Abstract
Antimicrobial dosing is currently attracting attention as a way to minimize the emergence of resistance. Three dose-based strategies have been advocated, each with shortcomings. Focus on killing susceptible cells overlooks resistant mutant subpopulations that may be present before treatment or generated during therapy; keeping therapeutic drug concentrations above the mutant prevention concentration (MPC; resistant mutant MIC) may be overly stringent; and dosage escalation modelling uses indirect estimates of resistant mutant subpopulation susceptibility (multiples of bulk population susceptibility, MIC) rather than direct estimates from MPC. The latter is significant because MPC and MIC are discordant with multiple pathogen isolates. Combining the strategies leads to MPC-based PK/PD thresholds (e.g. AUC24/MPC and t > MPC) for restricting resistant subpopulation enrichment and amplification. Using MPC-based thresholds to model dosing regimens that will restrict emergence of resistance requires generation of databases in which MPC is determined for many isolates.
Keywords: MPC, PK/PD indices, mutant selection window
Introduction
Antimicrobial resistance continues to make headlines, despite calls for restricting unnecessary antimicrobial use. Highly visible resistance problems, such as community-acquired methicillin-resistant Staphylococcus aureus,1,2 are generally caused by transmission of resistant pathogens from one person to another. Although transmission can often be limited by infection control methods, ultimately, we need to control its precursor, the acquisition of resistance. Several approaches have been proposed. The traditional strategy focuses on killing susceptible cells, as that should reduce the pool from which new mutants arise. More recently, emphasis has shifted to direct control of resistant subpopulations. They can selectively amplify under antimicrobial pressure and eventually dominate a population; at that point, antimicrobials and host defences may fail to control pathogen growth.
Although resistance is often considered an absolute term clinically, growth of many ‘resistant’ mutants can be inhibited by drug concentrations attainable therapeutically [such isolates have intermediate (dose-dependent) susceptibility]. The distinction between absolute clinical resistance, which may be used as an endpoint for modelling studies, and resistant mutants, which may be controlled by higher drug concentrations, is important for considering ways to halt the acquisition of resistance. Such considerations can lead to dosage increases3,4 or shifts from monotherapy to multidrug combination therapy.5 Below we sketch out three dosing strategies aimed at slowing the emergence of mutational resistance and then consider combining their best features. Induced phenotypic resistance, a situation in which all members of the pathogen population exhibit reduced susceptibility, is outside the present discussion.
Dosing to eradicate susceptible cells
In principle, reducing the bacterial load should suppress the acquisition of new mutants.6 Killing susceptible cells may also enable host defences to more effectively eliminate residual subpopulations of resistant mutants. Suitable antimicrobial doses are identified by measures of pathogen drug exposure that empirically correlate with the eradication of susceptible bacteria. For antimicrobials that are considered ‘time-dependent killers’, the pharmacodynamic term is time above MIC (t > MIC); for ‘concentration-dependent killers’, it is either area under the concentration–time curve in a 24 h period divided by MIC (AUC24/MIC) or maximal drug concentration divided by MIC (Cmax/MIC).7
As drug concentrations that kill susceptible cells may allow mutants to amplify,8–10 a dosing approach that addresses only bulk population susceptibility, which is measured by MIC, fails to consider resistant mutant subpopulations. Consequently, resistance can emerge during the eradication of susceptible cells (Figure 1), as seen with S. aureus colonizing the noses of tuberculosis patients.11 When a set of these patients was treated with rifampicin for tuberculosis, susceptible colonizing S. aureus was eradicated, but in 10% of the patients, rifampicin-resistant S. aureus was acquired. Thus, treatment strategies aimed only at eradicating susceptible cells may not block the acquisition of resistance.
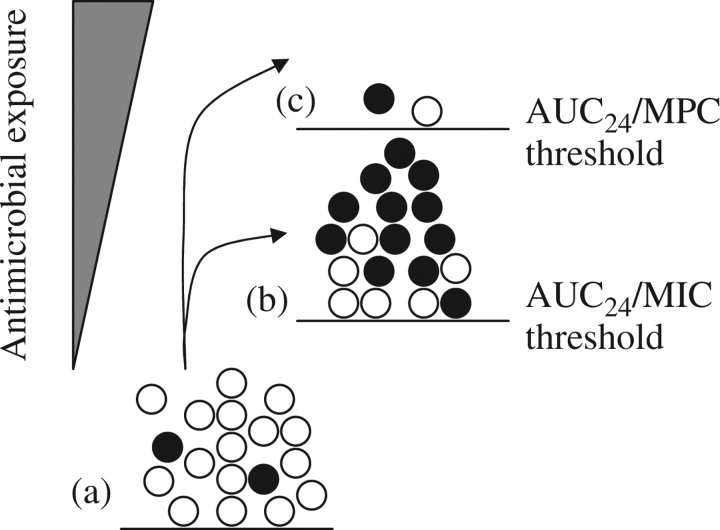
Figure 1.
