Abstract
Over the past decade, single-molecule fluorescence studies have elucidated the structure-function relationship of RNA molecules. The real-time observation of individual RNAs by single-molecule fluorescence has unveiled the dynamic behavior of complex RNA systems in unprecedented detail, revealing the presence of transient intermediate states and their kinetic pathways. This review provides an overview of how single-molecule fluorescence has been used to explore the dynamics of RNA folding and catalysis.
Keywords: RNA folding, RNA catalysis, single-molecule fluorescence
1. Introduction
RNA molecules are important biopolymers that show high functional versatility in living cells. In addition to their well-known function as an information carriers, they can play essential roles in catalysis and gene regulation [1]. Although RNA molecules make use of only four nucleobases, their ability to fold into a seemingly infinite number of dynamic structures is key for their functional diversity. The discovery of different functional aspects of RNA molecules has increased their potential in various applications in modern medicine and biotechnology [2-5].
Over two decades ago, it was first shown that RNA molecules have the capability to catalyze chemical reactions in the absence of proteins [6,7]. Subsequently, it was further recognized that RNA is a main player in two vital biological processes: splicing and translation. Pre-mRNA splicing is catalyzed by the spliceosome, a multi-megadalton ribonuclearprotein complex consisting of five RNAs and over 150 proteins [8,9]. In all living organisms, mRNA translation is catalyzed by the RNA components of the ribosome, another essential multi-megadalton ribonuclearprotein complex [10-12]. The ribosome is the largest and most abundant known ribozyme in nature.
In addition to its roles in catalysis, RNA molecules can also perform efficient regulatory roles in living organisms. Regulatory RNAs, such as micro-RNAs and small-interfering RNAs, can regulate gene expression by either direct translational suppression or by degradation of mRNA [13-15]. Riboswitches are regulatory RNAs that can regulate gene expression at both the transcriptional and the translational level, providing more evidence for the variety of RNA functions in living cells [16].
The folding of RNA into precise three-dimensional structures is essential for proper functioning of these molecules in nature. How a simple linear biopolymer like RNA adopts a functionally active tertiary structure is the crux of the RNA folding problem. Because of the rugged nature of their folding potential energy surfaces, RNAs can follow complex kinetic pathways and adopt multiple intermediate conformations before reaching the active state [17-29]. Interactions with metal ions and protein co-factors can influence the stability of these intermediates and cause changes in the RNA folding landscapes. UV melting, Isothermal Titration Calorimetry (ITC), Circular Dichroism (CD) and chemical footprinting are some important techniques that have been used traditionally to characterize RNA folding reactions [17-19,30-32]. However, all of these techniques deal with ensemble-averaged populations of a large number of molecules, which restricts the information that can be obtained if multiple conformations are present in solution. The emergence of single-molecule techniques has represented a tremendous advance in studies of RNA folding and catalysis, because it enables the direct observation of transient intermediates in isolated molecules, thereby resolving static and dynamic molecular heterogeneity and eliminating the requirement to synchronize all of the observed molecules to derive quantitative kinetic information. Thus, single-molecule spectroscopy provides insight into the functioning of complex biological systems by revealing the kinetic steps otherwise hidden in ensemble-averaged bulk experiments [25,33,34].
Since the first single-molecule RNA folding study a decade ago [35], single-molecule fluorescence has become a widely used technique to study the mechanisms of folding and catalysis in several RNA-based systems. The following sections review applications of single-molecule fluorescence that have significantly improved our understanding of RNA folding and catalysis. Since this review cannot include all the remarkable advances in the study of RNA folding and catalysis with single-molecule methods, we refer to these other reviews for further examples [26-29,36-44].
2. Experimental Approaches
Several single-molecule approaches have proven useful in adressing the structure-function relationships of RNA. For example, single-molecule force spectroscopy with optical tweezers has been used to study folding of the Tetrahymena ribozyme and the Adenosine riboswitch [45,46]. Here, we focus our attention to just one of the most widely used techniques for studying the structure and dynamics of RNA molecules: single-molecule fluorescence resonance energy transfer (smFRET).
2.1. Single molecule FRET
FRET is a powerful molecular ruler, which can be used to measure distances in the 2-10 nanometer range [47,48]. The combination of single-molecule detection with FRET has increased its potential in numerous applications in biological systems [49,50]. FRET is a distance-dependent, non-radiative energy transfer process that takes place between a donor and acceptor fluorophore linked to a biomolecule. Upon excitation of the donor fluorophore, the energy is transferred to the acceptor and increases its emission intensity while decreasing the emission intensity of the donor fluorophore. Energy transfer arises from dipole-dipole interactions between these fluorophores, and the efficiency of this process (E) is given by E = 1/(1+(R/R0)6), where R is the distance between the two dipoles and R0 is the Förster distance, at which 50% energy transfer efficiency is observed. R0 is typically 3-8 nm for most fluorophore-pairs. The FRET efficiency also depends on the orientation of the donor and acceptor fluorophores as well as the spectral overlap between the emission spectrum of the donor and the excitation spectrum of the acceptor molecule.
