Abstract
Magnetite nanoparticles (Chemicell SiMAG-TCL) were characterized by SQUID-relaxometry, susceptometry, and TEM. The magnetization detected by SQUID-relaxometry was 0.33% of that detected by susceptometry, indicating that the sensitivity of SQUID-relaxometry could be significantly increased through improved control of nanoparticle size. The relaxometry data were analyzed by the moment superposition model (MSM) to determine the distribution of nanoparticle moments. Analysis of the binding of CD34-conjugated nanoparticles to U937 leukemia cells revealed 60,000 nanoparticles per cell, which were collected from whole blood using a prototype magnetic biopsy needle, with a capture efficiency of >65% from a 750 µl sample volume in 1 minute.
Keywords: Magnetite, Nanoparticle, Magnetorelaxometry, SQUID detection, Susceptometry, Antibody-conjugation
We are developing SQUID (Superconducting Quantum Interference Device) relaxometry as a highly-sensitive platform for detecting and localizing superparamagnetic iron oxide nanoparticles specifically bound to cell-surface antigens (or other disease targets) in vivo. Preliminary in vitro experiments suggest that this technology will be capable of detecting just a few thousand cells located several centimeters from the sensors, enabling the early detection of cancer, or other diseases, in human subjects [1]. Currently we are investigating this methodology for the specific detection of breast cancer, transplant rejection, Alzheimer’s disease, and ovarian cancer. Further, as we develop other clinical applications of targeted magnetic nanoparticles, we are using SQUID-relaxometry as a tool for quantifying the binding of magnetic nanoparticles to cells. In this paper, we discuss the use of SQUID-relaxometry to evaluate the performance of a prototype magnetic biopsy needle [2,3] designed to capture and concentrate magnetically-labeled leukemia cells in vivo during bone marrow biopsy.
Magnetorelaxometry of nanoparticles (using SQUIDs or fluxgate magnetometers) is currently an active area of research [4–9]. To detect nanoparticles by relaxometry, the particles are first magnetized by a brief pulse of DC magnetic field, after which the sensors detect the decaying magnetization of the nanoparticles in zero field, as depicted in Fig. 1. Only those moments with relaxation times that fall within the measurement timescale (50 ms to 2 s, in our case) are detected. The magnetization of cell-bound nanoparticles relaxes by the Néel mechanism[10] (thermal fluctuations of the individual magnetic core orientations). At zero field, the Néel relaxation time constant is given by
| (1) |
Where τ0 is customarily taken to be 1 ns, K is the anisotropy energy density of the magnetic material, and V is the volume of the magnetic particle [10]. The relaxation time constant therefore depends strongly on the particle diameter, resulting in a very narrow range of particle diameters with relaxation times detectable within the timescale of the measurement. The magnetization of unbound magnetic particles in fluid also relaxes by Brownian rotation of the particle[11]. For sufficiently small particles (with hydrodynamic diameters less than a few hundred nanometers), Brownian relaxation is generally so fast that unbound particles are not detected, allowing the quantification of nanoparticle binding even in a large background of unbound particles [11,12].
Fig. 1.
The SQUID-relaxometry experiment. Particles that relax too quickly (during the 50 ms “dead time”) are not detected. Particles that relax too slowly (≫ 2 seconds) are also not detected, because SQUIDs only sense changes in magnetic flux.
When considering an ensemble of polydisperse nanoparticles, the sensitivity of the SQUID-relaxometry technique is strongly dependent on the distribution of nanoparticle properties, particularly those properties (particle volume and anisotropy energy density) that directly influence the Néel relaxation time. Because of this, our requirement for low polydispersity of these properties is more stringent than is required for magnetic nanoparticles used in other applications such as MRI or magnetic cell separation.
One aim of the present work is to characterize the nanoparticles we are currently using to determine what fraction of the iron oxide is actually detected by SQUID-relaxometry. For example, in order to evaluate the performance of the magnetic needle in collecting leukemia cells from a bone marrow biopsy, we must be able to calculate the total amount of magnetic material attached to the target cells (all of which is attracted to the needle) based on the magnetic moment measured by relaxometry. Furthermore, knowing the detectable fraction will allow us to determine what improvements in detection sensitivity will be possible through reduced polydispersity, an issue of critical importance to developing in vivo SQUID-relaxometry for clinical applications.
In this work, we characterize multi-core magnetite nanoparticles (Chemicell SiMAG-TCL), currently used in several applications in our laboratory, by two methods. One of these is SQUID-relaxometry (which is sensitive to only a narrow distribution of particles). The second method is SQUID-susceptometry (sensitive to particles with relaxation times up to ~200 s, i.e., all unblocked particles). Following the method of Chantrell [13], we interpret the magnetization curves (M vs. B) and relaxation curve (M vs. t) using a moment superposition model (MSM), which explicitly includes the distribution of nanoparticle moments. The nanoparticle characterization results are then applied to the quantitative analysis of in vitro experiments involving the binding of the nanoparticles to cells and the capture of nanoparticle-labeled cells by a prototype magnetic biopsy needle.
