Summary
Cancers result from large-scale deregulation of genes that lead to cancer pathophysiologies such as increase proliferation, decreased apoptosis, increased motility, increased angiogenesis, and others. Genes that influence proliferation and apoptosis are particularly attractive as therapeutic targets. To identify genes that influence these phenotypes, we have developed simple and rapid methods to measure apoptosis and cell proliferation using high content screening with YO-PRO®-1 and anti-BrdU staining of BrdU pulsed cells, respectively.
Keywords: Apoptosis, BrdU, cell cycle, cellomics, high content analysis, high content screening, image analysis, propidium iodide, siRNA, YO-PRO-1
1. Introduction
The genomic revolution has spawned a variety of global analysis methods to assess abnormalities in the genome, epigenome, and transcriptome that occur during carcinogenesis and cancer progression. Functional assessment of the genes revealed by these analyses is important both to identify potential therapeutic targets and/or to suggest molecular markers that might predict patient outcome. This requires efficient methods to measure cancer-related changes in cellular phenotypes that result from manipulation of the genes revealed by “-omic” analyses. Relevant phenotypic changes in cells involve cancer hallmarks such as genome instability, cell proliferation, apoptosis, angiogenesis, invasion, and metastasis (1).
Functional prioritization of aberrant genes is challenging because literally thousands of genes already have been implicated by profiling experiments (2). Our approach has been to use high content imaging to assess changes in two important cancer related end points—apoptosis and proliferation—during manipulation of candidate genes using siRNAs and/or gene-specific small molecule inhibitors (3,4). We have used this strategy in multiple cancer cell lines to evaluate genes that have been identified as potential therapeutic or diagnostic targets. Two high content screening methods employed by our lab are YO-PRO®-1 staining of cells to measure apoptosis and BrdU incorporation to measure proliferation (5,6). An example, using these techniques to evaluate a gene amplified and overexpressed in some ovarian cancers, will be used to demonstrate the evaluation of cellular phenotype after treatment with siRNA to this gene.
2. Materials
1. Cell lines testing positive for amplification and overexpression.
2. Cell lines negative for amplification and overexpression.
3. Tissue culture media selected for optimal growth of cells.
4. Lipofectamine 2000 (Invitrogen, Carlsbad, CA).
5. Optimem.
6. 1X Sterile PBS (phosphate-buffered saline).
7. Multichannel pipet (20–200 μL).
8. Aspirator.
9. Coulter Counter® (or hemocytometer—see Note 1).
10. Cuvet for Coulter Counter.
11. Isoton II Diluent (Beckman Coulter; Fullerton, CA).
12. Sterile disposable multichannel pipet basins.
13. YO-PRO-1 iodide 491/509 (Molecular Probes, Eugene, OR).
14. Hoechst 33342.
15. BrdU (bromodeoxyuridine).
16. Anti-BrdU monoclonal antibody (BD Biosciences, San Diego, CA).
17. Rabbit antimouse Alexa Fluor® 488 (Invitrogen).
18. PI (propidium iodide).
19. NaOH.
20. 70% ethanol.
21. Tween-20.
22. KineticScan HCS Reader (KSR; Cellomics, Inc., Pittsburg, PA).
3. Methods
The methods described next outline (1) seeding and treatment of cell lines, (2) staining cells with YO-PRO-1and Hoechst 33342, (3) incorporation of BrdU and antibody staining in proliferating cells, (4) scanning the plates using a high content screening system, and (5) analyzing the data.
3.1. Seeding and Treating Cells
The seeding and treating of cells with siRNA is described in Subheadings 3.1.1.–3.1.4. This includes (1) a description of criteria to select cell lines to test, (2) counting and seeding the cells onto a 96-well plate, and (3) treating cells in 96-well plate.
3.1.1. Selection of Cell Lines
In the example presented here we identified target genes by finding regions of the genome that are amplified (or lost) in ovarian cancer cell lines and tumors (7–9). These regions were identified based on DNA copy number changes of many different cell lines measured by comparative genomic hybridization arrays (10,11). Once amplified regions were identified, the genes in these regions were examined for overexpression. One of the amplified regions identified was chromosome 8q24 and is associated with poor outcome in ovarian cancers (9,12,13). Several genes were selected for testing using high-content analysis and one example is presented here. Once a gene was selected for testing in this system, cell lines were then selected based on either being positive or negative for the amplification and over expression of this gene. Four representative cell lines were selected for each category (see Note 2). For other applications cell lines would be selected based on the information required.
