Abstract
In order to withstand changes in their environment, bacteria have evolved mechanisms to sense the surrounding environment, integrate these signals and adapt their physiology to thrive under fluctuating conditions. Among these mechanisms, the ability of bacteria to exchange information between cells has become a dynamic field of interest for microbiologists over the past four decades. First described by Nelson et al.,1 this phenomenon often referred as either cell-cell communication, Quorum Sensing and/or AutoInduction involves the synthesis of small signal molecules called autoinducers. These signal molecules may be sensed by the bacterial population in the vicinity and induce regulation of gene expression. To date, three major communication systems have been described in bacteria. In this mini-review, we discuss the involvement of known communication systems in the transmission of information in the species Listeria monocytogenes. We will also discuss the latest findings on the role of communication in the regulation by Listeria monocytogenes of major adaptive strategies.
Key words: listeria, quorum sensing, agr system, communication, virulence, biofilm, regulation
In Gram-negative bacteria, signal molecules belong to the homoserine lactone (HSL) family. The system, elucidated in 1983, involves the synthesis by the enzyme LuxI of the signal molecule that diffuses freely.2 A cytoplasmic transcriptional regulator LuxR binds the HSL and affects transcription of target genes. In Gram positive bacteria, signal molecules are usually short peptides (linear or cyclic) processed by a transmembrane protein and other proteases.3 Once secreted, these autoinducers accumulate and interact with a two-component system. The signal is translocated from the outside to a cytoplasmic regulator via a phosphorylation cascade after binding of the autoinducer to the sensor histidine kinase. The phosphorylated regulator affects transcription of target genes. A third system has been described more recently.4 The signal molecule, called AutoInducer 2 (AI-2), is a family of furanone derivatives synthesized by LuxS. The AI-2 system has been proposed as an interspecies communication system as AI-2 production has been reported in both Gram negative and Gram-positive bacteria. Listeria monocytogenes is the causative agent of listeriosis.5 Its capacity to withstand stressful conditions, to colonize surfaces and grow as biofilm contribute to its ability to adapt and survive in many habitats.6
AI-2 System: The Metabolic Hypothesis
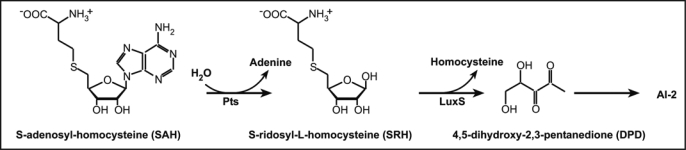
Fifteen years ago, the study of Vibrio harveyi a marine bacterium led to the elucidation of a new Quorum Sensing system based on the release of a furanone derivative called Auto Inducer 2 (AI-2).7 In this system, two enzymes, Pfs and LuxS catalyze the conversion of S-adenosyl homocystein (SAH), a toxic by-product of the Activated Methyl Cycle (AMC), into homocysteine and 4,5-dihydroxy-2,3-pentanedione (DPD). The latter spontaneously rearranges into various cyclic compounds commonly called AI-2 (Fig. 1).
Figure 1.
Synthesis of AI-2 through the Pfs and LuxS pathway.
As orthologs of luxS and pfs of the AI-2 production pathway have been identified in silico in the genome of most γ-, β- and ε-proteobacteria and firmicutes,7,8 AI-2 has been postulated to be a universal signal involved in bacterial interspecies communication.9 This is of a special interest in biofilm studies where cell-cell communication is postulated to play a key role. Both genes, pfs (lmo1494) and luxS (lmo1288) are present on the genome of Listeria monocytogenes (http://genolist.pasteur.fr/ListiList/) and two different studies reported that a luxS mutant shows a reduced AI-2 activity and an increased biofilm formation on stainless steel10 and on glass.11 Interestingly, complementation of the mutant strain with in vitro synthesized AI-2 has no effect on biofilm formation but the addition of exogenous S-ribosyl homocysteine (SRH) increased the number of attached cells. Moreover, it appears that L. monocytogenes luxS mutant, accumulates more SAH and SRH in its culture supernatant than the parental strains does.10 These results are to be correlated with those obtained on Neisseria meningitidis pfs and luxS mutants12 where growth defects are due to metabolic imbalance instead of the absence of AI-2 signal and favors the “metabolic hypothesis” to explain such a phenotype. so far, these experimental evidence and the lack of any known receptor for AI-2 in L. monocytogenes13 suggest that AI-2 is not a communication signal in the genus Listeria.
