Abstract
Molecules associated with the outer surface of living cells exhibit complex, non-Brownian patterns of diffusion. In this report, supported lipid bilayers were patterned with nanoscale barriers to capture key aspects of this anomalous diffusion in a controllable format. First, long-range diffusion coefficients of membrane-associated molecules were significantly reduced by the presence of the barriers, while short-range diffusion was unaffected. Second, this modulation was more pronounced for large molecular complexes than for individual lipids. Surprisingly, the quantitative effect of these barriers on long-range lipid diffusion could be accurately simulated using a simple, continuum-based model of diffusion on a nanostructured surface; we thus describe a metamaterial that captures the properties of the outer membrane of living cells.
The outer surface of cells presents a complex, nanostructured, yet fluid environment that controls the movement of signaling proteins. The lateral movement of many membrane biomolecules, including transmembrane or tethered proteins as well as lipids themselves, can be interpreted as being free and isotropic within compartments of the cell membrane measuring tens to hundreds of nanometers in scale1-6. These compartments are delineated by semi-permeable barriers that arise from interactions between the plasma membrane, underlying cytoskeleton, and associated proteins6-8. Fluctuations in these structures allow biomolecules to occasionally cross between compartments, allowing long-range, but comparatively slow, transport over the cell surface. More formally, transport along the membrane is an anomalous, non-Brownian process that can be characterized by two diffusion coefficients, one that describes short-range motion within an individual compartment and a second, smaller, effective diffusion coefficient that is associated with long-range motion over many barriers. The extent to which these values differ is dependent on the spacing and properties of the barriers as well as the diffusing molecule. Emerging models suggest significant impacts of this behavior on cell signaling2, 9, 10, but experimental systems for testing these hypotheses are not widely available. In this report, we capture this anomalous diffusion by nanopatterning supported lipid bilayers with barriers to lipid diffusion using a geometry that captures the semipermeable nature of those posed to be present in living cells. As is posed by models of these interactions, we aim to gain control over long-range diffusion, while maintaining local, isotropic diffusion associated with a membrane in the absence of such barriers. We demonstrate that these nanopatterned barriers give rise to different short- and long-range diffusion coefficients of lipids and membrane-associated proteins, and provide a quantitative model of this diffusion that suggests specific aspects of membrane structure at the sub-micrometer level.
The basic substrate supported lipid bilayer system consists of a phospholipid membrane in close association with an appropriate surface11, 12. A thin, subnanometer-thick layer of water separates the bilayer and substrate13-15, imparting lateral mobility to the membrane components. For reasons that are not well understood, bilayer formation is supported by a limited set of materials, notably silicon oxide-based substrates (glass, quartz, and silicon wafers, polydimethylsiloxane) and mica, which promote fusion of lipid vesicles into an extended, planar bilayer structure. Other materials such as metals, photoresists, or surface-adsorbed proteins are inert to vesicle fusion and provide a powerful method for directing the layout of supported lipid bilayers when micropatterned onto bilayer-compatible substrates16-18. Here, we extend this micropatterning approach to nanoscale dimensions in order to capture the scales of membrane compartments observed in living cells.
Glass substrates were patterned with linear arrays of barriers (Figure 1A) that are designed to restrict bilayer formation to the intervening regions of the surface. Barriers were spaced at either 125- or 250-nm center-to-center intervals (“spacing” in Figure 1A), dimensions that are associated with the size of membrane compartments on living cells. For this study, the barriers are specified to be 50 nm in width, the narrowest that could reliably and controllably be fabricated using our processing equipment (as described below). To capture the semi-permeable properties of their cell-based counterparts, gaps of 30, 40, and 50 nm length were interspersed throughout the barriers. Diffusing membrane molecules thus periodically encounter a break in the barrier, capturing the basic mechanism of the anchored-protein “picket fence” model, in which transmembrane proteins attached to cytoskeleton disrupt both leaflets of the membrane. These gaps were spaced such that the periodicity of each pattern is 500 nm (“length”). Gaps were staggered between lines, as opposed to being aligned at the same location on each barrier. An important aspect of this barrier layout is that while compartments observed on cell membranes are essentially closed-perimeter patches, the barriers used here break up the surface into parallel stripes, a geometry chosen to simplify the measurement of short- and long-range diffusion coefficients. Specifically, molecular motion parallel to the barriers is unhindered; in the absence of any impact of the barriers on local membrane mobility, measurement of long-range diffusion coefficients in this direction will be numerically equivalent to their short-range counterparts. The impact of these barriers on short-range and long-range diffusive behavior can thus be estimated by simultaneously measuring long-range diffusion along and across the barriers. Importantly, these long-range diffusion coefficients can be measured using standard fluorescence recovery after photobleaching (FRAP) techniques, offering simplified analysis and longer timescale observations than contemporary single particle tracking approaches, which require specialized, high temporal- and spatial-resolution equipment to reveal these dynamics.
