similar to a viral response when exposed to nanomaterials. Viruses are very similar in size range to the carbon nanomaterials used here, around 20 nm in diameter. FGFR, EGFR, and other RTK pathways have been implicated in viral response in numerous studies. An early step in viral infection is the targeting of the virus to cell surface receptors. Many viral receptors have been identified, including signaling receptors such as EGFR, chemokine receptor, platelet-derived growth factor receptor, fibroblast growth factor receptor, tumor necrosis factor receptor family, and various integrin receptors. Usually multiple receptors are targeted by the virus for binding, signaling, and entry. Virus also impinges upon the signal transduction pathway in the sense that their binding to the receptor perturbs the normal receptor-coupled signal transduction pathways. Many receptors, e.g., EGFR, are potent stimulators of the mitogen-activated protein kinase (MAPK) signaling pathway. Chronic stimulation of EGFR and of multiple steps in the MAPK signaling pathway is involved in multiple cellular processes, especially in the interaction between viruses and tyrosine kinase pathways.91 One interesting observation is the down-regulation of EGFR by >4-fold, which indicates that the nano-onion and nanotubes might serve as therapeutics for EGFR-overexpressing epithelial cancers, such as >20% of the breast cancer. This could be a very interesting use of the cytotoxicity of the carbon nanomaterials, which has been suggested28 and demonstrated by other groups.29 In addition to regulation of EGFR and FGFR1 expression, the overexpression of VEGF mRNA is also observed at both high dose experiments. The secretion of VEGF could be the cellular wound healing response to the addition of nanoparticles. In addition to the ability to activate epithelial proliferation, it may also be a last-resort cellular response to save the epithelial cells from apoptosis.92
Promoter analysis identified EGR1/Krox as one of the over-represented transcription regulatory elements on up-regulated genes in almost all experimental settings (Figure 5). In addition, with high dosage of treatment, additional transcription factors (ETS1 and IRF for MWCNTs, E2F and C/EBP-delta for MWCNOs) might be involved (Figure 5). In general, the profiles of enriched TREs are dramatically different in the individual experiments. For the down-regulated genes from MWCNT treatment, there are enrichment of GATA4, USF, and ELK1 at low dosage and COMP1 (cooperates with myogenic proteins 1) at higher dosage. For the carbon onions treatment, the lower dosage is correlated with enrichment of GATA4, USF, elk1, and Egr1/Knox in down-regulated genes and high dosage with enrichment of GATA1, HES1, PAX, and E2F1. The upstream events leading to the different expression patterns seem to be related to ERK and p38 MAPK activities and the induction of interferon signaling. These analyses suggest that the induction of the p38/ERK pathway and the type I IFN response are the upstream signaling events (see Figure 6 for the pathway analysis result illustration) responsible for changes in cellular transcription due to MWCNO and MWCNT treatment of cells. Indeed, the pathway responses shown here are similar to the response of human bronchial epithelial cells to combustion-derived metals.93
In summary, combined with the result from functional analysis, this study clearly showed that at high dosage, carbon particles can seriously impact the cellular functions in maintenance, growth, and differentiation. Of these two nanomaterials, MWCNTs appear to induce more stress on the cells than MWCNOs. Our data suggest that there is a qualitative difference in response to low dose and high dose treatment of carbon particles in human skin fibroblasts. Carbon tubes at high dose induced innate immune responses, whereas carbon onions did not. This indicates that cells respond differently according to the structures of nanomaterials. Our data also suggest that carbon atoms released from nanomaterials may participate in cell metabolic pathways. It is evident from our studies that carbon nanomaterials have a toxic effect on lung and skin cells. As little as 15 000 MWCNOs per cell and a few dozen MWCNTs per cell induced cell death in this study. Therefore, as this potentially revolutionary technology is further developed, specific heed must be given to minimizing unwanted effects upon both producers and consumers. The regulation of p38/ERK and the EGFR also opens the door that the carbon nano-onions and potentially other carbon nanomaterials can be exploited as a nanomedicine platform for cancer therapy, especially epithelially derived cancers.