In silico Investigation of Peptide-Based Communication Systems in the Genome of Listeria sp
Two communication systems have been extensively studied in Gram-positive bacteria. These are the development of competence of Bacillus subtilis associated with the autoinducer ComX,14 and the agr system first described in Staphylococcus aureus.15
In silico analysis indicates that orthologs of most of the competence genes of B. subtilis are present in the genome of Listeria monocytogenes EGD-e (http://genolist.pasteur.fr/ListiList/), but the gene coding for the inducer (comX), the transmembrane protein (comQ) and the two component system (comP/comA) are absent in the genome. Indeed so far, natural genetic transformation of L. monocytogenes has not been observed experimentally.16
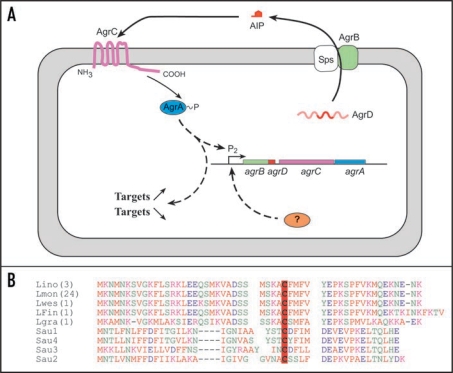
On the opposite, the four genes of the agr system are represented in the genome of Listeria sp. These four genes agrBDCA (Fig. 2) have a similar organization as the agrBDCA operon of Staphylococcus where agrB and agrD code for a transmembrane protein and the propeptide AgrD. AgrB is involved in the processing of AgrD into a cyclic autoinducer peptide (AIP) and its exportation. The genes agrC and agrA code for the sensor kinase and response regulator of a two-component system AgrC/AgrA.
Figure 2.
The agr system of L. monocytogenes. (A) Schematic of agr autoinduction. AgrB and Sps process AgrD into a secreted cyclic autoinducing peptide (AIP). The detection of AIP by the sensor kinase AgrC induces a phosphorylation cascade resulting in the activation of the regulator AgrA. (B) AgrD sequence alignments of Listeria and the four groups of staphylococcal AgrD. Lino: Listeria innocua; Lmon: Listeria monocytogenes; Lwes: Listeria welshimeri; LFin: Listeria monocytogenes Finland; Lgra: Listeria grayi. In brackets, number of genomes available online; identical sequences were observed in these genomes. In bold face and red-boxed, the conserved cystein residue critical for the thiolactone ring.
Comparison of the genomes available indicates a high level of conservation of the agr genes within the genus Listeria (Fig. 2). Unlike what is observed among isolates of Staphylococcus aureus where 4 classes of AIP are evidenced,17 sequence analysis of Listeria AgrD showed 99% identity in the species L. monocytogenes, L. innocua and L. welshimeri. AgrD is different in the species L. grayi. Putative AIPs are identical in L. monocytogenes, L. innocua and L. welshimeri but is different in L. grayi; these differences in autoinduction and AIP sensing are in accordance with evolution in the genus Listeria.18,19 Furthermore, in silico analysis failed to identify putative orthologs of the staphylococcal non-coding RNA regulator RNAIII in the genus Listeria and in ortholog systems of other Gram-positive bacteria. This suggests that, on an evolutionary point of view, RNAIII is an innovation specific to Staphylococcaceae.20
agr-Mediated Regulation of Virulence
The first studies on the role of the agr system in the regulation of virulence were performed by characterizing the virulence potential of a Tn1545 insertion21 and an in frame deletion agrA mutant.22 The authors reported that their respective mutants were not affected in their intracellular infectious cycle assayed in vitro on Caco-2, HepG-2 and Cos-1 cell lines and in macrophages.21,22 An increase in LD50 was observed with the Tn1545 insertion agrA mutant during infection of Swiss mice,21 however in-frame deletion of agrA did not affect virulence in a BALB/C mice infection assay.22 These results suggested that AgrA was not directly involved in the regulation of virulence-associated gene expression.