The power of single-molecule detection combined with the sensitivity of FRET has made single-molecule FRET (smFRET) one most widely-used real-time detection techniques to monitor structural dynamics, kinetics and thermodynamics of biopolymers. In order to achieve single-molecule sensitivity, it is critical to reduce all sources of background fluorescence [51,52]. To this aim, two techniques are primarily used: confocal and total internal reflection fluorescence (TIRF) microscopy. Both these techniques reduce background fluorescence by limiting the detection volume.
In confocal microscopy, the out-of-focus light transmitted to the detector is reduced by a small aperture in the image plane of the microscope, and detection is effectively limited to a small (femtoliter-size) volume, thus increasing the signal-to-noise ratio by suppressing the background signal. Confocal microscopy helps to detect freely diffusing molecules in solution [53]. However, the short dwell time of these molecules in the detection volume is the main limitation of this technique. In order to overcome this limitation, two main strategies have been developed during the past years: Immobilization of the sample in agarose or polyacrylamide gels [54] and the surface immobilization of molecules by specific interactions, such as a biotin-streptavidin bridge [35].
In contrast to confocal microscopy, TIRF microscopy aids in the detection of molecules by exciting with an evanescent wave generated by total internal reflection of a laser beam. The reduction of the excitation volume to a thin sheet at the interface between the slide and the solution decreases the background signal and the excessive photobleaching of molecules outside the detection volume. This technique facilitates the simultaneous detection of a large number of molecules simultaneously in a wide observation field with the use of a charge-coupled device (CCD) camera. These features increase the applicability of this technique by alleviating some of the disadvantages of confocal microscopy. However, the time resolution that can be obtained by current CCD cameras is still lower than that of the avalanche photodiodes (APD) and photomultipliers that are used in confocal fluorescence microscopy.
3. Single-molecule studies of RNA
The proper understanding of RNA folding dynamics is critical to the determination of its structure-function relationship. The presence of multiple kinetic pathways and intermediate RNA structures make this task challenging using ensemble-averaged experiments. However, single-molecule FRET has shown its ability to overcome many of these challenges by revealing transient intermediates on the folding pathway of various RNA molecules. As a result, smFRET has been widely used to elucidate the folding dynamics of various catalytic RNAs ranging from the small hairpin and VS ribozymes to large catalytic RNAs such as RNase P RNA and group II introns [25,33,34,55-60]. The following section summarizes some of the folding studies of biologically important small and large catalytic RNAs, as well as protein-mediated RNA folding in the assembly of the ribonucleoprotein (RNP) complexes.
3.1. Folding studies of small RNAs
Small ribozymes are critical model systems, which provide basic information about the folding dynamics of RNA. The hairpin ribozyme is one of the best-characterized small RNA enzymes [26,36,39,42,52,61,62]. This small ribozyme is derived from the self-cleaving negative strand of the satellite RNA in the tobacco ringspot virus [63]. In the rolling-circle replication of some viral RNAs, the hairpin ribozyme catalyzes the autocatalytic reaction to process multimeric products into unit-length satellite RNA genomes. The structure of the hairpin ribozyme consists of a four-way helical junction with two internal loops on adjacent arms that can dock on each other [25]. In the minimal version of this ribozyme, two of the helices can be removed to yield a two-way junction with docking internal loops A and B (Fig. 1a). The hairpin ribozyme can form an inactive extended conformation (undocked state) or an active bent conformation (docked state) containing tertiary interactions between loops A and B [64].
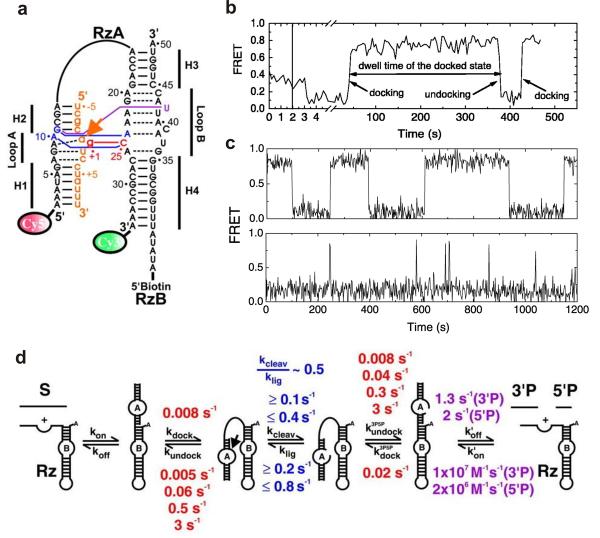
Fig. 1.
Single molecule FRET studies of the hairpin ribozyme. (a) The fluorophore-labeled two-way junction hairpin ribozyme binds with the substrate (orange). Terminal Cy3 and Cy5 fluorophores serve as the fluorescent donor-acceptor pair and biotin is used for surface immobilization through a biotin-streptavidin bridge. (b) A typical sm-FRET time trajectory of a single hairpin ribozyme. The FRET states at 0.8 and 0.2 represent the docked and the undocked states, respectively. (c) The FRET time traces of two different ribozyme molecules exhibit heterogeneous undocking kinetics and reveal the memory effect of the hairpin ribozyme. (d) The multistep reaction pathway of the hairpin ribozyme with the observed rate constants. From reference [33], X. Zhuang et. al., Science 296, 1473 (2002). Reprinted with permission from AAAS.
