Abstract
Almost all creatures have invented sophisticated mechanisms to adjust their developmental and metabolic processes to the changing light intensities in day and night. Recent findings suggest that one such mechanism is signaling via heterotrimeric G-proteins.1 The Trichoderma reesei (anamorph of Hypocrea jecorina) G-α subunit gene gna3 was found to be responsive to light and influenced by the light regulatory protein ENVOY.2 GNA3 significantly impacts regulation of cellulase gene expression only in light.1 While the exact mechanism of this regulation remains to be determined, first hints point to a regulation at the transcriptional level, since we observed light induced complex formation within the gna3 promotor. At least some of the components of this putatively regulatory protein complex also bind to the env1-promotor. These data indicate that the signal related by GNA3 is of light-dependent significance for H. jecorina and that the pathway of heterotrimeric G-protein signaling may be a target of the light perception machinery in fungi.
Key words: Trichoderma reesei, Hypocrea jecorina, heterotrimeric G-protein signaling, light response
Light perception and signaling as well as circadian rhythms and the effects of the respective signaling machineries on gene regulation and physiology have received much attention during the last decades.3–5 Among the many factors reported to be involved in triggering light responses the photoreceptor WHITE COLLAR-1 (WC-1)6 and the transcription factor WHITE COLLAR-2 (WC-2)7,8 have been shown to be the only essential components for light perception and blue light signaling in Neurospora crassa identified today. Another blue light photoreceptor of N. crassa, VIVID, is regulated by the WC-1-WC-2 complex and also forms a transient complex with WC-1, thus establishing a negative feedback loop leading to its own downregulation.9,10 By functioning as a clock associated negative regulator of the WC-1-WC-2 complex, VIVID also regulates other light responsive, clock controlled genes.
We have recently shown, that ENVOY, an ortholog of VIVID in the filamentous fungus Hypocrea jecorina (anamorph Trichoderma reesei), is not only involved in regulation of light-responsive genes,11 but also in regulation of cellulase gene expression (i.e. the cellobiohydrolases cbh1 and cbh2).2 Moreover, we found that ENVOY impacts transcription of the subtype III G-α subunit gene gna3. Although the nature of the signal transmitted by GNA3 is so far unknown, it became clear that the effect of activation of GNA3 was only observed in light.1
Fungal heterotrimeric G proteins have been shown to play a major role in signaling to various processes such as regulation of growth, reproduction, morphogenesis, virulence, secondary metabolite production and pathogenic development.12–17 However, crosstalk between heterotrimeric G-protein signaling pathways and light response has not yet been explored in fungi, although hints as to light influenced effects of G-proteins on growth or conidiation are available.18,19 Nevertheless, since such phenomena are known from mammals (reviewed in refs. 20 and 21) interconnected processes within these two important pathways are not without precedent.
Our findings raised the question how the light signal could be integrated with the GNA3 signal. In this respect, the occurrence of five copies of the recently discovered, putatively light related promotor motif EUM1,2 in the promotor of gna3 caught our attention. Hence we investigated the possibility that this light-dependent regulation is at least in part executed at the transcriptional level.
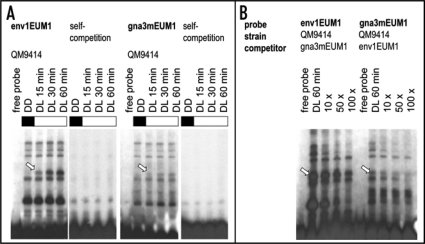
We used oligonucleotides derived from the env1-promotor, in which the motif was initially found and we analyzed the binding of proteins to this motif in electrophoretic mobility shift assays with cell free extracts of H. jecorina. All steps after harvesting the mycelium were performed in darkness with red safety light in order to prevent in vitro effects on complex formation.22 Using the wild-type strain QM9414, pregrown in darkness and illuminated thereafter with 1800 lux for 15, 30 or 60 minutes in Mandels Andreotti medium with 1% (w/v) glycerol as carbon source,1 we could indeed detect a protein complex appearing after the onset of illumination, which shows increasing signal strength with prolonged illumination (Fig. 1A). This effect occurs both with the oligonucleotide env1EUM1 which was derived from the env1-promotor as well as with the one comprising the five-fold EUM1 motif present in the gna3-promotor (gna3mEUM1). Concluding from the obvious resemblance of the protein complexes bound to these two probes, similar proteins could be the constituents of these complexes. To investigate this hypothesis, competition experiments with excess cold probes of the respective other promotor were performed using an experimental approach similar to that applied by He and coworkers,23 i.e., competition of complex formation with oligonucleotides derived from the promotor region of another gene comprising the common promotor motif of interest. Competition of a complex formed with the motif derived from the env1 promotor with the motif, as present in the gna3 promotor would thus suggest binding to both motifs. As shown in Figure 1B, complete competition of the light dependent complex occurred when env1EUM1 was added to the gna3-derived probe, whereas complex formation with the env1-derived probe was considerably weakened upon competition with the oligonucleotide derived from the gna3-promotor, but not abolished indicating stronger binding to the env1 promotor. While minor differences in complex formation suggest a certain variation of regulatory factors binding to these promotors, the majority of the complexes seem to be at work in both promotors.