Acknowledgments
We thank Mr. Kevin Peet and Ms. Lonnette Robinson for excellent administrative support. We thank Alistar McDonald for critical reading of the manuscript and Professor Marc Shuman and Professor Song Li for support. F. Chen was supported by NIH Grant R1CA95393-01, DOD BCRP BC045345 Grant, DARPA, UCSF Prostate Cancer SPORE award (NIH Grant P50 CA89520), by a DOE LDRD grant to A. P. Alivisatos/J. W. Gray, and by NIH P50 Grant CA112970 to J. W. Gray. This work was performed under the auspices of the U.S. Department of Energy, at the University of California/Lawrence Berkeley National Laboratory, under Contract No. DE-AC03-76SF00098.
Footnotes
Supporting Information Available: Details of materials and methods discussed, figures showing cell viability measurements and a scatter plot of normalized GeneChip data, and tables listing the top 20 genes in Figure 4. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Colvin VL. The potential environmental impact of engineered nanomaterials. Nat Biotechnol. 2003;21:1166–1170. doi: 10.1038/nbt875. [DOI] [PubMed] [Google Scholar]
- 2.Service RF. American Chemical Society meeting. Nanomaterials show signs of toxicity. Science. 2003;300:243. doi: 10.1126/science.300.5617.243a. [DOI] [PubMed] [Google Scholar]
- 3.Proffitt F. Nanotechnology. Yellow light for nanotech. Science. 2004;305:762. doi: 10.1126/science.305.5685.762b. [DOI] [PubMed] [Google Scholar]
- 4.Service RF. Nanotoxicology. Nanotechnology grows up. Science. 2004;304:1732–1734. doi: 10.1126/science.304.5678.1732. [DOI] [PubMed] [Google Scholar]
- 5.Curl RFRES, Kroto HW, O’Brien S, Heath JR. How the news that we were not the first to conceive of soccer ball C60 got to us. J Mol Graphics Modell. 2001;19:185–186. doi: 10.1016/s1093-3263(00)00107-8. [DOI] [PubMed] [Google Scholar]
- 6.Iijima S. Helical microtubules of graphitic carbon. Nature. 1991;354:56–58. [Google Scholar]
- 7.Iijima S, Ichihashi T. Single-shell carbon nanotubes of 1-nm diameter. Nature. 1993;363:603–605. [Google Scholar]
- 8.Kadish KM, Ruoff RS, editors. Fullerenes: Chemistry, Physics, and Technology. Wiley-Interscience; New York: 2000. [Google Scholar]
- 9.Dresselhaus MS, Dresselhaus G, Eklund PC. Science of Fullerenes and Carbon Nanotubes. Academic Press; New York: 1996. [Google Scholar]
- 10.Ajayan PM. Nanotubes from Carbon. Chem Rev. 1999;99:1787–1799. doi: 10.1021/cr970102g. [DOI] [PubMed] [Google Scholar]
- 11.Sugai T, et al. New Synthesis of High-Quality Double-Walled Carbon Nanotubes by High-Temperature Pulsed Arc Discharge. Nano Lett. 2003;3:769–773. [Google Scholar]
- 12.Sinnott SB, Andrews R. Carbon Nanotubes: Synthesis, Properties and Applications. Crit Rev Solid State Mater Sci. 2001;26:145–249. [Google Scholar]
- 13.Tan SJ, et al. Individual single-wall carbon nanotubes as quantum wires. Nature. 1997;386:474–477. [Google Scholar]
- 14.Colbert DT, Smalley RE. Fullerene nanobutes for molecular electronics. Trends Biotechnol. 1999;17:46–50. [Google Scholar]
- 15.White CT, Todorov TN. Quantum electronics: Nanotubes go ballistic. Nature. 2001;411:649–651. doi: 10.1038/35079720. [DOI] [PubMed] [Google Scholar]
- 16.Calvert P. Carbon Nanotubes. 1997:277–292. [Google Scholar]