To understand the correlation between structural dynamics and function of the hairpin ribozyme, Steven Chu, Nils Walter and coworkers performed the first smFRET experiments on the minimal hairpin ribozyme with the two-way junction [33]. The hairpin ribozyme was fluorophore labeled by attaching Cy3 and Cy5 dyes (FRET donor and acceptor, respectively) to the 3′ and 5′ ends of the RzA strand (Fig. 1a). A biotin was attached to the 5′ end of the RzB strand for surface immobilization via a biotin-streptavidin interaction. The fluorescence intensity from individual donor and acceptor fluorophores (Id and Ia) was detected using TIRF excitation and the apparent FRET efficiency calculated as FRET = Ia/(Ia + Id). Single-molecule FRET time trajectories reveal the structural dynamics of each molecule (Fig. 1b and c). The smFRET data show that the two ribozyme conformations (docked, FRET ∼ 0.9, and undocked, FRET ∼ 0.2) are in dynamic equilibrium. From the FRET trajectories, the kinetic rates of docking and undocking can be directly measured by determining the dwell time distributions in each state. The data showed a single rate of docking (0.008 s−1), and four different undocking rates were obtained, revealing the presence of four docked sub-populations, each with distinct undocking kinetic rates (Fig. 1d). Interestingly, a molecule in a given sub-population tends to remain in the same state (Fig. 1c) without switching between different docked states, revealing the presence of a strong memory effect in the folding pathway of the hairpin ribozyme. This behavior was also present in the natural form of the hairpin ribozyme in both docking and undocking [25]. The heterogeneous kinetics of the hairpin ribozyme were further explored by characterizing the effect of site-specific mutations and metal ion titrations on the transition state of the two-state folding reaction [55]. The metal ion titrations suggested the presence of a compact transition state, which can be stabilized by electrostatic interactions to a similar extent as the docked state, while the mutation analysis showed that the native tertiary contacts are, at most, only partially formed. The characterization of the transition state of the hairpin ribozyme extended the existing knowledge of RNA folding and suggested that the formation of a compact transition state without well-formed tertiary contacts may be a general phenomenon in elementary RNA folding reactions.
Further, smFRET combined with functional assays and kinetic simulations was used to study the impact of the essential functional groups located far from the cleavage site on both docking and undocking rate constants [56]. Similar to the wild type, all observed variants showed heterogeneous undocking kinetics with distinct undocking rate constants. However, heterogeneous docking kinetics were observed only in the variant with a modification at the domain junction. The persistence of molecular heterogeneity with memory effect for all variants under different buffer conditions confirms the previously identified unique nature of the hairpin ribozyme. Surprisingly, most modifications not only affect docking and undocking, but also significantly impact the internal chemistry rate constants over a substantial distance from the site of catalysis, revealing the presence of a network of coupled molecular motions that connects distant parts of the RNA with its reaction site.
Despite the simplicity of the hairpin ribozyme structure, smFRET revealed complex structural dynamics, providing useful information for in-depth understanding of the structure-function relationship. By enabling the discovery of previously unidentified complex behavior of the small ribozymes, these findings highlight the potential of smFRET over traditional ensemble-averaged experiments. Because of the modular nature of RNA molecules, studying small domains in isolation can be helpful to deepen our understanding of the folding dynamics of more complex large ribozymes [65-68].
3.2. Folding studies of large RNAs
The capability of smFRET to monitor the complex folding dynamics of large RNA enzymes was proven by numerous interesting applications. In the following sections, we highlight three examples: The Tetrahymena ribozyme, the catalytic domain of RNase P and the group II intron ribozyme.
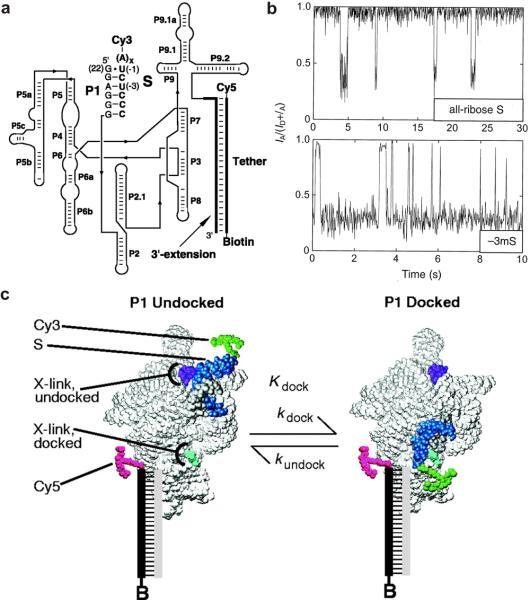
The Tetrahymena ribozyme is a large, multi-domain RNA enzyme that has been extensively characterized [6,69]. It was derived from a self-splicing group I intron and comprises a highly conserved catalytic core (P4 - P8) with additional conserved peripheral structural elements (Fig. 2a). The P4 - P6 domains have the ability to fold independently into the native tertiary structure in a magnesium-dependent manner [20,70-72]. As such, the complex structure of the Tetrahymena ribozyme, together with its P4 - P6 domain, provides a valuable model system to study the folding of large RNAs. Early smFRET studies showed that the P1 duplex reversibly docks into the preformed active core of the ribozyme to form the active structure [22]. A fluorophore-labeled ribozyme was used to monitor P1 docking and the overall folding of the Tetrahymena ribozyme in order to dissect the complex RNA folding pathway (Fig. 2a and 2c). Single-molecule FRET trajectories directly revealed a dynamic equilibrium between the docked (high FRET) and the undocked (low FRET) states (Fig. 2b). The analysis of folding transitions of the Tetrahymena ribozyme indicates the presence of multiple folding pathways (Fig. 2b, compare top and bottom traces). The folding transition state of the Tetrahymena ribozyme was probed by comparing the effects of mutations and denaturants on the docking rate (kdock) and equilibrium constant (Kdock) using smFRET [73]. Modification of eight groups in the P1 duplex that form tertiary interactions with the active core decreased the docking equilibrium (500-fold) to a greater extent than the docking rate constant (2-fold). The lower sensitivity of kdock to these modifications suggests that these tertiary interactions are not present in the transition state, suggesting an early transition state without tertiary contacts between P1 and the ribozyme core, as in the case of the hairpin ribozyme [55].
