Summary
MicroRNAs (miRNAs) play critical roles in the regulation of gene expression. However, since miRNA activity requires base pairing with only 6-8 nucleotides of mRNA, predicting target mRNAs is a major challenge. Recently, high-throughput sequencing of RNAs isolated by crosslinking immunoprecipitation (HITS-CLIP) has identified functional protein-RNA interaction sites. Here we use HITS-CLIP to covalently crosslink native Argonaute (Ago) protein-RNA complexes in mouse brain. This produced two simultaneous datasets—Ago-miRNA and Ago-mRNA binding sites—that were combined with bioinformatic analysis to identify miRNA-target mRNA interaction sites. We validated genome-wide interaction maps for miR-124, and generated additional maps for the 20 most abundant miRNAs present in P13 mouse brain. Ago HITS-CLIP provides a general platform for exploring the specificity and range of miRNA action in vivo, and identifies precise sequences for targeting clinically relevant miRNA-mRNA interactions.
Sophisticated mechanisms regulating RNA may explain the gap between the great complexity of cellular functions and the limited number of primary transcripts. Regulation by microRNAs (miRNAs) underscore this possibility, as miRNAs are each believed to directly bind to many mRNAs to regulate their translation or stability1,2, and thereby control a wide range of activities, including development, immune function and neuronal biology3-5. Many miRNAs are evolutionarily conserved, although others are species-specific (including human miRNAs not conserved in chimpanzees)6, consistent with roles ranging from generating cellular to organismal diversity.
Despite their biologic importance, determining miRNA targets is major challenge. The problem stems from the discovery that functional mRNA regulation requires interaction with as few as 6 nucleotides (nt) of miRNA seed sequence7. Such 6-mers are present on average every ∼4 kb, so that miRNAs could regulate a broad range of targets, but the full extent of their action is not known. Bioinformatic analysis has greatly improved the ability to predict bona fide miRNA binding sites8-10, principally by constraining searches for evolutionary conserved seed matches in 3′ UTR. Nonetheless, different algorithms produce divergent results with high false positive rates3,11,12. In addition, many miRNAs are present in closely related miRNA families, complicating interpretation of loss of function studies in mammals13,14, although such studies have been informative for several miRNAs3,15-17. miRNA overexpression or knockdown studies, most recently in combination with proteomic studies11,18, have led to the conclusion that individual miRNAs generally regulate a relatively small number of proteins at modest levels (< 2-fold), although the false-positive rate of target predictions remains high (∼up to 66%)11, and the data sets analyzed have been of limited size (∼5,000 proteins). Similar high false-positive rates have been observed when miRNAs were co-immunoprecipitated with Ago proteins19-23. A critical caveat common to all of these studies is their inability to definitively distinguish direct from indirect miRNA-target interactions. At the same time, as therapeutic antisense strategies become more viable17,24,25, knowledge of direct miRNA target sites has become increasingly important.
Recently, we developed HITS-CLIP to directly identify protein-RNA interactions in living tissues in a genome-wide manner26,27. This method28,29 uses UV-irradiation to covalently crosslink RNA-protein complexes that are in direct contact (∼single Angstrom distances) within cells, allowing them to be stringently purified. Partial RNA digestion reduces bound RNA to fragments that can be sequenced by high throughput methods, yielding genome wide maps and functional insights26,30. Recent X-Ray crystal structures of an Ago-miRNA-mRNA ternary complex31 suggest that Ago may make sufficiently close contacts to allow Ago HITS-CLIP to simultaneously identify Ago bound miRNAs and the nearby mRNA sites. Here we use Ago HITS-CLIP to define the sites of Ago interaction in vivo, decoding a precise map of miRNA-mRNA interactions in the mouse brain. This provides a platform that can establish the direct targets upon which miRNAs act in a variety of biologic contexts, and the rules by which they do so.
Ago RNA targets in the mouse brain
HITS-CLIP experiments rely on a means of purifying RNA binding proteins (RNABPs)26-29. To purify Ago bound to mouse brain RNAs, we UV-irradiated P13 neocortex and immunoprecipitated Ago under stringent conditions. After confirming the specificity of Ago immunoprecipitation (Fig. 1a), we radiolabeled RNA, further purified crosslinked Ago-RNA complexes by SDS-PAGE and nitrocellulose transfer, and visualized them by autoradiography. We observed complexes of two different modal sizes (∼110 kD and ∼130kD; Fig. 1b and Supplementary Fig. 1), suggesting that Ago (97kD) was crosslinked to two different RNA species. RT-PCR amplification revealed that the ∼110 kD band harbored ∼22 nt crosslinked RNAs and upper band both 22 nt and larger RNAs (Fig. 1c). These products were sequenced with high throughput methods26 and found to correspond to miRNAs and mRNAs, respectively (Supplementary Table 1), suggesting that Ago might be sufficiently close to both miRNA and target mRNAs to form crosslinks to both molecules in the ternary complex (Fig. 1d). Such a result would allow the search for miRNA binding sites to be constrained to both the subset of miRNAs directly bound by Ago-CLIP and to the local regions of mRNAs to which Ago crosslinked, potentially reducing the rate of false positive predictions of miRNA binding.