3.1.2. Cell Lines
The cell lines with the 8q24 amplification used for this example were CAOV4, HEY, OVCA432, and OVCAR-8. Four others, A2780, SKOV3, OV-90, and CAOV3 were also selected because they do not contain this amplification. CAOV3, CAOV4, OVCAR-8, and OV90 were purchased from American Type Culture Collection and the remaining cell lines were kindly donated by Gordon Mills, MD Anderson, Dallas, TX. The cell lines were maintained in L-15 (CAOV4), RPMI 1640 (HEY, OVCAR8, and A2780), MEM (OVCA432 and CAOV3), and M199/MCDB (OV90), supplemented with 10% fetal bovine serum (FBS). The media selected should be optimal for propagation of the cell line used.
The treatment used for the cells depends on the experimental goals, but this example, treatment with siRNA will be described. One of the important considerations for any treatment is to include a group of mock-treated cells. In our experiments, each condition is repeated in one of four wells to ensure reproducibility. If it is possible and convenient, randomizing the position in which, repeat wells are placed is a good practice, because of potential variability in the plate, edge effects, or any other unforeseen effects that might be dependent on the position of the cells in the plate.
3.1.3. Seeding Cells on a 96-Well Plate
The appropriate number of cells per well on a 96-well plate varies with the size of cells and the length of time from seeding to analysis. In this example, 5000 cells per well are plated in a 96-well plate (see Note 3). We have also used similar protocols in a 24-well plate and have found that 50,000–100,000 cells per well is optimal. Typically, the assays are performed within 48 h after seeding.
Adherent cells are dissociated from the culture dish surface with 0.25% trypsin EDTA and then the trypsin EDTA is inactivated with media containing 10% FBS. If the cells are sensitive to trypsin they can be spun down at this point and resuspended in media plus 10% FBS. Spin trypsin-sensitive cells down gently in a conical tube in a table-top centrifuge at approx 1200 RCF, remove media, and resuspend cell pellets into new media.
Approximately 100 μL of the cell mixture is then transferred to a Coulter Counter cuvet containing 10 mL isotone. A dilution of 101 is set on the Coulter Counter display. After the system is primed according to manufacturer's instructions, cells are counted and a calculation is made to determine the dilution of cells to obtain media containing 50,000 cells/mL. The Coulter Counter is then either put through a priming cycle so that another group of cells can be counted or, if counting is completed, the cleaning solution is put into the Coulter Counter cuvet and the system is cleaned to prevent future clogging. The cells are then diluted in the same media used for propagation. Before each step the cells should be gently, but thoroughly mixed to ensure accurate counting and even spreading in the wells of the plate. Approximately 10 mL of diluted cells are required per 96-well plate.
Once the dilution has been established the appropriate volume of diluted cells, depending on how many 96-well plates are required, is added to a sterile trough. The maximum volume for these receptacles is 55 mL. The multichannel pipet can be used to add 100 μL of diluted cells per well. The plates are then immediately placed at 37°C in 5% CO2 for incubation overnight. This will allow the cells to adhere to the plates and recover from the trypsin treatment.
3.1.4. siRNA Transfection
1. Label two sterile microcentrifuge tubes with the designation A or B.
2. In tube A add 10 μL of Opti-MEM+ X μL of 20 μM of siRNA stock (see Table 1 for amount of siRNA oligonucleotide to add).
3. In tube B add 10 μL of Opti-MEM+ X μL of 1 μg/μL Lipofectamine 2000.
4. Incubate each tube at room temperature for 5 min.
5. Add the contents of tube B to tube A. Incubate the mixture at room temperature for 20 min.
6. Approximately 5 min before the 20 min incubation is over, replace culture media with 100 μL of Opti-MEM in each well of a 96-well plate and with 500 μL in each well of a 24-well plate.
7. Add the tube A + B siRNA-Lipofectamine 2000 mixture gradually and gently to wells containing Opti-MEM. For each well of a 96-well plate, add 20 μL of the mixture. For each well of a 24-well plate, add 100 μL of the mixture.
8. Incubate plate at 37°C in a CO2 incubator for 2–4 h.
9. Replace Opti-MEM with complete culture media.
Table 1.
Recommended Volumes of 20 μM Stock siRNA and 1 μg/μL Lipofectamine 2000 for Different Final Concentrations of siRNA
| siRNA final concentration |
|||
|---|---|---|---|
| Culture format | 40 (nM) | 80 (nM) | 120 (nM) |
| 24-well (μL/well) | 1.25 | 2.5 | 3.75 |
| 96-well (μL/well) | 0.25 | 0.5 | 0.75 |
A positive control should also be added to ensure the assay is performed properly. In this example, cells were treated with paclitaxel, causing both apoptosis and proliferation effects as expected. After treatment, cells are incubated for an appropriate period of time to ensure that both the siRNA has had time to downregulate the gene of interest and any resulting downstream effects have occurred. Initial testing is generally required to establish a range of concentrations of siRNA or times relevant for the analysis. Additional cells are treated with the siRNA so that some can be processed for RT-PCR and Western blots to verify knockdown of the transcript and protein of interest. After cells are incubated sufficiently long, the protocol for identifying changes in apoptosis and/or proliferation can begin.