Fig. 2.
Single molecule FRET studies of the Tetrahymena ribozyme. (a) The fluorophore-labeled structure of the Tetrahymena ribozyme. The Cy3 and Cy5 fluorophores serve as the fluorescent donor and acceptor, respectively. The biotin is used for surface immobilization of the ribozyme complex through a biotin-streptavidin bridge. (b) FRET time trajectories of the Tetrahymena ribozyme showing P1 docking and undocking. The FRET states at ∼0.9 and ∼0.3 correspond to the docked and undocked states, respectively. (c) A model showing the docking and undocking of the P1 duplex of the Tetrahymena ribozyme. The p1 duplex between the 3′ end of the ribozyme (gray) and S (blue) reversibly docks into the rest of the ribozyme (light gray). From reference [22], X. Zhuang et. al., Science 288, 2048 (2000). Reprinted with permission from AAAS.
Other studies have used single-molecule force spectroscopy to study the unfolding from the native state to a fully extended single stranded RNA [45]. This technique uses focused laser beams to manipulate biological molecules immobilized between two separated beads in solution [74,75]. The resulting mechanical unfolding trajectories showed the presence of eight intermediates with discrete kinetic barriers, demonstrating the complex folding behavior of this catalytic RNA.
The catalytic domain of the RNase P RNA is a well-characterized large RNA enzyme. It catalyzes the maturation reaction of tRNA in bacteria and eukaryotes by cleaving the 5′ leader sequence of precursor tRNA [7,76]. The ensemble analysis of this large catalytic RNA describes the two-step folding pathway consisting of three main conformational states: the unfolded state (U), the intermediate state (I), and the folded native state (N) [77]. Magnesium-dependent, smFRET-mediated folding analysis of RNase P RNA revealed the existence of at least two previously unidentified folding intermediates [57]. Further, FRET time trajectories of individual RNA enzymes indicated the presence of five subpopulations based on three FRET states as well as dynamics between different states. Collectively, the observed heterogeneity and slow conformational transitions indicate the early folding of this catalytic RNA through a series of intermediates.
More recently, the folding of the Bacillus stearothermophlilus RNase P catalytic domain was studied by a specially designed single-molecule, non-equilibrium periodic [Mg2+]-jump method [58]. This approach allows probing of the regions of the free energy landscape to understand the early folding pathways of a large RNA. According to the observed FRET trajectories, [Mg2+] jumps from 0.01 mM to 0.1 mM show two-state behavior and jumps from 0.01 mM to 0.4 mM show complex multistate folding behavior. Before the jump, RNA molecules in low and high FRET states show dynamics in two distinct regions of the energy landscape that are separated by a high-energy barrier. Structural changes involved in crossing this barrier that did not change FRET values but changed their dynamics were identified. The observed slow dynamics between different conformations helped reveal the presence of a long memory effect of folding in this catalytic RNA. This specially-designed smFRET approach in addition to previous smFRET data collectively elucidate complex folding dynamics of the RNase P RNA by characterizing undisclosed folding intermediates and the free energy landscape of this RNA enzyme.
Group II intron ribozymes rank among the largest protein-free, multidomain ribozymes found in nature. Their catalytic mechanism resembles that of nuclear splicing in eukaryotes [78]. The ribozyme's secondary structure consists of six domains (D1-D6) that radiate from a central core, but only nucleotides involved in tertiary interactions are highly conserved [79]. The minimal active form is the D135 ribozyme, derived from the group II intron Sc.ai5γ of Saccharomyces cerevisiae, which consists of the necessary components for catalysis and provides a model system to study group II introns. The unique folding pathway of the group II intron ribozyme consists of an initial slow compaction step of domain I (D1), followed by rapid folding of the remaining domains onto the D1 scaffold to form the native state (N) [80]. Previous studies have described the two-step folding pathway for this D135 ribozyme that contains one obligatory intermediate (U ↔ I ↔ N) at high salt conditions [81]. The fast folding step has been recently characterized by smFRET experiments [34]. Because of its large size, the D135 ribozyme was fluorophore-labeled and biotinylated by hybridizing fluorophore-labeled and biotinylated DNA strands with the ribozyme (Fig. 3c). Then, the whole complex was immobilized on a microscope slide through a biotin-streptavidin interaction and detected using TIRF-based smFRET (Fig. 3a).