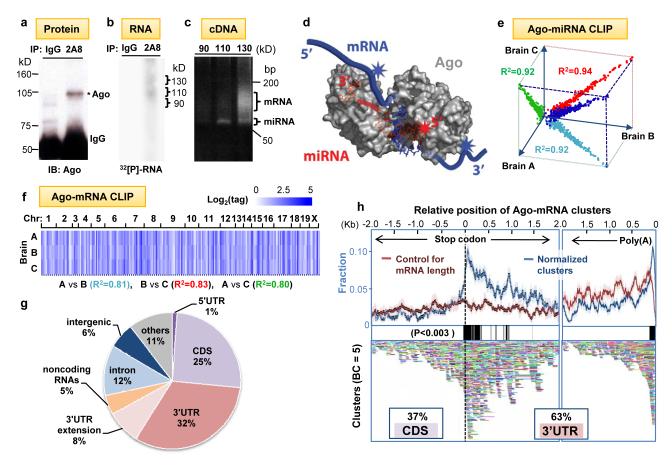
Fig. 1. Argonaute HITS-CLIP.
(a) Immunoblot (IB) analysis of Ago immunoprecipitates (IP) from P13 mouse neocortex using pre-immune IgG as a control or anti-Ago monoclonal antibody 2A8 were blotted with 7G1-1* antibody (Supplementary Methods). (b) Autoradiogram of 32P-labeled RNA crosslinked to mouse brain Ago purified by IP. RNA-protein complexes of ∼110 kD and ∼ 130 kD are seen with 2A8 but not control IP. (c) PCR products amplified after linker (36 nt) ligation to RNA products excised from (b). Products from the 110 kD RNA-protein complex were ∼22 nt miRNAs, and those from 130 kD complexes were predominantly mRNAs. (d) Illustration showing proposed interpretation of data in (c). Ago (drawn based on structure 3F73 in PDB)31 binds in a ternary complex to both miRNA and mRNA, with sufficiently close contacts to allow UV-crosslinking to either RNA; mRNA tags will be in the immediate vicinity of miRNA binding sites. (e-f) Reproducibility of all Ago-miRNA tags (e; graphed as log2(normalized miRNA frequency) per brain) or all tags within Ago-mRNA clusters (f; see Supplementary Figs. 2-3). We estimated that sequencing depth was near saturation (Supplementary Fig. 16). (g) Location of reproducible Ago-mRNA tags (tags in clusters; BC≥2) in the genome. Annotations are from RefSeq; ”others” are unannotated EST transcripts, non-coding RNAs are from lincRNAs50 or FANTOM3. (h) Top panel: The position of robust Ago-mRNA clusters (BC = 5) in transcripts is plotted relative to the stop codon and 3′ end (presumptive poly(A) site, as indicated). Data is plotted as normalized density relative to transcript abundance for Ago-mRNA clusters (blue) or control clusters (red), (standard deviations are shown in light colors; see Supplementary Methods). Regions with significant enrichment relative to control are indicated with black bars (> 3 standard deviations; P<0.003). Cluster enrichment ∼1kb downstream from stop codon appears to be due to a large number of transcripts with ∼1kb 3′ UTRs (data not shown). Bottom panel: All individual clusters (BC=5) are shown (each is a different color).
To differentiate robust from non-specific or transient Ago-RNA interactions, we compared the results from biologic replicate experiments done with two different monoclonal antibodies (Supplementary Figs. 1-3). The background was further reduced by in silico random CLIP, a normalization algorithm that accounted for variation in transcript length and abundance (Supplementary Figs. 4-5). The set of Ago-crosslinked miRNAs and mRNAs were highly reproducible. Among biologic triplicates or among 5 replicates done with two antibodies, the Pearson correlation coefficient was R2 > 0.9 and > 0.84, respectively, for Ago-miRNA CLIP (Fig. 1e, Supplementary Fig. 2) and R2 ≥ 0.8 and ≥ 0.65, respectively, for Ago-mRNA CLIP (Fig. 1f, Supplementary Fig. 3). We identified 454 unique miRNAs crosslinked to Ago in mouse brain, with Ago-miR-30e being the most abundant species (14% of total tags; Supplementary Fig. 2); these results were consistent with previous estimates assessed by cloning frequency32 or bead-based cytometry33, although the correlation with published results (R2 = 0.2 - 0.32; Supplementary Fig. 6) was not as high as among our biologic replicates. These discrepancies might be due to differences in the ages of brain used, regulation of Ago-mRNA interactions, and/or increased sensitivity allowed by stringent CLIP conditions and consequent improved signal:noise. To facilitate the analysis of large numbers of Ago-mRNA CLIP tags (∼1.5 × 106 unique tags; Supplementary Table 1), we analyzed overlapping tags (clusters)26, which were normalized by in silico random CLIP and sorted by biologic complexity26 (“BC”; a measure of reproducibility between biologic replicates; see Supplementary Figs. 5, 7). 1,463 robust clusters (BC=5; i.e. harboring CLIP tags in all five biologic experiments using both antibodies) mapped to 829 different brain transcripts, and 990 clusters had at least 10 tags (Supplementary Fig. 7).