3.2. Staining Cells With YO-PRO-1 and Hoechst
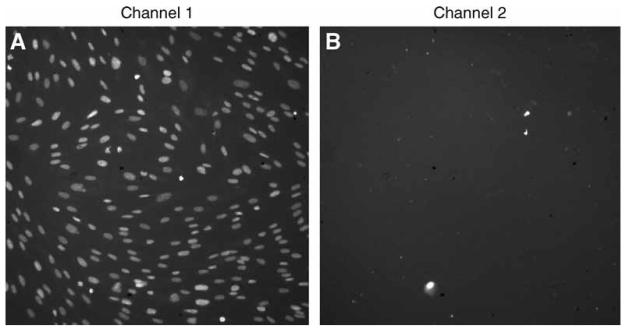
YO-PRO-1 iodide permits analysis of apoptotic cells without interfering with cell viability (14). Cells stained with YO-PRO-1 are counter stained with Hoechst 33342, a nucleic acid stain permeant in both live and dead cells, whereas YO-PRO-1 is only permeant in cells that are beginning to undergo apoptosis (15–17). Therefore, apoptotic cells fluoresce both green and blue, whereas live cells fluoresce only blue (see Fig. 1A,B). This section describes staining of cells with YO-PRO-1 iodide to measure apoptosis and imaging the plates using the KSR.
Fig. 1.
Images generated by the KineticScan® HCS Reader. Cells were plated on 96-well plates, transfected with siRNA, incubated for 48 h, and then stained with Hoechst and YO-PRO®-1. Plates were transported to the KineticScan for image generation, then automated analysis was performed on the collected images. Each nuclei imaged by the KSR is identified with the Cell Health Profiling BioApplication software in (A) the blue channel by Hoechst staining or (B) the green channel by YO-PRO-1 staining. Measurements of intensity and area, among others, are then made using the Cell Health Profiling algorithm. (Please see the companion CD for the color version of this figure.)
3.2.1. Staining Cells With YO-PRO®-1
After incubation with the siRNA or other reagents is completed the cells are then stained with YO-PRO-1 and Hoechst 33342 (see Note 4). Approximately 10 mL of the appropriate media containing these DNA stains is required per 96-well plate. The following steps are performed to complete the staining of the cells.
1. Warm the appropriate media to 37°C in a water bath. Use the same media that was used to propagate the cells.
2. Thaw YO-PRO-1 and Hoechst.
3. Transfer 10 mL of prewarmed media per 96-well plate to a 50-mL conical tube.
4. Add 20 μL of 10 mg/mL YO-PRO-1 per 10 mL media.
5. Add 20 μL of 10mg/mL Hoechst per 10 mL media. This will make a 2X solution for staining.
6. In a laminar flow hood transfer the media containing YO-PRO-1 and Hoechst into a solution basin for transfer into 96-well plates.
7. Using a multichannel pipet, gently transfer 100 μL of the dye plus media mixture into each well.
8. Rock the plate gently to mix and then transfer back into a 37°C incubator with 5% CO2 for 30 min (see Note 5).
9. After 30 min the plate is immediately imaged using the Cellomic's KSR or the ArrayScan.
3.2.2. Imaging 96-Well Plates
Plates are immediately imaged using the KSR system. This high content analysis system scans plates by imaging a designated number of fields in each well of a 96-well plate with user designated filter sets. The XF100 filter set is used for this protocol, although any filter set that allows the visualization of FITC and Hoechst can be used. The user also designates the wells to be imaged. In this assay, two images are collected per field, one in the blue channel image (Hoechst) and one in the green channel image (YO-PRO-1). Before beginning the scan, plates should be examined using an inverted light microscope to assess the density of cells. The density of the cells will help determine how many fields per well should be imaged with a selected objective. Typically, the ×10 objective is used and 5–10 fields per well are imaged. If the cell density is low up to 30 fields can be imaged in each channel.
Images are acquired by the KSR and then analyzed using the Cellomics, Inc. BioApplication® software, in two separate steps. This is in contrast to the ArrayScan, in which these two processes are performed simultaneously. Image acquisition is executed as specified by the manufacturer of the instrument (Cellomics, Inc.). Briefly, the instrument and light source are turned on 10 min before the end of the last incubation of when using the YO-PRO1 staining protocol. The user then double clicks on the Cellomics, Inc. KineticScan icon and types in a username and password. The Cell Health Profiling® protocol is selected for this assay and modified, as needed, according the manufacturer's instructions. The main modifications required are (1) the length of exposure for each channel, dependent on the intensity of staining, and (2) the wells to be imaged. Because the images are analyzed after they are collected, it is important to ensure they are saved to disk in the final dialog box of the scanning software. After the scan is completed the plate is removed. The images are then analyzed as discussed in Subheading 3.4. Results from a typical experiment are graphed (see Fig. 2).
