Abstract
There is increasing evidence for epistatic interactions between gene products (e.g. KIR) encoded within the Leukocyte Receptor Complex (LRC) with those (e.g. HLA) of the Major Histocompatibility Complex (MHC), resulting in susceptibility to disease. Identification of such associations at the DNA level requires comprehensive knowledge of the genetic variation and haplotype structure of the underlying loci. The LRC haplotype project aims to provide this knowledge by sequencing common LRC haplotypes.
Keywords: haplotypes; killer immunoglobulin-like receptor; leucocyte receptor complex; polymorphism, single nucleotide
The leucocyte receptor complex (LRC) on chromosome 19q13.4 encodes multiple clusters of natural killer receptors and related molecules belonging to the immunoglobulin superfamily (1, 2). These include the killer immunoglobulin-like receptors (KIR), the leucocyte immunoglobulin-like receptors (LILR) and the leucocyte-associated immunoglobulin-like receptors. Of these, KIRs are the most widely studied receptor because of their high levels of polymorphism, giving rise to multiple haplotypes, and because of the increasing appreciation of their role in human health and disease (3, 4).
Following the example of sequencing haplotypes from the major histocompatibility complex region (5-7), the LRC Haplotype Project aims to determine the complete DNA sequences of multiple haplotypes, with initial focus on the KIR and LILR clusters and flanking framework loci (spanning approximately 1.0 megabase). Depending on the presence/absence of certain KIR genes, the resulting haplotypes can be subdivided into two groups: A and B (8), and representatives of both groups will be analysed. The resources generated as part of this project will be made publicly available from the project web site (http://www.sanger.ac.uk/HGP/Chr19/LRC/) and will include clones, contig maps, genomic sequences, annotation and a catalogue of all single nucleotide polymorphisms (SNPs) and deletion/insertion polymorphisms.
The LRC Haplotype Project web site, shown in Figure 1, includes a colour-coded table of haplotypes linked to the ImMunoGeneTics (IMGT)/human leucocyte antigen (HLA) database (http://www.ebi.ac.uk/imgt/hla/) where appropriate. At the time of writing, data were available from two consanguineous cell lines (PGF, COX). Although both these cell lines are homozygous for HLA, KIR typing suggests that COX carries both group A and B haplotypes, while PGF has two different A haplotypes. It is our intention to sequence sufficient clones from each cell line to resolve these haplotypes.
Figure 1.
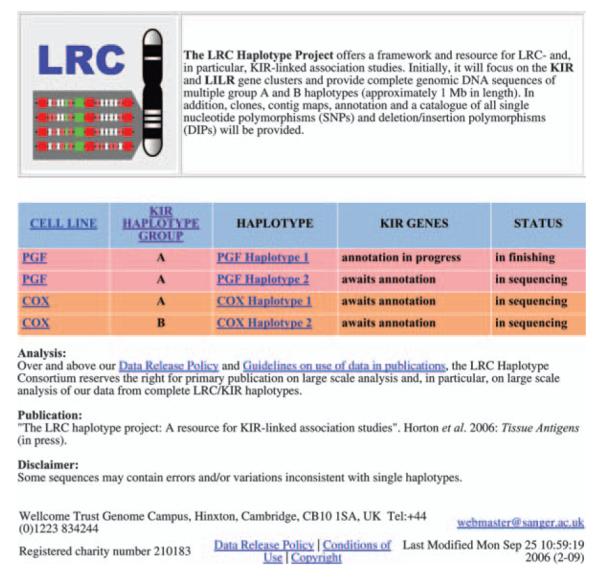
Web site of the LRC Haplotype Project (http://www.sanger.ac.uk/HGP/Chr19/LRC/) showing a colour-coded haplotype table and the accompanying side menu at bottom left.