- 17.Iijima S. Direct observation of the tetrahedral bonding in graphitized carbon black by high-resolution electronmicroscopy. J Cryst Growth. 1980;50:675–683. [Google Scholar]
- 18.Ugarte D. Onion-like graphitic particles. Carbon. 1995;33:989–993. [Google Scholar]
- 19.Sano N, Wang H, Chhowalla M, Alexandrou I, Amaratunga GAJ. Nanotechnology: Synthesis of carbon “onions” in water. Nature (London) 2001;414:506–507. doi: 10.1038/35107141. [DOI] [PubMed] [Google Scholar]
- 20.Sano N, et al. Properties of carbon onions produced by an arc discharge in water. J Appl Phys. 2002;92:2783–2788. [Google Scholar]
- 21.Lange H, et al. Nanocarbon production by arc discharge in water. Carbon. 2003;41:1617–1623. [Google Scholar]
- 22.Chen B, Selegue JP, Wijeratne L, Bom D, Meier MS. Proc Carbon ‘02, Int Conf Carbon. 2002. Characterization of carbon nano-onions by using flow field-flow fractionation; p. 22.27.75. [Google Scholar]
- 23.Choi MS, Kim Y-J, Yi J-H, Altman I, Pikhitsa P. US Pat Appl Publ. :10. ((S. Korea). U.S., 2004) [Google Scholar]
- 24.Brabec C, Hirsch A. PCT Int Appl. Vol. 19. Siemens Aktieng-esellschaft; Germany: 2003. WO. [Google Scholar]
- 25.Kajiura H, Shiraishi S, Negishi E, Ata M, Yamada A. Jpn Kokai Tokkyo Koho. Vol. 13. Sony Corp.; Japan Jp: 2001. [Google Scholar]
- 26.Maynard AD, et al. Exposure to carbon nanotube material: aerosol release during the handling of unrefined single-walled carbon nanotube material. J Toxicol Environ Health, Part A. 2004;67:87–107. doi: 10.1080/15287390490253688. [DOI] [PubMed] [Google Scholar]
- 27.Silva VM, Corson N, Elder A, Oberdorster G. The Rat Ear Vein Model For Investigating In Vivo Thrombogenicity Of Ultrafine Particles (Ufp) Toxicol Sci. 2005 doi: 10.1093/toxsci/kfi142. [DOI] [PubMed] [Google Scholar]
- 28.Sayes CM, et al. The differential cytotoxicity of water-soluble fullerenes. Nano Lett. 2004;4:1881–1887. [Google Scholar]
- 29.Shi Kam NW, O’Connell M, Wisdom JA, Dai H. Carbon nanotubes as multifunctional biological transporters and near-infrared agents for selective cancer cell destruction. Proc Natl Acad Sci US A. 2005;102:11600–11605. doi: 10.1073/pnas.0502680102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yinghuai Z, et al. Substituted carborane-appended water-soluble single-wall carbon nanotubes: new approach to boron neutron capture therapy drug delivery. J Am Chem Soc. 2005;127:9875–9880. doi: 10.1021/ja0517116. [DOI] [PubMed] [Google Scholar]
- 31.Burlaka AP, et al. Catalytic system of the reactive oxygen species on the C60 fullerene basis. Exp Oncol. 2004;26:326–327. [PubMed] [Google Scholar]
- 32.Nelson MA, et al. Effects of acute and subchronic exposure of topically applied fullerene extracts on the mouse skin. Toxicol Ind Health. 1993;9:623–630. doi: 10.1177/074823379300900405. [DOI] [PubMed] [Google Scholar]
- 33.Ali SS, et al. A biologically effective fullerene (C60) derivative with superoxide dismutase mimetic properties. Free Radical Biol Med. 2004;37:1191–1202. doi: 10.1016/j.freeradbiomed.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 34.Rancan F, et al. Cytotoxicity and photocytotoxicity of a dendritic C(60) monoadduct and a malonic acid C(60) tris-adduct on Jurkat cells. J Photochem Photobiol, B. 2002;67:157–162. doi: 10.1016/s1011-1344(02)00320-2. [DOI] [PubMed] [Google Scholar]
