Abstract
Mitochondria exist as dynamic networks that are constantly remodeled through the opposing actions of fusion and fission proteins. Changes in the expression of these proteins alter mitochondrial shape and size, and may promote or inhibit the propagation of apoptotic signals. Using mitochondrially targeted EGFP or DsRed2 to identify mitochondria, we observed a short, distinctly tubular mitochondrial morphology in postnatal cortical neurons in culture and in retinal ganglion cells in vivo, whereas longer, highly interconnected mitochondrial networks were detected in cortical astrocytes in vitro and non-neuronal cells in the retina in vivo. Differential expression patterns of fusion and fission proteins, in part, appear to determine these morphological differences as neurons expressed markedly high levels of Drp1 and OPA1 proteins compared to non-neuronal cells. This finding was corroborated using optic tissue samples. Moreover, cortical neurons expressed several splice variants of Drp1 including a neuron-specific isoform which incorporates exon 3. Knockdown or dominant negative interference of endogenous Drp1 significantly increased mitochondrial length in both neurons and non-neuronal cells, but caused cell death only in cortical neurons. Conversely, depletion of the fusion protein, Mfn2, but not Mfn1, caused extensive mitochondrial fission and cell death. Thus, Drp1 and Mfn2 in normal cortical neurons not only regulate mitochondrial morphology, but are also required for cell survival. The present findings point to unique patterns of Drp1 expression and selective vulnerability to reduced levels of Drp1 expression/activity in neurons, and demonstrate that the regulation of mitochondrial dynamics must be tightly regulated in neurons.
Keywords: apoptosis, mitochondrial dynamics, mitochondrial fusion, mitochondrial fission, neuronal cell death, dynamin related protein-1
Introduction
Mitochondria are essential for maintaining neuronal viability and neuronal function because of their ability to provide ATP, regulate calcium homeostasis and control Fe/S cluster biogenesis (DiMauro and Schon, 2008). Consistent with this critical role, abnormalities in mitochondrial structure and function are closely associated with a wide array of neurological diseases and nervous system injury (Chan, 2004; Lin and Beal, 2006). Recent evidence demonstrates that mitochondria are highly dynamic structures that display significant variability in size, shape and distribution. In addition to being highly motile structures, mitochondria can change size and shape by undergoing fission and fusion. These opposing processes may play an important role in mediating the activity-dependent regulation of mitochondrial distribution in neurons (Li et al., 2004; Sung et al., 2008), an activity that has recently been linked to creating or maintaining synaptic contacts (Li et al., 2008). In addition, alterations in mitochondrial fission and fusion have been associated with neuronal degeneration induced by diverse neurotoxic stresses (Barsoum et al., 2006; Jahani-Asl et al., 2007; Yuan et al., 2007; Poole et al., 2008; Yang et al., 2008; Deng et al., 2008). Thus, the regulation of mitochondrial dynamics may be critical for maintaining neuronal health, connectivity and function (Knott et al., 2008).
Changes in mitochondrial shape, size and function result from a shift in the steady state balance between fission and fusion, and are carried out by a well conserved cellular machinery comprised of dynamin-related GTPases (Chen and Chan, 2005). Three such GTPases, Mfn1, Mfn2, and OPA1 regulate fusion in mammalian cells (Cipolat et al., 2004; Chen et al., 2005). Conversely, dynamin-related protein 1 (Drp1) and Fis1 have been identified as components of the mammalian mitochondrial fission machinery (Smirnova et al., 2001; James et al., 2003; Yoon et al., 2003). The importance of fusion and fission proteins to the biology of neurons is underscored by the occurrence of several neurodegenerative diseases that result from mutations in these genes. Mutations in Mfn2 cause the autosomal dominant disease Charcot-Marie-Tooth (CMT) type 2A, a peripheral neuropathy of motor and sensory neurons (Zuchner et al., 2004). Mutations in OPA1 promote dominant optic atrophy (DOA), a commonly inherited form of optic nerve degeneration (Alexander et al., 2000; Delettre et al., 2000). CMT4A is caused by mutations in GDAP1, a protein implicated in mitochondrial fission (Niemann et al., 2005). Finally, a mutation in the fission gene Drp1 has recently been identified in a newborn child with abnormal brain development and optic atrophy (Waterham et al., 2007).
Despite the emerging relationship between the regulation of mitochondrial dynamics and the maintenance of neuronal structure and function, very little is known regarding the cellular machinery that regulates mitochondrial fusion and fission in healthy neurons or how it responds to disease and stress. Also, it is not clear whether our current understanding of mitochondrial dynamics in non-neuronal cells is directly applicable to neurons. Therefore, in the present study, we determined if the regulation of mitochondrial dynamics differed between neurons and non-neuronal cells. The present findings demonstrate that neurons express unique patterns of fission/fusion proteins and that they are vulnerable to either reduced levels/activity of the fission protein Drp1 or conversely, the fusion protein Mfn2, but not Mfn1. Thus, mitochondrial dynamics must be tightly regulated in neurons to maintain function and viability.
Materials and Methods
Animals and cell culture
Primary cultures of postnatal cortical and hippocampal neurons from newborn mice (day 0) and embryonic telencephalic neurons from day 14.5 mouse embryos were prepared as previously described (Xiang et al., 1996; Johnson et al., 1999) and maintained for 3 or 4 days prior to experimental manipulations unless otherwise specified. Neuronal viability was determined based on morphological criteria as previously described (Xiang et al., 1996); nuclear morphology (Hoechst 33258 staining) or the overall morphology of neurons visualized with EGFP fluorescence following lentivirus infection. Mouse embryonic fibroblasts (MEF), mouse astrocytes and HeLa (a human cervical carcinoma cell line) cells were prepared and maintained as previously described (Uo et al., 2005; Uo et al., 2007). Adult spinal cord progenitor cells were prepared and maintained as a mixture of neurospheres and attached cells in DMEM/F12 (1:1) supplemented with 2 mM glutamine, 1% N2 supplements, 5 μg/ml heparin, 20 ng/ml EGF, and 20 ng/ml FGF-2 as previously described (Petit et al., 2007).
Immunoblotting
Brains were freshly obtained from 6 month-old C57BL/6 mice following cervical dislocation. Superficial layers of superior colliculus (bilateral) were dissected out using a small spatula, optic nerves were separated at the chiasm and eye cup, and retinas were cleanly removed from dissected eye cups, and then each was placed in tubes containing ice-cold lysis buffer (50 mM Tris-HCl, pH 7.4; 150 mM NaCl; 5 mM EDTA; 1% Triton X-100; 100 μg/mL PMSF; 1% protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO) and 1% phosphatase inhibitor cocktail I and II (Sigma-Aldrich, St. Louis, MO)). Samples of tissue from 10 mice each were pooled. All tissue was initially broken down with a tube-fitted pestle then further sheared by pulling through successively smaller gauge needle syringes (up to 26G). Lysate was boiled for 5 min in a 95°C water bath then centrifuged for 10 min at 16,000g. The protein concentration of the supernatant was quantified with a BCA assay (Pierce Biotechnology, Rockford, IL). Protein extracts from cultured cells were prepared in SDS-PAGE sample buffer (10 % glycerol, 62.5 mM Tris-HCl, pH 6.8, 2% SDS). Protein samples were subjected to gel electrophoresis using 10 μg protein/lane on 4–15% SDS gradient polyacrylamide gels unless otherwise specified, followed by immunoblotting as previously described (Uo et al., 2005; Uo et al., 2007).
Immunofluorescence
Cultured cells were fixed in 4% paraformaldehyde (PFA) in 0.1 M phosphate buffer, pH 7.4 (PB), permeabilized with 0.2% Triton X-100, and processed for immunostaining as described (Xiang et al., 1996). Fluorescent microscopic images were captured on an Axiovert 200 inverted microscope (Carl Zeiss Microimaging, Thornwood, NY) equipped with a cooled CCD camera (SensiCam, Cooke Corp, Auburn Hills, MI). Images to be directly compared were captured and processed in an identical manner with Slidebook™ imaging software (Intelligent Imaging Innovations, Denver, CO) and Adobe Photoshop (version 7.0.1. or CS2 9.0.2).
For in vivo samples, ten days after LV-Mito-EGFP injection, mice were deeply anesthetized with 300 mg/kg sodium pentobarbital, then perfused transcardially with 4% PFA in PB; eyes were dissected out, superior cut made, cornea and lens removed before placing the eyecup in 4% PFA for 1 h. Eyecups were washed in PB, retinas dissected out and vitreous removed using angled jeweler’s forceps. Retinas were cryoprotected by overnight incubation in 30% sucrose and subjected to three consecutive freeze-thaw cycles in 30% sucrose before immunolabeling. Retinas were rinsed 3 times (10 min each) in 0.1M Tris buffer (TB, pH 7.4) and then blocked for 1h in 5% donkey serum (Jackson ImmunoResearch Laboratories, West Grove, PA) and 0.4% Triton X-100 (Sigma-Aldrich, St. Louis, MO) in TB (block solution). Tissue was incubated overnight with primary antibodies against NeuN and neurofilament-200 (neurofilament-H/NF-H) in the block solution. Retinas were then rinsed 3 times (10 min each) in TB, incubated for 30 min in block solution then for 2h in Cy5 or RedX-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) diluted in blocking solution. After final rinsing (3 times in TB), wholemount retinas were mounted on SuperFrost slides (Fisher Scientific, Fairlawn, NJ) and coverslipped in gelvitol. High magnification confocal stacks of retinas were taken using a Bio-Rad Radiance 2000 confocal microscope. Image stacks were processed using Volocity software (Improvision, Ltd., Waltham, MA).
Dilutions of primary antibodies and their sources are provided in Table 1.
Table 1.