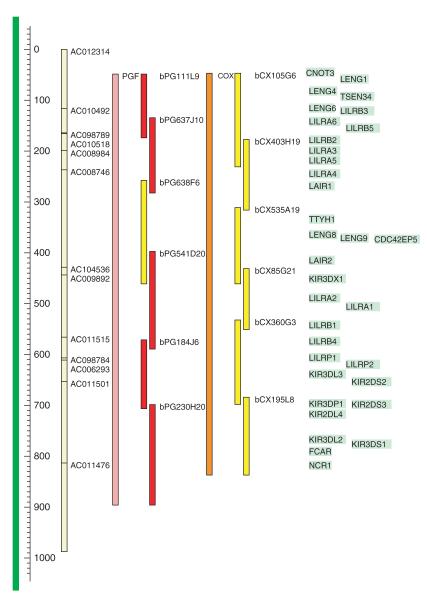
A side menu on the web site allows access to pages for the following, among others: Links (including IPD-KIR http://www.ebi.ac.uk/ipd/kir; and Vega http://vega.sanger.ac.uk databases), Progress map, Help, Sequence and Variation data, which will be populated during the project. The Progress map provides detailed (clickable) clone and contig information for each haplotype. Further haplotypes will be added as and when available. The sequencing progress to date, shown in Figure 2, is colour coded and illustrates the targeted LRC region between the genes CNOT3 and NCR1. The individual BACs from the CHORI-501 library (for PGF) and CHORI-502 (for COX) can be requested from BACPAC resources (http://www.chori.org/bacpac/). The consortium aims to update the web site regularly.
Figure 2.
Tiling path of the human LRC. The map includes from left to right: scale bar (dark green) and scale in kilobases; current reference sequence (pale yellow) spanning 13 clones from AC012314 to AC011476 (NCBI36 chr19 59300001..60300000); PGF (pale red) and COX (orange) mapped contig positions and landmark LRC gene symbols (black text on pale-green background). The BAC clones have been mapped along the LRC and are colour-coded according to their current sequencing status: finished in red and pre-finishing in yellow.
The resource described here represents a general framework for LRC-linked, and especially KIR-linked, association studies. The LRC Consortium welcomes future collaborations involving the analysis of further haplotypes and disease association studies. While the data are made available in good faith before publication, some sequences may contain errors and/or variations inconsistent with single haplotypes. The consortium reserves the right to primary publication of large-scale analysis of the data and, in particular, large-scale analysis of the complete LRC haplotypes.
Acknowledgments
R. H., P. C., M. M. M., J. G. S., S. S., S. P., H. S., J. H., J. R. and S. B. were supported by the Wellcome Trust. J. A. T., R. W. and J. T. were supported by the Medical Research Council. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health under contract number NO1-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organization imply endorsements by the U.S. Government. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- 1.Wende H, Colonna M, Ziegler A, Volz A. Organization of the leukocyte receptor cluster (LRC) on human chromosome 19q13.4. Mamm Genome. 1999;10:154–60. doi: 10.1007/s003359900961. [DOI] [PubMed] [Google Scholar]
- 2.Kelley J, Walter L, Trowsdale J. Comparative genomics of natural killer cell receptor gene clusters. PLoS Genet. 2005;1:129–39. doi: 10.1371/journal.pgen.0010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–14. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 4.Bashirova AA, Martin MP, McVicar DW, Carrington M. The killer immunoglobulin-like receptor gene cluster: tuning the genome for defense. Annu Rev Genomics Hum Genet. 2006;7:277–300. doi: 10.1146/annurev.genom.7.080505.115726. [DOI] [PubMed] [Google Scholar]
- 5.Allcock RJ, Atrazhev AM, Beck S, et al. The MHC haplotype project: a resource for HLA-linked association studies. Tissue Antigens. 2002;59:520–1. doi: 10.1034/j.1399-0039.2002.590609.x. [DOI] [PubMed] [Google Scholar]
- 6.Stewart CA, Horton R, Allcock RJ, et al. Complete MHC haplotype sequencing for common disease gene mapping. Genome Res. 2004;14:1176–87. doi: 10.1101/gr.2188104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Traherne JA, Horton R, Roberts AN, et al. Genetic analysis of completely sequenced disease-associated MHC haplotypes identifies shuffling of segments in recent human history. PLoS Genet. 2006;2:e9. doi: 10.1371/journal.pgen.0020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson MJ, Torkar M, Haude A, et al. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci U S A. 2000;97:4778–83. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]