Figure 1.
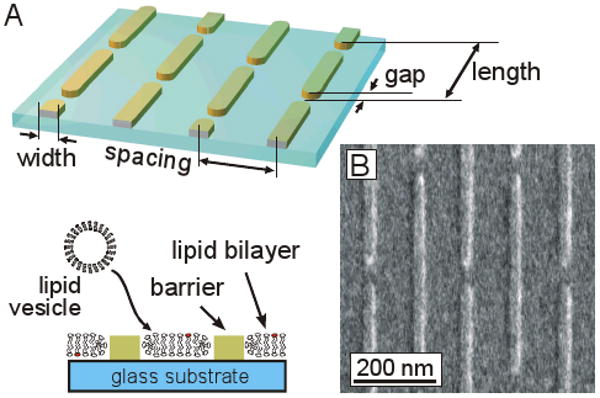
Nanopatterning of supported lipid bilayers. (A) A metal lift-off method was used to define parallel barriers on glass. These barriers are designed to disrupt supported lipid bilayers that will subsequently formed on these substrates. (B) Electron micrograph of a resultant nanopatterned surface.
An electron-beam based lift-off process was used to create these barriers. Coverslips were spin-coated with a lower resist layer of 25kDa PMMA, an upper resist layer of 950kDa PMMA (together ∼ 200 nm), and finally a charge dispersion layer of Aquasave (Mitsubishi Rayon Co., New York, NY). PMMA was patterned using an FEI XL-30 Scanning Electron Microscope fitted with a Nabity Pattern Generator and developed using standard methods. A 4 nm-thick layer (unless otherwise specified) of either Cr or Ti was then deposited using an electron beam evaporation system, and then the PMMA and overlying metal were finally removed in acetone. An electron micrograph of a typical surface is shown in Figure 1B, demonstrating the successful patterning of these substrates. Individual patterns covered an 80 μm × 80 μm region, each containing a specific combination of spacing and gap. Small unilamellar vesicles (SUVs) of egg phosphatidylcholine (egg PC, Avanti Polar Lipids) supplemented with 1 molar % Texas Red 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (TR-DHPE, Molecular Probes), at a concentration of 5 mg/ml in water, were prepared by extrusion through 50-nm pore polycarbonate membranes (Avanti Polar Lipids) using standard techniques19. Supported lipid bilayers were formed by exposing the nanopatterned substrates to SUVs for 5 minutes, followed by extensive rinsing with PBS and then deionized water. Imaging of substrates was carried out in phosphate buffered saline (pH 7.4).
Anisotropic diffusion of nanopatterned lipid bilayers
Fluorescence recovery after photobleaching methods were used to examine diffusion of TR-DHPE lipids, as illustrated in Figure 2A. This example contains Ti barriers spaced 250 nm center-to-center and oriented vertically in this image, interspersed with 50 nm gaps. Bilayers on the patterned regions appeared uniform by fluorescence microscopy but of slightly less intensity than the surrounding areas, consistent with the presence of a series of sub-diffraction-scale barriers that interrupt lipid bilayer formation. The left image shows a roughly 40 μm photo bleach spot created in the bilayer, while the right image was collected 30 seconds later. The initially round photobleach spot becomes elongated, illustrating the anisotropic properties of this surface and faster long-range diffusion in the vertical direction. At later timepoints, no evidence of the initial photobleach pattern remained, indicating that there is essentially no immobile fraction of TR-DHPE lipids.
Figure 2.