List of antibodies used
| Antigen | Application | Antibody | Dilution | Source |
|---|---|---|---|---|
| Drp1 | WB, IF | clone 8 | 2000/500 | BD Transduction Laboratories™ |
| Fis1 | WB | 3491 | 500 | BioVision |
| Mfn1 | WB | 110–58853 | 1000 | Novus Biologicals, Inc |
| Mfn2 | WB | M6319 | 2000 | Sigma-Aldrich |
| OPA1 | WB | #9661 | 2000 | BD Transduction Laboratories™ |
| NeuN | IF | MAB377 | 500 | Millipore |
| Neurofilament 200 | IF | N4142 | 500 | Sigma-Aldrich |
| HSP60 | IF | H-300 | 2000 | Santa Cruz Biotechnology |
| β-Actin | WB | AC-15 | 20000 | Sigma-Aldrich |
| β-Tubulin III | IF | TUJ | 1000 | Aves Labs Inc. |
| TuJ1 | 1000 | Covance | ||
| FLAG | WB, IF, IP | M2 | 2000 | Sigma-Aldrich |
| Bcl-xL | IP | 54H6 | 100 | Cell Signaling Technology |
| MYC-Tag | WB | 9B11 | 2000 | Cell Signaling Technology |
| SUMO-1 | WB | S8070 | 2000 | Sigma-Aldrich |
WB: Western blot; IF: immunofluorescence, IP: immunoprecipitation
Analysis of Drp1 binding to Bcl-xL and Drp1 SUMOylation
For co-immunoprecipitation (co-IP) studies, at 24 hr after transfection with Lipofectamine 2000 reagent (Invitrogen), subconfluent HEK 293 FT cells on 6-well multidish (Nunc) were lysed in the co-IP buffer (10 mM HEPES-KOH, pH 7.5, 1 mM EDTA, 150 mM NaCl, 1% Triton X-100) supplemented with proteinase inhibitors. Lysates were divided into two parts, and subjected to immunoprecipitation with 2 μg of either Bcl-xL antibody (54H6, Cell Signaling Technology) or normal Rabbit IgG (Invitrogen) bound to ImmunoPure® Immobilized Protein G (Pierce). To detect Drp1 SUMOylation, HEK 293 FT cells were lysed in RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% sodium deoxycholate, 0.1% SDS, 1% Triton X-100) containing protease inhibitors 24 hr after transfection. The resultant lysates were incubated with anti-FLAG M2 agarose (Sigma-Aldrich). Beads were washed 4 times with the respective buffers used for extraction and boiled with 2x SDS-PAGE sample buffer. The eluted proteins were resolved by SDS-PAGE gels, followed by Western blot analysis using appropriate antibodies.
Plasmid construction and lentivirus production
Human pcDNA3-Drp1 for transient expression of wild-type Drp1 (Smirnova et al., 1998) was kindly provided by Dr Richard Youle (NIH). It is important to note that this cDNA lacks exon 3, 16 and 17. Expression constructs for FLAG-tagged wild-type (Drp1WT) and dominant-negative mutant Drp1 (Drp1K38A) were created as follows. Using pcDNA3-Drp1 as a template, a cDNA fragment encoding N-terminally FLAG-tagged partial Drp1 protein was prepared by PCR with forward primer: GGA TCC GCC ACC ATG GAT TAC AAG GAT GAC GAC GAT AAG GAG GCG CTA ATT CCT GTC ATA AAC and reverse primer: AAA TCC ACA AGT GTC AAA TTG ACA ACG (BamHI site is singly underlined and the sequence encoding FLAG tag was dashed). The amplified fragment contains the HindIII site and this was digested with BamHI and HindIII. Subsequently, the resultant fragment and the HindIII-XhoI fragment of pcDNA3-Drp1 were introduced together into the BamHI-XhoI site of the pcDNA3 vector (Invitrogen) to produce pcDNA3 FLAG-Drp1WT. An expression construct for the dominant negative mutant of Drp1, pcDNA3 FLAG-Drp1K38A, was created using the QuikChange XL Site-Directed Mutagenesis Kit (Stratagene). For lentivirus-mediated expression of Drp1WT and Drp1K38A, DNA fragments were liberated from the respective pcDNA3 FLAG-Drp1 vectors by digestion with NdeI and XhoI. These fragments were inserted into the NdeI-XhoI site of the pSL6 IRES-GFP vector (Uo et al., 2007) to produce lentiviral transfer vectors co-expressing EGFP and either FLAG-tagged Drp1WT or Drp1K38A from the same transcripts.
Each cDNA encoding a variable splicing variant of human Drp1 was created using pcDNA3 FLAG-Drp1WT as a parental plasmid. Human Drp1 cDNA encoding transcript variant 1 (NM012062.2) which lacks exon 3 but contains exon 16 and 17 was purchased from Origene (Rockville, MD) and used as a template for PCR in some cases. Primers with DNA sequence corresponding to exon 3 were used for PCR to introduce the exon 3 sequence into the human Drp1 cDNA. Further information about the cloning methods and primers is available upon request.
For expression of the MYC-tagged Bcl-xL protein, a cDNA fragment encoding human Bcl-xL was liberated from the pCMVTag2 Bcl-xL plasmid (Uo et al., 2007) by digestion with BamHI and XhoI. The resultant fragment was inserted into the same site of the pCMVTag3 plasmid (Stratagene, La Jolla, CA) to produce pCMVTag3 Bcl-xL.
For lentivirus-mediated expression of a mitochondrially-targeted form of EGFP protein, a DNA fragment was liberated from the pMito-EGFP plasmid (Uo et al., 2007) by digestion with NheI and NotI, followed by trimming to obtain blunt-ended fragments. These fragments were inserted into the blunt-ended XhoI site of the pSL6 vector (originally provided by Dr. W.R. Osborne, University of Washington) to produce pSL6 Mito-EGFP. pSL6 Mito-DsRed2 was created using the same strategy for expression of a mitochondrially-targeted form of DsRed2 protein.
The pre-made lentiviral pLKO.1 plasmid expressing shRNA for mouse Drp1 (Sigma-Aldrich: Clone ID: NM_152816.1-1101s1c1), Mfn1 (NM_024200.2-1521s1c1) and Mfn2 (NM_133201.1-2295s1c1) were used for knockdown of expression of mouse Drp1, Mfn1 and Mfn2, respectively. The MISSION Non-Target shRNA Control Vector was used to express control shRNA that does not target mouse genes. In the case of Mfn1 and Mfn2 knockdown, the puromycin-resistant gene in pLKO.1 plasmid was replaced by cDNA encoding EGFP. The MISSION Non-Target shRNA Control Vector was processed in the same manner.
The concentrated virus stocks were titered by transducing HT1080 cells with limiting dilutions of the stock and by assessing expression of fluorescence proteins through flow cytometry or quantifying proviral DNA by real-time PCR. Stable knockdown of human Drp1 in HeLa cells was achieved by using pREP4 vector (Invitrogen) to which a U6 promoter-driven shRNA sequence for human Drp1 was subcloned from the pSHAG-1 (Cold Spring Harbor Laboratory, Cold Spring, NY)-based shRNA expression system (kindly provided by Dr. R. J. Youle, NIH) according to the published protocol (Lee et al., 2004).
Reverse transcription (RT)-PCR based identification of Drp1 splice variants
Total RNA was isolated from the specified cells in culture using the RNeasy kit (Qiagen, Inc., Valencia, CA) according to the manufacturer’s protocols. cDNA synthesis and target gene amplification were performed using the OneStep RT-PCR kit (Qiagen) with the following primers for analysis of inclusion/exclusion of exon 3 (forward primer, P1: GTG GGA AGA GCT CAG TGC TGG AAA GC; reverse primer, P2: CTT GTC GAA TTT CAT CAA AAT CTG TGT AAA G) and for that of exon 16 and 17 (forward primer, P3: CTT AGT GGC AAT TGA GCT AGC GTA TATC; reverse primer, P4: GTT GTT GGT TCC TGA CCA CCG TCT CCA ATC). Each primer binding site is shown in Figure 3A. RT was carried out at 55°C for 30 min followed by a 15 min denaturation step at 94°C. This was followed by non-quantitative PCR with 35 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s. 100 ng of total RNA was used in each reaction mixture (20 μl).
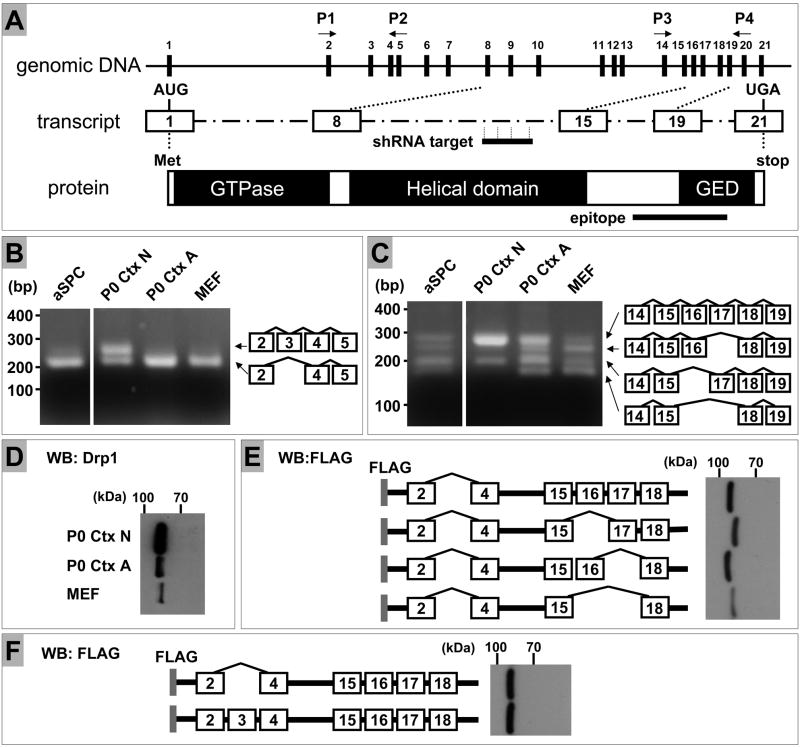
Figure 3. Analysis of expression of Drp1 splicing variants in mouse cells.
(A) Schematic representation of genomic organization, transcript and protein of mouse Drp1. Numbering of exons is based on the sequence of transcript variant “a” (GenBank accession No., NM152816). shRNA target sequence resides in the region corresponding to exon 11, whereas the peptide sequence used to raise the Drp1 antibody we used encompasses the region corresponding to exon 18–20. Thus, all of the splicing variants and the respective proteins can be targeted by our shRNA sequence and recognized by the antibody used in this study, respectively.
(B) Total RNA was prepared from adult spinal cord neural progenitor cells (aSPC), primary neuronal cultures derived from postnatal day 0 cortex (P0 Ctx N), astrocyte cultures from postnatal day 0 cortex (P0 Ctx A) and mouse embryonic fibroblasts (MEF). Each sample was subjected to RT-PCR analysis using primers P1 and P2 for amplification of exon 2–5. The resultant reaction mixtures were resolved on 2.0 % agarose gels with markers (100 bp PCR Molecular Ruler, Bio-Rad. Hercules, CA). The structure of each splicing variant was confirmed by direct sequencing.
(C) Total RNA was prepared and subjected to RT-PCR analysis using primers P3 and P4 for amplification of exon 14–19, followed by agarose gel analysis as described above in (B). The aSPC and other samples were applied to the same agarose gel. The resultant photo images, which contained unrelated samples, were cut and rearranged to facilitate the comparison.
(D) Protein samples prepared as described in Fig. 2A were subjected to SDS polyacrylamide gel electrophoresis using a constant 7.5% gel followed by Western blot analysis. Drp1 protein from MEF migrated faster than those from P0 cortical neurons (P0 Ctx N) and astrocytes (P0 Ctx A).