Materials and methods
Nanoparticles
SiMAG-TCL (lot 0808/07) (Chemicell GmbH, Berlin, Germany) are 100 nm diameter particles, each comprised of approximately 20 individual Fe3O4 cores embedded in a silica matrix. The particle surface is functionalized with carboxyl groups to enable binding to antibodies. The nanoparticles were imaged by transmission electron microscopy (Tecnai G2 F30 at 300kV, FEI Corporation, Hillsboro, Oregon, USA) as shown in Fig. 2. The nanoparticles are supplied in sterile, deionized water at a concentration of 50 mg[solids]/ml, with 8.5 × 1011 particles/mg. The iron concentration of the stock solution was determined destructively by dissolving in acid, forming the phenanthroline/Fe2+ complex, and then quantifying the concentration of a known dilution spectrophotometrically [14]. The concentration was found to be 27 mg[Fe]/ml, confirming that the nanoparticles are 75 wt.% Fe3O4 (54 wt.% Fe).
Fig. 2.
Transmission electron micrograph of SiMAG-TCL. Under the measurement conditions, the porous silica shells on the iron oxide particles provide very little contrast. The observed structures are the magnetic cores, most of which appear to have diameters in the 5–20 nm range.
SQUID-relaxometry
Detection of nanoparticles by relaxometry was performed using a seven-channel low-temperature SQUID array (BTi 2004, 4D-Neuroimaging, San Diego, CA) originally designed for magnetoencephalography. Second order gradiometers with a baseline of 4 cm are used to reject background magnetic fields due to distant sources, allowing the measurements to be performed in an unshielded environment. Due to RF interference, the sensitivity of the system is currently limited to ~10−12 T/√Hz. The seven gradiometer coils are located at the bottom of the liquid He dewar, 1.9 cm from the outer dewar surface, arranged with six in a circle of 2.15 cm radius and one at the center. For in vitro measurements on small samples, the sample is located at a distance z ≈ 2.8–3.5 cm below the bottom of the center coil. The samples are uniformly magnetized (parallel to the center gradiometer axis) using a 60 cm-square Helmholtz array (L = 16 mH, B/I = 0.107 mT/A) powered by a 3 kW current-regulated supply. The decaying magnetization is sampled at a rate of 1 kHz (beginning 50 ms after switching off the magnetizing pulse) and digitized using a National Instruments PXI8336 16-channel digitizer and LabVIEW 8.5.1 acquisition software (National Instruments, Austin, TX). Our standard measurement protocol is to apply a 3.8 mT field for 0.3 s and then acquire data for 2 s (see Fig. 1), with 10 repetitions, subsequently averaged, to improve signal-to-noise. Magnetization curves (M vs. B) are obtained, using the same apparatus and timing, by varying the applied field between 0 and 4.0 mT.
Data analysis is performed using the Multi-Source Analysis program, written in our lab using MATLAB (The MathWorks Inc., Natick, MA). After signal averaging, background data (acquired with no sample) is subtracted from the sample data. The seven relaxation curves are fit by a logarithmic function and an exponential function to determine the magnetic field amplitude at each sensor position (by extrapolation to the time at which the magnetizing pulse is switched off) [1]. In order to solve the inverse problem, we fit the spatial dependence of the magnetic field by modeling the sample as a single magnetic dipole, which allows us to determine the location (x, y, z) and magnetic moment (mz) of the source. The least-squares fit is performed using the Levenberg-Marquardt algorithm. A multiple dipole model may be used to determine the spatial coordinates and moments for multiple discrete sources using n different sample positions – equivalent to a sensor array with 7n elements.
To measure the desired Néel relaxation of the nanoparticles, the particles must be immobilized. In the case of cell samples, the antibody-conjugated nanoparticles are immobilized by the binding of the antibodies to receptors on the cell surface. In the case of unconjugated nanoparticles, 10–20 µl of stock nanoparticle solution is applied to a Q-tips cotton swab (Unilever, Trumball, CT) and allowed to dry in air.
SQUID-susceptometry
Magnetic characterizations were performed using an MPMS-7 SQUID magnetometer system (Quantum Design, San Diego, CA, USA). DC Magnetization curves were acquired by equilibrating the sample at the measurement temperature in zero field, then incrementally increasing the field and pausing 100 seconds at each field before measurement. Five sequential measurements were taken at each field, a mean of those measurements calculated, and the three values with the lowest deviation from the mean retained. The mean of these three values was then reported as the moment. Zero Field Cooled (ZFC) curves were determined by cooling the sample in the absence of a magnetic field to 5 K, then slowly warming in a 1 mT field. After thermally equilibrating at a target temperature, a series of five measurements was taken and the values were processed as described previously for magnetization curves to obtain a reported magnetization value. AC susceptibility curves were performed by applying a series of 0.2 mT AC magnetic fields (f = 0.1 – 1000 Hz) to a thermally equilibrated sample. The in-phase (χ’) and out-of-phase (χ”) AC responses of the sample were digitized and recorded.