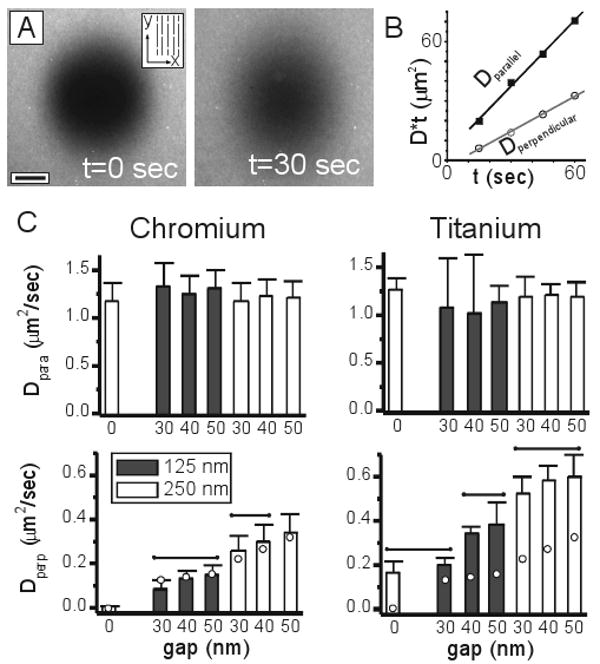
Anomalous diffusion captured on nanopatterned surfaces. (A) Anisotropic diffusion of TR-DHPE lipids on a surface containing barriers spaced 250 nm apart, and interspersed with 50 nm gaps. A 40 μm photo bleach spot was created on this surface, and images were collected immediately and 30 seconds after this process. Barriers are aligned vertically in this image, as indicated in the inset schematic. Scale bar = 10 μm. (B) Measurement of long-range diffusion coefficients. Dparallel provides a measure of short-range, unhindered diffusion in the lipid bilayer, while Dperpendicular captures the influence of barriers on long-range diffusion. (C) Diffusion coefficients as a function of barrier spacing, gap size, and barrier material. Data are mean ± s.d., and were compared by ANOVA methods, using Tukey's criteria, α=0.05, n = 4-12. No differences were detected between data of Dparallel (top row). For data of Dperpendicular (bottom row), groups within which no significant differences were detected are indicated by the overbars. Circles indicated values of Dperpendicular predicted using a continuum model (see text).
Long-range diffusion coefficients parallel and perpendicular to the barriers were calculated from images capturing the recovery process using a 2-dimensional Fourier transform method20, 21. Briefly, if C(x,y,t) represents the 2-dimensional concentration profile of a diffusing species (i.e., an image of a fluorescent marker) at some time, t, and Ĉ(u, v, t) is the Fourier transform of this data, then Ĉ(u,v,t) = Ĉ(u,v,0) * exp(−4π2((Dxt)u2+(Dyt)v2)), where Ĉ(u,v,0) represents the transform of data collected at time t=0, and Dx and Dy are long-range diffusion coefficients along two primary, independent axes. Non-linear curve fitting was used to estimate values of Dx*t and Dy*t that provide the best fit between the transformed data at t=0 and each of the subsequent timepoints; diffusion coefficients were calculated as the slope of these values with respect to time. Notably, information representing the photobleach spot was associated with low wavenumber components of the transforms. Curve fitting was restricted to these components both to limit the fitting procedure on the photobleach spot dynamics and to reject the influence of the immobile barriers, which were represented at much higher wavenumbers. The y-axis was assigned to be along the barriers, such that Dparallel ≡ Dy and Dperpendicular ≡ Dx. Values of D*t extracted from the timeseries from which the images in Figure 2A were taken are presented in Figure 2B and are linear along both directions, indicating that long-range diffusion appears as a purely Brownian process along these two independent axes with Dparallel being larger than Dperpendicular, providing a quantitative verification of anisotropic long-range diffusion. Diffusion coefficients measured on all surfaces in these two directions are presented in Figure 2C and discussed below.