Ago-mRNA HITS-CLIP tags were enriched in transcribed mRNAs (Fig. 1g). The pattern of tags mirrored the results of functional assays with miRNAs34, which show no biologic activity when seed sites are present in the 5′ UTR (1% Ago-mRNA tags), and high efficacy in 3′ UTRs (40% tags including 8% within 10kb downstream of annotated transcripts, regions likely to have unannotated 3′ UTRs26,35). In addition, an extensive set of tags were identified in other locations, including coding sequence (CDS; 25%), a location for which there is emerging evidence of miRNA regulation36-40, introns (12%), and non-coding RNAs (4%), suggesting that these sites may provide new insights into miRNA biology. Within mRNAs, Ago-mRNA clusters were highly enriched in the 3′ UTR (∼60%; Supplementary Fig. 8), especially around stop codons (with a peak ∼50 nt downstream) and at the 3′ end of transcripts (∼70 nt upstream of presumptive poly(A) sites; P<0.003, Fig. 1h), consistent with bioinformatic observations from microarray data34. Taken together, this data suggested that Ago-mRNA clusters might be associated with functional binding sites.
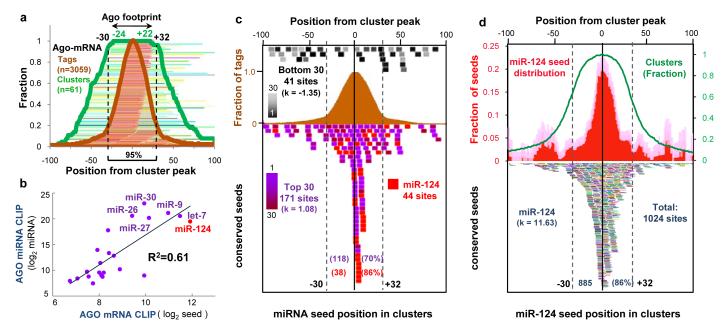
To examine the relationship between Ago-mRNA clusters and potential sites of miRNA action, we first performed an unbiased search for all 6-8 nt sequence motifs within Ago-mRNA clusters using linear regression analysis (Supplementary Methods). The 6 most enriched motifs corresponded to seed sequences present within the most frequently crosslinked miRNAs in Ago-miRNA CLIP (Ago-miRNAs; Supplementary Table 2). The most significant match corresponded to the seed sequence of miR-124, a well-studied brain-specific miRNA (P = 8.3 × 10-58; since miR-124 is only the 8th most frequently crosslinked Ago-miRNA, this data may indicate over-representation of miR-124 seed matches in the genome (Supplementary Table 2-3), or contributions from currently unknown rules of Ago binding). To more precisely define the region of mRNA complexed with Ago, we examined the width of 61 robust Ago-mRNA clusters (BC = 5; total tags > 30) relative to their peaks (determined by cubic spline interpolation; Supplementary Methods). We found that Ago bound within 45-62 nt of cluster peaks ≥ 95% of the time (Fig. 2a), and we defined this region as the average Ago-mRNA footprint.
Fig. 2. Distribution of mRNA tags correlates with seed sequences of miRNAs from Ago CLIP.
(a) Ago-mRNA cluster width and peaks. The peaks of 61 robust clusters (BC = 5, peak height >30, with single peaks) were determined, and the position of tags (brown lines and fraction plotted as brown graph) and width of individual clusters (green lines and fraction plotted as green graph) are shown relative to the peaks (Supplementary Methods). The minimum region of overlap of all clusters (100%) was within -24 and +22 nt of cluster peaks, and ≥ 95% were within -30 and +32 nt, suggesting that the Ago footprint on mRNA spans 62 nt (or, more stringently, 46 nt). (b) A linear regression model was used to compare miRNA seed matches enriched in the stringent Ago footprint region with the frequency of miRNAs experimentally determined by Ago-miRNA HITS-CLIP (see also Supplementary Fig. 9). (c) The position of conserved core seed matches (position 2-7) from the top 30 Ago-bound miRNAs (independent of peak height; purple colors, including miR-124, red; or, bottom 30 miRNAs, expressed at extremely low levels in brain (Supplementary Fig. 2); black to gray colors) are plotted relative to the peak of 134 robust clusters (BC = 5, peak height >30). 118 of 171 seed matches are within the Ago 62 nt footprint (d) The position of conserved miR-124 seed matches (bottom panel; each is represented by a different color) were plotted relative to the peak position of all Ago-mRNA clusters (BC ≥ 2). Top panel; distribution of mir-124 seed matches (plotted relative to cluster peak, normalized to number of clusters; red graph); pale color indicates standard deviation. Excess kurtosis (k) indicates that seed sites are present in a sharp peak relative to a normal distribution.