(E) HEK 293 FT cells were transiently transfected with plasmids harboring FLAG-tagged human Drp1 cDNA with all combinations of inclusion/exclusion of exon 16 and 17. All of these cDNAs lack exon 3. Protein extracts from transfectants were subjected to Western blot using a constant SDS 7.5% polyacrylamide gel to detect FLAG-tagged proteins. The DNA sequences corresponding to exon 16 and 17 encode 26 and 11 amino acids, respectively. Exclusion of exon 16 yielded a faster migrating form as expected, whereas inclusion or exclusion of exon 17 did not provide separable migration patterns under these conditions. Since neuronal Drp1 is expected to be predominantly the form that includes both exon 16 and 17 (C), exclusion of exon 16, not exon 17, can explain the faster moving Drp1 from MEF relative to the neuronal Drp1.
(F) FLAG-tagged human Drp1 cDNAs containing both exon 16 and 17 with/without exon 3 were used in the same way as described in (E). The DNA sequence corresponding to exon 3 encodes 13 amino acids. Inclusion or exclusion of exon 3 did not provide separable migration patterns under these conditions.
Intracellular localization of Drp1 splice variants
Intracellular localization of the Drp1 splice variants was evaluated by expressing them via transient transfection using Lipofectamine 2000 (Invitrogen). HEK 293 FT cells were plated in four-well multidishes (Nunc) at a subconfluent density and transfected with 0.2 μg of pMito-EGFP plasmid and 0.8 μg of a FLAG-tagged Drp1 variant plasmid as previously described (Uo et al., 2005; Uo et al., 2007). Postnatal cortical neurons in four-well multidishes (4 × 105 cells per well) were transfected one day after plating with 1 μg of a FLAG-tagged Drp1 variant plasmid. 24 and 48 hr after transfection, HEK 293 FT cells and neurons were fixed, respectively and processed for FLAG immunostaining to visualize the intracellular distribution of the exogenously expressed Drp1 variants.
Intraocular Injection
To label mitochondria in retinal cells, lentiviral vector carrying Mito-EGFP was injected intraorbitally in 10 month old DBA/2 mice anesthetized with Avertin (1.3% tribromoethanol and 0.8% tert-amyl alcohol) placed supine in a plasticine support mold. A 32G Hamilton syringe attached to a microinjection device was used to deliver 5×103 transduction units (TU) (1μL of 5×103 TU/μl) of the virus to the mouse eye posterior chamber by entering from the nasal orientation and just posterior to the ora serrata. Injections took place over 2 min and injections were made in both eyes.
Results
Mitochondrial shape, size and distribution were evaluated in neurons and non-neuronal cells in culture by expressing a mitochondrially targeted form of EGFP or DsRed2. In postnatal cortical neurons mitochondria displayed short, tubular shaped morphology and were randomly distributed throughout the neurites. Mitochondria were highly concentrated in the soma but retained their short tubular stature (Fig. 1Aa). Similar mitochondrial morphological features were observed in hippocampal and cerebellar granule neurons in culture (data not shown). In marked contrast to neurons, Mito-EGFP and Mito-DsRed2 positive structures displayed a reticular network in primary astrocytes (Fig. 1Ab) and other non-transformed or transformed non-neuronal cells including mouse embryonic fibroblasts (MEF: Fig. 1Ac) and HeLa cells (Supplemental Figure 1).
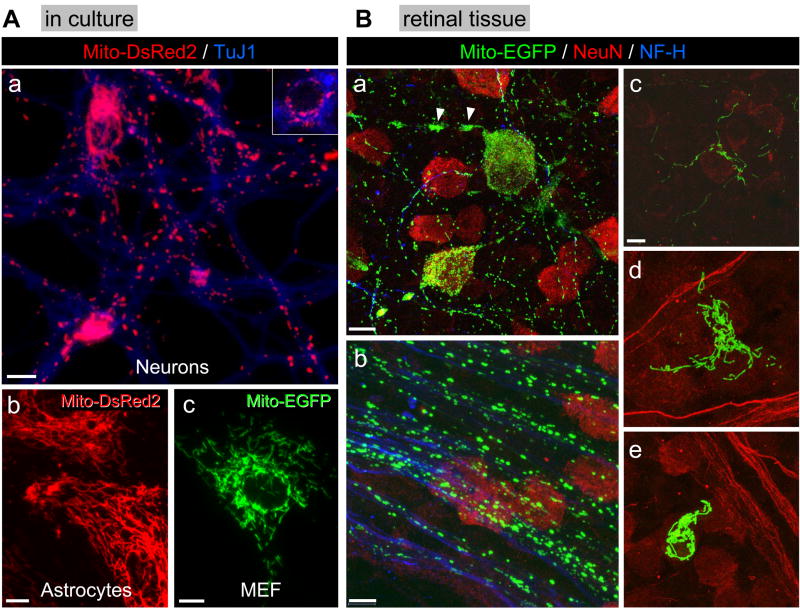
Figure 1. Mitochondrial morphology in neurons and non-neuronal cells in vitro and in vivo.
A) Primary cultures of (a) postnatal mouse cortical neurons, (b) postnatal mouse cortical astrocytes and (c) mouse embryonic fibroblasts (MEF) were infected with lentivirus expressing mitochondrially targeted EGFP (10 MOI) or DsRed2 (10 MOI) 24 hr after plating. Cells were fixed 48 hr after infection. Neurons in (a) were immunostained for β-tubulin III (TuJ1 antigen), a neuronal marker. The gamma factor for (a) was set at 0.4 using Slidebook software in order to bring up the MitoDsRed2 fluorescence intensity in the neurites. An inset using a lower fluorescence intensity depicts mitochondrial morphology in the soma of neurons. Scale bar (a–c) = 10 μm. The images are representative of four independent experiments.
B) Mito-EGFP lentivirus (5×103 TU) was injected into the posterior chamber of the eye where it was taken up by several cell types. (a) NeuN-positive retinal ganglion cells expressing Mito-EGFP show small, punctate or tubular mitochondria well-dispersed throughout the neuronal architecture including the cell soma. An exception is seen in the axon initial segment where mitochondria tend to aggregate and form clusters of EGFP-positive mitochondria (arrowheads: see Supplemental Figure 2 for more detail). (b) Retinal ganglion cell axons immunopositive for neurofilament-200 (NF-H) chain, an axonal marker, show a wide range of EGFP-positive mitochondrial morphology. The larger puncta may represent individual or clusters of mitochondria. (c) NeuN-negative cells present in the nerve fiber layer, above the retinal ganglion cells, also display highly elongated mitochondria. (d) A NeuN-negative cell in the retinal nerve fiber layer has large numbers of elongated mitochondria. (e) Another NeuN-negative cell, possibly a macrophage, also has a large concentration of EGFP-positive elongated mitochondria. The images are representative of three independent experiments. Scale bars: 20 μm in (a), 10 μm in (b–e). Note: in (d) and (e), neurofilament-200 staining is also presented in red, not in blue, together with NeuN staining.
Mitochondrial shape was also evaluated in vivo to validate the results obtained with cultured cells. This was accomplished by infecting retinal ganglion neurons and glia in the retina with lentivirus expressing Mito-EGFP. Retina was chosen because it provided a straightforward approach for viral injection (posterior chamber) and the soma and axons are easily visualized in the well characterized lamellar structure of the retina. The soma of retinal ganglion cells identified based on NeuN positivity were packed with Mito-EGFP-positive mitochondria but they retained the short, tubular structure that was observed in cultured cortical neurons (Fig. 1Ba). Interestingly, in a rare case of degenerating retinal ganglion cells the mitochondria appear fused and clustered around the nucleus (Supplemental Figure 2). Neurofilament-200 positive fibers in the optic nerve representing axons of retinal ganglion cells also displayed short, tubular mitochondria that were randomly dispersed along the entire length of the processes (Fig. 1Bb). In contrast, NeuN-negative cells displayed an interconnected network of Mito-EGFP positive mitochondria (Fig. 1Bc–e). These structures were considerably longer than those observed in neurons and they appeared to extend into cellular extensions of these non-neuronal cells.
Next, we evaluated expression levels of proteins that regulate mitochondrial fusion and fission to determine if there is a molecular basis to account for these differences. Expression levels of each fission/fusion protein examined were very similar among neurons in culture irrespective of their developmental age (E14.5 vs. postnatal) or location (cortical vs. hippocampal) (Fig. 2A). Striking differences emerged from a comparison among neurons, astrocytes and mouse embryonic fibroblasts (MEF). Drp1 and OPA1 levels in neurons were markedly higher than those in non-neuronal cells.
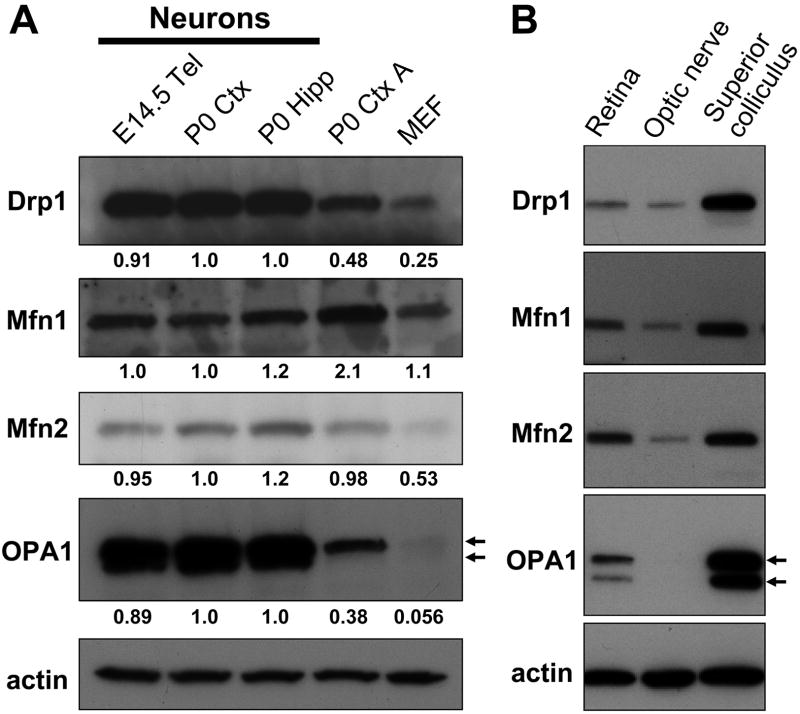
Figure 2. Comparative analysis of expression of mitochondrial fusion/fission proteins among various cultured cells and tissues.