Lipids exhibit free diffusion parallel to barriers
Long-range diffusion coefficients of TR-DHPE measured parallel to the barriers, Dparallel, were independent of barrier spacing, gap, and composition (Figure 2C, top row, as compared by ANOVA with Tukey's criterion, α=0.05) and similar to that on unpatterned regions of the substrates, indicating that mobility along this direction is essentially unhindered. These measurements are thus representative of the short-range diffusion of molecules away from the barriers. We note that during traversal of a gap in the barrier, individual lipid molecules will experience a short period of time in which transport in the direction parallel to the barriers will be limited. However, these gaps represent a small fraction of the total accessible surface; any effect of this limited mobility on long-range diffusion along the barrier direction is likely smaller than that detectable using our analyses.
Chromium barriers provide fine control over long-range diffusion
The behavior of lipids on our nanopatterned surfaces varied as a function of barrier material. We first focus on the surfaces containing chromium barriers, with later comparison to those with titanium.
On the chromium-patterned surfaces of all combinations of gap and spacing, long-range diffusion measured perpendicular to the barriers was significantly reduced in comparison to that parallel to these structures (Figure 2B, bottom row, ANOVA with Tukey's criterion, α=0.05). As expected, decreasing the spacing between barriers or the opportunity for a molecule to diffuse across (around) a barrier significantly reduces long-range diffusion.
Surprisingly, these results could be accurately predicted using a simple continuum model, based on the diffusion equation ∂C(x,y,t)/∂t = Df *∇2C(x,y,t) applied to a perforated, 2-dimensional slab capturing the geometry defined in Figure 1A and based on a single, isotropic diffusion coefficient, Df. The diamonds in the bottom row of Figure 2C indicate the long-range diffusion coefficients predicted by this model, using on a free diffusion coefficient of Df = 1.1 μm2/sec, typical of that observed on an unpatterned bilayer. The good agreement over all geometries (again, limited to the chromium barrier surfaces) suggests that: 1) chromium structures completely disrupt the bilayer and, 2) the physical properties of the membrane can be treated as an isotropic and homogenous material surrounding these barriers. This was surprising, since interrupting the lipid bilayer structure with a metal barrier or other edge would, at the simplest conceptual level, likely expose the hydrophobic core to water. In response to such a high-energy edge, lipids locally rearrange to balance the energy of membrane deformation against that of exposing the hydrophobic tails and forming a “hairpin” suggested in the schematic in the lower part of Figure 1A; such a structure has not been directly observed, but is supported by secondary measurements of membrane properties and molecular dynamics simulations22. The impact of such deformations on lipid partitioning and mobility are not well understood, particularly in the context of a substrate-supported lipid bilayer interfacing with a material barrier. Notably, previous studies have provided mixed results into even the spatial influence of such an edge region on diffusion, ranging from sub-micrometer to multi-micrometer distances23-26; in particular, Howland and colleagues26 identified a non-fluid phase and lipid-free moat in fluid bilayers surrounding a change in surface chemistry.
Assuming a similar, dramatic change in diffusivity, namely that of a non-fluid boundary region at the bilayer-barrier interface, our observations of diffusion parallel to the barriers suggests that any such boundary layer must be much less than half of the distance between the smallest barrier spacing, thus less than 33-nm. Furthermore, the good agreement between our experimental measurements and the continuum model suggest that in the assumption of a non-fluid boundary layer, such a region must be small compared to the smallest gap spacing (30 nm), or on the order of 10s of nanometers. Molecular dynamics simulations by Jiang et al. 22 suggest that, topologically, the hairpin configuration can be accommodated within a small, 10-nm region encompassing the bilayer edge, supporting the idea that any boundary layer on our nanopatterned surfaces may indeed be this thin. As an alternative to the presence of a single, non-fluid boundary layer, the bilayer edge on our surfaces may be composed of a potentially wider region with properties that vary in terms of diffusivity in a more gradual fashion. We note that the impact of such regions may be simply diluted to below the level of detectability in our experiments by a large fraction of membrane that does behave as a homogenous, isotropic medium.