Within Ago-mRNA footprints (11,118 clusters; BC≥2) we found a high correlation between the frequency of Ago-miRNAs and the frequency of their seed matches (Fig. 2b and Supplementary Figure 9). The seed matches were near the peaks of 134 robust clusters (Fig. 2c; Supplementary Table 3 and Supplementary Figure 13), and their distribution was leptokurtic (with a higher and sharper peak than a normal distribution; excess kurtosis (k) = 1.08, versus 0 in a normal distribution). As a control, seed matches for a “negative” group of miRNAs (the lowest ranks from the Ago-miRNA list) were uniformly distributed across these clusters (Fig. 2c; k = -1.35, versus -1.2 in uniform distribution). Taken together, these results indicate that the Ago-mRNA footprint is rich in and may predict miRNA binding sites with enhanced specificity over purely bioinformatic approaches (at least a 3-fold improvement in false-positives; Supplemental Methods).
Ago HITS-CLIP and miR-124
To further explore the relationship between the Ago-mRNA footprint and miRNA binding we focused on miR-124 sites. There was a marked enrichment of conserved miR-124 seed matches in Ago-mRNA clusters (BC ≥ 2; Fig. 2d, estimated false-positive rate of 13%; Supplementary Methods). 86% of the predicted miR-124 binding sites were present within the Ago footprint region, again in a tight peak region showing leptokurtic distribution (Fig. 2d; k = 11.63). While some predicted seeds outside of the Ago footprint might correspond to false positives, we noted small secondary peaks at ∼ +/- 50 nt outside the Ago footprint (Fig. 2d), suggesting the possibility of cooperative secondary miR-124 binding sites in some transcripts, consistent with prior data34. Relative to more stringent analyses (Fig. 2C), our analysis at this threshold (BC ≥ 2) was more sensitive and sufficiently specific such that we used it for subsequent analyses (Supplementary Figure 7).
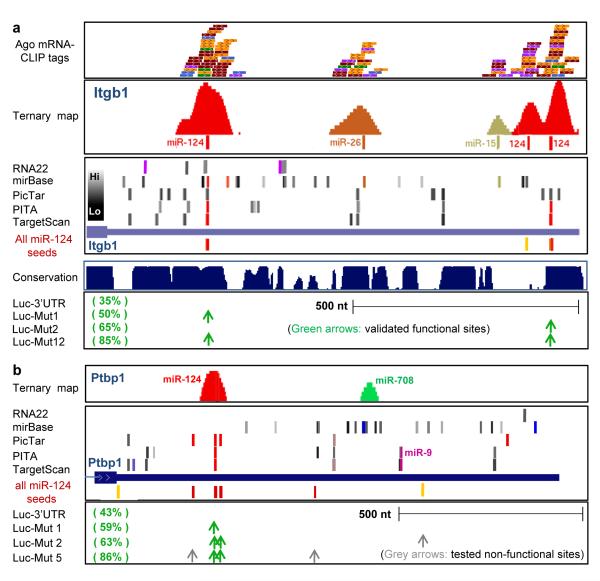
We searched published examples of miR-124 regulated transcripts for Ago-mRNA clusters harboring miR-124 seeds within the predicted 62 nt Ago footprint (referred to as Ago-miR-124 ternary clusters). We identified such ternary clusters in 5 of 5 transcripts in which miR-124 seed sites had been well defined by functional studies (including mutagenesis of seed sequences in full length 3′UTR; Fig. 4 and Supplementary Fig. 10). In each of these transcripts, there were many predicted miRNA target sites in the 3′ UTR relative to the small number of Ago-mRNA ternary clusters found, suggesting that there may be a significant number of false positive predictions from bioinformatic algorithms (Fig. 3 and Supplementary Fig. 10). For example the 3′ UTR of Itgb1 mRNA has ∼50 predicted miRNA target sites, including two miR-124 sites, but only 5 Ago-mRNA ternary clusters (Fig. 3a). Using the Ago footprint to predict which miRNAs bound at these sites (Ago ternary map; Supplementary Methods) we identified three as miR-124 sites, one of which was not predicted computationally because the seed sequence is not conserved (Fig. 3a); similar observations were made in the Ctdsp1 3′ UTR (Supplementary Fig. 10). Previous luciferase assays demonstrated that miR-124 suppression of Itgb1 (to 35% control levels) was partially reversed (to 85% control levels) by mutating both of the two predicted seed sequences41; our observation of an Ago-miR-124 ternary cluster at this third non-conserved site may explain the partial rescue. Conversely, in the Ptbp1 3′ UTR, the absence of any Ago footprint at a predicted miR-9 seed site was consistent with prior studies which found this site to be non-functional42 (Fig. 3b). Additionally, Ptbp1 has seven predicted miR-124 seed sites, of which five were previously tested and only 2 found to be functional in luciferase assays42; only these 2 sites harbored Ago-miR-124 ternary clusters (Fig. 3b).
Fig. 4. Meta-analysis of Ago-mRNA clusters in large-scale screens of miR-124 regulated targets.