Nanoparticle-antibody conjugation
Nanoparticles were conjugated by the following method, suggested by Andrew Wang [15]. Ten mg of SiMAG-TCL nanoparticles were aliquoted into 15 ml conical tubes (Greiner Bio-One, San Diego, CA) and brought to a total volume of 10 ml with double distilled water. N-hydroxysulfosuccinimide (Sulfo-NHS) (Pierce, Rockford, Il) and 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC) (Pierce, Rockford, Il) were prepared fresh at a concentration of 25 mg/ml each in separate tubes with double distilled water. One hundred microliters of the EDC and Sulfo-NHS were each added to the nanoparticles and incubated at room temperature on a LabQuake shaker (LabIndustries, Inc., Berkeley, CA) for 20 minutes. Nanoparticles were brought to pH 8.0 with 50 mM NaHCO3 (Sigma-Aldrich, St. Louis, MO), 50 µg of anti-CD34 antibody (BD Biosciences, San Jose, CA) was added, and the mixture was incubated at room temperature on a LabQuake shaker for 2 hours. The antibody-nanoparticle mixture was centrifuged at 7,500 RCF for 30 minutes at 4°C. The supernatant was removed and 10 ml of double distilled water was added to the pelleted NPs. The centrifugation parameters were repeated once more and the supernatant was removed. The remaining pellet was resuspended in a total volume of 240 µl PBS/ (Gibco-BRL, Rockville, MD) 0.5% fetal bovine serum (FBS) (HyClone, Logan, UT). CD34-conjugated nanoparticles were stored at 4°C for up to 2 days. The magnetic moment from 20 µl of the antibody-conjugated solution was found to be (1.4 ± 0.1) × 10−7 A-m2, equivalent to 7 × 1011 nanoparticles.
In vitro magnetic labeling of cells
U937 cells were purchased commercially from American Type Culture Collection (ATCC, Manassas, VA) and cultured in RPMI supplemented with 10% FBS (v/v) (HyClone, Logan, UT), 1% penicillin streptomycin (v/v) (Gibco-BRL, Rockville, MD) and 4 µg/mL ciprofloxacin (Bayer, West Haven, CT) at 37ºC with 5% CO2. The cells were harvested and washed using PBS (Gibco-BRL, Rockville, MD) and counted using a hemocytometer (Hausser Scientific, Horsham, PA).
Magnetic biopsy needle
The magnetic needle prototype [2] consists of a 1 mm diameter stainless steel rod, at the end of which are two cylindrical NdFeB magnets (1 mm diameter, 2 mm length) separated by a stainless steel spacer (2 mm length), all contained within a tight-fitting polyimide tube. The needle assembly is then inserted into a polyimide sheath, which is designed to fit through a standard bone marrow biopsy tool to allow the tip of the magnetic needle to sample the bone marrow in vivo during a biopsy procedure.
For the in vitro tests reported here, U937 cells in RPMI media (described above) were diluted in donor whole blood and anti-coagulated in 10U/ml of heparin (Becton-Dickinson, San Jose, CA) resulting in a total sample volume of 750 µl (contained in a 1.5 ml vial). CD34-conjugated nanoparticles (20 µl, 7 × 1011 particles) were then added, and the sample was incubated for 60 minutes at 4°C on a nutator (Becton-Dickinson, San Jose, CA). To capture labeled cells, the magnetic needle tip, covered by the polyimide sheath, was inserted in the center of the cell-spiked blood sample and held in position for 1 minute. The sheath was then removed and the material collected on the sheath was resuspended in 200 µl of RPMI media and assessed by SQUID relaxometry and light microscopy. For microscopy, glass slide preparations were made using a Cyto-centrifuge (Shandon, Pittsburgh, PA) and stained with Prussian blue by TriCore Reference Laboratories (Albuquerque, NM).