Effect of PK/PD thresholds on major susceptible and minor resistant pathogen populations treated with a lethal agent. (a) Unchallenged bacterial population contains a majority of susceptible cells (open circles) and a small subpopulation of resistant mutants (filled circles). (b) When drug exposure passes an AUC24/MIC threshold above which mutants, but not susceptible cells, can survive and proliferate, enrichment and amplification of resistant mutant subpopulations occur. (c) When drug exposure exceeds an AUC24/MPC threshold above which resistant mutants and susceptible cells are both killed, acquisition of resistance fails to occur (a few pathogen cells may persist, as shown). With fluoroquinolones, AUC24/MPC of 20–70 restricts emergence of resistance.31
Drug concentrations above mutant prevention concentration (MPC)
A second approach involves direct measurement of resistant mutant subpopulation susceptibility (MPC).12 MPC is estimated as the drug concentration that blocks growth when 1010 cells are applied to agar12 or tested in liquid medium13 (MIC determination uses 104–105 cells). Large inocula ensure the presence of mutant subpopulations; consequently, MPC estimates resistant subpopulation susceptibility. If antimicrobial concentrations are kept above MPC throughout therapy, mutant subpopulation amplification will be inhibited. How and when resistant subpopulations are generated has no influence on this approach, which holds for both bacteriostatic and bactericidal agents. MPC has been measured for fluoroquinolones, linezolid, macrolides, β-lactams, vancomycin and daptomycin (the major failure to measure MPC has been for rifampicin with Escherichia coli).14–17 The strategy of maintaining drug concentrations above MPC suffers from being more stringent than necessary for antimicrobials that kill resistant pathogens. For example, with a local S. aureus infection of rabbits, levofloxacin concentrations need to be above MPC for only 20% of the dosing interval to restrict mutant amplification.8
Dose escalation modelling
A third approach involves mathematical modelling of data obtained with escalating doses to identify the susceptible population-based drug exposure (AUC24/MIC) that blocks amplification of resistant mutant subpopulations and kills most susceptible cells.18 Monte Carlo simulation is then used to estimate the fraction of patients for which a given dose will achieve the restrictive value of AUC24/MIC.
A key assumption with this method is that dominant population susceptibility (MIC) is proportional to resistant subpopulation susceptibility (MPC) among many different clinical isolates, thereby allowing particular multiples of MIC to approximate mutant subpopulation susceptibility. Although proportionality is by definition true for any single isolate, experimental measurements indicate that MPC and MIC are discordant when multiple isolates are surveyed.19–21 Lack of proportionality may arise from patient isolates harbouring mutations that confer little increase in MIC, but a large increase in MPC and vice versa.22–24 Thus, MIC-based PK/PD thresholds are likely to be inherently less accurate for suppressing selective amplification of resistant mutant subpopulations than those based on direct measurement of resistant subpopulation susceptibility (e.g. AUC24/MPC). The dose escalation modelling also requires testing enough cells to assure that amplification of the least susceptible, single-step mutant is suppressed.18,25
Combined approach
As problems with the dose escalation modelling approach derive from using bulk population susceptibility rather than mutant subpopulation susceptibility, an improvement would be to use an MPC-based threshold (for example, AUC24/MPC) rather than an MIC-based one (for example, AUC24/MIC) as a target parameter for blocking resistant subpopulation proliferation. The same conclusion is drawn from a slightly different argument. Empirical relationships between drug exposure and efficacy identify AUC24/MIC for some agents and t > MIC for others, as the PK/PD index that relates to bulk pathogen survival.7 These indices for susceptible cells should apply to resistant subpopulations if MPC is substituted for MIC, as MPC correlates with the MIC of the least-susceptible single (next)-step mutant.26 Then, AUC24/MPC and t > MPC would be thresholds for preventing resistant mutant subpopulations from proliferating (see also Olofsson et al.27). In principle, Cmax could be substituted for AUC24.8,28 However, this would require more complex population pharmacokinetic expressions, as AUC24 includes a consideration of dosage frequency, which is absent from raw Cmax measurements.