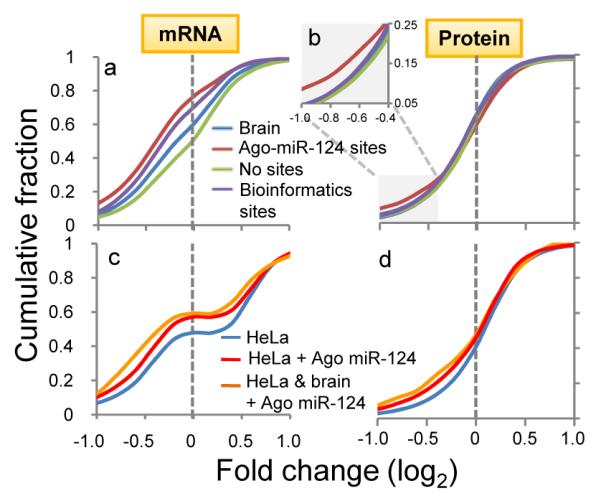
(a) Transcripts with predicted (conserved) miR-124 seeds (purple line) showed miR-124 suppression relative to all transcripts expressed in brain and cell lines (blue line) or those with no miR-124 seed sequences (green line). Transcripts with Ago-miR-124 ternary clusters (containing both miR-124 seed sequences and Ago-mRNA CLIP tags; red line) showed further miR-124 suppression. (b) Similar results were seen when analyzing miR-124 dependent protein suppression (identified by SILAC)11, with discrimination by the presence of Ago-miR-124 ternary clusters especially evident where there were smaller numbers of transcripts showing larger changes (log2<-0.4; inset). (c) Transcripts expressed in miR-124 transfected HeLa cells that harbor new Ago-miR-124 clusters (red line; or a subset of transcripts also harboring Ago-miR-124 clusters in mouse brain; yellow line), compared with previous analysis7 of regulated transcripts in miR-124 transfected HeLa cells. (d) As in (c), plotted for predicted protein levels, compared with prior data11.
Fig. 3. Ago-miRNA ternary clusters in validated miR-124 mRNA targets.
(a) Ago—mRNA CLIP tags (top panel: raw tags, one color per biologic replicate: second panel: ternary map of Ago-mRNA normalized clusters around top predicted miRNA sites) compared with predicted miRNA sites (using indicated algorithms) for the 3′ UTR of Itgb1 (third panel, see Supplementary Methods; colors indicate predicted top 20 miRNAs as in Fig. 5; grey bars indicate miRNAs ranking below the top 20 (per heat-map) in Ago-miRNA CLIP). All predicted miR-124 6-8 mer seeds (conserved (red) or non-conserved (yellow) sites) are shown. Bottom panel shows data from luciferase assays in which mutagenesis of predicted miR-124 seeds at the indicated positions had the indicated effects on restoring from miR-124 mediated suppression (35% of baseline luciferase levels)41. (b) Ago-miRNA ternary maps compared with previously reported functional data for Ptbp142; see Supplementary Fig. 10 for maps of Ctdsp144 and Vamp344.
To extend these observations, we compiled brain-expressed transcripts from a meta-analysis of five published microarray experiments which identified transcripts downregulated following miR-124 overexpression in HeLa and other cell lines (Supplementary Methods)7,11,21,42,43. Brain-expressed transcripts that had predicted 3′ UTR miR-124 seeds were also suppressed at the mRNA and protein level by miR-124 overexpression (Fig. 4), consistent with previous experiments11,18. However, transcripts with Ago-miR-124 ternary clusters had a significantly greater tendency to be downregulated at the mRNA and protein level (P<0.01, Kolmogorov-Smirnov test, Fig. 4). Ago-miR-124 ternary clusters had much greater predictive value (true positive rate = 73%) and specificity (92.5%) than analysis of conserved seed sequences alone (Supplementary Fig. 11). We validated these studies experimentally by examining Ago-mRNA clusters that appeared de novo in HeLa cells (which do not express endogenous miR-124) after miR-124 transfection. Applying this data to the meta-analysis, mRNAs whose 3′ UTRs harbored new AgomiR-124 ternary clusters after miR-124 transfection showed an even greater enrichment in miR-124-dependent changes in transcript (Fig. 4c) and protein (Fig. 4d) levels (P<0.01, Kolmogorov-Smirnov test).
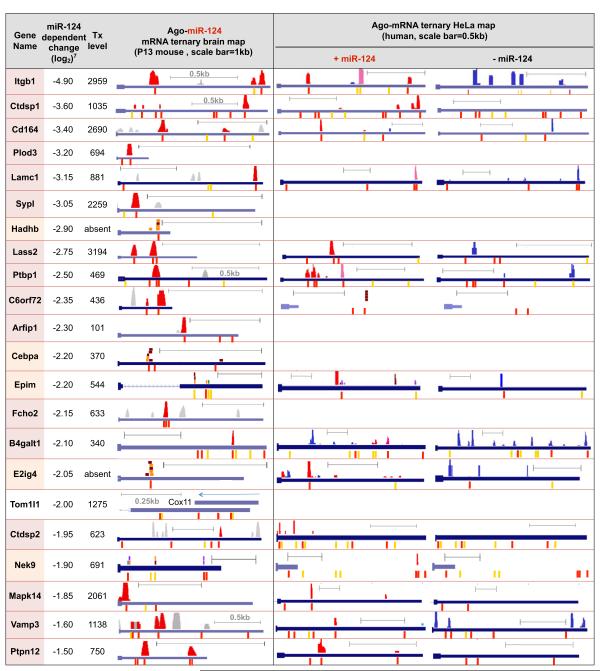
We next examined Ago-miR-124 ternary clusters present in validated individual transcripts identified from among 168 candidate miR-124 regulated transcripts7. These targets were previously analyzed by Hannon and colleagues44 using a rigorous 3-part strategy to experimentally validate 22 of them (although miR-124 binding sites were not generally defined). 16 of 22 harbored Ago-miR-124 ternary clusters in the 3′ UTR (Table 1), and in 5 additional transcripts with low expression levels, Ago-mRNA CLIP-tags were identified at predicted miR-124 seed sites. For transcripts of even moderate abundance (with normalized probe intensities ≥700; average for P13 brain transcripts ∼1,255; Supplementary Methods), we identified all 10/10 predicted targets (Table 1). These data indicate that identifying Ago-miR-124 ternary clusters markedly enhances the sensitivity and specificity of detecting bona fide miR-124 targets.