Results and discussion
Characterization of SiMAG TCL
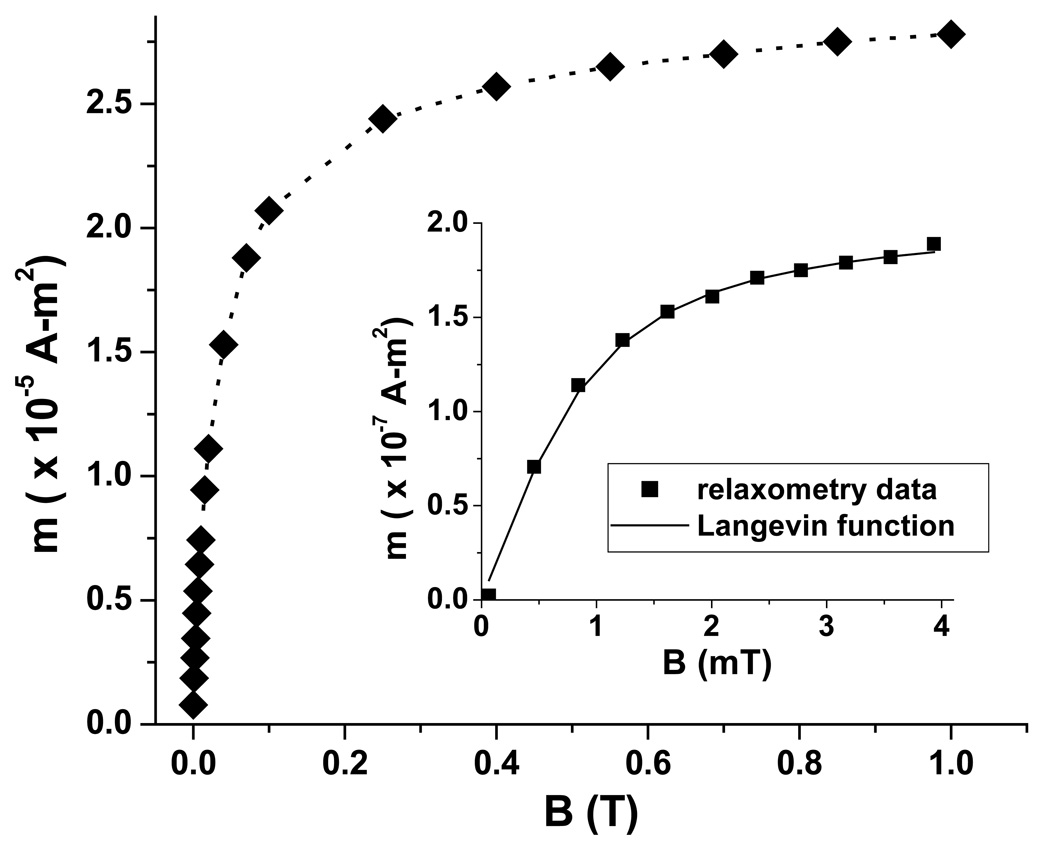
The zero-field cooled magnetization curve of 0.53 mg of immobilized SiMAG-TCL nanoparticles, acquired by susceptometry at 1 mT, indicates that the average blocking temperature is 260 K (data not shown). Fig. 3 shows the magnetization curve (M vs. B), also acquired by susceptometry (◆), at 250 K, just below the blocking temperature. From these data, the average saturation magnetization was determined to be 72.9 A-m2/kg[Fe3O4], somewhat lower than the bulk value (~92 A-m2/kg).
Fig. 3.
Magnetization curves of SiMAG-TCL particles acquired by susceptometry (0.53 mg, ◆) and relaxometry (1 mg, ■). The saturation magnetization detected by relaxometry is two orders of magnitude lower than that of susceptometry, because only a fraction of the nanoparticles exhibit detectable relaxation times.
The room-temperature magnetization curve acquired by relaxometry (from 1 mg of particles) is shown in Fig. 3 (inset). By fitting the data with a simple Langevin function, the magnetic moment μobs of the nanoparticle cores detectable by relaxometry is estimated to be 9.7 × 10−18 A-m2. The fit also yields an apparent saturation magnetization of 0.24 A-m2/kg[Fe3O4], which is only 0.33% of the value determined by susceptometry. This result clearly demonstrates that there is a wide distribution of Néel relaxation times, due to the distribution of core diameters, such that only a small fraction of the magnetite cores are detectable by relaxometry.
Using our standard magnetizing field of 3.8 mT (used in the cell experiments described below), the relaxometry-observed magnetization reaches 89% of the saturation value. Under these conditions, the observed magnetic moment from 1 mg of nanoparticles (8.5 × 1011 100-nm multi-core particles, each containing 8.8 × 10−19 kg of Fe3O4) is 1.7 × 10−7 A-m2. Using the saturation magnetization determined by susceptometry, we calculate that the total magnetic moment from 1 mg of particles is actually 5.8 × 10−5 A-m2. As we analyze the results of nanoparticle-cell binding experiments, these data allow us to convert the magnetic moment measured by relaxometry into other useful experimental parameters, namely the total number of bound nanoparticles, the total quantity of iron oxide, and the actual magnetic moment per cell.
If we could separate the relaxometry-observable cores from the others, we would presumably measure by relaxometry a saturation magnetization very similar to that measured by susceptometry for the whole ensemble. Using the susceptometry value (72.9 A-m2/kg[Fe3O4]), the fitted value of μobs implies that the mass of a relaxometry-observed core is ~1.3 × 10−19 kg. Assuming that the cores are spherical and have the same density as bulk magnetite (5240 kg/m3), we then estimate that the diameter of the cores observed by relaxometry is ~37 nm, larger than the majority of cores visible in the TEM image (Fig. 2) and larger than the predicted value of 23 nm [1], which was calculated using K = 1.34 × 104 J/m3, the value for bulk magnetite. To achieve the same relaxation time (~1 s), the product KV must be the same; thus, the larger observed core size suggests that K is smaller than the bulk value, in this case by approximately a factor of 4.