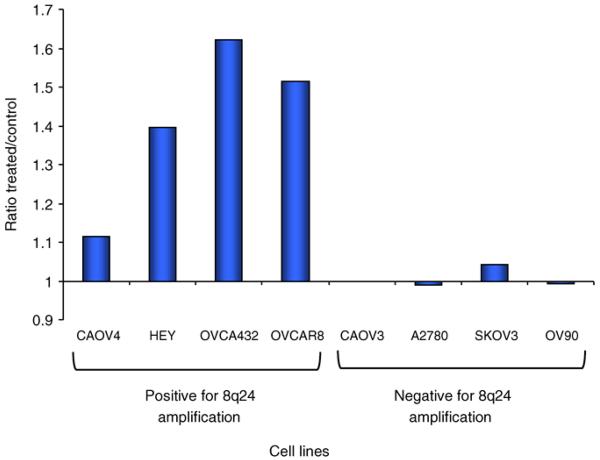
Fig. 2.
Results of YO-PRO®-1 staining in ovarian cancer cells. Amplification of 8q24 (amplified in 50% of ovarian cancers) is associated with poor survival. siRNA was developed against a gene in this region and used to transfect ovarian cancer cells, both negative and positive for amplification of this region. Staining with YO-PRO-1 shows that treatment with siRNA against this selected gene was demonstrated to induce apoptosis in four overexpressing cell lines compared to lipofectamine treated cells, whereas not inducing significant apoptosis in three of the four negative cell lines. (Please see the companion CD for the color version of this figure.)
3.3. Staining Cells With Incorporated BrdU
The protocol used for examining cell proliferation is based on protocols typically used for flow cytometry (18). Cells are pulsed with a 1 mM solution of BrdU before they are fixed and stained for BrdU and DNA content. Only cells that are actively replicating DNA incorporate BrdU into the newly synthesized DNA, providing a measurement of the number of cells in S-phase during the pulse. Because BrdU is a derivative of uridine that replaces thymidine in replicating DNA, it provides an indication of the rate of cell proliferation. The duration of the pulse with BrdU can vary depending on the proliferation rate of the cells. The typical time for incubation with media containing BrdU is 30 min, however if the cells proliferate rapidly less time is required. In this protocol a mouse monoclonal anti-BrdU antibody is used and its location is visualized with an antimouse AlexaFluor®488 antibody (see Fig. 3B). Cells are counterstained with propidium iodide to provide an indication of total DNA content and for object identification (see Fig. 3A).
Fig. 3.
Images generated by the KineticScan® HCS Reader. Cells were plated on 96-well plates, transfected with siRNA, incubated for 48 h, and then fixed. They were then stained with propidium iodide and the monoclonal antibody for BrdU followed by an antimouse AlexaFluor®488. Plates were transported to the KineticScan for image generation, then automated analysis was performed on the collected images as described in the text. Each nuclei imaged by the KSR is identified with the Cell Health Profiling Bioapplication Software in (A) the red channel by PI staining or (B) the green channel by AlexaFluor 488 staining. This staining indicates the cells that underwent DNA replication during the pulse with BrdU.
3.3.1. Pulsing Cells With BrdU and Fixing
1. Add BrdU directly to cell media to achieve a final concentration of 10 μM (see Note 6).
2. Pulse for 30 min. If the cells proliferate more rapidly or slowly than typical tissue culture cells the length of pulse can be determined empirically and adjusted accordingly. For example, when using rapidly proliferating cells the pulse can be as short as 5 min to obtain labeling of a significant percentage of the cells. However, for most cells 30 min is optimal. Place cells at 37°C in 5% CO2 during the pulse.
3. Remove media. It is preferable to use a multichannel pipet for this step, because it is gentler than an aspirator and fewer cells are lost.
4. Add enough cold 70% ETOH to cover cells. With a 96-well plate 100 μL is sufficient. The 70% ethanol should be stored at −20°C.
5. Place cells covered, at 4°C until ready to stain. The incubation should be at least 1–2 h, but we have stored cells in 70% ethanol for several days and have obtained adequate results.
3.3.2. Staining Cells With Incorporated BrdU
1. Remove 70% ETOH and allow cells to air-dry for 1–2 min. Cells should not be allowed to dry completely.
2. Add 0.07 N NaOH to denature the DNA and incubate at RT for 3 min. This will allow the incorporated BrdU to be accessible to an anti-BrdU antibody.