In the latest report on the role of the agr system in the virulence of L. monocytogenes, the virulence potential of an in-frame agrD deletion mutant was investigated.23 These authors evidenced internalin A-dependent differences invasion when in vitro assays were performed with exponential phase-grown inocula while differences were not significant when inocula were grown to stationary phase, the experimental condition used in the previous studies. Bioluminescent in vivo imaging confirmed a virulence defect of ΔagrD mutant when exponentially grown cells were injected intravenously. It is striking to notice how critical is the procedure of inoculum preparation (exponential versus stationary) on the outcome of these in vitro and in vivo virulence assays.
agr-Mediated Biofilm Formation
The growth of ΔagrA mutants was similar to the growth of the parental strain at temperatures ranging from 4°C to 43°C,21,22 in the presence of 9% NaCl and 0.025% H2O2 thus demonstrating the absence of pleiotropic effects.22
However, in our laboratory we have demonstrated that in-frame deletion of agrA affects the ability of L. monocytogenes to adhere to glass, polystyrene and stainless steel. For example, adhesion to glass slides was decreased by 62% in comparison to the parental strain EGD-e. Strikingly, a similar phenotype was observed with an agrD in-frame deletion mutant.24 This is indirect evidence that AgrD is the precursor of an auto-inducing peptide (AIP). However, this AIP has yet to be isolated and sequenced. Nevertheless, the presence of a conserved cystein residue within the sequence of AgrD (Fig. 2) confirms that cyclic auto-inducing peptides can derive from this pro-peptide. This critical conserved cystein residue is observed in AIPs of the ortholog agr systems of S. aureus,25 Enterococcus faecalis26 and Lactobacillus plantarum.27
ΔagrA and ΔagrD mutants were also affected during growth as biofilm within the first 24 h of incubation under static conditions in a microtiter plate assay24 and under dynamic conditions in a flow-cell model of growth.28 Similar conclusions were reached by other authors.23
Spatiotemporal expression of agr expression of a GFP reporter strain was followed in situ during biofilm development under batch and dynamic conditions.28 Surprisingly, GFP expression, recorded with a confocal scanning laser microscope, depended on the growth conditions. It was low under static conditions. Unorganized multicellular layers of cells were observed, only a few expressing agr (less than 1% of the total number of cells in a 48 h biofilm). Under dynamic conditions, agr expression increased from 15% during adhesion to 80% in flow-cell mature biofilms. The agr expression was mainly recorded at the apex of the biofilm, in a complex network of cells organized in chains that surrounded ball-shaped microcolonies.
The agr System and the Regulation of Transcription
In order to refine our understanding of the significance of agr signaling in the physiology of L. monocytogenes, it is of importance to determine the targets of AgrA. A recent study gives some clues on the genes that might be regulated by this response regulator.23 These authors compared global gene expression profiles of an agrD deletion mutant and its parental strain L. monocytogenes EGD-e. During exponential phase, 55 genes and 66 genes respectively were up and downregulated in the mutant. In our laboratory, similar results were observed with an agrA deletion mutant of L. monocytogenes EGD-e (Piveteau P, personal communication). The number of genes differentially regulated was higher during stationary phase; the number of genes up and downregulated respectively was 345 and 325 respectively.23 All categories of genes were affected by the mutation. The transcription of a rather large number of genes (58) coding for regulatory proteins was affected.23 These preliminary results suggest first of all that the agr system plays a major role in the adaptation of L. monocytogenes to environmental conditions, and secondly, that AgrA is probably involved in a complex regulatory network. However, it is necessary to know whether these expression differences are a direct consequence of a low abundance of AgrA in the ΔagrD background or rather an indirect consequence due to a disturbance within a complex regulatory network.
Autoinduction, Communication and Quorum Sensing: Where Does agr Stand?
Although there is no doubt that the agr system is indeed a communication system based on autoinduction, whether it is dedicated to Quorum Sensing (QS) remains on open question. QS, first proposed by Fuqua et al. (1994)29 can be defined as “the phenomenon whereby the accumulation of signaling molecules in the surrounding environment enables a single cell to assess the number of bacteria (cell density) so that the population as a whole can make a coordinated response.”30 Experimental evidence indicate that the transcription of the agr genes is lower in the agrA and agrD in-frame deletion mutants compared to the parental strain EGD-e thus confirming that the agr system is autoregulated.24 However, during growth of L. monocytogenes in liquid cultures, the agr operon is transcribed to a similar level during mid exponential and early stationary phase,21,24 suggesting that the agr system is not a mechanism to assess cell density in order to proceed in coordinated behavior of the whole population.