Conclusions
Halting the selective amplification of resistant mutant subpopulations requires more stringent antimicrobial therapies than required to clear susceptible infections. For several reasons, values of AUC24/MPC and t > MPC are expected to be preferred PK/PD thresholds for defining dosing regimens that block mutant subpopulation proliferation. This expectation must now be confirmed with animal infection models. The application of these expressions to large patient populations requires a standard methodology for the measurement of MPC16,25,29,30 and then generation of extensive databases of population MPC.
Funding
The work summarized was supported by NIH grants AI035257, AI073491 and AI068014.
Transparency declarations
None to declare.
References
- 1.Klevens R, Morrison M, Nadle J, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. J Am Med Assoc. 2007;298:1763–71. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 2.Zetola N, Francis J, Nuermberger E, et al. Community-acquired methicillin-resistant Staphylococcus aureus: an emerging threat. Lancet Infect Dis. 2005;5:275–86. doi: 10.1016/S1473-3099(05)70112-2. [DOI] [PubMed] [Google Scholar]
- 3.Goldstein F, Garau J. 30 years of penicillin-resistant S. pneumoniae: myth or reality? Lancet. 1997;350:233–4. doi: 10.1016/s0140-6736(05)62222-2. [DOI] [PubMed] [Google Scholar]
- 4.Dunbar L, Wunderink R, Habib M, et al. High-dose, short-course levofloxacin for community-acquired pneumonia: a new treatment paradigm. Clin Infect Dis. 2003;37:752–60. doi: 10.1086/377539. [DOI] [PubMed] [Google Scholar]
- 5.Fox W, Elklard G, Mitchison D. Studies on the treatment of tuberculosis undertaken by the British Medical Research Council Tuberculosis Units, 1946–1986, with relevant subsequent publications. Int J Tuberc Lung Dis. 1999;3:S231–79. [PubMed] [Google Scholar]
- 6.Stratton C. Dead bugs don't mutate: susceptibility issues in the emergence of bacterial resistance. Emerg Infect Dis. 2003;9:10–6. doi: 10.3201/eid0901.020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craig WA. Pharmacodynamics of antimicrobials: general concepts and applications. In: Ambrose P, editor. Antimicrobial Pharmacodynamics in Theory and Clinical Practice. New York: Marcel Dekker; 2002. pp. 1–22. [Google Scholar]
- 8.Cui J, Liu Y, Wang R, et al. The mutant selection window demonstrated in rabbits infected with Staphylococcus aureus. J Infect Dis. 2006;194:1601–8. doi: 10.1086/508752. [DOI] [PubMed] [Google Scholar]
- 9.Lipsitch M, Levin B. The population dynamics of antimicrobial chemotherapy. Antimicrob Agents Chemother. 1997;41:363–73. doi: 10.1128/aac.41.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gumbo T, Louie A, Deziel M, et al. Pharmacodynamic evidence that ciprofloxacin failure against tuberculosis is not due to poor microbial kill but to rapid emergence of resistance. Antimicrob Agents Chemother. 2005;49:3178–81. doi: 10.1128/AAC.49.8.3178-3181.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Cui J, Wang R, et al. Selection of rifampicin-resistant Staphylococcus aureus during tuberculosis therapy: concurrent bacterial eradication and acquisition of resistance. J Antimicrob Chemother. 2005;56:1172–5. doi: 10.1093/jac/dki364. [DOI] [PubMed] [Google Scholar]
- 12.Dong Y, Zhao X, Domagala J, et al. Effect of fluoroquinolone concentration on selection of resistant mutants of Mycobacterium bovis BCG and Staphylococcus aureus. Antimicrob Agents Chemother. 1999;43:1756–8. doi: 10.1128/aac.43.7.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quinn B, Hussain S, Malik M, et al. Daptomycin inoculum effects and mutant prevention concentration with Staphylococcus aureus. J Antimicrob Chemother. 2007;60:1380–3. doi: 10.1093/jac/dkm375. [DOI] [PubMed] [Google Scholar]
- 14.Zhao X, Drlica K. Restricting the selection of antibiotic-resistant mutants: measurement and potential uses of the mutant selection window. J Infect Dis. 2002;185:561–5. doi: 10.1086/338571. [DOI] [PubMed] [Google Scholar]