3. Remove NaOH and add 100–200 μL of PBS to neutralize the base.
4. Mix anti-BrdU with PBS/Tween at a 1/100 dilution. For one 96-well plate, at least 5 mL of PBS/Tween is required, containing 50 μL of the mouse anti-BrdU antibody.
5. Gently add 50 μL of anti-BrdU dilution per well to cells (see Note 7). Incubate 45 min at room temperature.
6. Wash twice with PBS/Tween. Do this step gently so that the cells remain attached to the plate.
7. Mix the antimouse AlexaFluor 488 antibody in PBS/Tween at 1/500. For a 96-well plate use 5 mL PBS/Tween and then add 10 μL of the antimouse antibody AlexaFluor 488. Protect the mixture from light.
8. Add 50 μL to each well, and incubate for 30–60 min, in the dark, at room temperatu.
9. Wash three times with PBS/Tween.
10. Incubate cells for 5 min in 0.5 μg/mL PI in PBS.
11. Wash cells 1X with PBS. Add 200 μL PBS, seal with clear and adhesive plate sealing film, and either scan immediately or place the plate at 4°C until ready to scan.
3.3.3. Imaging 96-Well Plates
The KSR® system scans plates by imaging a designated number of fields in each well of the plate, with a user designated objective and filter set, in a similar manner to the YO-PRO-1 staining previously described in Subheading 3.2.2. Because the cells in this assay are fixed the plate can be stored at 4°C for several days or imaged immediately. In this particular assay, two images will be collected per field, one red channel image (PI) and one green channel image (AlexaFluor 488). Before beginning the scan, as described in Subheading 3.2.2., the plates should be examined using an inverted light microscope to assess the density of cells to determine the number of fields imaged per well. In this protocol, the XF93 filter set and ×10 objective are typically used and 5–10 fields are imaged unless the density is low. With lower cell densities, a greater number of fields can be imaged. The software should be set so that the first channel is set for Texas Red and used for object identification. This is because PI stained nuclei can be visualized in this channel and all cells are stained with PI.
As described in Subheading 3.2.2. images are acquired by the KSR and then analyzed using a Cellomics Bioapplication as a separate step. Cell health profiling is the bioapplication used for this assay and also, modified as needed according the manufacturer's instructions as in the previous protocol. Typical images acquired using this protocol are shown (see Fig. 3A,B). After the scan is complete the software is used to eject the plate. The images are then analyzed as described in the next section.
3.4. Processing of the Acquired Images
Described below are the two different methods we use for (1) analyzing the plate stained using YO-PRO-1 and (2) the plate stained using the anti-BrdU and secondary antibodies. When staining with YO-PRO-1, the essential information is the percentage of the total number of cells stained with YO-PRO-1 over a certain threshold of staining intensity. In addition, the intensity of the Hoechst staining in this assay can also be used as an indicator of apoptosis, because nuclei condense while undergoing apoptosis. This causes a greater average intensity of fluorescence within the apoptotic nuclei.
When cells are stained with PI and anti-BrdU, the fluorescence intensity for both of the dyes is important. This is because the total DNA content and the amount of DNA that has incorporated BrdU in each nucleus are both important pieces of information for the final analysis.
3.4.1. Image Analysis of Hoechst and YO-PRO 1 Stained Cells
The Cell Health Profiling Bioapplication (Cellomics, Inc.) is used to analyze the images obtained with the KSR, using manufacturer's instructions, with a few modifications. It is important to have enough cells to obtain statistically significant results, but not so many cells that they become clumped and do not allow the image analysis software to adequately identify individual nuclei (see Note 8). For initial optimization of the image analysis software, the Hoechst stained nuclei images from channel 1 are used to define individual nuclei.
In this part of the optimization it is important to identify all of the nuclei that are stained with Hoechst and to distinguish individual nuclei. In order to start this process a single typical image of Hoechst stained nuclei is selected from the collection of images in an experiment. In order to optimize both object selection and discrimination, the user can alter several settings within the software. There are many examples of this type of optimization in the documentation provided by the developer of the software (Cellomics, Inc.).
Briefly, the first parameter optimized in the image analysis software, allows all the present nuclei to be identified as an object for analyses in addition to the area of the object in which the algorithm will be applied. The algorithm we use the most frequently for this purpose is the Isodata threshold object identification method, which selects a certain percentage of the brightest pixels in an image. If nuclei are not brightly stained the fixed threshold object identification method can be used. In optimizing the Isodata threshold algorithm, we apply numbers between −0.2 and −0.8, depending on the intensity and uniformity of the Hoechst staining of the nuclei. Larger values result in the rejection of dimmer pixels in the image. The optimal setting for this algorithm is ultimately determined empirically by using several different numbers, and each time a new number is chosen, the algorithm is applied to the selected image. Once a value is chosen, several images should be tested with this algorithm to insure uniform results. More details are found in manufacturer's protocol entitled “ArrayScan HCS Reader: Cell Health Profiling BioApplication Guide.” This is either provided with the software or can be obtained online at www.clubhcs.com after registering.