- 35.Oberdorster E. Manufactured nanomaterials (fullerenes, C60) induce oxidative stress in the brain of juvenile largemouth bass. Environ Health Perspect. 2004;112:1058–1062. doi: 10.1289/ehp.7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frampton MW, et al. Effects of exposure to ultrafine carbon particles in healthy subjects and subjects with asthma. Res Rep–Health Eff Inst. 2004 discussion 49–63, 1–47. [PubMed] [Google Scholar]
- 37.Block ML, et al. Nanometer size diesel exhaust particles are selectively toxic to dopaminergic neurons: the role of microglia, phagocytosis, and NADPH oxidase. FASEB J. 2004;18:1618–1620. doi: 10.1096/fj.04-1945fje. [DOI] [PubMed] [Google Scholar]
- 38.Lam CW, James JT, McCluskey R, Hunter RL. Pulmonary toxicity of single-wall carbon nanotubes in mice 7 and 90 days after intratracheal instillation. Toxicol Sci. 2004;77:126–134. doi: 10.1093/toxsci/kfg243. [DOI] [PubMed] [Google Scholar]
- 39.Monteiro-Riviere NA, Nemanich RJ, Inman AO, Wang YY, Riviere JE. Multiwalled carbon nanotube interactions with human epidermal keratinocytes. Toxicol Lett. 2005;155:377–384. doi: 10.1016/j.toxlet.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 40.Shvedova AA, et al. Exposure to carbon nanotube material: assessment of nanotube cytotoxicity using human keratinocyte cells. J Toxicol Environ Health, Part A. 2003;66:1909–1926. doi: 10.1080/713853956. [DOI] [PubMed] [Google Scholar]
- 41.Warheit DB, et al. Comparative pulmonary toxicity assessment of single-wall carbon nanotubes in rats. Toxicol Sci. 2004;77:117–125. doi: 10.1093/toxsci/kfg228. [DOI] [PubMed] [Google Scholar]
- 42.Jia G, et al. Cytotoxicity of Carbon Nanomaterials: Single-Wall Nanotube, Multi-Wall Nanotube, and Fullerene. 2005;39:1378–1383. doi: 10.1021/es048729l. [DOI] [PubMed] [Google Scholar]
- 43.Goodman CM, McCusker CD, Yilmaz T, Rotello VM. Toxicity of gold nanoparticles functionalized with cationic and anionic side chains. Bioconjugate Chem. 2004;15:897–900. doi: 10.1021/bc049951i. [DOI] [PubMed] [Google Scholar]
- 44.Lockman PR, Koziara JM, Mumper RJ, Allen DD. Nanoparticle surface charges alter blood-brain barrier integrity and permeability. J Drug Targeting. 2004;12:635–641. doi: 10.1080/10611860400015936. [DOI] [PubMed] [Google Scholar]
- 45.Kirchner C, et al. Cytotoxicity of colloidal CdSe and CdSe/ZnS nanoparticles. Nano Lett. 2005;5:331–338. doi: 10.1021/nl047996m. [DOI] [PubMed] [Google Scholar]
- 46.Cunningham MJ, Magnuson SR, Falduto MT, Balzano L, Resasco DE. Investigating the toxicity of nanoscale materials by gene expression profiling: A systems biology approach. American Chemical Society Annual Meeting Presentation; 2005. [Google Scholar]
- 47.Andrews R, Jacques D, Qian D, Rantell T. Multiwall carbon nanotubes: synthesis and application. Acc Chem Res. 2002;35:1008–1017. doi: 10.1021/ar010151m. [DOI] [PubMed] [Google Scholar]
- 48.Wronski R, Golob N, Grygar E, Windisch M. Two-color, fluorescence-based microplate assay for apoptosis detection. Bio-techniques. 2002;32:666–668. [PubMed] [Google Scholar]
- 49.Ding LH, et al. Gene expression profiles of normal human fibroblasts after exposure to ionizing radiation: a comparative study of low and high doses. Radiat Res. 2005;164:17–26. doi: 10.1667/rr3354. [DOI] [PubMed] [Google Scholar]