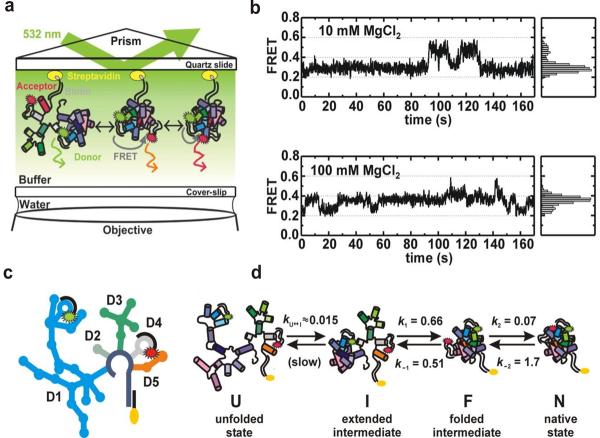
Fig. 3.
Single molecule FRET studies of group II intron ribozymes. (a) Experimental setup of the total internal reflection fluorescence spectroscopy (TIRF) based single molecule FRET. Fluorophore-labeled ribozyme construct is immobilized on a quartz slide through a biotin-streptavidin linkage. (b) Single molecule time trajectories and histograms at different Mg2+ concentrations showing the presence of three different FRET states (∼0.25, 0.4 and 0.6). (c) A schematic structure of the Cy3 and Cy5 fluorophore-labeled D135-L14 ribozyme containing five domains (D1, D2, D3, D4 and D5). (d) Folding pathway of the D135-L14 ribozyme consisting of four different states: the unfolded state (U), the extended-intermediate state (I), the folded-state (F), and the native state (N). Reprinted with permission from reference [34], M Steiner et. al., PNAS 105, 13853 (2008). Copyright (2008) National Academy of Sciences, U.S.A.
The single-molecule data revealed three distinct structural conformations with a previously unidentified on-pathway folding intermediate (Fig. 3b and d). Further, the observed fast folding rates between different states indicated that these conformations are connected by small activation barriers. FRET time trajectories showed that the native conformation occurs only transiently at 20 mM or higher Mg2+ concentrations. However, the binding of the substrate with the ribozyme stabilizes the high FRET conformation, suggesting this structure acts as a catalytically active conformation in vitro. Interestingly, it was found that increasing [Mg2+] lead to an increase in structural dynamics. The fraction of dynamic molecules versus [Mg2+] yields a dissociation constant that coincides with the values for bulk folding and cleavage. This result links the structural dynamics of D135 to its function, posing the interesting possibility that domain motion contributes to successful reactivity.
3.3. Protein-mediated RNA folding
RNA is a structurally and functionally flexible biomolecule, which often requires specific protein assistance to form functionally active conformations in vivo. Protein factors can promote RNA folding into the native conformation by either enhancing the structural stability or by chaperoning the folding process [82,83]. RNA-protein complexes (RNPs) require numerous protein factors to coordinate the assembly of these large structures for specific biological function. The capability of single-molecule fluorescence to dissect the reaction pathways by direct observation of the folding processes paved the way for its use in characterizing the assembly of large RNA-protein complexes.
In order to explore the assembly of the telomerase RNP complex through stepwise protein-mediated RNA folding, Zhuang and coworkers performed single-molecule fluorescence experiments using telomerase from Tetrahymena thermophila as a model system [84]. The telomerase RNP complex solves the end replication problem by adding telomeric DNA to the termini of the linear chromosomes. The complete telomerase complex contains three main components: telomerase RNA, telomerase reverse transcriptase (TERT) and protein cofactors. Telomerase RNA and TERT are key components for telomerase activity. However, in vivo telomerase biogenesis requires RNA-binding proteins to promote the specific assembly pathway [85]. Therefore, the real-time observation of the Tetrahymena telomerase RNP assembly was detected using the telomerase RNA, TERT and the La-motif protein p65 (Fig. 4a) [84]. In the experimental set-up, the fluorophore-labeled telomerase RNA was immobilized on a surface via a biotin-streptavidin interaction, and RNA folding was monitored in the presence or the absence of the TERT and the holoenzyme p65 using smFRET (Fig. 4a).
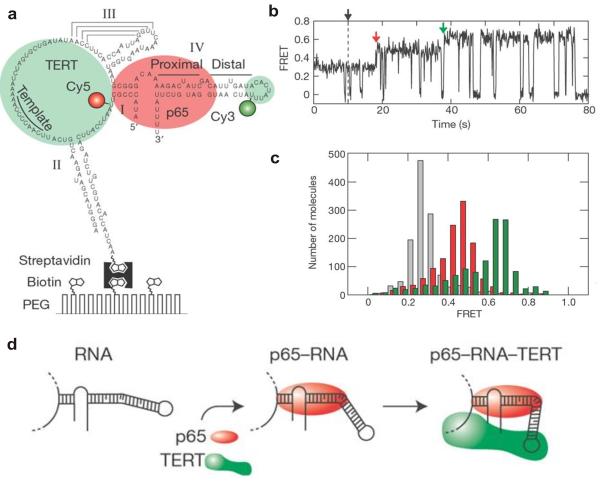
Fig. 4.