Table 1. Ago-miR-124 ternary maps in brain and transfected HeLa cells.
Left panel: Comparison of 22 validated miR-124 targets44 with Ago-miR-124 ternary clusters in brain transcripts (BC ≥ 2); miR-124 clusters are shown graphically above each 3′ UTR, as in Fig. 3, and corresponding conserved (red) or non-conserved (yellow) miR-124 seeds below) were identified in 16 of 22 validated transcripts (gene names colored red). In 5 additional transcripts (names colored orange) clusters were below normalization threshold, but individual Ago-mRNA CLIP tags are shown. Tom1l1 had no detectable Ago binding, although notably a brain-expressed antisense transcript (Cox11) is present. Previously reported changes in miR-124 dependent transcript levels from miR-124 transfected HeLa cells7, and levels of the same transcript (“tx”) in mouse brain (normalized probe intensities) are shown. Right panels: Columns represent Ago-miR-124 ternary clusters in HeLa cells transfected with either miR-124 or a control microRNA (“-miR-124”). 17 de novo Ago-miR-124 ternary clusters (red; present only after miR-124 transfection) are shown, together with clusters present in both control and miR-124 transfected cells (purple), or present in control HeLa cells alone (blue).
We examined this set of 22 targets44 for Ago-mRNA clusters that appeared in the 3′ UTR after miR-124 transfection (Table 1). Remarkably, from among many potential miR-124 seed sites, 17 de novo Ago-miR-124 ternary clusters appeared after transfection and 14 of 17 were at precisely the positions predicted from brain Ago miR-124 maps. Genome-wide, a total of 526 of new Ago-miR-124 clusters were detected in transcripts expressed in HeLa cells and mouse brain. These results confirmed that the Ago ternary map identifies functional sites of miRNA regulation.
Predicting miRNA functional networks
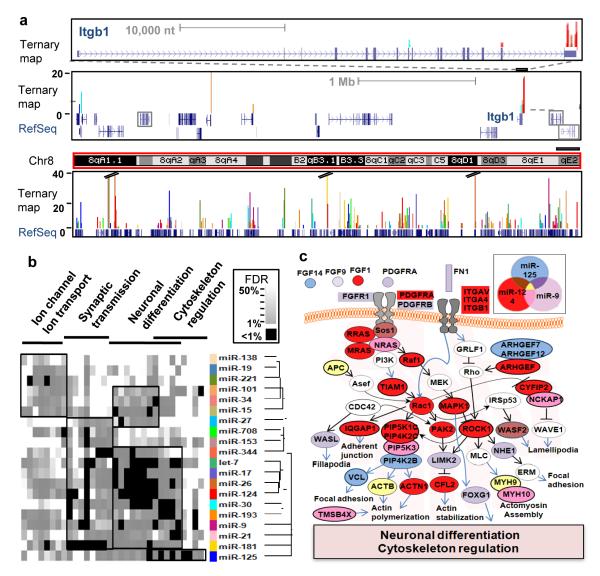
Based on the robust correlation between previously validated miR-124 functional sites and Ago HITS-CLIP, we examined brain Ago-mRNA clusters to predict binding maps for the 20 most abundant Ago-miRNAs (Supplementary Method). These maps (Fig. 5a) show that Ago binds to target transcripts at very specific sites: on average there are only 2.6 Ago-mRNA clusters (BC ≥ 2) per Ago regulated transcript (2.12 per 3′ UTR) and each miRNA binds an average of only 655 targets (Supplementary Figs. 7 and 13). To explore the potential of Ago HITS-CLIP maps to define miRNA regulated transcripts, we examined the functions encoded by the predicted targets of these 20 miRNAs using gene ontology (GO) analysis; comparison of these results with predictions made using GO analysis of TargetScan predictions (Supplementary Fig. 14) demonstrates that the false discovery rate (FDR) and ‘quality’ of the protein network deteriorate substantially when the Ago-mRNA map is not used. Target predictions from the Ago HITS-CLIP map suggest that diverse neuronal functions are regulated by different sets of miRNAs (Fig. 5b). The largest set of miRNA associated functions, “neuronal differentiation” illustrates interwoven but distinct pathways predicted to be regulated by three miRNAs expressed in neurons (Fig. 5c). The Ago-RNA ternary map corresponds remarkably well with the current view of miR-124, miR-125 and miR-9 biology, including actions to promote neurite outgrowth and differentiation by inhibiting Ago-miR-124 targets (Fig. 5c; discussed in Supplemental Fig. 15).