- 50.Dolbeare F, Gratzner H, Pallavicini MG, Gray JW. Flow cytometric measurement of total DNA content and incorporated bromodeoxyuridine. Proc Natl Acad Sci US A. 1983;80:5573–5577. doi: 10.1073/pnas.80.18.5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang XL, Fan CH, Zhu HS. Photoinduced cytotoxicity of malonic acid [C(60)]fullerene derivatives and its mechanism. Toxicol in Vitro. 2002;16:41–46. doi: 10.1016/s0887-2333(01)00102-3. [DOI] [PubMed] [Google Scholar]
- 52.Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 53.Honda K, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 54.Harada H, et al. Absence of the type I IFN system in EC cells: transcriptional activator (IRF-1) and repressor (IRF-2) genes are developmentally regulated. Cell. 1990;63:303–312. doi: 10.1016/0092-8674(90)90163-9. [DOI] [PubMed] [Google Scholar]
- 55.Weier HU, George CX, Greulich KM, Samuel CE. The interferon-inducible, double-stranded RNA-specific adenosine deaminase gene (DSRAD) maps to human chromosome 1q21.1–21.2. Genomics. 1995;30:372–375. doi: 10.1006/geno.1995.0034. [DOI] [PubMed] [Google Scholar]
- 56.Luster AD, Unkeless JC, Ravetch JV. Gamma-interferon transcriptionally regulates an early-response gene containing homology to platelet proteins. Nature. 1985;315:672–676. doi: 10.1038/315672a0. [DOI] [PubMed] [Google Scholar]
- 57.Luster AD, Jhanwar SC, Chaganti RS, Kersey JH, Ravetch JV. Interferon-inducible gene maps to a chromosomal band associated with a (4; 11) translocation in acute leukemia cells. Proc Natl Acad Sci US A. 1987;84:2868–2871. doi: 10.1073/pnas.84.9.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clauss IM, et al. Chromosomal localization of two human genes inducible by interferons, double-stranded RNA, and viruses. Cytogenet Cell Genet. 1990;53:166–168. doi: 10.1159/000132920. [DOI] [PubMed] [Google Scholar]
- 59.Porter AC, et al. Interferon response element of the human gene 1988, 6–16. EMBO J. 1988;7:85–92. doi: 10.1002/j.1460-2075.1988.tb02786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chebath J, Merlin G, Metz R, Benech P, Revel M. Interferon-induced 56 000 Mr protein and its mRNA in human cells: molecular cloning and partial sequence of the cDNA. Nucleic Acids Res. 1983;11:1213–1226. doi: 10.1093/nar/11.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lafage M, et al. The interferon- and virus-inducible IFI-56K and IFI-54K genes are located on human chromosome 10 at bands q23–q24. Genomics. 1992;13:458–460. doi: 10.1016/0888-7543(92)90272-t. [DOI] [PubMed] [Google Scholar]
- 62.Zhu H, Cong JP, Shenk T. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc Natl Acad Sci US A. 1997;94:13985–13990. doi: 10.1073/pnas.94.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Niikura T, Hirata R, Weil SC. A novel interferon-inducible gene expressed during myeloid differentiation. Blood Cells Mol Dis. 1997;23:337–349. doi: 10.1006/bcmd.1997.0151. [DOI] [PubMed] [Google Scholar]
- 64.Horisberger MA, et al. cDNA cloning and assignment to chromosome 21 of IFI-78K gene, the human equivalent of murine Mx gene. Somat Cell Mol Genet. 1988;14:123–131. doi: 10.1007/BF01534397. [DOI] [PubMed] [Google Scholar]
- 65.Melen K, et al. Human MxB protein, an interferon-alpha-inducible GTPase, contains a nuclear targeting signal and is localized in the heterochromatin region beneath the nuclear envelope. J Biol Chem. 1996;271:23478–23486. doi: 10.1074/jbc.271.38.23478. [DOI] [PubMed] [Google Scholar]