Protein-mediated RNA folding in assembly of the telomerase RNP complexes. (a) The Cy3 (donor) and Cy5 (acceptor) fluorophore-labeled full length telomerase RNA molecule immobilized on a surface via a biotin-streptavidin linkage. (b) smFRET time trajectory showing the assembly of the RNP complex after the addition of p65 and TERT (black arrow). The assembly pathway consists of two steps: P65-induced FRET transition (red arrow) and complete assembly of the p65-RNA-TERT ternary complex (green arrow). (c) FRET histogram showing the distribution of the FRET states in the absence of protein (grey), the presence of 10 nM p65 (red) and the presence of both 10 nM p65 and 32 nM TERT1-516 (green). (d) A schematic representation of RNA folding observed in real-time assembly of the telomerase RNP complex. Reprinted by permission from Macmillan Publishers Ltd: [Nature] [84], copyright (2007).
Telomerase RNA showed a single FRET distribution centered at 0.29 in the absence of proteins. Addition of the holoenzyme p65 shifted the observed FRET value toward the higher value of 0.46, indicating a p65-induced conformational change of RNA that brings the ends of stem I and stem IV into closer proximity. However, further characterization of the p65-induced intermediates by mutational studies revealed that the p65-induced structural changes occur mainly around the conserved GA bulge in stem IV. Upon addition of purified TERT into the p65-RNA complex, the FRET value shifted further to a high value of 0.65 (Fig. 4b and c). This indicates the formation of the p65-RNA-TERT ternary complex with a close association of the helix IV and stem I.
To further understand the RNA folding process, a mixture of proteins containing both p65 and TERT was introduced to the immobilized RNAs. The majority (74%) of the resulting FRET trajectories showed a two-step pathway with an intermediate FRET state at 0.46, revealing the hierarchical telomerase assembly pathway directed by stepwise protein-mediated RNA folding. In the absence of p65, FRET traces of the TERT-RNA complex did not exhibit the previously observed high FRET state (∼0.65), but the addition of p65 into the TERT-RNA complex was able to rescue the high FRET state. This provided strong evidence for the p65-induced RNA conformation as an essential intermediate during the assembly pathway. Using smFRET, the authors were able to observe and characterize the hierarchical assembly pathway of the Tetrahymena telomerase RNP complex (Fig. 4d).
3.4. RNA catalysis
Numerous studies have explored the catalytic nature of RNA using ensemble-averaged techniques [86]. Compared to these methods, smFRET offers the potential to dissect the reaction pathway by revealing transient intermediates and minor subpopulations making a significant contribution to characterize catalytic RNAs. During the last decade, single-molecule techniques have become widely used to explore the numerous catalytic RNAs ranging from small ribozymes such as the hairpin ribozyme to large catalytic RNAs such as the Tetrahymena ribozyme [22,33,56,87]. Here, we summarize some of the fruitful applications of smFRET to illustrate its contribution to a greater understanding of RNA catalysis.
The hairpin ribozyme reversibly cleaves its substrate into two products through a cleavage pathway consisting of substrate binding, interdomain docking or folding of the ribozyme-substrate complex into a catalytically active docked state, cleavage of the substrate, interdomain undocking or unfolding of the ribozyme-product complex, and release of the products from the ribozyme. In order to dissect this multi-step cleavage reaction, single-molecule fluorescence was applied to both the minimal and natural forms of the hairpin ribozyme [33,87]. The first smFRET study performed with the minimal form of the hairpin ribozyme containing a two-way junction exhibited three distinct FRET states corresponding to the docked, undocked and substrate-free or unbound states during the cleavage reaction [33]. An increase of the substrate-free state population over time indicates the cleavage and rapid dissociation of the cleaved products from the undocked state. In addition to confirming the previously proposed cleavage reaction pathway by ensemble experiments, this study was able to reveal kinetic rate constants of this reaction for the individual conformational changes. Based on these observed rate constants, the conformational transitions between docked and undocked states and the equilibrium constants of the reversible cleavage step are recognized as the rate-limiting points of the overall cleavage reaction of the two-way junction ribozyme. Furthermore, the apparent heterogeneity in the undocking rate constants demonstrates the heterogeneous cleavage kinetics of these catalytic RNAs.
The internal cleavage and ligation reactions were directly observed for the first time by the single-molecule fluorescence studies conducted with the natural form of the hairpin ribozyme containing a four-way junction [87]. According to the resulting FRET trajectories, molecules can remain stably in the docked state or display rapid docking and undocking transitions representing the ligated and cleaved forms of the ribozyme, respectively. These distinct structural dynamics between the cleaved and ligated forms enable the recognition of multiple cycles of internal cleavage and ligation of a single ribozyme. Further, the analysis of chemical rate constants indicates a stronger equilibrium bias toward the ligated state. Compared to the minimal hairpin ribozyme, the natural form of this ribozyme shows a different rate-limiting mechanism. The cleavage step accelerates the undocking of the four-way junction ribozyme and exhibits a faster undocking rate than the ligation rate. As a result, the overall cleavage reaction rate of the four-way junction ribozyme is primarily limited at the internal cleavage step. In addition to previously observed heterogeneity of undocking, the natural form of the hairpin ribozyme showed heterogeneity in rate constants of both the folding and the unfolding steps of the ribozyme. The observed kinetic heterogeneity in smFRET studies elucidates the fast and the slow phases of the cleavage pathway of the hairpin ribozyme.