In order to select individual nuclei, optimizing the image processing parameters of background correction, object smooth factor, and object segmentation in this software can minimize the effect of clumping on the final analysis. Instructions and examples of how changing these parameters can affect the image analysis can also be found in the manufacturer's protocol. In general these parameters are optimized using images from both a negative and positive control and once parameters are optimized they are tested using images from a subset of the experimental wells to ensure that visual observations are reflected in the results of the image analysis.
For measurements of apoptosis, the desired information is the percentage of cells stained with YO-PRO-1, because this provides a measurement of the portion of cells that have become permeable to the nucleic acid stain. The cell health-profiling algorithm allows measurements of staining intensity in both the nucleus (circ) and the cytoplasm (ring) (see Fig. 4). Because YO-PRO-1 is a nucleic acid stain, measurements of staining intensity are only required for the nucleus, so the algorithm is set with a zero ring width. Once acceptable parameters are selected for the image analysis software that adequately identifies individual nuclei using channel 1, then image selection parameters in channel 2 are modified so that only stained nuclei are selected for analysis. In general, the only parameter that needs to be modified here is the average intensity threshold. The threshold should be set using both negative and positive controls so that only clearly stained nuclei are selected. This number will be reflected in the final analysis as the percentage selected. It is this number that is compared between control and experimental wells.
Fig. 4.
Example of image analysis performed by the Cell Health Profiling BioApplication. The image is identified in channel 1 and a circle is placed around the nucleus. The area within this circle is termed circ. Using the image in channel 2, a second circle is drawn a specified number of pixels from the first circle (circ) and this is termed ring. Separate measurements of fluorescent intensity can be obtained for both of these regions of the cell. Because the stains that are used in this protocol are nucleic acid specific, the analysis of the region termed ring is not necessary.
3.4.2. Image Analysis of PI and BrdU Antibody Stained Cells
The image analysis software that we use to analyze these images is also the Cell Health Profiling Bioapplication. The same basic tenets outlined in Subheading 3.4.1. for identifying individual nuclei apply in this section. The only difference is that total intensity measurements are important, so it is necessary to ensure that the entire nucleus is identified and circled. In this case initial optimization is done using the PI stained nuclei from channel 1. If some cells are clumped and the software is unable to identify them as separate objects, they can be removed from the final analysis by altering the object selection parameters in channel 1. Parameters that can be changed to eliminate these include total intensity, area, and shape. Nucleus area is the most reliable parameter to vary in our hands, because this is relatively constant from individual cell to individual cell and if the cells are clumped this number increases as a function of the number of nuclei in the clump.
In this particular protocol, all of the objects identified and selected in channel 1 should be selected and identified in channel 2. This is because the total intensity of anti-BrdU antibody staining in channel 2 is the important measurement, not the presence or absence of staining. The negative controls for anti-BrdU antibody staining should include wells that have been stained with only the secondary antibody to access the intensity of the nonspecific staining. The total intensity measurement of these nuclei should be extremely low and significantly different from positively stained nuclei. Total intensity measurements are obtained for both the PI and the anti-BrdU antibody staining, and these are plotted to obtain information about cell proliferation (see Fig. 5). As an alternative, the percentage of cells in S-phase, during the BrdU pulse can be used as an indication of cellular proliferation. This would require object selection in channel 2 as in Subheading 3.4.1.
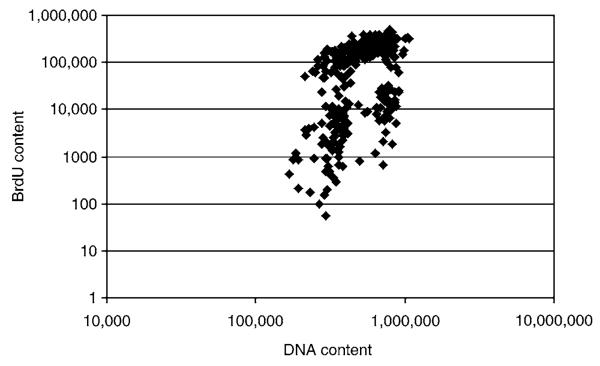
Fig. 5.
Measurement of cell proliferation after treatment with siRNA. Cells were plated on 96-well plates, treated, pulsed with BrdU, fixed, and then stained with anti-BrdU and PI. Plates were transported to the KSR for image collection and then automated analysis was performed on the collected images. This is a typical scatter plot of BrdU staining intensity vs PI intensity. This is used for calculating the number of cells in G0/G1, S, and G2/M phases.