To gain a better, structural understanding of these nanopatterned bilayers, we used atomic force microscopy to examine the topology of supported membranes formed on glass patterned with chromium barriers. Bilayers of egg PC + 1% TR-DHPE were examined in contact mode using a MikroMasch CSC38 cantilever (hydrophilic, 10.0 nm tip radius, 30° tip cone angle) with a force of 5 nN. Figure 3A shows a micrometer-scale scan of a nanopatterned bilayer on a substrate containing relatively tall (>10 nm) barriers, chosen to simplify identification with respect to the lipids. The barriers are visible as the tall, diagonal lines pointing to the lower right of the image, while small defects in the bilayer are observable as occasional dark pits dispersed throughout the image. Figure 3B shows a higher-resolution scan taken perpendicular to a series of three successive barriers. This scan also includes a bilayer defect (at ∼240 nm), providing a zero-height reference. From scans such as this, we measure a bilayer thickness of about 3 nm, which likely reflects a slight compression of the bilayer under the tip forces. We were not able to detect large deviations in the bilayer structure at the bilayer-barrier interfaces, between the barriers, or around the defects. Thus, this topological data is consistent with our FRAP measurements that the bilayers are effectively homogenous to within a small distance of the bilayer-barrier interface.
Figure 3.
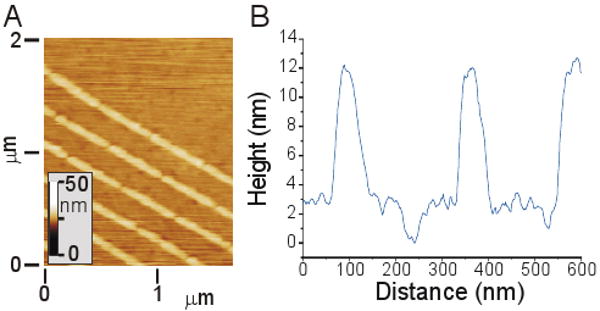
Atomic Force Microscopy of bilayer nanostructure. (A) Micrometer-scale scan of an egg PC bilayer formed on a substrate containing tall (∼12 nm) chromium barriers. (B) Higher magnification scan, illustrating bilayer topology between a series of barriers. A bilayer defect is shown at ∼240 nm in this scan.
Modulation of diffusion by clustering
An emerging model regarding the impact of this anomalous diffusion on cell signaling is that oligomerization of membrane proteins decreases their ability to cross the barriers, causing them to become localized to the site of clustering. We explored the ability of our surfaces to capture this process using supported lipid bilayers containing the ganglioside GM1 which were subsequently oligomerized using cholera toxin subunit B (CTB); pentamers of CTB bind up to five GM1 molecules, yielding a large, membrane-associated structure of roughly 30 nm2 footprint on the membrane27, as opposed to the ∼ 1 nm2 area associated with individual TR-DHPE lipids. Specifically, lipid vesicles composed of 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine (DOPC) supplemented with 1 % TR-DHPE and 0.1 % GM1 (Avanti) were prepared by extrusion and used to form bilayers on surfaces containing linear arrays of chrome barriers spaced 250 nm center-to-center and interspersed with 30 nm gaps. The resultant surfaces were blocked with bovine serum albumin (20 μg/ml in PBS for 30 minutes) to reduce non-specific protein binding, and subsequently incubated with fluorescently-labeled CTB (Alexa-488 conjugate, Invitrogen) at a concentration of 10 μg/ml. Binding of CTB was specific for GM1, as no adsorption of this protein to the bilayer and barriers was observed on surfaces in which GM1 was omitted.
Prior to the addition of CTB, diffusion of TR-DHPE was similar to that reported in Figure 2. CTB was then added to these surfaces. In all experiments, diffusion of CTB along the barriers was observed to be effectively Brownian (a linear relation between Dparallel*t and imaging interval, t), but varied considerably between experiments (including controls carried out on unpatterned glass) ranging from Dparallel = 0.02 to 0.12 μm2/sec. No immobile fraction of CTB was detected on these surfaces. In comparison to motion along the barriers, diffusion perpendicular to these structures was highly restricted. Specifically, CTB/GM1 complexes exhibited a Dperpendicular / Dparallel ratio of typically ∼ 0.02, as compared to 0.2 for TR-DHPE, demonstrating that these large structures have less ability to cross the nanoscale barriers than the smaller TR-DHPE lipids, and demonstrating the basic phenomenon posed to enhance the activity of protein clusters on the surface of living cells.