(A) Protein samples were prepared from primary neuronal cultures derived from embryonic day 14.5 telencephalon (E14.5 Tel), postnatal day 0 cortex (P0 Ctx) or hippocampus (P0 Hipp), astrocyte cultures from postnatal day 0 cortex (P0 Ctx A) and mouse embryonic fibroblasts (MEF). Expression of mitochondrial fusion (Mfn1, Mfn2 and OPA1) and fission (Drp1) GTPase proteins in each sample was evaluated by Western blot analysis. The intensity of each band corresponding to the specified protein was quantitated using ImageJ 1.41o software (National Institutes of Health, Bethesda, MD) and normalized against actin. For OPA1, the summed intensity of two major bands (indicated by arrows) was used for quantitation. The value for the respective protein in each sample was then normalized to that in postnatal cortical neuronal extracts. Similar results were obtained for P0 Ctx N and MEF in at least three independent experiments.
(B) Protein samples were prepared from retina, optic nerve and superior colliculus dissected out from 6 month old C57BL/6 mice and subjected to Western blot analysis as described in (A). The data is representative of two independent experiments.
Expression of mitochondrial fission/fusion proteins in vivo was also evaluated for mouse retina, optic nerve and the superior colliculus to correlate with the in vivo mitochondrial morphology studies performed above with retinal ganglion cells and optic nerve. Drp1 and OPA1 levels were highly elevated in the superior colliculus relative to retina and optic nerve in 6 month old C57BL/6 mice (Fig. 2B). In contrast, Mfn1 and Mfn2 levels were not elevated as much in the superior colliculus relative to retina and optic nerve. Since the superior colliculus is a highly laminated structure containing neuronal cell bodies, the demonstration of elevated Drp1 and OPA1 expression in this tissue is consistent with data obtained with postnatal neurons in vitro. The high levels of Drp1 may account for the shorter mitochondrial units observed in neurons and suggest that the constitutive regulation of mitochondrial dynamics in neurons may be weighted toward increased fission compared to non-neuronal cells.
The presence and respective functions of multiple splicing variants related to the OPA1 gene are well documented in various cell types and tissues including brain (Song et al., 2007; Akepati et al., 2008). Similarly, Drp1 is known to express multiple splicing variants (Yoon et al., 1998; Smirnova et al., 1998; Howng et al., 2004). However, little is known regarding their precise tissue/cell type-specific expression and their functional variations. Thus, we examined whether neurons express selective subsets of Drp1 variants. First, we carried out an initial RT-PCR screen of total RNA extracted from different cell types using primers specific for each variable exon (Fig. 3A). As previously reported, we confirmed that various cell types frequently skip exon 3, 16 and 17 singly or in combination, while all the other exons are always included in the transcripts (data not shown). In cultured cortical neurons Drp1 transcripts were expressed in two forms with or without exon 3 included, while this exon was always excluded in astrocytes, MEF and adult spinal cord neural progenitor cells (Fig. 3B). Thus, inclusion of exon 3, originally reported as being brain-specific (Yoon et al., 1998), is unequivocally specific to post-mitotic differentiated neurons. According to currently available literature (Smirnova et al., 1998; Howng et al., 2004) and the public database, (http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene), the total number of exons in the human Drp1 gene is considered to be 20, based on the exon structure of transcript variant 1 (GenBank accession No., NM012062) which lacks the sequence corresponding to exon 3. However, our BLAST search on the database (human expressed sequence tags) indeed confirmed the presence of transcripts containing the exon 3-sequence in human brain tissues including cerebellum (DC305528), amygdala (DC315924), hippocampus (DC334877) and substantia nigra (DC336964). Thus, exon 3-containing Drp1 is likely to be expressed as a functional protein, and we propose that inclusion of exon 3 may yield a functionally discrete form of the Drp1 protein. Unique features of neuronal Drp1 were further exemplified by preferential inclusion of both exon 16 and 17 (Fig. 3C). Interestingly, the exclusion of exon 17 was never observed in neurons. Conversely, the other cell types tested here provided expression patterns with all possible combinations of inclusion/exclusion of exon 16 and 17 with some preference for inclusion of each exon (Fig. 3C).
Since neurons express longer Drp1 isoforms compared with other cell types, we re-defined the migration patterns of Drp1 from different cell types on a constant 7.5% SDS-PAGE gel. Obviously, the majority of Drp1 from MEF migrated faster than Drp1 from postnatal cortical neurons and astrocytes (Fig. 3D). This difference is most likely due to exclusion of exon 16 in MEF based on the migration patterns of multiple FLAG-tagged variants of human Drp1 (Fig. 3E). FLAG-tagged variants containing or lacking exon 3 migrated to the same position (Fig. 3F). Thus, under the conditions tested here, neuronal variants with or without exon 3 should co-migrate and give rise to a single immunoreactive band just as we observed for astrocyte Drp1 which lacks exon 3.
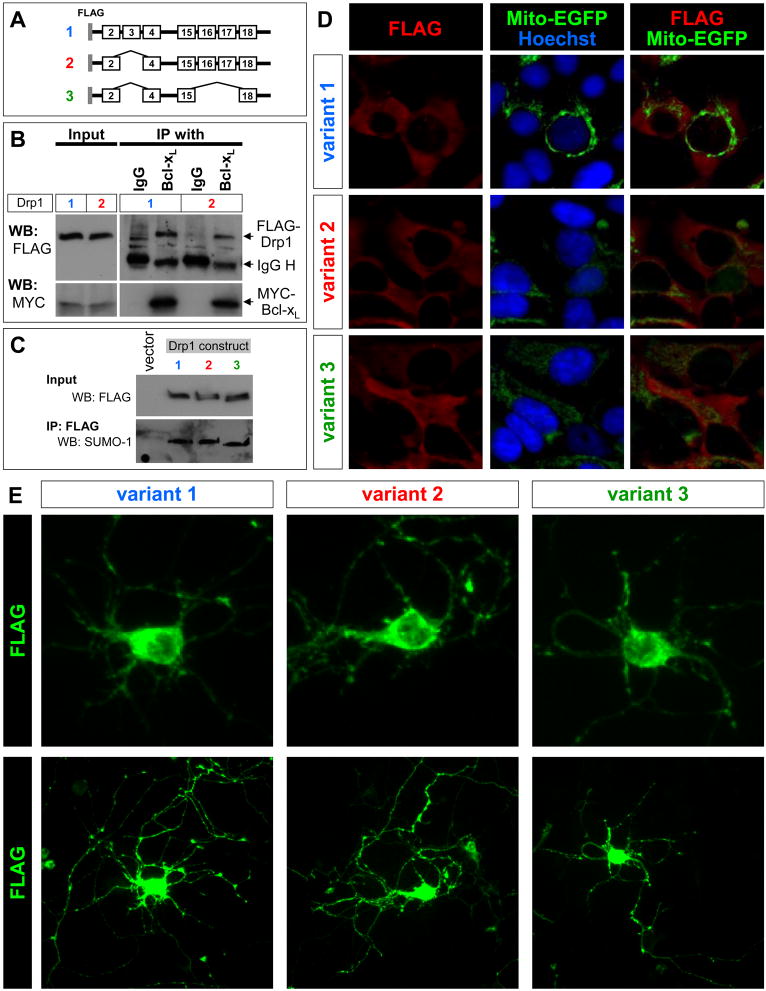
Drp1 function is regulated through association with other proteins including Bcl-xL as well as diverse post-translational modifications such as SUMOylation (Wasiak et al., 2007; Li et al., 2008; Santel and Frank, 2008). These modes of regulation may be altered by inclusion of exon 3 and/or exon 16/17. We initiated studies to test these possibilities using Drp1 constructs shown in Fig. 4A. Overexpression of Drp1 variants in HEK 293 FT cells showed that inclusion of exon 3 in Drp1 is not a determinant of binding to Bcl-xL (Fig. 4B). Furthermore, we found that the presence or absence of exon 3, 16 and 17 did not affect the SUMOylation of Drp1 (Fig. 4C). Similarly, we also determined that the presence or absence of exon 3, 16 and 17 did not effect the intracellular localization of Drp1 in HEK 293 FT cells (Fig. 4D) or postnatal cortical neurons (Fig. 4E).
Figure 4. Drp1 variants display similar properties in binding to Bcl-xL, SUMOylation and intracellular localization.
(A) Schematic representation of the Drp1 constructs used for (B)–(D). Note the variations in the inclusion/exclusion of exon 3 and exon 16/17.
(B) HEK 293 FT cells were transiently co-transfected with a MYC-tagged Bcl-xL plasmid and one of the Drp1 variant plasmids, as shown in (A), as indicated by the numbers. Lysates were prepared 24 hr after transfection and subjected to immunoprecipitation (IP) with anti-Bcl-xL antibody followed by Western blot analysis (WB) using anti-FLAG and anti-MYC tag antibodies to detect the exogenously expressed Drp1 and Bcl-xL proteins, respectively. The positions of IgG heavy chains (IgG H) are also indicated. Normal rabbit IgG was used as a negative control for the IP.
(C) HEK 293 FT cells were transiently transfected with one of the Drp1 variant plasmids, as shown in (A), as indicated by the numbers. Lysates were prepared 24 hr after transfection and were subjected to IP with anti-FLAG antibody followed by Western blot analysis (WB) using anti-SUMO-1 to detect SUMOylated Drp1. The bands recognized by SUMO-1 antibody were also detected by FLAG antibody (data not shown).
(D) HEK 293 FT cells and (E) postnatal cortical neurons were transiently transfected with one of the plasmids shown in (A). Intracellular distribution of the exogenously expressed Drp1 was determined by FLAG immunostaining 24 (D) and 48 hr (E) after transfection. Mitochondria in HEK 293 FT cells were visualized by co-transfection with the Mito-EGFP plasmid. Upper and lower panels show the distribution of each splice variant in the soma and neurites in the same cell, respectively (E).