In situ expression also gave results that are not in agreement with the QS paradigm; heterogeneity of agr expression during biofilm growth28 does not fit either with the definition of QS. On the contrary, our results corroborate with two theories presented more recently. The theory of Diffusion Sensing (DS) postulates that autoinduction enables the cell to evaluate the diffusion and mixing of autoinducers and other molecules in the cell's environment.31 The second theory, Efficiency sensing (ES), is an elegant attempt to explain autoinduction and signaling in complex environments thus unifying QS and DS in a global theory of cell-cell communication.32 The latter theory of ES is the most convincing in regard to our experimental results.
Conclusions
Information exchange and signaling are central to the adaptation of bacteria to environmental conditions. In the genus Listeria, AI-2 does not fit with the definition of a signaling molecule. So far, the agr system is the only communication system described in the genus Listeria. It is based on autoinduction but evidence suggests that Quorum Sensing does not apply to this communication system. Major adaptive responses are regulated through agr autoinduction; these include virulence and biofilm formation. It is tempting to propose that AgrA may be involved in a complex regulatory network in charge of coordinating gene expression for an optimal adaptation of Listeria monocytogenes and Listeria sp. to environmental conditions. A synthetic approach will have to be developed to decipher such a network.
Footnotes
Previously published online as a Communicative & Integrative Biology E-publication: http://www.landesbioscience.com/journals/cib/article/8610
References
- 1.Nealson KH, Platt T, Hastings JW. Cellular control of the synthesis and activity of the bacterial luminescent system. J Bacteriol. 1970;104:313–322. doi: 10.1128/jb.104.1.313-322.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engebrecht J, Nealson K, Silverman M. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell. 1983;32:773–781. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- 3.Dunny GM, Leonard BAB. Cell-cell communication in gram-positive bacteria. Ann Rev Microbiol. 1997;51:527–564. doi: 10.1146/annurev.micro.51.1.527. [DOI] [PubMed] [Google Scholar]
- 4.Surette MG, Miller MB, Bassler BL. Quorum sensing in Escherichia coli, Salmonella typhimurium and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Nat Acad Sci USA. 1999;96:1639–1644. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vazquez-Boland JA, Kuhn M, Berche P, Chakraborty T, Dominguez-Bernal G, Goebel W, et al. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev. 2001;14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gray MJ, Freitag NE, Boor KJ. How the bacterial pathogen Listeria monocytogenes mediates the switch from environmental Dr. Jekyll to pathogenic Mr. Hyde. Inf Immun. 2006;74:2505–2512. doi: 10.1128/IAI.74.5.2505-2512.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bassler BL, Wright M, Silverman MR. Multiple signaling systems controlling expression of luminescence in Vibrio harveyi—sequence and function of genes encoding a 2nd sensory pathway. Mol Microbiol. 1994;13:273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- 8.Sun JB, Daniel R, Wagner-Dobler I, Zeng AP. Is autoinducer-2 a universal signal for interspecies communication: a comparative genomic and phylogenetic analysis of the synthesis and signal transduction pathways. BMC Evol Biol. 2004;4 doi: 10.1186/1471-2148-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schauder S, Shokat K, Surette MG, Bassler BL. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol Microbiol. 2001;41:463–476. doi: 10.1046/j.1365-2958.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- 10.Challan Belval S, Gal L, Margiewes S, Garmyn D, Piveteau P, Guzzo J. Assessment of the roles of LuxS, S-ribosyl homocysteine and autoinducer 2 in cell attachment during biofilm formation by Listeria monocytogenes EGD-e. Appl Env Microbiol. 2006;72:2644–2650. doi: 10.1128/AEM.72.4.2644-2650.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sela S, Frank S, Belausov E, Pinto R. A mutation in the luxS gene influences Listeria monocytogenes biofilm formation. Appl Env Microbiol. 2006;72:5653–5658. doi: 10.1128/AEM.00048-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heurlier K, Vendeville A, Halliday N, Green A, Winzer K, Tang CM, et al. Growth deficiencies of Neisseria meningitidis pfs and luxS mutants are not due to inactivation of quorum sensing. J Bacteriol. 2009;191:1293–1302. doi: 10.1128/JB.01170-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rezzonico F, Duffy B. Lack of genomic evidence of AI-2 receptors suggests a non-quorum sensing role for luxS in most bacteria. BMC Microbiol. 2008;8:154. doi: 10.1186/1471-2180-8-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magnuson R, Solomon J, Grossman AD. Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell. 1994;77:207–216. doi: 10.1016/0092-8674(94)90313-1. [DOI] [PubMed] [Google Scholar]