Barrier composition influences bilayer structure
While our analyses suggest that the use of chromium as a barrier material provides a straightforward, elegant approach to nanopatterning bilayers, a very different result was observed on barriers of titanium. As shown in Figure 2C, measured values of Dperpendicular were higher than that observed on chromium and/or predicted by the diffusion equation model for all pattern geometries. Moreover, significant diffusion was observed across the barriers with zero gap size. This result suggests that unlike chromium, titanium does not completely inhibit formation of an overlying supported bilayer, allowing at least partial connectivity between regions separated by this material. Notably, barrier composition is not usually given much consideration in studies of patterned bilayers, chosen more often on the basis of ease of fabrication or other experimental consideration such as durability. However, the barriers used in this study are much smaller than those typically used to direct lipid diffusion, and indeed the width of these barriers approaches that of the individual vesicles from which the planar structure are formed. At these scales, vesicles interacting with the barrier are likely to also interact with the glass substrate on either side of the barrier, or with a planar bilayer forming on that surface, leading to a continuous bilayer over a structure. In support of this idea, diffusion was not observed over Ti barriers measuring 100 nm in width and thus twice that of the expected diameter of the extruded vesicles (data not shown). Furthermore, recent studies have demonstrated that lipids can interact with bulk titanium oxide surfaces, forming structures that suggest lipid-substrate interactions that are weaker than those on silicon oxide yet stronger than on other materials28, 29. In addition, a recent report by Zhou and colleagues30 showed that lipid bilayers can form over individual carbon nanotubes as part of a new sensor design.
As a further demonstration of the formation of bilayers over Ti structures, we patterned surfaces with a square grid layout of 50 nm-wide barriers on glass, spaced at 500 nm intervals. Lipid bilayers formed on these surfaces exhibited complete recovery after photobleaching (Figure 4A), demonstrating an intact structure over the barriers. Moreover, long-range diffusion coefficients over these barriers were reduced to roughly half of that on unpatterned substrates, suggesting an alternative approach to creating semi-permeable barriers. However, we also found that these structures were very fragile. Figure 4A was collected using a relatively shallow photobleach (short exposure) of the TR-DHPE lipids. Deeper photobleach exposures resulted in fragmentation of the bilayer along the barriers (often outside of the photobleach region) as illustrated in Figure 4B. While lipids could diffuse within the individual corrals, the long-range diffusion was interrupted at random edges, leading to segmentation of the surface into corrals that were connected to each other, but separate from other groups. One explanation of this bilayer fracture is that exposure of the bilayers to light induces contraction or loss of bilayer material, a process that has been described using UV wavelengths31, 32; we speculate that photobleaching of the Texas Red fluorophores at their respective excitation wavelength may produce a similar effect here. Alternatively, local heating of the barriers may be responsible for these effects. In either case, the partial continuity of the bilayer at the barrier interface, or additional bending of the membrane at these locations, may explain the propensity of the bilayer to break at these edges. The onset of behavior was highly variable between surfaces of identical preparation, further highlighting the fragile nature of this configuration. This fragility limits the use of Ti barriers in mimicking membrane/cytoskeleton interactions, but may find utility as a simple method of dynamically patterning lipid bilayers.
Figure 4.
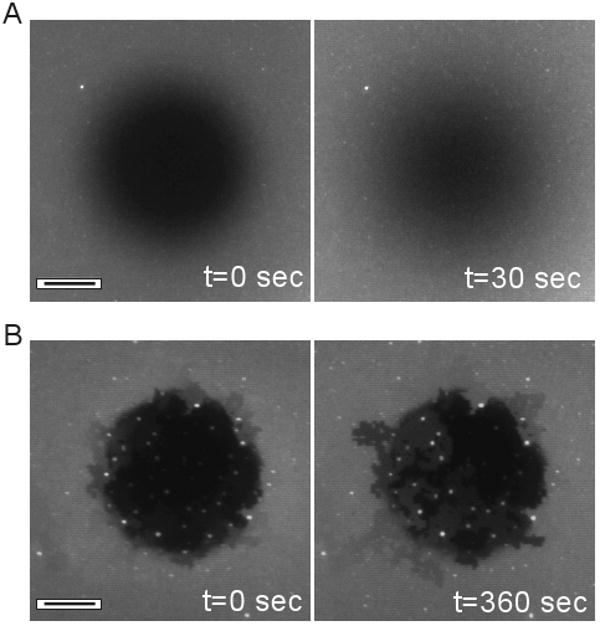
Diffusion over Ti barriers. Substrates were patterned with square grids of Ti barriers spaced 500 nm apart. (A) Recovery of a photobleach spot, demonstrating continuity of the lipid bilayers over the Ti barriers. (B) Longer exposure of the bilayer to the photobleach illumination induces separation of membrane patches from each other along the Ti barriers. Scale bars = 10 μm.