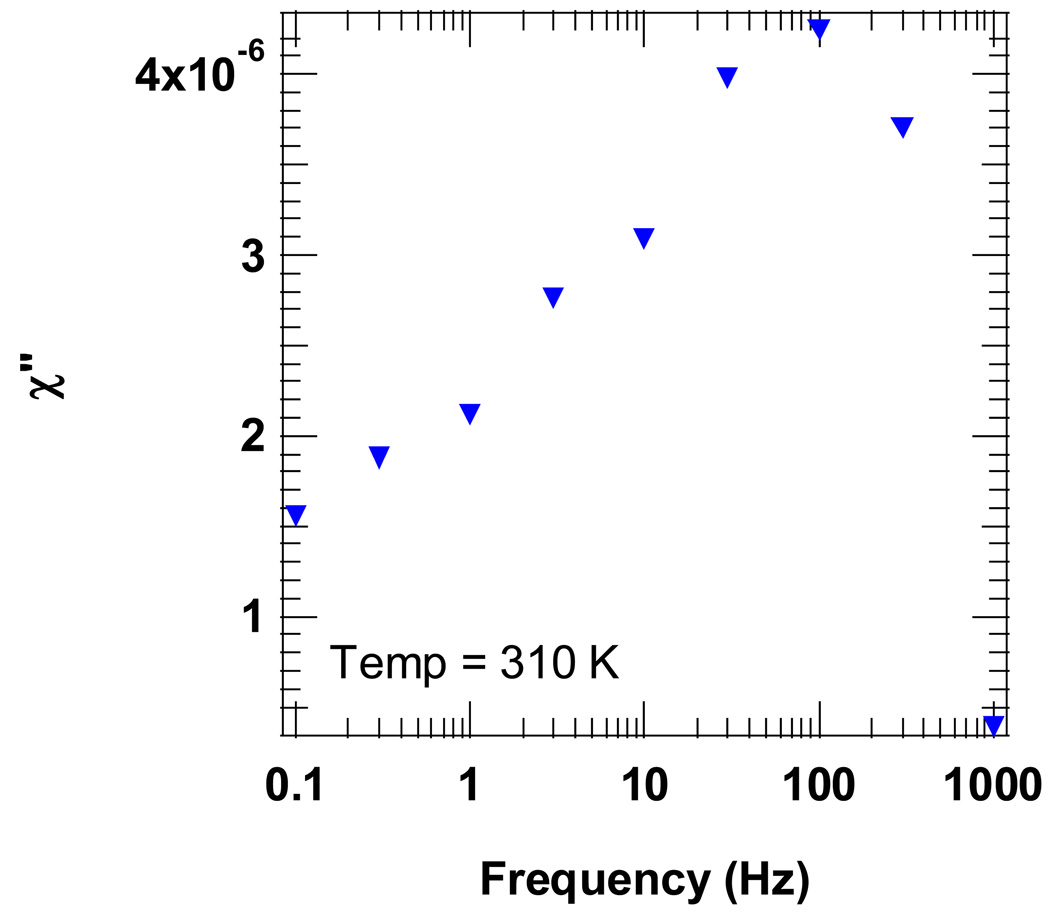
The imaginary component of the AC magnetic susceptibility (χ”) directly probes the relaxation times of the nanoparticle ensemble [16]. As shown in Fig. 4, there is a distinct peak in χ” at f = 100 Hz indicating that the average relaxation time, given by (2πfpeak)−1, is approximately 1.6 ms. Thus, the relaxation time of the average particle is too short (i.e., the diameter is too small) to be detected by our relaxometry method, due to the 50 ms “dead time” after the magnetizing pulse (see Fig. 1). Thus, the AC magnetic susceptibility provides further evidence that most of the nanoparticle cores in the sample are too small to be detected by relaxometry.
Fig. 4.
The frequency dependence of the AC susceptibility of immobilized SiMAG-TCL nanoparticles directly probes the distribution of Néel relaxation times. The loss peak centered at 100 Hz indicates that the volume-averaged relaxation time is ~1.6 ms (= (2πfpeak)−1). While there is significant loss in the 0.1–1 Hz range (corresponding to the SQUID-relaxometry timescale), the majority of magnetite cores relax too quickly to be detected by our relaxometry method.