- 66.Bonnevie-Nielsen V, Larsen ML, Frifelt JJ, Michelsen B, Lernmark A. Association of IDDM and attenuated response of 2′,5′-oligoadenylate synthetase to yellow fever vaccine. Diabetes. 1989;38:1636–1642. doi: 10.2337/diab.38.12.1636. [DOI] [PubMed] [Google Scholar]
- 67.Bonnevie-Nielsen V, et al. Variation in antiviral 2′,5′-oligoadenylate synthetase (2′5′AS) enzyme activity is controlled by a single-nucleotide polymorphism at a splice-acceptor site in the OAS1 gene. Am J Hum Genet. 2005;76:623–633. doi: 10.1086/429391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Whiteheart SW, et al. SNAP family of NSF attachment proteins includes a brain-specific isoform. Nature. 1993;362:353–355. doi: 10.1038/362353a0. [DOI] [PubMed] [Google Scholar]
- 69.Lemons PP, Chen D, Bernstein AM, Bennett MK, Whiteheart SW. Regulated secretion in platelets: identification of elements of the platelet exocytosis machinery. Blood. 1997;90:1490–1500. [PubMed] [Google Scholar]
- 70.Ravichandran V, Chawla A, Roche PA. Identification of a novel syntaxin- and synaptobrevin/VAMP-binding protein, SNAP-23, expressed in nonneuronal tissues. J Biol Chem. 1996;271:13300–13303. doi: 10.1074/jbc.271.23.13300. [DOI] [PubMed] [Google Scholar]
- 71.Mansour SJ, Herbrick JA, Scherer SW, Melancon P. Human GBF1 is a ubiquitously expressed gene of the sec7 domain family mapping to 10q24. Genomics. 1998;54:323–327. doi: 10.1006/geno.1998.5563. [DOI] [PubMed] [Google Scholar]
- 72.Walczak H, et al. TRAIL-R2: a novel apoptosis-mediating receptor for TRAIL. EMBO J. 1997;16:5386–5397. doi: 10.1093/emboj/16.17.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McCarthy JV, Ni J, Dixit VM. RIP2 is a novel NF-kappaB-activating and cell death-inducing kinase. J Biol Chem. 1998;273:16968–16975. doi: 10.1074/jbc.273.27.16968. [DOI] [PubMed] [Google Scholar]
- 74.Sibilia M, et al. The EGF receptor provides an essential survival signal for SOS-dependent skin tumor development. Cell. 2000;102:211–220. doi: 10.1016/s0092-8674(00)00026-x. [DOI] [PubMed] [Google Scholar]
- 75.Opferman JT, et al. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature. 2003;426:671–676. doi: 10.1038/nature02067. [DOI] [PubMed] [Google Scholar]
- 76.Boise LH, et al. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- 77.Senechal K, Halpern J, Sawyers CL. The CRKL adaptor protein transforms fibroblasts and functions in transformation by the BCR-ABL oncogene. J Biol Chem. 1996;271:23255–23261. doi: 10.1074/jbc.271.38.23255. [DOI] [PubMed] [Google Scholar]
- 78.Wakasugi K, Schimmel P. Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science. 1999;284:147–151. doi: 10.1126/science.284.5411.147. [DOI] [PubMed] [Google Scholar]
- 79.Seshadri T, Campisi J. Repression of c-fos transcription and an altered genetic program in senescent human fibroblasts. Science. 1990;247:205–209. doi: 10.1126/science.2104680. [DOI] [PubMed] [Google Scholar]
- 80.Chuang CF, Ng SY. Functional divergence of the MAP kinase pathway. ERK1 and ERK2 activate specific transcription factors. FEBS Lett. 1994;346:229–234. doi: 10.1016/0014-5793(94)00480-3. [DOI] [PubMed] [Google Scholar]