In summary, we demonstrate that nanopatterning of supported lipid bilayers with hard materials, particularly chromium, provides a powerful and predictable method for capturing the anomalous, non-Brownian diffusion of membrane lipids and proteins observed on the surface of living cells. We note that these barriers retard long-range diffusion by a mechanism that is slightly different than that posed to operate in living cells. Specifically, most models describing the semi-permeable nature of such barriers point to time-dependent fluctuations between the membrane, cytoskeleton, and proteins that link or are otherwise associated with these structures; at any given position along the edge of a compartment, molecules can cross to an adjoining region if at that moment there is a relatively large separation between these components. However, the dynamics of these fugitive interactions have not been observed directly, and would be very difficult to replicate, much less control, in a model system. Our approach trades some replication of this specific mechanism for the strengths of contemporary lithographic processes, which are in the ability to define barriers with great precision and control over geometry. These methods will provide a powerful platform for testing and comparing various models of anomalous diffusion, including their functional impacts on cell signaling.
Acknowledgments
We gratefully thank Evren U. Azeloglu for assistance with AFM imaging of the supported bilayers. This study was supported in part by the National Institutes of Health through the NIH Roadmap for Medical Research (PN2 EY016586). Its contents do not necessarily represent the official views of the NIH or the National Eye Institute. This work was also partially supported by the Nanoscale Science and Engineering Initiative of the National Science Foundation under NSF Award Number CHE-0641523 and by the New York State Office of Science, Technology, and Academic Research (NYSTAR).
References
- 1.Kusumi A, Sako Y, Yamamoto M. Biophysical Journal. 1993;65(5):2021–40. doi: 10.1016/S0006-3495(93)81253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kusumi A, Suzuki K, Koyasako K. Current Opinion in Cell Biology. 1999;11(5):582–90. doi: 10.1016/s0955-0674(99)00020-4. [DOI] [PubMed] [Google Scholar]
- 3.Ritchie K, Iino R, Fujiwara T, Murase K, Kusumi A. Molecular Membrane Biology. 2003;20(1):13–18. doi: 10.1080/0968768021000055698. [DOI] [PubMed] [Google Scholar]
- 4.Ritchie K, Spector J. Biopolymers. 2007;87(9999):95–101. doi: 10.1002/bip.20801. [DOI] [PubMed] [Google Scholar]
- 5.Fujiwara T, Ritchie K, Murakoshi H, Jacobson K, Kusumi A. Journal of Cell Biology. 2002;157(6):1071–81. doi: 10.1083/jcb.200202050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morone N, Fujiwara T, Murase K, Kasai RS, Ike H, Yuasa S, Usukura J, Kusumi A. J Cell Biol. 2006;174(6):851–862. doi: 10.1083/jcb.200606007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ritchie K, Kusumi A. Sub-Cellular Biochemistry. 2004;37:233–45. doi: 10.1007/978-1-4757-5806-1_7. [DOI] [PubMed] [Google Scholar]
- 8.Kusumi A, Sako Y. Current Opinion in Cell Biology. 1996;8(4):566–574. doi: 10.1016/s0955-0674(96)80036-6. [DOI] [PubMed] [Google Scholar]