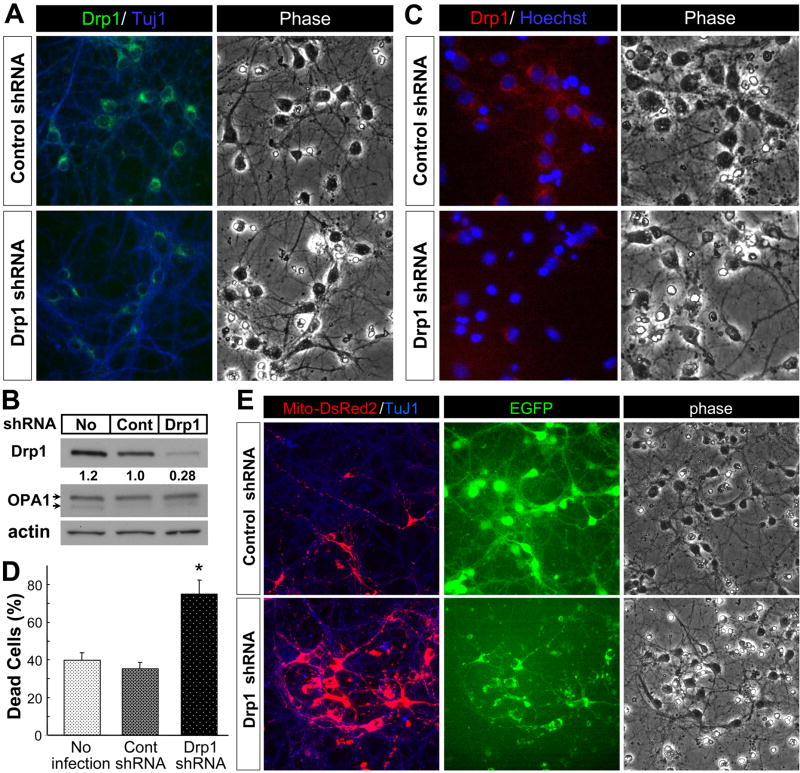
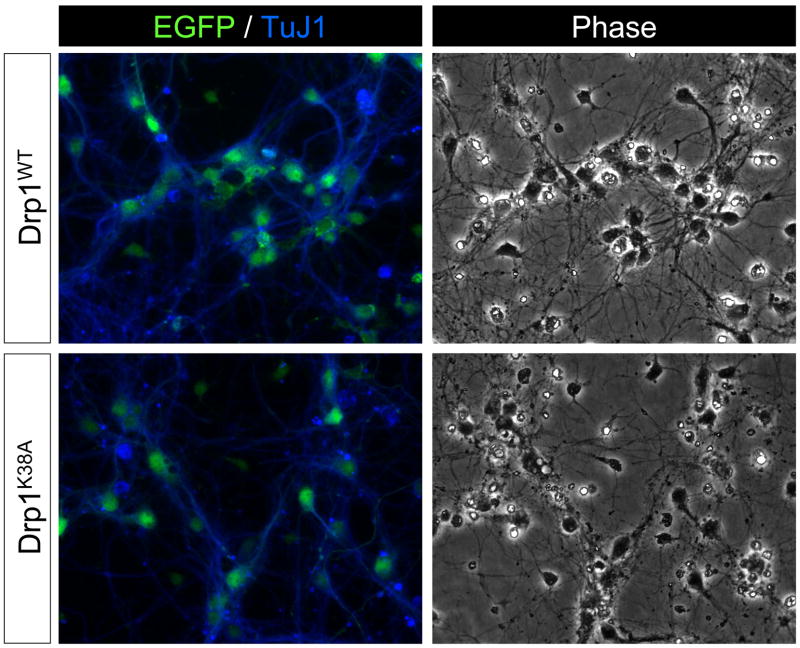
The observation that neurons displayed a unique pattern of Drp1 expression prompted us to determine if neuronal Drp1 plays a distinct role in regulating mitochondrial morphology and cell viability. To this end, the expression of Drp1 was knocked down by using lentivirus expressing Drp1 shRNA. Drp1 shRNA significantly reduced Drp1 expression in postnatal cortical neurons relative to neurons infected with lentivirus expressing control shRNA (Fig. 5A). The reduction in Drp1 protein mediated by the shRNA was confirmed by Western blot analysis, which showed no effect on OPA1 levels (Fig. 5B) and by immunostaining (Fig. 5C). Drp1 knockdown induced a significant increase in neuronal cell death as evidenced by the accumulation of neurons displaying nuclear condensation (Hoechst stain: Fig. 5C), highly shrunken cell bodies (phase contrast image: Fig. 5C, E) and the overall loss of neuronal arborization in Drp1 shRNA-expressing neurons as revealed by EGFP fluorescence (Fig. 5E EGFP). When quantitated, there was a two-fold increase in the number of dying neurons induced by Drp1 shRNA treatment relative to non-infected and control shRNA lentivirus-infected neurons (Fig. 5D). Mitochondria in the processes of neurons expressing Drp1 shRNA were significantly longer than those observed in neurons expressing control shRNA (Fig. 5E), consistent with a reduction in mitochondrial fission. Also, there was increased accumulation of Mito-DsRed2 fluorescence throughout and, in particular, near and in the soma of the neurons expressing Drp1 shRNA compared to those expressing control shRNA. To corroborate the effects obtained by suppressing Drp1 expression with Drp1 shRNA we also evaluated the viability of neurons in response to blocking Drp1 activity by expressing a dominant negative form of Drp1 (Drp1K38A). The K38A mutation changes a critical lysine in the G1 consensus motif of the GTPase domain into an alanine, thereby inhibiting GTP binding to Drp1 (Smirnova et al., 1998). This mutation is sufficient to inhibit mitochondrial fission in a dominant-negative fashion in mammalian cells (Smirnova et al., 2001). Postnatal cortical neurons over-expressing wild-type Drp1 (Drp1WT) displayed healthy cell bodies, and their neurites looked smooth and continuous (Fig. 6), a characteristic associated with healthy neurons (Xiang et al., 1996). In marked contrast, neurons infected with Drp1K38A lentivirus displayed a significant degree of cellular damage. Their cell bodies were shrunken and their neurites were thinner, fragmented in appearance and generally less abundant (Fig. 6, phase-contrast image). These results, consistent with those obtained by Drp1 shRNA, suggest that the deleterious effect of Drp1 knockdown (Fig. 5) is not due to an off-target effect of the Drp1 shRNA.
Figure 5. Drp1 shRNA reduces Drp1 protein levels and induces neuronal cell death.
(A) Primary cultures of postnatal mouse cortical neurons were infected with control shRNA (10 MOI) or Drp1 shRNA (10 MOI) lentivirus 24 hr after plating. Cells were fixed five days after infection and immunostained for Drp1 or β-tubulin III (TuJ1 antigen). The data is representative of three independent experiments.
(B) Primary cultures of postnatal mouse cortical neurons were infected as described in (A) above. Five days later protein extracts were prepared as described in the methods and expression of mitochondrial fission (Drp1) and fusion (OPA1) proteins was evaluated by Western blot analysis. Drp1 expression levels were quantitated and normalized against actin as described in Fig. 2A and presented as values relative to the level in control shRNA. The data is representative of two independent experiments. No, no infection; Cont, control shRNA lentivirus; Drp1, Drp1 shRNA lentivirus
(C) Primary cultures of postnatal mouse cortical neurons were infected as described in (A) above and five day later the cells were fixed, immunostained for Drp1 and stained with Hoechst 33258 to depict nuclear morphology. The data is representative of three independent experiments.
(D) The percentage of damaged neurons displaying shrunken, condensed nuclei based on Hoechst 33258 DNA staining was determined five days after infection in ten random fields per condition (approximately 1500 cells counted in total per condition). The data is representative of three independent experiments. *, significantly different from all other conditions (p < 0.0005, one-way ANOVA using Tukey’s post hoc test).
(E) Primary cultures of postnatal mouse cortical neurons were co-infected with Mito-DsRed2 lentivirus (10 MOI) and either control shRNA or Drp1 shRNA lentivirus (10 MOI) that co-express EGFP 24 hr after plating. Cells were fixed five days after infection and immunostained for β-tubulin III (TuJ1 antigen), a neuronal marker. EGFP fluorescence represents neurons expressing control/Drp1 shRNA. Representative data are shown from three independent experiments.
Figure 6. Dominant negative Drp1 induces neuronal cell death.
Primary cultures of postnatal mouse cortical neurons were infected with Drp1WT (10 MOI) or Drp1K38A (10 MOI) lentivirus 24 hr after plating. The lentivirus expressing either form of Drp1 also co-expressed EGFP from an internal ribosomal entry site. Cells were fixed five days after infection and immunostained for β-tubulin III (TuJ1 antigen), a neuronal marker. Representative data are shown from four independent experiments.
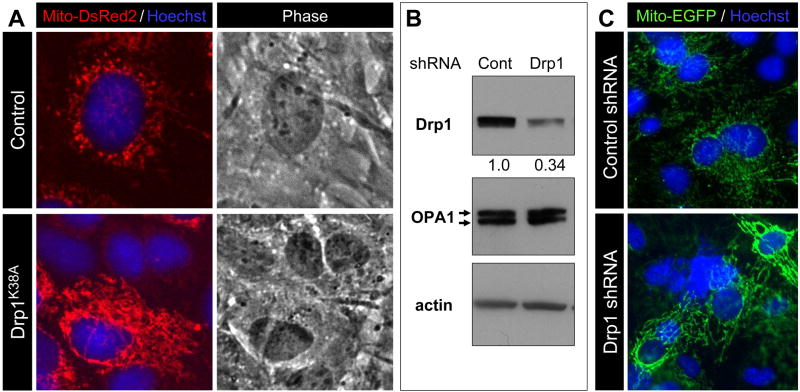
In contrast to its detrimental effect on neuronal viability, suppression of Drp1 activity/expression in MEF (Fig. 7A, C) and HeLa cells (Supplemental Figure 1) showed no evidence of damage or cell death (data not shown) despite clear evidence of reduced Drp1 expression (Fig. 7B) and increased mitochondrial fusion (Fig. 7A, C). Moreover, in HeLa cells Drp1 overexpression showed no effect on mitochondrial morphology or cell viability, either (Supplemental Figure 1A). Thus, the adverse effects associated with Drp1 suppression on cellular integrity and viability appear to occur specifically in neurons. These findings suggest that constitutively high levels of Drp1 are a prerequisite for cell viability in neurons.
Figure 7. Drp1 shRNA reduces Drp1 protein levels and increases mitochondrial fusion but does not cause cell death in mouse embryonic fibroblasts.
(A) MEF were co-infected with Mito-DsRed2 lentivirus (2 MOI) and with either control lentivirus expressing EGFP (10 MOI) or dominant negative Drp1K38A lentivirus co-expressing EGFP (10 MOI) 24 hr after plating. Cells were fixed five days after infection and stained with Hoechst 33258 to depict nuclear morphology. Under the infection conditions used, the infection efficiency with Drp1K38A lentivirus and its control virus was 100% (EGFP fluorescence not shown). Representative data are shown from two independent experiments.
(B) MEF were infected with lentivirus expressing control shRNA (10 MOI) or Drp1 shRNA (10 MOI) 24 hr after plating. Expression of mitochondrial fission (Drp1) and fusion (OPA1) proteins in each sample was evaluated five days after infection by Western blot analysis. The intensity of Drp1 bands was quantitated and normalized against β-actin as described in Figure 2A and presented as values relative to the level in control shRNA. Representative data are shown from two independent experiments.
(C) MEF were co-infected with Mito-EGFP lentivirus (2 MOI) and either control shRNA or Drp1 shRNA lentivirus (10 MOI) 24 hr after plating. Cells were fixed five days after infection and stained with Hoechst 33258 to depict nuclear morphology. The lentivirus used in this study did not express an infection marker (EGFP), so viability was judged in those cells displaying a high degree of mitochondrial fusion that was never observed in naïve MEF expressing only Mito-EGFP or those co-infected with Mito-EGFP and control shRNA lentivirus. No evidence of damage or cell death was observed in cells expressing extensive mitochondrial fusion even five days after infection based on the morphology of Hoechst stained nuclei. Representative data are shown from two independent experiments.