Fig. 5. Ago-miRNA ternary maps.
(a) Genome-wide views of Ago-miRNA ternary maps for the top 20 miRNAs from Ago HITS-CLIP (colors represent individual miRNA targets as indicated in (b)) are shown for the Itgb1 gene (top panel), the local gene region (middle panel; all transcripts are expressed in P13 brain except those outlined in grey boxes), showing tags in neighboring 3′ UTRs, and for all of chromosome 8 (bottom panel). (b) Heat map derived from gene ontology (GO) analysis of transcripts identified as targets of each of the top 20 miRNAs. Tree shows the hierarchical clustering of miRNAs based on GO (Supplementary Methods). Significant clusters are outlined with black boxes (see Supplementary Fig. 12). (c) Ago HITS-CLIP targets are shown for the most significant pathways (neuronal differentiation/cytoskeleton regulation; based on FDR; Fig. 5b) for miR-124, miR-9 and miR-125 in mouse brain. Actin cytoskeleton pathways are shown based on KEGG database (http://www.genome.jp/kegg/).
Discussion
Ago-miRNA-mRNA ternary maps identify functionally relevant miRNA binding sites in living tissues, and were developed in the context of several recent studies. Crystallographic structures of Ago miRNA-mRNA ternary complexes31 demonstrated close contacts between all three molecules, consistent with the ability of CLIP, which requires close protein-RNA contacts28,29, to detect both Ago miRNA and mRNA interactions. The development of HITS-CLIP set the stage for generating and analyzing genome-wide RNA-protein maps in the brain26 and cultured cells30. High throughput experiments and bioinformatic analysis together generated genome-wide predictions of miRNA seed sequences, particularly of miR-124. These studies demonstrated that miR-124 simultaneously represses hundreds of transcripts7,11,21,42-44, and provided a genome-wide “gold standard” with which to compare Ago HITS-CLIP data. This allowed estimates specificity, false positive and false negatives rates (∼93%, ∼13-27% and ∼15-25%, respectively; Supplementary Methods) that, although limited to one Ago-miRNA-mRNA dataset, indicate that experimental Ago HITS-CLIP data outperformed bioinformatic predictions alone (Fig. 4; Supplementary Fig. 11).
Although we used seed-driven approaches to validate targets, not all Ago binding need be constrained by these rules. 27% of Ago-mRNA clusters have no predicted seed matches among the top 20 Ago-miRNAs families. Such orphan clusters might bind other miRNAs, or miRNAs that follow other rules of binding, such as wobble or bulge nucleotides40,45,46 (Chi & Darnell, unpublished observations). Orphan clusters also provide another means of estimating the false negative rate (∼15%; Supplementary Methods), which compares favorably with previous studies in which false negative rates were between 50% and 70%11,18,20.
Ago HITS-CLIP resolves some roadblocks that have arisen in efforts to understand miRNA action. It has been difficult to discriminate direct from indirect actions of miRNAs, and to extrapolate miRNA overexpression studies in tissue culture to organismal miRNA action. Target RNAs have previously been identified by immunoprecipitation, microarray analysis21,44, and reporter validation assays, with the concern that low stringency immunoprecipitation of non-crosslinked RNA-protein complexes47, including Ago-miRNAs48, may purify indirect targets. This has spurred interest in efforts to explore miRNA-target identification by covalently crosslinking, using formaldehyde or 4-thio-uridine modified RNA in culture to identify transcripts complexed with Ago, miRNAs and additional proteins48,49. HITS-CLIP offers a clear means of identifying direct Ago targets and identifying specific interaction sites, which in turn offers the possibility of specifically targeting miRNA activity.
Ago-HITS-CLIP compliments bioinformatic approaches to miRNA target identification by restricting the sequence space to be analyzed to the ∼45-60 nt Ago footprint. For highly conserved 3′ UTRs, such as those of the RNABPs Ptbp2, Nova1, and Fmr1, many miRNA sites are predicted using algorithms that rely on sequence conservation, but each has only one Ago-mRNA CLIP cluster (Supplementary Fig. 10). In fact, miRNA selectivity is very high (Fig. 4) such that on average transcripts have between 1-3 major Ago binding sites in a single tissue (Fig. 3 and Supplementary Figs. 10, 13-14). Ago-mRNA binding sites themselves have no apparent sequence preference (data not shown), suggesting that accessibility may rely on additional RNABPs. Such a mechanism, which may be assessed by overlaying HITS-CLIP maps of different RNABPs26, could provide a means of dynamically regulating miRNA binding and regulation1.