Figure 1.
Characterization of protein complex-binding to EUM1 within the env1 or gna3-promotor. (A) EMSA analysis with annealed and labeled oligonucleotide derived from the env1 or gna3 promotor (env1EUM1F 5′ GAT CTC TTG TCC CTT TAC TCT GTG CTC TCT CTA CCT GCC T 3′; env1EUM1R 5′ GAT CAG GCA GGT AGA GAG AGC ACA GAG TAA AGG GAC AAG A 3′; gna3mEUM1F 5′ GAT CGA CTC GTT GCT GTG CTG TGC TGT GCT GTG CTG TGC TGT 3′ gna3mEUM1R 5′ GAT CAC AGC ACA GCA CAG CAC AGC ACA GCA CAG CAA CGA GTC 3′; lower case letters indicate bases added for labeling) and 30 µg of H. jecorina cell free extracts.24,25 The wild-type strain QM9414 was pregrown in darkness and harvested after illumination for the time indicated. (B) Competition experiments with cell free extracts prepared as described above after 60 minutes of illumination with 10-fold, 50-fold or 100-fold excess of cold competitor. Arrows point at the light-dependent protein complex found binding to both the env1 and the gna3-promotor. Experiments were carried out with both probes on the same gel and in case of self-competitions with 120-fold excess, uncompeted protein extracts were loaded on the same gel as control and exposed to similar signal strength. All experimental procedures after harvesting the mycelia were performed in complete darkness.
This finding is especially intriguing, given the fact that gna3 is influenced by ENVOY.2 Although ENVOY is no functional ortholog of N. crassa VIVID,2 a feedback mechanism involving BLR1 and BLR2, the H. jecorina orthologous of N. crassa White Collar 1 and 2 could be at work. Such a feedback could impact not only complex formation at EUM1 within the env1-promotor but also within other promotors bearing this motif, which would explain the results for gna3. Further analyses are warranted to clarify, whether also post-transcriptional regulation and modification/activation of GNA3 occur in a light dependent manner and thus contribute to the integration of the two signal transduction pathways of light response and heterotrimeric G-protein signaling. First indications in this respect come from the suggested involvement of an RGS-protein in the regulatory effect of GNA3, which is only observed in light.1
Although the finding that a G-α subunit, representing the first intracellular target of an environmental signal, is regulated by light is intriguing in itself, the question as to the benefit of this regulation for the fungus remains. The information if the fungus grows on the surface (light) or under the surface of its substrate (darkness) is crucial for the decision whether to initiate sporulation (which would not be wise when growing underground) or to start efforts to escape the harmful effects of light by inducing protective or repair mechanisms or by entering the substrate. Consequently it seems necessary to receive a detailed review of the current environmental condition, a task to be executed for example by the membrane spanning G-protein coupled receptors and their cognate heterotrimeric G-proteins. Since their regulatory output is obviously different in light and darkness, light dependent regulation of G-proteins might serve the function to determine the significance of the signal to be transduced according to light or darkness, thus economically providing energy for the processes most urgently needed under the respective condition.
Acknowledgements
This work was supported by FWF (Austrian Science Fund)-grant P20004 to Monika Schmoll. Monika Schmoll is recipient of an APART fellowship of the Austrian Academy of Sciences at the Institute of Chemical Engineering, Vienna University of Technology.