- 15.Firsov A, Smirnova M, Lubenko I, et al. Testing the mutant selection window hypothesis with Staphylococcus aureus exposed to daptomycin and vancomycin in an in vitro dynamic model. J Antimicrob Chemother. 2006;58:1185–92. doi: 10.1093/jac/dkl387. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez J, Cebrian L, Lopez M, et al. Mutant prevention concentration: comparison of fluoroquinolones and linezolid with Mycobacterium tuberculosis. J Antimicrob Chemother. 2004;53:441–4. doi: 10.1093/jac/dkh119. [DOI] [PubMed] [Google Scholar]
- 17.Drlica K. The mutant selection window and antimicrobial resistance. J Antimicrob Chemother. 2003;52:11–7. doi: 10.1093/jac/dkg269. [DOI] [PubMed] [Google Scholar]
- 18.Jumbe N, Louie A, Leary R, et al. Application of a mathematical model to prevent in vivo amplification of antibiotic-resistant bacterial populations during therapy. J Clin Invest. 2003;112:275–85. doi: 10.1172/JCI16814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcusson L, Olofsson S, Lindgren P, et al. Mutant prevention concentration of ciprofloxacin for urinary tract infection isolates of Escherichia coli. J Antimicrob Chemother. 2005;55:938–43. doi: 10.1093/jac/dki136. [DOI] [PubMed] [Google Scholar]
- 20.Drlica K, Zhao X, Blondeau J, et al. Low correlation between minimal inhibitory concentration (MIC) and mutant prevention concentration (MPC) Antimicrob Agents Chemother. 2006;50:403–4. doi: 10.1128/AAC.50.1.403-404.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tam V, Louie A, Deziel M, et al. The relationship between quinolone exposures and resistance amplification is characterized by an inverted U: a new paradigm for optimizing pharmacodynamics to counterselect resistance. Antimicrob Agents Chemother. 2007;51:744–7. doi: 10.1128/AAC.00334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, Zhao X, Drlica K. Selection of Streptococcus pneumoniae mutants having reduced susceptibility to levofloxacin and moxifloxacin. Antimicrob Agents Chemother. 2002;46:522–4. doi: 10.1128/AAC.46.2.522-524.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drlica K, Zhao X, Wang JY, et al. An anti-mutant approach for antimicrobial use. In: Fong I, Drlica K, editors. Antimicrobial Resistance and Implications for the 21st Century. New York City: Springer; 2008. pp. 371–400. [Google Scholar]
- 24.Bast D, Low D, Duncan C, et al. Fluoroquinolone resistance in clinical isolates of Streptococcus pneumoniae: contributions of type II topoisomerase mutations and efflux on levels of resistance. Antimicrob Agents Chemother. 2000;44:3049–54. doi: 10.1128/aac.44.11.3049-3054.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen G, Zhao X, Drlica K, et al. Mutant prevention concentration for ciprofloxacin and levofloxacin with Pseudomonas aeruginosa. Int J Antimicrob Agents. 2006;27:120–4. doi: 10.1016/j.ijantimicag.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 26.Zhao X, Drlica K. Restricting the selection of antibiotic-resistant mutants: a general strategy derived from fluoroquinolone studies. Clin Infect Dis. 2001;33(Suppl 3):S147–56. doi: 10.1086/321841. [DOI] [PubMed] [Google Scholar]
- 27.Olofsson S, Marcusson L, Komp-Lindgren P, et al. Selection of ciprofloxacin resistance in Escherichia coli in an in vitro kinetic model: relation between drug exposure and mutant prevention concentration. J Antimicrob Chemother. 2006;57:1116–21. doi: 10.1093/jac/dkl135. [DOI] [PubMed] [Google Scholar]
- 28.Rybak MJ. Pharmacodynamics: relation to antimicrobial resistance. Am J Infect Control. 2006;34(Suppl 1):S28–45. doi: 10.1016/j.ajic.2006.05.227. [DOI] [PubMed] [Google Scholar]
- 29.Blondeau J, Zhao X, Hansen G, et al. Mutant prevention concentrations (MPC) for fluoroquinolones with clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother. 2001;45:433–8. doi: 10.1128/AAC.45.2.433-438.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Metzler K, Hansen G, Hedlin P, et al. Comparison of minimal inhibitory and mutant prevention concentrations of 4 fluoroquinolones: methicillin-susceptible and -resistant Staphylococcus aureus. Int J Antimicrob Agents. 2004;24:161–7. doi: 10.1016/j.ijantimicag.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 31.Drlica K, Zhao X. Mutant selection window hypothesis updated. Clin Infect Dis. 2007;44:681–8. doi: 10.1086/511642. [DOI] [PubMed] [Google Scholar]