By simultaneously generating binding maps for multiple miRNAs, Ago HITS-CLIP offers a new approach to understanding combinatorial control of target RNA expression. Both the FDR rate and evidence for such miRNA target protein networks deteriorate substantially when predicted maps are generated without experimental Ago HITS-CLIP data (Supplementary Fig. 15). At the same time, analysis of a single miRNA, miR-124, demonstrated that its expression not only induced Ago to bind miR-124 sites, but reduced or precluded Ago binding to sites occupied in untransfected cells (Table 1), perhaps reflecting competition between a limited capacity for miRNA binding on a given 3′ UTR. Such Ago occlusion has important mechanistic, experimental and clinical implications, where studies manipulating miRNA levels are envisioned.
Methods summary
Ago HITS-CLIP was performed in biologic replicate as described26,27 (using monoclonal antibody 2A8 or 7G1-1* as described in Supplementary Methods). High-throughput sequencing was performed with an Illumina Genome Analyzer.
Microarrays. Affymetrix exon arrays (MoEx 1.0 ST) were used to measure transcript abundance in P13 mouse brain and data was analyzed with Affymetrix Power Tools.
Bioinformatics analysis used the UCSC genome browser, miRBASE, BioPython, Scipy and GoMiner, as described in Supplementary Methods.
Supplementary Material
Acknowledgments
The authors thank members of the Darnell laboratory for discussion, J. Fak for help with exon arrays, C. Zhang for bioinformatic discussions, and G. Dunn, D. Licatalosi, C. Marney, M. Frias, T. Eom, J. Darnell, M. Yano and C. Zhang for critical review of the manuscript. Thanks to Z. Mourelatos for generously supplying the 2A8 antibody and communicating unpublished results, to G. Hannon for helpful discussions, and to Scott Dewell for help with high throughput sequencing. This work was supported in part by grants from the NIH (R.B.D.), the Cornell/Rockefeller/Sloan-Kettering Tri-Institutional Program in Computational Biology and Medicine (S.W.C.) and MD-PhD Program (J.B.Z.). R.B.D. is an Investigator of the Howard Hughes Medical Institute.
References
- 1.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 2.Sharp PA. The centrality of RNA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 6.Berezikov E, et al. Diversity of microRNAs in human and chimpanzee brain. Nat Genet. 2006;38:1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- 7.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 8.Krek A, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 9.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 10.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38(Suppl):S8–13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 11.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bentwich I. Prediction and validation of microRNAs and their targets. FEBS Lett. 2005;579:5904–5910. doi: 10.1016/j.febslet.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 13.Ventura A, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- 16.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 17.van Rooij E, Liu N, Olson EN. MicroRNAs flex their muscles. Trends Genet. 2008;24:159–166. doi: 10.1016/j.tig.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 18.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 19.Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 2007;4:76–84. doi: 10.4161/rna.4.2.4640. [DOI] [PubMed] [Google Scholar]
- 20.Easow G, Teleman AA, Cohen SM. Isolation of microRNA targets by miRNP immunopurification. RNA. 2007;13:1198–1204. doi: 10.1261/rna.563707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hendrickson DG, Hogan DJ, Herschlag D, Ferrell JE, Brown PO. Systematic identification of mRNAs recruited to argonaute 2 by specific microRNAs and corresponding changes in transcript abundance. PLoS ONE. 2008;3:e2126. doi: 10.1371/journal.pone.0002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L, et al. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol Cell. 2007;28:598–613. doi: 10.1016/j.molcel.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammell M, et al. mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nat Methods. 2008;5:813–819. doi: 10.1038/nmeth.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Rooij E, Marshall WS, Olson EN. Toward microRNA-based therapeutics for heart disease: the sense in antisense. Circ Res. 2008;103:919–928. doi: 10.1161/CIRCRESAHA.108.183426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wheeler TM, Lueck JD, Swanson MS, Dirksen RT, Thornton CA. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J Clin Invest. 2007;117:3952–3957. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ule J, et al. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 28.Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 29.Jensen KB, Darnell RB. CLIP: crosslinking and immunoprecipitation of in vivo RNA targets of RNA-binding proteins. Methods Mol Biol. 2008;488:85–98. doi: 10.1007/978-1-60327-475-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeo GW, et al. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009 doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, et al. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 34.Grimson A, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14:872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A. 2008;105:14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lal A, et al. p16(INK4a) translation suppressed by miR-24. PLoS ONE. 2008;3:e1864. doi: 10.1371/journal.pone.0001864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen WF, Hu YL, Uttarwar L, Passegue E, Largman C. MicroRNA-126 regulates HOXA9 by binding to the homeobox. Mol Cell Biol. 2008;28:4609–4619. doi: 10.1128/MCB.01652-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 41.Yu JY, Chung KH, Deo M, Thompson RC, Turner DL. MicroRNA miR-124 regulates neurite outgrowth during neuronal differentiation. Exp Cell Res. 2008;314:2618–2633. doi: 10.1016/j.yexcr.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X, Wang X. Systematic identification of microRNA functions by combining target prediction and expression profiling. Nucleic Acids Res. 2006;34:1646–1652. doi: 10.1093/nar/gkl068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karginov FV, et al. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci U S A. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- 46.Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kirino Y, Mourelatos Z. Site-specific crosslinking of human microRNPs to RNA targets. RNA. 2008;14:2254–2259. doi: 10.1261/rna.1133808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 50.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.