- 15.Recsei P, Kreiswirth B, Oreilly M, Schlievert P, Gruss A, Novick RP. Regulation of exoprotein gene-expression in Staphylococcus-aureus by agr. Mol Gen Genet. 1986;202:58–61. doi: 10.1007/BF00330517. [DOI] [PubMed] [Google Scholar]
- 16.Borezee E, Msadek T, Durant L, Berche P. Identification in Listeria monocytogenes of MecA, a homologue of the Bacillus subtilis competence regulatory protein. J. Bacteriol 2000;182:5931–5934. doi: 10.1128/jb.182.20.5931-5934.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright JS, Traber KE, Corrigan R, Benson SA, Musser JM, Novick RP. The agr radiation: an early event in the evolution of Staphylococci. J Bacteriol. 2005;187:5585–5594. doi: 10.1128/JB.187.16.5585-5594.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buchrieser C. Biodiversity of the species Listeria monocytogenes and the genus Listeria. Microb Infect. 2007;9:1147–1155. doi: 10.1016/j.micinf.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Schmid MW, Ng EYW, Lampidis R, Emmerth M, Walcher M, Kreft J, et al. Evolutionary history of the genus Listeria and its virulence genes. Syst Appl Microbiol. 2005;28:1–18. doi: 10.1016/j.syapm.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 20.Wuster A, Babu MM. Conservation and evolutionary dynamics of the agr cell-to-cell communication system across firmicutes. J Bacteriol. 2008;190:743–746. doi: 10.1128/JB.01135-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Autret N, Raynaud C, Dubail I, Berche P, Charbit A. Identification of the agr locus of Listeria monocytogenes: role in bacterial virulence. Infect Immun. 2003;71:4463–4471. doi: 10.1128/IAI.71.8.4463-4471.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams T, Bauer S, Beier D, Kuhn M. Construction and characterization of Listeria monocytogenes mutants with in-frame deletions in the response regulator genes identified in the genome sequence. Infect Immun. 2005;73:3152–3159. doi: 10.1128/IAI.73.5.3152-3159.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riedel CU, Monk IR, Casey PG, Waidmann MS, Gahan CG, Hill C. AgrD-dependent quorum sensing affects biofilm formation, invasion, virulence and global gene expression profiles in Listeria monocytogenes. Mol Microbiol. 2009 doi: 10.1111/j.1365-2958.2008.06589.x. [DOI] [PubMed] [Google Scholar]
- 24.Rieu A, Weidmann S, Garmyn D, Piveteau P, Guzzo J. agr system of Listeria monocytogenes EGD-e: role in adherence and differential expression pattern. Appl Env Microbiol. 2007;73:6125–6133. doi: 10.1128/AEM.00608-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji G, Beavis R, Novick RP. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 26.Nakayama J, Chen S, Oyama N, Nishiguchi K, Azab EA, Tanaka E, et al. Revised model for Enterococcus faecalis fsr quorum-sensing system: the small open reading frame fsrD encodes the gelatinase biosynthesis-activating pheromone propeptide corresponding to staphylococcal agrd. J Bacteriol. 2006;188:8321–8326. doi: 10.1128/JB.00865-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sturme MH, Nakayama J, Molenaar D, Murakami Y, Kunugi R, Fujii T, et al. An agr-like two-component regulatory system in Lactobacillus plantarum is involved in production of a novel cyclic peptide and regulation of adherence. J Bacteriol. 2005;187:5224–5235. doi: 10.1128/JB.187.15.5224-5235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rieu A, Briandet R, Habimana O, Garmyn D, Guzzo J, Piveteau P. Listeria monocytogenes EGD-e biofilms: no mushrooms but a network of knitted chains. Appl Env Microbiol. 2008;74:4491–4497. doi: 10.1128/AEM.00255-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.West SA, Diggle SP, Buckling A, Gardner A, Griffins AS. The social lives of microbes. Ann Rev Ecol Evol Syst. 2007;38:53–77. [Google Scholar]
- 31.Redfield RJ. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 2002;10:365–370. doi: 10.1016/s0966-842x(02)02400-9. [DOI] [PubMed] [Google Scholar]
- 32.Hense BA, Kuttler C, Muller J, Rothballer M, Hartmann A, Kreft JU. Does efficiency sensing unify diffusion and quorum sensing? Nat Rev Micobiol. 2007;5:230–239. doi: 10.1038/nrmicro1600. [DOI] [PubMed] [Google Scholar]