- 81.Hipskind RA, Baccarini M, Nordheim A. Transient activation of RAF-1, MEK, and ERK2 coincides kinetically with ternary complex factor phosphorylation and immediate-early gene promoter activity in vivo. Mol Cell Biol. 1994;14:6219–6231. doi: 10.1128/mcb.14.9.6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hodge C, et al. Growth hormone stimulates phosphorylation and activation of elk-1 and expression of c-fos, egr-1, and junB through activation of extracellular signal-regulated kinases 1 and 2. J Biol Chem. 1998;273:31327–31336. doi: 10.1074/jbc.273.47.31327. [DOI] [PubMed] [Google Scholar]
- 83.Lim CP, Jain N, Cao X. Stress-induced immediate-early gene, egr-1, involves activation of p38/JNK1. Oncogene. 1998;16:2915–2926. doi: 10.1038/sj.onc.1201834. [DOI] [PubMed] [Google Scholar]
- 84.Liang Q, et al. The transcription factor GATA4 is activated by extracellular signal-regulated kinase 1- and 2-mediated phosphorylation of serine 105 in cardiomyocytes. Mol Cell Biol. 2001;21:7460–7469. doi: 10.1128/MCB.21.21.7460-7469.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kerkela R, Pikkarainen S, Majalahti-Palviainen T, Tokola H, Ruskoaho H. Distinct roles of mitogen-activated protein kinase pathways in GATA-4 transcription factor-mediated regulation of B-type natriuretic peptide gene. J Biol Chem. 2002;277:13752–13760. doi: 10.1074/jbc.M105736200. [DOI] [PubMed] [Google Scholar]
- 86.Galibert MD, Carreira S, Goding CR. The Usf-1 transcription factor is a novel target for the stress-responsive p38 kinase and mediates UV-induced Tyrosinase expression. EMBO J. 2001;20:5022–5031. doi: 10.1093/emboj/20.17.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baldassare JJ, Bi Y, Bellone CJ. The role of p38 mitogen-activated protein kinase in IL-1 beta transcription. J Immunol. 1999;162:5367–5373. [PubMed] [Google Scholar]
- 88.O’Rourke J, Yuan R, DeWille J. CCAAT/enhancer-binding protein-delta (C/EBP-delta) is induced in growth-arrested mouse mammary epithelial cells. J Biol Chem. 1997;272:6291–6296. doi: 10.1074/jbc.272.10.6291. [DOI] [PubMed] [Google Scholar]
- 89.Ohtake F, et al. Modulation of oestrogen receptor signaling by association with the activated dioxin receptor. Nature. 2003;423:545–550. doi: 10.1038/nature01606. [DOI] [PubMed] [Google Scholar]
- 90.Matikainen T, et al. Aromatic hydrocarbon receptor-driven Bax gene expression is required for premature ovarian failure caused by biohazardous environmental chemicals. Nat Genet. 2001;28:355–360. doi: 10.1038/ng575. [DOI] [PubMed] [Google Scholar]
- 91.Braun-Falco M, Eisenried A, Buning H, Ring J. Recombinant adeno-associated virus type 2-mediated gene transfer into human keratinocytes is influenced by both the ubiquitin/proteasome pathway and epidermal growth factor receptor tyrosine kinase. Arch Dermatol Res. 2005;296:528–535. doi: 10.1007/s00403-005-0547-y. [DOI] [PubMed] [Google Scholar]
- 92.Alavi A, Hood JD, Frausto R, Stupack DG, Cheresh DA. Role of Raf in vascular protection from distinct apoptotic stimuli. Science. 2003;301:94–96. doi: 10.1126/science.1082015. [DOI] [PubMed] [Google Scholar]
- 93.Wu W, et al. Activation of the EGF receptor signaling pathway in human airway epithelial cells exposed to metals. Am J Physiol. 1999;277:L924–931. doi: 10.1152/ajplung.1999.277.5.L924. [DOI] [PubMed] [Google Scholar]