Footnotes
Previously published online as a Communicative & Integrative Biology E-publication: http://www.landesbioscience.com/journals/cib/article/8223
References
- 1.Schmoll M, Schuster A, do Nascimento Silva R, Kubicek CP. The G-alpha protein GNA3 of Hypocrea jecorina (anamorph Trichoderma reesei) regulates cellulase gene expression in the presence of light. Eukaryot Cell. 2009 doi: 10.1128/EC.00256-08. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmoll M, Franchi L, Kubicek CP. Envoy, a PAS/LOV domain protein of Hypocrea jecorina (Anamorph Trichoderma reesei), modulates cellulase gene transcription in response to light. Eukaryot Cell. 2005;4:1998–2007. doi: 10.1128/EC.4.12.1998-2007.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunlap JC, Loros JJ. The neurospora circadian system. J Biol Rhythms. 2004;19:414–424. doi: 10.1177/0748730404269116. [DOI] [PubMed] [Google Scholar]
- 4.Herrera-Estrella A, Horwitz BA. Looking through the eyes of fungi: molecular genetics of photoreception. Mol Microbiol. 2007;64:5–15. doi: 10.1111/j.1365-2958.2007.05632.x. [DOI] [PubMed] [Google Scholar]
- 5.Idnurm A, Heitman J. Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol. 2005;3:95. doi: 10.1371/journal.pbio.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ballario P, Vittorioso P, Magrelli A, Talora C, Cabibbo A, Macino G. White collar-1, a central regulator of blue light responses in Neurospora, is a zinc finger protein. EMBO J. 1996;15:1650–1657. [PMC free article] [PubMed] [Google Scholar]
- 7.Linden H, Macino G. White collar 2, a partner in blue-light signal transduction, controlling expression of light-regulated genes in Neurospora crassa. EMBO J. 1997;16:98–109. doi: 10.1093/emboj/16.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Talora C, Franchi L, Linden H, Ballario P, Macino G. Role of a white collar-1-white collar-2 complex in blue-light signal transduction. EMBO J. 1999;18:4961–4968. doi: 10.1093/emboj/18.18.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heintzen C, Loros JJ, Dunlap JC. The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell. 2001;104:453–464. doi: 10.1016/s0092-8674(01)00232-x. [DOI] [PubMed] [Google Scholar]
- 10.Schwerdtfeger C, Linden H. VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. EMBO J. 2003;22:4846–4855. doi: 10.1093/emboj/cdg451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schuster A, Kubicek CP, Friedl MA, Druzhinina IS, Schmoll M. Impact of light on Hypocrea jecorina and the multiple cellular roles of ENVOY in this process. BMC Genomics. 2007;8:449. doi: 10.1186/1471-2164-8-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bolker M. Sex and crime: heterotrimeric G proteins in fungal mating and pathogenesis. Fungal Genet Biol. 1998;25:143–156. doi: 10.1006/fgbi.1998.1102. [DOI] [PubMed] [Google Scholar]
- 13.D'Souza CA, Alspaugh JA, Yue C, Harashima T, Cox GM. Perfect JR, et al. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol Cell Biol. 2001;21:3179–3191. doi: 10.1128/MCB.21.9.3179-3191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Souza CA, Heitman J. Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol Rev. 2001;25:349–364. doi: 10.1111/j.1574-6976.2001.tb00582.x. [DOI] [PubMed] [Google Scholar]
- 15.Mukherjee PK, Latha J, Hadar R, Horwitz BA. Role of two G-protein alpha subunits, TgaA and TgaB, in the antagonism of plant pathogens by Trichoderma virens. Appl Environ Microbiol. 2004;70:542–549. doi: 10.1128/AEM.70.1.542-549.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reithner B, Brunner K, Schuhmacher R, Peissl I, Seidl V, Krska R, et al. The G protein alpha subunit Tga1 of Trichoderma atroviride is involved in chitinase formation and differential production of antifungal metabolites. Fungal Genet Biol. 2005;42:749–760. doi: 10.1016/j.fgb.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 17.Zeilinger S, Reithner B, Scala V, Peissl I, Lorito M, Mach RL. Signal transduction by Tga3, a novel G protein alpha subunit of Trichoderma atroviride. Appl Environ Microbiol. 2005;71:1591–1597. doi: 10.1128/AEM.71.3.1591-1597.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivey FD, Hodge PN, Turner GE, Borkovich KA. The G alpha i homologue gna-1 controls multiple differentiation pathways in Neurospora crassa. Mol Biol Cell. 1996;7:1283–1297. doi: 10.1091/mbc.7.8.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kays AM, Rowley PS, Baasiri RA, Borkovich KA. Regulation of conidiation and adenylyl cyclase levels by the Galpha protein GNA-3 in Neurospora crassa. Mol Cell Biol. 2000;20:7693–7705. doi: 10.1128/mcb.20.20.7693-7705.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arshavsky VY, Lamb TD, Pugh EN., Jr G proteins and phototransduction. Annu Rev Physiol. 2002;64:153–187. doi: 10.1146/annurev.physiol.64.082701.102229. [DOI] [PubMed] [Google Scholar]
- 21.Okano T, Fukada Y. Phototransduction cascade and circadian oscillator in chicken pineal gland. J Pineal Res. 1997;22:145–151. doi: 10.1111/j.1600-079x.1997.tb00316.x. [DOI] [PubMed] [Google Scholar]
- 22.Froehlich AC, Liu Y, Loros JJ, Dunlap JC. White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science. 2002;297:815–819. doi: 10.1126/science.1073681. [DOI] [PubMed] [Google Scholar]
- 23.He Q, Liu Y. Molecular mechanism of light responses in Neurospora: from light-induced transcription to photoadaptation. Genes Dev. 2005;19:2888–2899. doi: 10.1101/gad.1369605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stangl H, Gruber F, Kubicek CP. Characterization of the Trichoderma reesei cbh2 promoter. Curr Genet. 1993;23:115–122. doi: 10.1007/BF00352009. [DOI] [PubMed] [Google Scholar]
- 25.Zeilinger S, Mach RL, Kubicek CP. Two adjacent protein binding motifs in the cbh2 (cellobiohydrolase II-encoding) promoter of the fungus Hypocrea jecorina (Trichoderma reesei) cooperate in the induction by cellulose. J Biol Chem. 1998;273:34463–34471. doi: 10.1074/jbc.273.51.34463. [DOI] [PubMed] [Google Scholar]