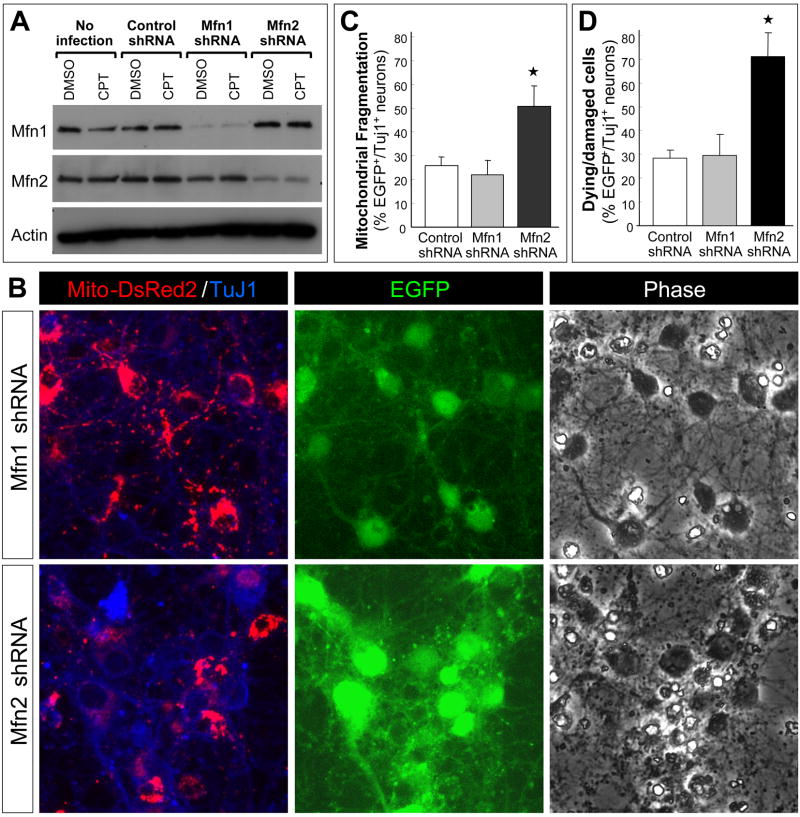
Western blot data for the proteins regulating mitochondrial dynamics and the results obtained with suppression of Drp1 expression (shRNA) and activity (K39A) suggest that the ratio of fusion/fission proteins is really important for maintenance of neuronal mitochondrial morphology and viability. To further evaluate this concept and to extend the results obtained with shifting the balance of fusion/fission toward fusion (Drp1 suppression), we determined if shifting the balance of mitochondrial dynamics towards fission also compromised the viability of postnatal cortical neurons. We have shown that both the Mfn1 and Mfn2 proteins are expressed in postnatal cortical neurons (Fig. 2A). Apoptotic induction by the DNA-damaging agent camptothecin did not alter expression levels of these proteins (Fig. 8A). Neurons infected with lentivirus expressing Mfn1 or Mfn2 shRNA showed significant and selective knockdown of their respective target protein (Fig. 8A). Mfn1 knockdown did not appear to change mitochondrial morphology and did not induce fission or a loss of viability (Fig. 8B, D). In contrast, Mfn2 knockdown resulted in a significant extent of fission (Fig. 8B, C) and neuronal cell death (Fig. 8D). Fission was so extensive in Mfn2 shRNA-expressing neurons that mitochondria were barely detectable in the processes (Fig. 8B). These findings demonstrate that maintaining a proper ratio of fusion/fission proteins is essential for sustaining mitochondrial morphology and viability in postnatal cortical neurons.
Figure 8. Mfn2 knockdown promotes extensive mitochondrial fission and loss of neuronal viability.
(A) Primary cultures of postnatal mouse cortical neurons were used without infection or infected with control shRNA, Mfn1 shRNA or Mfn2 shRNA lentivirus (10 MOI) 24 hr after plating. Neurons were treated with camptothecin (CPT: 5 μM) or DMSO (control) 48 after infection. Protein extracts were prepared, as described in the methods, 12 hr after treatment when neurons are committed to undergo apoptosis in a p53- and Bax-dependent manner. Expression of mitochondrial fusion proteins, Mfn1 and Mfn2, was evaluated by Western blot analysis.
(B) Neurons were co-infected with Mito-DsRed2 lentivirus (2 MOI) and either Mfn1 shRNA or Mfn2 shRNA lentivirus (10 MOI) that co-expresses EGFP 24 hr after plating. Cells were fixed two days after infection and immunostained for β-tubulin III, a neuronal marker (TuJ1 antigen). Representative data are shown from five independent experiments.
(C) and (D) Neurons were co-infected with Mito-DsRed2 lentivirus (2 MOI) and either control shRNA, Mfn1 shRNA or Mfn2 shRNA lentivirus (10 MOI) 24 hr after plating. Cells were fixed two days after infection and stained with Hoechst 33258 to depict nuclear morphology. Infected cells and neurons were identified based on the fluorescence of EGFP, co-expressed by the shRNA lentivirus used, and β-tubulin III (TuJ1) immunoreactivity, respectively. The fraction (%) of EGFP+/Tuj1+ neurons displaying (C) mitochondrial fragmentation (mitochondria less than 2μm in size) or (D) cell death (nuclear condensation and fragmentation based on Hoechst staining) was determined in three independent experiments by an observer that was blind to the treatments. The data reflects the average ± S.D. and is representative of three independent experiments. *, significantly different from all other conditions (p < 0.0005, one-way ANOVA using Tukey’s post hoc test).
Discussion
A growing body of evidence suggests that alterations in the regulation of mitochondrial fusion/fission are involved in acute brain injuries and chronic neurodegenerative diseases. However, the molecular basis for the extensive changes in mitochondrial fusion/fission that occur in neurons in response to specific types of injuries has not been elucidated. Moreover, it is not understood how mitochondrial fusion/fission events are constitutively regulated in developing or healthy differentiated neurons. In the present study, we carried out a comparative analysis of mitochondrial shape, the expression pattern of proteins that regulate mitochondrial fusion/fission and the functional consequences of interfering with the activity/expression of the fission protein, Drp1, in neuronal and non-neuronal cells. The results demonstrate that: (1) differentiated neurons in vitro and in vivo possess a short, distinctly tubular mitochondrial morphology, whereas non-neuronal cells exhibit longer, highly interconnected mitochondrial networks; (2) differential expression of Drp1 may, in part, account for those differences in mitochondrial shape and (3) differentiated neurons require high levels of Drp1 for maintenance of their viability.
Differentiated neurons exhibit a unique expression pattern of mitochondrial fusion/fission proteins
A similar pattern of highly elevated levels of Drp1 and OPA1 expression was observed in both cultured postnatal cortical neurons and the neuron rich superior colliculus relative to non-neuronal cells or glia-rich elements. The characteristic morphology and distribution of mitochondria was also conserved between cortical neurons in vitro and retinal ganglion cells in vivo. These findings suggest that cortical neurons in culture may be an appropriate and useful model for evaluating the effects of regulating Drp1 and OPA1 expression/activity on neuronal viability and function.
Although the present study demonstrated elevated Drp1 levels in neurons, it may not be the absolute levels of Drp1 alone, but rather the ratio of Drp1 levels to the level of mitofusin proteins, that dictates the relative rate of fission and fusion. Indeed, the ratio of Drp1 to Mfn1 and Mfn2 were very similar among neurons, while they were much lower for astrocytes and MEF (Drp1/Mfn1 ratio was 0.23 for both astrocytes and MEF; Drp1/Mfn2 ratios were 0.49 and 0.47 for astrocytes and MEF respectively, when the ratios of Drp1/Mfn1 and Drp1/Mfn2 in postnatal cortical neurons are set to 1.0: see Fig. 2A). The differential expression between neurons vs. non-neuronal cells (astrocytes and MEF) in culture was generally conserved in vivo, too, as retina and optic nerve, which are both rich in glial components, express relatively higher levels of Mfn proteins while superior colliculus, which is neuron rich, expresses relatively higher levels of Drp1 and OPA1.
Collectively, the Drp1/Mfn ratios observed suggest that neurons are more heavily weighted toward fission compared to non-neuronal cells. This may be necessary to maintain a small, tubular, “motile mitochondrial unit” enabling the highly dynamic mitochondrial transport required for neurons. This implies that neurons may be very sensitive to dysfunction in the mitochondrial fission machinery. Neurons also expressed high levels of OPA1 in vitro and in vivo. In addition, while our blots were not intended to fully resolve OPA1 proteins, it was clear that OPA1 isoforms were differentially expressed in neurons compared with non-neuronal cells. Large amounts of OPA1 might be necessary for the high degree of mitochondrial matrix remodeling that is required for mitochondrial maturation (Olichon et al., 2003; Griparic et al., 2004). Alternatively, high expression levels of OPA1 may normally serve to regulate cristae junction opening and cytochrome c release thereby providing anti-apoptotic activity (Yamaguchi et al., 2008).
Suppression of Drp1 leads to neuronal cell death
Excessive mitochondrial fission has increasingly been associated with mitochondrial dysfunction and the execution of apoptosis (Youle and Karbowski, 2005). Conversely, chemical or genetic interference with the mitochondrial fission machinery is reported to inhibit apoptotic mitochondrial fragmentation and delays cell death (Youle and Karbowski, 2005; Cassidy-Stone et al., 2008). Moreover, until very recently, human genetic studies had only identified mutations in fusion proteins such as Mfn2 and OPA1, which cause defects in the nervous system, Charcot-Marie Tooth Syndrome (Zuchner et al., 2004) and Autosomal Dominant Optic Atrophy (Alexander et al., 2000; Delettre et al., 2000), respectively. Therefore, inhibition of mitochondrial fission has been considered beneficial for mitochondrial function as well as protective against apoptosis-inducing stress. Contrary to this notion, in the present study, we found that suppression of Drp1 activity/expression can induce neuronal cell death, while Drp1 overexpression has no effect. Our observation, however, is consistent with a recent series of studies reporting: (1) the discovery of a dominant-negative mutation in the Drp1 gene in a newborn child with abnormal brain development (Waterham et al., 2007); (2) loss-of-function mutations found in the fission protein, GDAP1, that lead to Charcot-Marie-Tooth disease (Niemann et al., 2005); (3) genetic evidence that overexpression of mitochondrial fission proteins phenotypically counteracts disease-associated mitochondrial fusion and confers protection in a Drosophila model of Parkinson’s disease (Poole et al., 2008; Yang et al., 2008; Deng et al., 2008), and (4) the association of reduced levels of the Drp1 protein with abnormal mitochondrial morphology and distribution in fibroblasts from sporadic Alzheimer’s disease patients (Wang et al., 2008a). Moreover, Drp1-dependent fission is required for delivery of mitochondria to dendritic protrusions, vesicle clusters and synapses. Conversely, defects in the fission machinery lead to a loss of dendritic spines and synapses (Li et al., 2004; Li et al., 2008). From these results and our findings, we propose that neurons are more sensitive to excessive mitochondrial fusion than non-neuronal cells and a shift toward increased fusion may result in cellular dysfunction and ultimately cell death.