3.5. Data Analysis
Analysis of data is different for each type of staining and will be described separately.
3.5.1. Analysis of YO PRO 1 Stained Cells
As stated in Subheading 3.4.1. the important measurement is the percentage of cells stained by YO-PRO-1. These numbers are obtained by opening the files using Cellomics, Inc. vHCS View software. For this protocol, the parameters that are typically downloaded into an Excel spreadsheet include the total number of Hoechst stained nuclei, the average intensity of the Hoechst staining, and the percentage of nuclei selected in channel 2. Once these numbers are downloaded into Excel they can be graphed using a program such as GraphPad Prism® to allow curve fitting and basic statistical analysis. Standard deviations are determined for the experimental results using the averaged values from four independent wells. High standard deviations for an experiment might indicate that the image analysis software was not optimized for this application, high background, or uneven staining. If the variability is high, individual images should also be examined for evidence of software or hardware failure.
3.5.2. Analysis of Anti-BrdU Antibody Stained Cells
As stated in Subheading 3.4.2. it is important to obtain fluorescent intensity measurements for both PI staining and the AlexaFluor 488 staining of individual nuclei. The data is accessed using Cellomics, Inc. vVHS view software and instead of downloading average values for each well, intensity measurement both channels in each nucleus are downloaded. From these measurements, a bivariate distribution of DNA content (PI staining) Vs BrdU content (AlexaFluor 488 staining) can be generated on a scatter plot and analyzed for the proportion of cells in G1, S, and G2/M phases of the cell cycle (see Fig. 5). This is performed as described in ref. 19. A simple Student t-test can also be used to assess if there is a significant difference between the BrdU staining in the control vs the experimental.
Acknowledgments
The authors thank Dr. Gordon Mills for supplying many of the cell lines used in these studies.
Footnotes
A hemocytometer can also be used to count cells, but the process is longer and subject to greater variability than the results obtained with the Coulter Counter.
Cell lines must be selected based on the question that is being answered. For this application the question was whether or not the presence or absence of an amplification of a specific chromosomal amplification had an effect on cell proliferation and apoptosis. Cell lines were chosen that contain this amplification (positive) and cell lines that do not contain this amplification (negative). The experiment was then to knock down expression of a particular gene in this region with siRNA to determine if it plays an important role in cell proliferation or apoptosis. Because cancer is associated with the deregulation of these processes, it is important to understand the effect of overexpression of specific genes in this region.
When using larger cells, such as lung or skin fibroblasts, we typically reduce the number to 2500–3000 of cells per well in a 96-well plate. With this particular protocol, only the nucleus is used in the image analysis, therefore, 3000 cells should be a good starting point when evaluating larger cells in this protocol. Before doing this protocol on a large scale with a particular cell line it is a good practice to plate various numbers of cells in the wells of a 96-well plate, spanning 3000, and then check cell growth at the time-point they will be imaged in the final assay. It is important to avoid clumping of cells as much as possible, and this is achieved by mixing well before plating and keeping the density low enough so individual cells can be clearly differentiated from each other. This becomes important when performing image analysis, because the program analyzes individual nuclei. When cells are very close together the image analysis software might recognize several nuclei as one nucleus, impacting the final results.
Because the Hoechst is visualized in the blue channel and the YO-PRO-1 is visualized in the green channel, PI can also be added as a nucleic acid stain to identify necrotic cells. This would be visualized in yet a third channel. The only modification to the protocol is to add PI at a final concentration of 1 μg/mL to the staining mixture containing YO-PRO-1 and Hoechst. The filter set used would then be XF93 and the Texas Red channel would be added to the analysis.
Staining of the nuclei with YO-PRO-1 occurs almost immediately, but a short amount of time is required to obtain the desired intensity of staining with Hoechst.
During this step the media should not be changed because this could affect the growth rate of the cells. Growth media becomes “conditioned” after cells have been grown in it for a period of time, because it contains extracellular factors and metabolites secreted from the cells in the media, in addition to the original components of the media. These extracellular factors might include growth factors or other molecules that effect the proliferation of cells. We typically make our standard solution as 1 mM BrdU in PBS and add 1 μL to the 100 μL of media in each well of a 96-well plate.
If a 24-well plate is used add 100 μL and rock the plate gently back and forth on a rocker so that all cells are exposed to the antibody mixture. This can also be applied to the secondary antibody.