The Tetrahymena ribozyme, derived from a self-splicing group I intron, catalyzes the cleavage of the substrate by guanosine. The catalytic reaction of this self-splicing RNA enzyme was observed by single-molecule fluorescence with addition of guanosine to the surface-immobilized ribozyme-substrate complex [22]. After rapid binding of guanosine, the cleavage of the substrate was detected by the disappearance of fluorescence signal due to release of the Cy3-labeled cleavage product. The identical cleavage rate constants obtained from both the single-molecule and ensemble measurements indicate the validity of the single-molecule technique to study RNA catalysis.
3.5. The ribosome and single-molecule fluorescence
The ribosome is a complex molecular machine that catalyzes protein synthesis in all living cells. In bacteria, this large ribonucleoprotein complex is composed of two subunits, the small 30S subunit and the large 50S subunit, that assemble around an mRNA template to form a complete 70S ribosome. During translation, tRNAs recognize the information encoded in the mRNA template and convert it into a specific protein with the help of the ribosome. There are three tRNA binding sites at the subunit interface of the ribosome: the aminoacyl site (A site), the peptidyl site (P site), and the exit site (E site) [88].
Protein synthesis consists of three main steps: translation initiation, elongation and termination. Translation elongation is an extensively studied process that contains several important steps [89,90]. First, the ternary complex, containing aminoacyl tRNA (aa-tRNA), GTP and elongation factor Tu (Ef-Tu), binds with the A site and forms base pairs at two regions; The anticodon loop of the aa-tRNA base pairs with the mRNA codon at the A site within the 30S subunit, and the 3′CCA terminal residues base pair with the conserved ribosomal RNA (rRNA) within the 50S subunit. Correct codon-anticodon recognition triggers GTP hydrolysis by EF-Tu. After hydrolysis, the 3′ end of the A site aa-tRNA dissociates from EF-Tu(GDP) and resides in the peptidyltransferase center to promote peptide bond formation. Then the peptide attached to the P site tRNA is transferred to the A site tRNA, and the mRNA-tRNA complex translocates to reset the ribosome for the next round of protein synthesis.
Cryo-electron microscopy and chemical footprinting studies have identified two tRNA conformations on the ribosome: the classical state and the hybrid state [91]. In the classical state, each tRNA is bound to the A and P sites on both the 30S and 50S subunits (A/A-P/P). In the hybrid state, tRNAs remain bound to the A and P sites at the 30S subunit, while the 3′CCA ends of the P- and A-site tRNA are bound to the E and P sites of the 50S subunit, respectively (A/P-P/E). This suggests, high spatial and time resolution smFRET studies identified a new tRNA configuration in which only deacylated-tRNA adopts a hybrid state (A/A-P/E). Therefore, tRNAs exhibit dynamic exchange between three metastable configurations during translocation process [92].
In order to gain more information about the translation process and to determine the dynamics of the tRNAs on the ribosome, Steven Chu, Joseph Puglisi and coworkers performed TIRF-based smFRET experiments using surface-immobilized ribosomes [89,90]. In the experimental set-up, a single ribosome initiated with Cy3-labeled aa-tRNA (fMet-tRNAfMet) in the P site was first bound to biotinylated mRNA, and the whole complex was then immobilized on a microscope slide by a biotin-streptavidin bridge. Cy5-labeled EF-Tu-GTP-aa-tRNA ternary complexes were then delivered to the immobilized ribosomes, and the FRET change was observed over time. smFRET time trajectories showed the rapid change from low to high FRET (∼0.75), followed by dynamic fluctuations to an intermediate FRET value (Fig. 5a). Stable occupancy of the high FRET state indicates the complete accommodation of aa-tRNA into the A site and peptide bond formation. Consistent with previous data, the complete accommodation of aa-tRNA requires ∼93 ms [89].
Fig. 5.
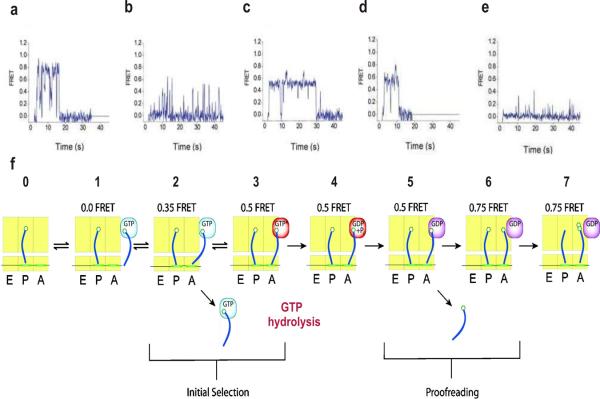
Detection of tRNA selection and dynamics in translation by single molecule FRET. Top panel shows FRET time traces obtained after stopped-flow delivery of EF-Tu-GTP-Phe-tRNAPhe (Cy5) to surface-immobilized ribosome complex containing fMet-tRNAfMet (Cy3) in the P site at different conditions. (a) Cognate aa-tRNA delivery. (b) Cognate aa-tRNA delivery in the presence of tetracycline. (c) Cognate aa-tRNA delivery in the presence of nonhydrolyzable GTP analog (GDPNP). (d) Cognate aa-tRNA delivery in the presence of kirromycin. (e) Near-cognate aa-tRNA delivery. (f) A model of tRNA selection at the A site of the ribosome. A, P and E sites of the ribosome are shown in yellow rectangles, and tRNAs are shown in blue. Reprinted by permission from Macmillan Publishers Ltd: [Nat Struct Mol Biol],[89] copyright (2004).