- 9.Kusumi A, Ike H, Nakada C, Murase K, Fujiwara T. Seminars in Immunology. 2005;17(1):3–21. doi: 10.1016/j.smim.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 10.Ike H, Kosugi A, Kato A, Iino R, Hirano H, Fujiwara T, Ritchie K, Kusumi A. Chemphyschem. 2003;4(6):620–6. doi: 10.1002/cphc.200300670. [DOI] [PubMed] [Google Scholar]
- 11.Brian AA, McConnell HM. Proceedings of the National Academy of Sciences of the United States of America. 1984;81(19):6159–63. doi: 10.1073/pnas.81.19.6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sackmann E. Science. 1996;271(5245):43–8. doi: 10.1126/science.271.5245.43. [DOI] [PubMed] [Google Scholar]
- 13.Bayerl TM, Bloom M. Biophysical Journal. 1990;58(2):357–362. doi: 10.1016/S0006-3495(90)82382-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heine DR, Rammohan AR, Balakrishnan J. Molecular Simulation. 2007;33(4):391–397. [Google Scholar]
- 15.Johnson SJ, Bayerl TM, McDermott DC, Adam GW, Rennie AR, Thomas RK, Sackmann E. Biophysical Journal. 1991;59(2):289–294. doi: 10.1016/S0006-3495(91)82222-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boxer SG. Current Opinion in Chemical Biology. 2000;4(6):704–709. doi: 10.1016/s1367-5931(00)00139-3. [DOI] [PubMed] [Google Scholar]
- 17.Groves JT, Boxer SG. Accounts of Chemical Research. 2002;35(3):149–157. doi: 10.1021/ar950039m. [DOI] [PubMed] [Google Scholar]
- 18.Groves JT, Ulman N, Boxer SG. Science. 1997;275:651–653. doi: 10.1126/science.275.5300.651. [DOI] [PubMed] [Google Scholar]
- 19.Kung LA, Kam L, Hovis JS, Boxer SG. Langmuir. 2000;16(17):6773–6776. [Google Scholar]
- 20.Smith BA, Clark WR, McConnell HM. PNAS. 1979;76(11):5641–5644. doi: 10.1073/pnas.76.11.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsay T, Jacobson K. Biophys J. 1991;60(2):360–368. doi: 10.1016/S0006-3495(91)82061-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang FY, Bouret Y, Kindt JT. Biophys J. 2004;87(1):182–192. doi: 10.1529/biophysj.103.031054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mossman KD, Campi G, Groves JT, Dustin ML. Science. 2005;310(5751):1191–3. doi: 10.1126/science.1119238. [DOI] [PubMed] [Google Scholar]
- 24.van Oudenaarden A, Boxer SG. Science. 1999;285:1046–1048. doi: 10.1126/science.285.5430.1046. [DOI] [PubMed] [Google Scholar]
- 25.Radler J, Strey H, Sackmann E. Langmuir. 1995;11:4539–4548. [Google Scholar]
- 26.Howland MC, Sapuri-Butti AR, Dixit SS, Dattelbaum AM, Shreve AP, Parikh AN. Journal of the American Chemical Society. 2005;127(18):6752–6765. doi: 10.1021/ja043439q. [DOI] [PubMed] [Google Scholar]
- 27.Merritt EA, Sarfaty S, Akker FVD, L'Hoir C, Martial JA, Hol WGJ. Protein Science. 1994;3(2):166–175. doi: 10.1002/pro.5560030202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rossetti FF, Bally M, Michel R, Textor M, Reviakine I. Langmuir. 2005;21(14):6443–6450. doi: 10.1021/la0509100. [DOI] [PubMed] [Google Scholar]
- 29.Yuan J, Parker ER, Hirst LS. Langmuir. 2007;23(14):7462–7465. doi: 10.1021/la701033a. [DOI] [PubMed] [Google Scholar]
- 30.Zhou X, Moran-Mirabal JM, Craighead HG, McEuen PL. Nature Nanotechnology. 2007;2(3):185–190. doi: 10.1038/nnano.2007.34. [DOI] [PubMed] [Google Scholar]
- 31.Yee CK, Amweg ML, Parikh AN. J Am Chem Soc. 2004;126(43):13962–13972. doi: 10.1021/ja047714k. [DOI] [PubMed] [Google Scholar]
- 32.Smith AM, Huser T, Parikh AN. Journal of the American Chemical Society. 2007;129(9):2422–2423. doi: 10.1021/ja065626p. [DOI] [PubMed] [Google Scholar]