Further studies will be necessary to determine whether mitochondrial fusion achieved through manipulation of other fission or fusion proteins (e.g., Mfn overexpression) can also cause neuronal damage as seen with Drp1 suppression. This will determine whether the detrimental effect of Drp1 suppression is mediated solely through Drp1’s function as a fission protein or through Drp1-specific functions that are distinct from its mitochondrial fission activity and mediated through protein-protein interactions such as the Drp1-Bcl-xL interaction recently reported (Li et al., 2008).
Although Drp1 overexpression had no effect on mitochondrial morphology or neuron viability, increased net fission activity resulting from Mfn2 suppression precipitated excessive mitochondrial fission and neuronal cell death. Surprisingly, Mfn1 knockdown did not affect mitochondrial morphology or neuronal viability suggesting that, of the two fusion proteins, Mfn2 plays a central role in the constitutive regulation of mitochondrial dynamics in neurons. Nonetheless, as Mfn1 expression is clearly abundant in postnatal cortical neurons, its function in mitochondrial dynamics in healthy and damaged neurons remains to be elucidated. The differential outcomes associated with Drp1 overexpression vs. Mfn2 suppression, both purported to increase fission activity, also remains to be explained.
Evidence for a potential neuron-specific function of Drp1
A key finding in the present study is the demonstration that the exon 3-containing Drp1 species is expressed specifically in neurons. Inclusion of exon 3 inserts an additional amino acid sequence in the GTPase domain. Notably, in dynamin A, the parental protein of Drp1, from Dictyostelium discoideum, inclusion of exon 3 inserts 13 amino acids into the loop connecting the β2 and β2A sheets of the GTPase domain. This loop of dynamin A is exposed to the solvent and has been proposed as an interaction site for a subset of proteins (Niemann et al., 2001). By analogy, the corresponding loop encoded by exon 3 in neuronal Drp1 may provide the neuron-specific form of Drp1 with unique activities or modify its distribution, binding partners or stability. Despite the potential importance of exon 3 inclusion, this form of Drp1 has not been used often for overexpression studies. As summarized in Supplemental Table 1, it has primarily been used for non-neuronal cells and neuroblastoma cells (Yoon et al., 1998; Wang et al., 2008a; Wang et al., 2008b) but not for differentiated neurons,. Thus, a Drp1 construct containing exon 3 has not been appropriately tested in differentiated neurons. Our initial findings suggest that inclusion of exon 3 in Drp1 is not a principal determinant of Drp1 binding to Bcl-xL. Moreover, exon 3 (and exons 16/17) is not required for SUMOylation of Drp1 and does not appear to modify the intracellular localization of Drp1 in HEK 293 FT cells or postnatal cortical neurons. Additional studies will be needed to fully appreciate any unique functions or binding interactions conferred by inclusion of exon 3 and exons 16/17.
Given the observation that exon 3 is excluded in spinal cord neural progenitor cells as shown in the present study and also in Neuro2A neuroblastoma cells (T.U. and R.S.M., unpublished observation), it will be important to determine when and how the neuron-specific splicing event incorporating exon 3 is switched on during neuronal development and if it is essential for any specific step of neuronal commitment, differentiation and maturation. A complete functional characterization of the neuron-specific isoform of Drp1 is essential for understanding how Drp1 contributes to the regulation of neuronal mitochondrial morphology and viability and also to neuron specific physiological functions such as synaptic transmission (Li et al., 2008).
Summary
The present findings indicate that postmitotic differentiated neurons display unique patterns of Drp1 expression compared to non-neuronal cells. Furthermore, we suggest that Drp1-dependent mitochondrial fission is essential for the maintenance of mitochondrial integrity in neurons and that any impairment in the Drp1-mediated fission process promotes changes in mitochondrial morphology consistent with increased mitochondrial fusion and subsequent loss of neuronal viability. In addition, the findings of this study clearly indicate that a significant shift in the balance of fusion/fission protein expression toward fission can also have deleterious effects on neuronal survival. Thus, the regulation of mitochondrial dynamics must be tightly regulated in neurons, but the contributions of individual fusion/fission proteins to mitochondrial dynamics as well as cell viability appear to be unique and specific.
Supplementary Material
Supplemental Figure 1. Suppression of Drp1 activity/expression induces extensive mitochondrial fusion with no cytopathic effects in HeLa cells.
(A) HeLa cells were transiently co-transfected with pMito-EGFP plasmid and a plasmid for expression of either Drp1WT or Drp1K38A. pCDNA3 plasmid was used as a vector control. Cells were fixed 48 hr later and processed for EGFP imaging (green). Nuclei were counterstained with Hoechst 33258 (blue).
(B) HeLa cells were transfected with a pREP4-based control (no shRNA sequence) or a human Drp1 shRNA construct (Lee et al., 2004). Cells were re-fed 24 hr later with fresh media containing 200 μg/ml hygromycin B for 2 days followed by 10 days in media containing 75 μg/ml hygromycin B for the selection of stable transfectants. Cells were then fixed and processed for immunostaining for Drp1 (red) and HSP60 (green), a mitochondrial marker, and for nuclear staining with Hoechst 33258 (blue).
Supplemental Figure 2. Short and punctate morphology is a normal feature of mitochondria in healthy retinal ganglion cells.
Images of Mito-EGFP fluorescence and NeuN immunoreactivity are shown for healthy retinal ganglion cells (A: the same image as for Fig. 1Ba) and a sick retinal ganglion cell (B). The great majority of retinal ganglion cells infected with Mito-EGFP lentivirus displayed a short and punctate mitochondrial morphology as shown in (A). In sharp contrast, mitochondrial morphology was dramatically different in sick cells. Sick ganglion cells were identified based on the swollen soma which is readily seen in the NeuN images and the relatively condensed nucleus (the area with less intense NeuN staining). In these very rare cases of sick retinal ganglion cells mitochondria appeared elongated/fused and clustered around the nucleus while being lost from the processes. These results indicate that although the short and punctate mitochondrial morphology in these retinal ganglion cells looks fragmented when compared to the mitochondrial morphology in non-neuronal cells, it represents a normal and rather neuron-specific mitochondrial morphology in retinal ganglion cells and is not due to any pathological condition that might have been caused by virus-mediated expression of Mito-EGFP. Of note, healthy neurons often displayed aggregated mitochondrial clusters in the area of the proximal axon called the “axon initial segment” (arrowheads). This was observed with a high incidence and, therefore, appears to be a normal feature of mitochondrial distribution in healthy retinal ganglion cells. Bars = 20 μm.
Acknowledgments
The authors wish to acknowledge members of the Morrison lab for thoughtful discussions of the data. We also thank Cody Wyles for technical assistance and Bonita Lee and Chelsey Johnson for genotyping. The authors also thank Dr. Richard J. Youle (NIH) for providing molecular tools for the Drp1 studies. This work was supported by grants from the National Institutes of Health NS35533 and NS056031 to R.S.M., a grant from the Mitochondrial Research Guild at Children’s Hospital and Research Center to R.S.M, Glaucoma Research Foundation Catalyst for a Cure initiative to P.J.H. and by a grant to support the viral core facility at the Fred Hutchinson Cancer Center (DK 56465).