Magnetic Needle Results
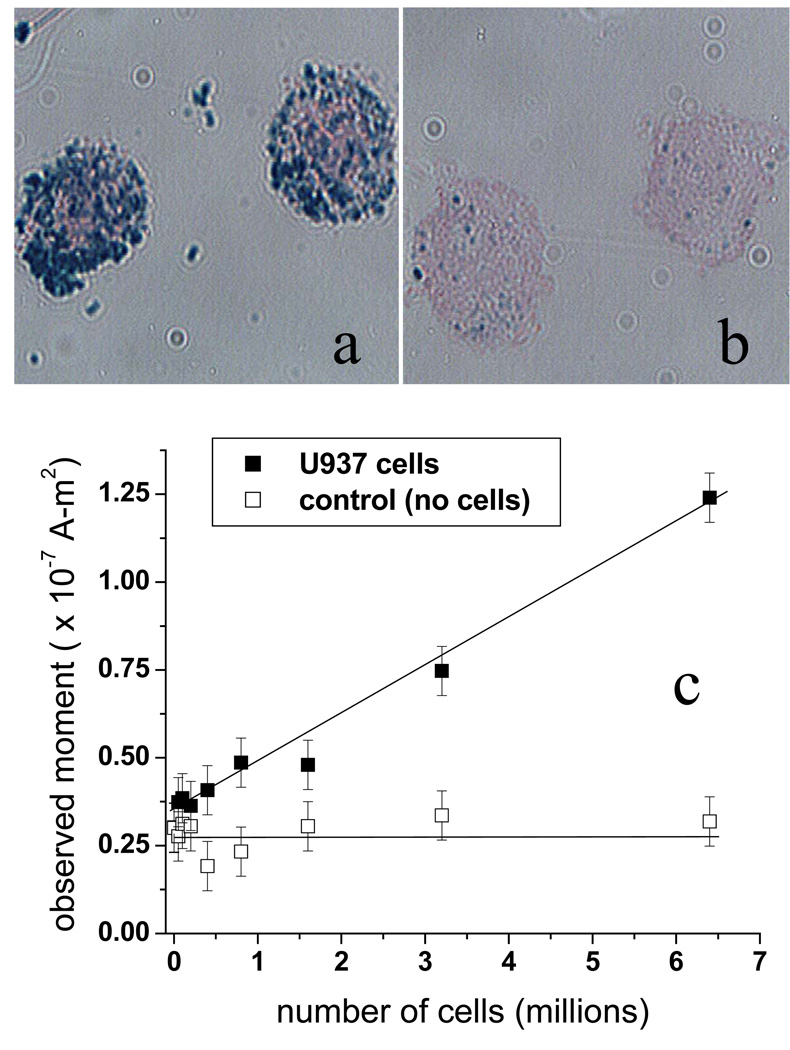
Preliminary in vitro tests of the magnetic needle biopsy technique utilized U937 cells labeled with CD34-conjugated SiMAG-TCL nanoparticles [3]. U937 is a human leukemic monocyte lymphoma cell line, which was chosen for these experiments due to its high cell-surface expression of CD34, a stem cell antigen expressed by several types of human leukemia blast cells [17]. Fig. 5 shows photomicrographs of U937 cells (~20 µm in diameter) incubated in PBS/0.5% FBS for 60 minutes with either CD34-conjugated nanoparticles (Fig. 5a) or nanoparticles that were not conjugated to antibodies (Fig. 5b). Prussian blue staining of the iron reveals significant binding of the CD34-conjugated nanoparticles to the cells and very little non-specific binding of the unconjugated nanoparticles. Previously, we determined that ~7 × 1011 nanoparticles are sufficient to completely label up to 10 million U937 cells [3]. The observed magnetic moment from 10 million labeled cells was found to be (1.20 ± 0.37) × 10−7 A-m2, after subtraction of the background signal (typically 0.2 – 0.4 × 10−7 A-m2) obtained without cells. The uncertainty is the standard deviation of four repeated measurements.
Fig. 5.
Photomicrographs of U937 cells incubated with (a) CD34-conjugated nanoparticles and (b) nanoparticles without antibodies. Prussian blue staining of the iron shows the location of the nanoparticles. (c) Magnetic needle capture of U937 cells spiked into normal blood and then labeled with CD34-conjugated nanoparticles. (In the control samples, no cells were added) The captured material was resuspended in RPMI media and measured by SQUID-relaxometry.
According to the analysis above, 1.20 × 10−7 A-m2 is equivalent to 6.0 × 1011 bound 100-nm particles, or ~85% of the 7 × 1011 nanoparticles used in the incubation. The observed moment per U937 cell is then 1.20 ± 0.4 × 10−14 A-m2, equivalent to 60,000 nanoparticles per cell. The number of nanoparticles that will theoretically bind to a single cell is limited by steric hindrance to 64,000, assuming a single random-close-packed layer of antibody-coated nanoparticles (modeled as spheres of diameter ~140 nm) covering a 20 µm-diameter spherical cell. This result agrees quantitatively with the observed magnetic moment per cell, consistent with Fig. 5a, which shows nearly complete coverage of the cell.
Fig. 5c shows the results of an in vitro experiment to test the ability of the magnetic biopsy needle to collect magnetically-labeled U937 cells from blood. U937 cells were first added to normal blood and then incubated with 7 × 1011 CD34-labeled nanoparticles for 60 min. The magnetic needle was then inserted in the sample vials for 1 min. to collect the labeled cells. A control experiment was also performed using the same mixtures of blood and cell suspension fluid containing no U937 cells. The magnetic moment of the material collected by the magnetic needle was found to be proportional to the number of U937 cells originally added to the sample. The highest concentration sample contained 6.4 ± 1.6 million cells in 750 µl; the material collected on the needle from this sample gave an observed magnetic moment of 9 × 10−8 A-m2, after subtraction of the non-specific background moment determined from the control experiment. Using the observed moment per cell obtained from the previous experiment (1.20 ± 0.37 × 10−14 A-m2), we calculate that 7.5 ± 2.3 million labeled cells were collected. Considering the uncertainties, we then estimate that the magnetic needle captured between 65% and 100% of the labeled cells.