This is the step in which the even distribution of the cells makes the most difference. It is important, especially for the YO-PRO-1 staining, that individual nuclei can be distinguished. This is because have an accurate count of the total number of nuclei affects the accuracy of the results. The most important information obtained from this analysis is the percentage of nuclei stained with YO-PRO-1, indicating the number of cells undergoing apoptosis. The percentage obtained might be artificially high, if clumps of cells are not distinguished from one another. The image analysis Cell Health Profiling Bioapplication Software has some features that allow it to distinguish individual nuclei in clumps, but it limited.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat. Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- 3.Silva J, Chang K, Hannon GJ, Rivas FV. RNA-interference-based functional genomics in mammalian cells: reverse genetics coming of age. Oncogene. 2004;23:8401–8409. doi: 10.1038/sj.onc.1208176. [DOI] [PubMed] [Google Scholar]
- 4.Singer O, Yanai A, Verma IM. Silence of the genes. Proc. Natl Acad. Sci. USA. 2004;101:5313–5314. doi: 10.1073/pnas.0401209101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dolbeare F, Gratzner H, Pallavicini MG, Gray JW. Flow cytometric measurement of total DNA content and incorporated bromodeoxyuridine. Proc. Natl Acad. Sci. USA. 1983;80:5573–5577. doi: 10.1073/pnas.80.18.5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wronski R, Golob N, Grygar E, Windisch M. Two-color, fluorescence-based microplate assay for apoptosis detection. Biotechniques. 2002;32:666–668. [PubMed] [Google Scholar]
- 7.Kiechle M, Jacobsen A, Schwarz-Boeger U, Hedderich J, Pfisterer J, Arnold N. Comparative genomic hybridization detects genetic imbalances in primary ovarian carcinomas as correlated with grade of differentiation. Cancer. 2001;91:534–540. [PubMed] [Google Scholar]
- 8.Patael-Karasik Y, Daniely M, Gotlieb WH, et al. Comparative genomic hybridization in inherited and sporadic ovarian tumors in Israel. Cancer Genet. Cytogenet. 2000;121:26–32. doi: 10.1016/s0165-4608(00)00224-7. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki S, Moore DH, 2nd., Ginzinger DG, et al. An approach to analysis of large-scale correlations between genome changes and clinical endpoints in ovarian cancer. Cancer Res. 2000;60:5382–5385. [PubMed] [Google Scholar]
- 10.Collins C, Volik S, Kowbel D, et al. Comprehensive genome sequence analysis of a breast cancer amplicon. Genome Res. 2001;11:1034–1042. doi: 10.1101/gr.174301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinkel D, Segraves R, Sudar D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat. Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 12.Iwabuchi H, Sakamoto M, Sakunaga H, et al. Genetic analysis of benign, low-grade, and high-grade ovarian tumors. Cancer Res. 1995;55:6172–6180. [PubMed] [Google Scholar]
- 13.Lapuk A, Volik S, Vincent R, et al. Computational BAC clone contig assembly for comprehensive genome analysis. Genes Chromosomes Cancer. 2004;40:66–71. doi: 10.1002/gcc.20016. [DOI] [PubMed] [Google Scholar]
- 14.Idziorek T, Estaquier J, De Bels F, Ameisen JC. YOPRO-1 permits cytofluorometric analysis of programmed cell death (apoptosis) without interfering with cell viability. J. Immunol. Methods. 1995;185:249–258. doi: 10.1016/0022-1759(95)00172-7. [DOI] [PubMed] [Google Scholar]
- 15.Daly JM, Jannot CB, Beerli RR, Graus-Porta D, Maurer FG, Hynes NE. Neu differentiation factor induces ErbB2 down-regulation and apoptosis of ErbB2-overexpressing breast tumor cells. Cancer Res. 1997;57:3804–3811. [PubMed] [Google Scholar]
- 16.Estaquier J, Idziorek T, Zou W, et al. T helper type 1/T helper type 2 cytokines and T cell death: preventive effect of interleukin 12 on activation-induced and CD95 (FAS/APO-1)-mediated apoptosis of CD4+ T cells from human immunodeficiency virus-infected persons. J. Exp. Med. 1995;182:1759–1767. doi: 10.1084/jem.182.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Estaquier J, Tanaka M, Suda T, Nagata S, Golstein P, Ameisen JC. Fas-mediated apoptosis of CD4+ and CD8+ T cells from human immunodeficiency virus-infected persons: differential in vitro preventive effect of cytokines and protease antagonists. Blood. 1996;87:4959–4966. [PubMed] [Google Scholar]
- 18.Gray JW, Dolbeare F, Pallavicini MG, Beisker W, Waldman F. Cell cycle analysis using flow cytometry. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1986;49:237–255. doi: 10.1080/09553008514552531. [DOI] [PubMed] [Google Scholar]
- 19.Dolbeare F, Selden JR. Immunochemical quantitation of bromodeoxyuridine: application to cell-cycle kinetics. Methods Cell Biol. 1994;41:297–316. [PubMed] [Google Scholar]