Abbreviations
- Drp1
dynamin related protein-1
- Mfn
mitofusin
- OPA1
optic atrophy-1
- Fis1
fission-1
- PFA
paraformaldehyde
- RT
reverse transcription
- MEF
mouse embryonic fibroblast
- MOI
multiplicity of infection
- TU
transduction unit
- PB
phosphate buffer
- TB
Tris-Base buffer
- shRNA
short hairpin RNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akepati VR, Muller EC, Otto A, Strauss HM, Portwich M, Alexander C. Characterization of OPA1 isoforms isolated from mouse tissues. J Neurochem. 2008;106:372–383. doi: 10.1111/j.1471-4159.2008.05401.x. [DOI] [PubMed] [Google Scholar]
- Alexander C, Votruba M, Pesch UE, Thiselton DL, Mayer S, Moore A, Rodriguez M, Kellner U, Leo-Kottler B, Auburger G, Bhattacharya SS, Wissinger B. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat Genet. 2000;26:211–215. doi: 10.1038/79944. [DOI] [PubMed] [Google Scholar]
- Barsoum MJ, Yuan H, Gerencser AA, Liot G, Kushnareva Y, Graber S, Kovacs I, Lee WD, Waggoner J, Cui J, White AD, Bossy B, Martinou JC, Youle RJ, Lipton SA, Ellisman MH, Perkins GA, Bossy-Wetzel E. Nitric oxide-induced mitochondrial fission is regulated by dynamin-related GTPases in neurons. Embo J. 2006;25:3900–3911. doi: 10.1038/sj.emboj.7601253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Stone A, Chipuk JE, Ingerman E, Song C, Yoo C, Kuwana T, Kurth MJ, Shaw JT, Hinshaw JE, Green DR, Nunnari J. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev Cell. 2008;14:193–204. doi: 10.1016/j.devcel.2007.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PH. Mitochondria and neuronal death/survival signaling pathways in cerebral ischemia. Neurochem Res. 2004;29:1943–1949. doi: 10.1007/s11064-004-6869-x. [DOI] [PubMed] [Google Scholar]
- Chen H, Chan DC. Emerging functions of mammalian mitochondrial fusion and fission. Hum Mol Genet. 2005;14:R283–289. doi: 10.1093/hmg/ddi270. Spec No. 2. [DOI] [PubMed] [Google Scholar]
- Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- Cipolat S, Martins de Brito O, Dal Zilio B, Scorrano L. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc Natl Acad Sci U S A. 2004;101:15927–15932. doi: 10.1073/pnas.0407043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delettre C, Lenaers G, Griffoin JM, Gigarel N, Lorenzo C, Belenguer P, Pelloquin L, Grosgeorge J, Turc-Carel C, Perret E, Astarie-Dequeker C, Lasquellec L, Arnaud B, Ducommun B, Kaplan J, Hamel CP. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet. 2000;26:207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- Deng H, Dodson MW, Huang H, Guo M. The Parkinson’s disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc Natl Acad Sci U S A. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMauro S, Schon EA. Mitochondrial disorders in the nervous system. Annu Rev Neurosci. 2008;31:91–123. doi: 10.1146/annurev.neuro.30.051606.094302. [DOI] [PubMed] [Google Scholar]
- Griparic L, van der Wel NN, Orozco IJ, Peters PJ, van der Bliek AM. Loss of the intermembrane space protein Mgm1/OPA1 induces swelling and localized constrictions along the lengths of mitochondria. J Biol Chem. 2004;279:18792–18798. doi: 10.1074/jbc.M400920200. [DOI] [PubMed] [Google Scholar]
- Howng SL, Sy WD, Cheng TS, Lieu AS, Wang C, Tzou WS, Cho CL, Hong YR. Genomic organization, alternative splicing, and promoter analysis of human dynamin-like protein gene. Biochem Biophys Res Commun. 2004;314:766–772. doi: 10.1016/j.bbrc.2003.12.172. [DOI] [PubMed] [Google Scholar]
- Jahani-Asl A, Cheung EC, Neuspiel M, MacLaurin JG, Fortin A, Park DS, McBride HM, Slack RS. Mitofusin 2 protects cerebellar granule neurons against injury-induced cell death. J Biol Chem. 2007;282:23788–23798. doi: 10.1074/jbc.M703812200. [DOI] [PubMed] [Google Scholar]
- James DI, Parone PA, Mattenberger Y, Martinou JC. hFis1, a novel component of the mammalian mitochondrial fission machinery. J Biol Chem. 2003;278:36373–36379. doi: 10.1074/jbc.M303758200. [DOI] [PubMed] [Google Scholar]
- Johnson MD, Kinoshita Y, Xiang H, Ghatan S, Morrison RS. Contribution of p53-dependent caspase activation to neuronal cell death declines with neuronal maturation. J Neurosci. 1999;19:2996–3006. doi: 10.1523/JNEUROSCI.19-08-02996.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott AB, Perkins G, Schwarzenbacher R, Bossy-Wetzel E. Mitochondrial fragmentation in neurodegeneration. Nat Rev Neurosci. 2008;9:505–518. doi: 10.1038/nrn2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Jeong SY, Karbowski M, Smith CL, Youle RJ. Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol Biol Cell. 2004;15:5001–5011. doi: 10.1091/mbc.E04-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Chen Y, Jones AF, Sanger RH, Collis LP, Flannery R, McNay EC, Yu T, Schwarzenbacher R, Bossy B, Bossy-Wetzel E, Bennett MV, Pypaert M, Hickman JA, Smith PJ, Hardwick JM, Jonas EA. Bcl-xL induces Drp1-dependent synapse formation in cultured hippocampal neurons. Proc Natl Acad Sci U S A. 2008;105:2169–2174. doi: 10.1073/pnas.0711647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Okamoto K, Hayashi Y, Sheng M. The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell. 2004;119:873–887. doi: 10.1016/j.cell.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Niemann A, Ruegg M, La Padula V, Schenone A, Suter U. Ganglioside-induced differentiation associated protein 1 is a regulator of the mitochondrial network: new implications for Charcot-Marie-Tooth disease. J Cell Biol. 2005;170:1067–1078. doi: 10.1083/jcb.200507087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann HH, Knetsch ML, Scherer A, Manstein DJ, Kull FJ. Crystal structure of a dynamin GTPase domain in both nucleotide-free and GDP-bound forms. Embo J. 2001;20:5813–5821. doi: 10.1093/emboj/20.21.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olichon A, Baricault L, Gas N, Guillou E, Valette A, Belenguer P, Lenaers G. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J Biol Chem. 2003;278:7743–7746. doi: 10.1074/jbc.C200677200. [DOI] [PubMed] [Google Scholar]
- Petit A, Sellers DL, Liebl DJ, Tessier-Lavigne M, Kennedy TE, Horner PJ. Adult spinal cord progenitor cells are repelled by netrin-1 in the embryonic and injured adult spinal cord. Proc Natl Acad Sci U S A. 2007;104:17837–17842. doi: 10.1073/pnas.0703240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AC, Thomas RE, Andrews LA, McBride HM, Whitworth AJ, Pallanck LJ. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci U S A. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santel A, Frank S. Shaping mitochondria: The complex posttranslational regulation of the mitochondrial fission protein DRP1. IUBMB Life. 2008;60:448–455. doi: 10.1002/iub.71. [DOI] [PubMed] [Google Scholar]
- Smirnova E, Shurland DL, Ryazantsev SN, van der Bliek AM. A human dynamin-related protein controls the distribution of mitochondria. J Cell Biol. 1998;143:351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Chen H, Fiket M, Alexander C, Chan DC. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J Cell Biol. 2007;178:749–755. doi: 10.1083/jcb.200704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung JY, Engmann O, Teylan MA, Nairn AC, Greengard P, Kim Y. WAVE1 controls neuronal activity-induced mitochondrial distribution in dendritic spines. Proc Natl Acad Sci U S A. 2008;105:3112–3116. doi: 10.1073/pnas.0712180105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uo T, Kinoshita Y, Morrison RS. Neurons exclusively express N-Bak, a BH3 domain-only Bak isoform that promotes neuronal apoptosis. J Biol Chem. 2005;280:9065–9073. doi: 10.1074/jbc.M413030200. [DOI] [PubMed] [Google Scholar]
- Uo T, Kinoshita Y, Morrison RS. Apoptotic actions of p53 require transcriptional activation of PUMA and do not involve a direct mitochondrial/cytoplasmic site of action in postnatal cortical neurons. J Neurosci. 2007;27:12198–12210. doi: 10.1523/JNEUROSCI.3222-05.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Su B, Fujioka H, Zhu X. Dynamin-like protein 1 reduction underlies mitochondrial morphology and distribution abnormalities in fibroblasts from sporadic Alzheimer’s disease patients. Am J Pathol. 2008a;173:470–482. doi: 10.2353/ajpath.2008.071208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Su B, Siedlak SL, Moreira PI, Fujioka H, Wang Y, Casadesus G, Zhu X. Amyloid-beta overproduction causes abnormal mitochondrial dynamics via differential modulation of mitochondrial fission/fusion proteins. Proc Natl Acad Sci U S A. 2008b;105:19318–19323. doi: 10.1073/pnas.0804871105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasiak S, Zunino R, McBride HM. Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J Cell Biol. 2007;177:439–450. doi: 10.1083/jcb.200610042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham HR, Koster J, van Roermund CW, Mooyer PA, Wanders RJ, Leonard JV. A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med. 2007;356:1736–1741. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]
- Xiang H, Hochman DW, Saya H, Fujiwara T, Schwartzkroin PA, Morrison RS. Evidence for p53-mediated modulation of neuronal viability. J Neurosci. 1996;16:6753–6765. doi: 10.1523/JNEUROSCI.16-21-06753.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi R, Lartigue L, Perkins G, Scott RT, Dixit A, Kushnareva Y, Kuwana T, Ellisman MH, Newmeyer DD. Opa1-mediated cristae opening is Bax/Bak and BH3 dependent, required for apoptosis, and independent of Bak oligomerization. Mol Cell. 2008;31:557–569. doi: 10.1016/j.molcel.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ouyang Y, Yang L, Beal MF, McQuibban A, Vogel H, Lu B. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc Natl Acad Sci U S A. 2008;105:7070–7075. doi: 10.1073/pnas.0711845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y, Pitts KR, Dahan S, McNiven MA. A novel dynamin-like protein associates with cytoplasmic vesicles and tubules of the endoplasmic reticulum in mammalian cells. J Cell Biol. 1998;140:779–793. doi: 10.1083/jcb.140.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y, Krueger EW, Oswald BJ, McNiven MA. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol Cell Biol. 2003;23:5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, Karbowski M. Mitochondrial fission in apoptosis. Nat Rev Mol Cell Biol. 2005;6:657–663. doi: 10.1038/nrm1697. [DOI] [PubMed] [Google Scholar]
- Yuan H, Gerencser AA, Liot G, Lipton SA, Ellisman M, Perkins GA, Bossy-Wetzel E. Mitochondrial fission is an upstream and required event for bax foci formation in response to nitric oxide in cortical neurons. Cell Death Differ. 2007;14:462–471. doi: 10.1038/sj.cdd.4402046. [DOI] [PubMed] [Google Scholar]
- Zuchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J, Parman Y, Evgrafov O, Jonghe PD, Takahashi Y, Tsuji S, Pericak-Vance MA, Quattrone A, Battaloglu E, Polyakov AV, Timmerman V, Schroder JM, Vance JM. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Suppression of Drp1 activity/expression induces extensive mitochondrial fusion with no cytopathic effects in HeLa cells.
(A) HeLa cells were transiently co-transfected with pMito-EGFP plasmid and a plasmid for expression of either Drp1WT or Drp1K38A. pCDNA3 plasmid was used as a vector control. Cells were fixed 48 hr later and processed for EGFP imaging (green). Nuclei were counterstained with Hoechst 33258 (blue).
(B) HeLa cells were transfected with a pREP4-based control (no shRNA sequence) or a human Drp1 shRNA construct (Lee et al., 2004). Cells were re-fed 24 hr later with fresh media containing 200 μg/ml hygromycin B for 2 days followed by 10 days in media containing 75 μg/ml hygromycin B for the selection of stable transfectants. Cells were then fixed and processed for immunostaining for Drp1 (red) and HSP60 (green), a mitochondrial marker, and for nuclear staining with Hoechst 33258 (blue).
Supplemental Figure 2. Short and punctate morphology is a normal feature of mitochondria in healthy retinal ganglion cells.
Images of Mito-EGFP fluorescence and NeuN immunoreactivity are shown for healthy retinal ganglion cells (A: the same image as for Fig. 1Ba) and a sick retinal ganglion cell (B). The great majority of retinal ganglion cells infected with Mito-EGFP lentivirus displayed a short and punctate mitochondrial morphology as shown in (A). In sharp contrast, mitochondrial morphology was dramatically different in sick cells. Sick ganglion cells were identified based on the swollen soma which is readily seen in the NeuN images and the relatively condensed nucleus (the area with less intense NeuN staining). In these very rare cases of sick retinal ganglion cells mitochondria appeared elongated/fused and clustered around the nucleus while being lost from the processes. These results indicate that although the short and punctate mitochondrial morphology in these retinal ganglion cells looks fragmented when compared to the mitochondrial morphology in non-neuronal cells, it represents a normal and rather neuron-specific mitochondrial morphology in retinal ganglion cells and is not due to any pathological condition that might have been caused by virus-mediated expression of Mito-EGFP. Of note, healthy neurons often displayed aggregated mitochondrial clusters in the area of the proximal axon called the “axon initial segment” (arrowheads). This was observed with a high incidence and, therefore, appears to be a normal feature of mitochondrial distribution in healthy retinal ganglion cells. Bars = 20 μm.