Approximating the magnetic needle as a single dipole of strength of 4.72 × 10−3 A-m2 (based on a surface field of 0.47 T for each of the two magnets), we calculate (using Eq. 16 from Ref. 2) that the collection range of the needle is 4.8 mm for a collection time of 1 min. in blood at room temperature. The calculation assumes that each 20-µm diameter cell carries a magnetic moment of 4 × 10−12 A-m2 (~60,000 particles per cell). The magnetic moment used here is the actual magnetic moment (calculated from the relaxometry-observed moment), because all of the magnetic material on the cells is attracted by the needle. The volume of fluid corresponding to the 4.8 mm collection radius is 463 µl, which is 62% of the 750 µl sample volume. This theoretical result agrees reasonably well with the captured fraction determined from the relaxometry measurements, considering that this range calculation does not take into account cooperative magnetic or hydrodynamic effects (e.g., chain formation, convection) which are known to enhance the rate of collection [18].
Moment Superposition Model analysis
It is somewhat surprising that the relaxometry data in Fig. 3 can be fit rather well by a simple Langevin function (i.e., by assuming a single value of µ), as there is clearly a wide distribution of moments present in the sample. The moment superposition model (MSM) [13] was developed to treat the case of a distribution of non-identical, non-interacting, superparamagnetic particles and has been used by other workers to interpret relaxation curves (M vs. t) acquired by magnetorelaxometry [4–9]. Such a model may also be used to calculate the shape of the magnetization curve, M vs. B, as we show below.
Similar to Chantrell [13], we model the magnetization observed by relaxometry in terms of a distribution f(μ) of N dipole moments:
| (2) |
where L is the Langevin function, tpulse is the duration of the magnetizing pulse, t is the time after the end of the magnetizing pulse, and the Néel relaxation time τ is taken to be
| (3) |
following Ref. 4. HK is the anisotropy field, with HK = 2K/Ms. We assume a log-normal distribution of dipole moments
| (4) |
The mean moment m’ and the variance s2 are calculated from m and σ2 in the standard way.
To fit the relaxometry data (shown in Fig. 3, inset), we constrained Ms to the value determined by susceptometry (3.82 × 105 A-m), assuming the bulk density of magnetite. Then the value of m was adjusted manually, and σ and HK were allowed to vary to obtain the best fit. The value of m was adjusted until the resulting fit satisfied the experimentally-determined constraint that the observable magnetization calculated from Eq. 2 (for t=50 ms, B=3.8 mT) should be 0.33% of the maximum possible magnetization. The best fit satisfying the constraint yielded the following parameters: m’ = 5.7 × 10−18 A-m2, s = 2.0 × 10−18 A-m2, and HK = 9.6 mT. The fit is not shown, because it is virtually indistinguishable from the simple Langevin function fit in Fig. 3. However, compared to the simple Langevin fit, the MSM fit results in a larger value for the average observed moment (μobs = 1.67 × 10−17 A-m2) and a lower value of K (1.8 × 103 J/m3).
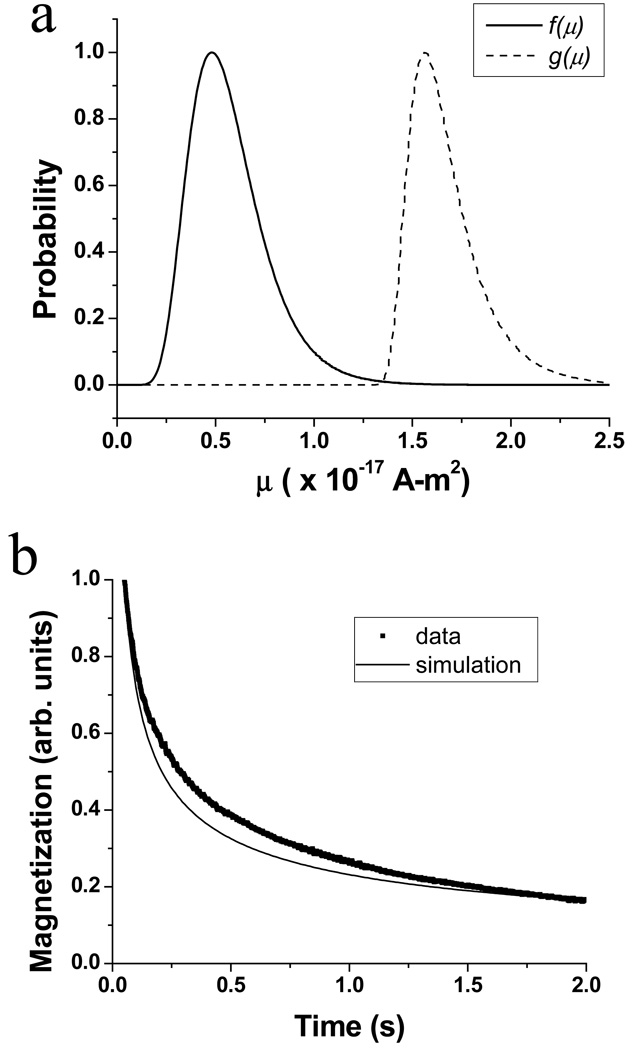
Based on the fit of the magnetization curve, the resulting distribution of moments f(μ) (solid line) and the distribution of observable magnetization g(μ) (dashed line) are shown in Fig. 6a. The moments contributing to the observable magnetization are those with short enough relaxation times (at 3.8 mT) that they become magnetized in 0.3 s, and long enough relaxation times (at zero field) that the magnetization is not completely relaxed at 50 ms. Qualitatively, these distributions agree with the TEM and AC susceptibility results, which indicate that the mean moment is smaller than μobs. Using the fitted value of HK, we calculate that a moment of 7.6 × 10−18 A-m2 would give rise to the 100 Hz peak observed in Fig. 4, in reasonable agreement with the peak of f(μ) in Fig. 6a. However, the typical particle size (15 nm) observed in the TEM (Fig. 2) corresponds to a moment which is an order of magnitude smaller.
Fig. 6.
(a) Distribution of moments f(μ) (solid line) and distribution of observable magnetization g(μ) (dashed line) determined by MSM analysis of the relaxometry data in Fig. 3. (b) Relaxation curves determined experimentally (■) and by simulation (line) using Eq. 2 with the same parameters as in (a).
In Fig. 6b, the relaxation curve (M vs. t, with B = 3.8 mT) was calculated using Eq. 2 and the fit parameters determined above (solid line). The calculated curve shows a somewhat faster decay than the experimental data (■). Together, these results indicate that further refinements to the model will be required to simultaneously account for all measured data. Currently, we are considering the inclusion of the anisotropy energy terms in the calculation of the Langevin function, and we are exploring different functional forms for the B-dependence of the Néel time constant and the moment distribution.
Conclusions
Characterization of magnetite nanoparticles (Chemicell SiMAG-TCL) showed that the magnetization detected by SQUID-relaxometry was 0.33% of that detected by susceptometry, indicating that the sensitivity of SQUID-relaxometry could be significantly increased through improved control of nanoparticle size. We also demonstrated that SQUID-relaxometry and susceptometry provide complementary information, which is useful for quantitatively analyzing nanoparticle-cell binding experiments and for evaluating the results obtained by moment superposition model analysis.
Acknowledgments
Senior Scientific, LLC, acknowledges the support of the National Institutes of Health SBIR program. JEJ acknowledges the support of the Howard Hughes Medical Institute Medical Research Training Fellowship program. This work was performed, in part, at the Center for Integrated Nanotechnologies, a U.S. Department of Energy, Office of Basic Energy Sciences user facility. Sandia National Laboratories is a multi-program laboratory operated by Sandia Corporation, a Lockheed-Martin Company, for the U. S. Department of Energy under Contract No. DE-AC04-94AL85000.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Flynn ER, Bryant HC. Phys. Med. and Biol. 2005;50:1273. doi: 10.1088/0031-9155/50/6/016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryant HC, Sergatskov DA, Lovato DA, et al. Phys. Med. Biol. 2007;52:4009. doi: 10.1088/0031-9155/52/14/001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaetao JE, Lovato DM, Adolphi NL, et al. manuscript in preparation. [Google Scholar]
- 4.Eberbeck D, Hartwig S, Steinhoff U, et al. Magnetohydrodynamics. 2003;39:77. [Google Scholar]
- 5.Romanus E, Berkov DV, Prass S, et al. Nanotechnology. 2004;14:1251. doi: 10.1088/0957-4484/14/12/003. [DOI] [PubMed] [Google Scholar]
- 6.Eberbeck D, Wiekhorst F, Steinhoff U, et al. J. Phys.: Condens. Matter. 2006;18:S2829. [Google Scholar]
- 7.Enpuku K, Tanaka T, Matsuda T, et al. J. Appl. Phys. 2007;102:054901. [Google Scholar]
- 8.Ludwig F, Heim E, Schilling M. J. Appl. Phys. 101;2007:113909. [Google Scholar]
- 9.Ludwig F, Heim E, Schilling M, et al. J. Appl. Phys. 2008;103:07A314. [Google Scholar]
- 10.Néel L. Adv. Phys. 1955;4:191. [Google Scholar]
- 11.Kötitz R, Weitschies W, Trahms L, et al. J. Magn. Magn. Mater. 1999;201:102. [Google Scholar]
- 12.Chemla YR, Grossman HL, Poon Y, et al. Proc. Natl. Acad. Sci. 97;2000:14268. doi: 10.1073/pnas.97.26.14268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chantrell RW, Hoon SR, Tanner BK. J. Magn. Mater. 1983;38:133. [Google Scholar]
- 14.ASTM E394-00, Standard Test Method for Iron in Trace Quantities Using the 1,10-Phenanthroline Method. 2000 [Google Scholar]
- 15.Wang A. Fayetteville, AR, USA: Ocean NanoTech, LLC; private communication. [Google Scholar]
- 16.Chung SH, Hoffmann A, Bader SD, et al. Appl. Phys. Lett. 2004;85:2971. [Google Scholar]
- 17.Basso G, Lanza F, Orfao A, et al. J. Biol. Regul. Homeos. Agents. 2001;15:68. [PubMed] [Google Scholar]
- 18.Mikkelsen C, Hansen M, Bruus H. J. Magn. Magn. Mater. 2005;293:578. [Google Scholar]