To further characterize this process, known inhibitors that block specific steps of the reaction pathway were linked with single-molecule fluorescence experiments [89]. In the presence of tetracycline, which inhibits A site tRNA delivery, the resulting FRET traces showed frequent excursions to the low FRET state (∼0.35) with only rare excursions to the high FRET state. This indicates that the tetracycline does not completely inhibit initial binding, but rather prevents later steps of the tRNA selection (Fig. 5b). The non-hydrolysable GTP analog, GDPNP, was able to efficiently stall aa-tRNA selection before GTP hydrolysis, and a stable FRET value of 0.5 was exhibited after rapid transition through the 0.35 FRET state (Fig. 5c). Kirromycin, which inhibits the structural transition of EF-Tu to the GDP-bound conformation, showed similar FRET traces to the GDPNP-stalled ribosomes, indicating its ability to stall the aa-tRNA selection step after GTP hydrolysis (Fig. 5d).
To investigate the fidelity of aa-tRNA selection and proofreading, mRNA at the A site was programmed with a near cognate (CUU) or a non-cognate (AAA) codon, instead of a cognate codon (UUU) [89]. FRET traces with the near cognate codon exhibited transient occupancy of the 0.35 FRET state with rare transitions to high FRET states (Fig. 5e). However, the non-cognate codon effectively eliminated the occurrence of the codon recognition state (0.35 FRET state) on the observation time scale. Comparison of smFRET data indicates a ∼6-fold preference of cognate aa-tRNAs over near cognate aa-tRNA in the initial selection step and a ∼24-fold preference in the proofreading step. Based on smFRET data, the overall error frequency of aa-tRNA selection of ∼7.1×10−3 was obtained indicating the high fidelity of this process. Overall, these studies provide more details about the translation process by direct observation of the step-wise movement of aa-tRNAs and their dynamics on the ribosome (Fig. 5f).
Single-molecule fluorescence enables direct observation of the ribosomal intersubunit dynamics in real time [93]. The dynamics of the ribosome constructed with fluorescently labeled subunits (30S subunit labeled with acceptor-Cy5 on S6 or S11 protein and 50S subunit labeled with donor-Cy3 on L9 protein) were detected using TIRF-based smFRET experiments. According to the smFRET traces, the pre-translocation ribosome fluctuates between the rotated (FRET ∼0.4) and non-rotated (FRET ∼0.56) conformations, which correspond to the hybrid and classical states, respectively. In contrast, the post-translocation ribosome remains mainly fixed in the non-rotated classical state.
Translation-elongation was also characterized using a different labeling strategy. The main goal of this study was to determine the real-time dynamic coupling between the ribosome and tRNAs using smFRET [94]. The L1 stalk of the ribosome is a universally conserved and highly dynamic part of the 50S subunit, which interacts with the central fold domain of the deacylated tRNA. To monitor the direct correlation of the ribosome and tRNA dynamics, the ribosomal protein L1 was labeled with Cy5, and Cy3 was incorporated into the tRNAPhe. The post-translocation-elongation complex (POST-1) was assembled with OH-tRNAfMet in the E site, fMet-Phe-(Cy3)tRNAPhe in the P site and an empty A site. To characterize pre-translocation, POST1 reacted with EF-Tu(GTP)Lys-tRNALys to generate the pre-translocation complex (PRE-1), leaving the newly deacylated OH-(Cy3)tRNAPhe in the P site and newly formed fMet-Phe-Lys-tRNALys in the A site. smFRET time trajectories exhibited two FRET states, the low FRET state ∼0.21 and the high FRET state ∼0.84. This indicates that the pre-translocation complex fluctuates between a non-ratcheted state (tRNAs in the classical configuration without the L1 stalk-tRNA interaction) and a ratcheted state (tRNAs in an intermediate hybrid configuration with the L1 stalk-tRNA interaction). However, the binding of EF-G forms POST-2 containing deacylated OH-(Cy3)tRNAPhe in the E site, fMet-Phe-Lys-tRNALys in the P site and an empty A site and shifts the equilibrium towards the high FRET state, indicating the formation of the ratcheted state. Collectively, these smFRET data describe the association of the L1 stalk with tRNA throughout the translocation process, suggesting the ability of L1 stalks to direct tRNA movements during translocation [94].
More recently, the dynamics of the L1 stalk of the ribosome were further studied by attaching fluorescent labels directly to the ribosome, at protein L1 and protein L33 of the 50S subunit [95]. smFRET data revealed the movement of the L1 stalk relative to the body of the 50S subunit between at least three distinct conformational states: an open conformation (FRET ∼0.25) in which L1 stalk is positioned away from the body of the 50S subunit, a half-closed conformation (FRET ∼0.4) in which L1 stalk moves inwards to interact with deacylated tRNA bound in the E/E classical state, and a closed conformation (FRET ∼0.55) that results in the inward excursion of the L1 stalk by interacting with the deacylated tRNA in the P/E hybrid state. Therefore, the movement of the L1 stalk promotes the remodeling of the ribosomal E site based on the positions and acylation states of tRNA bound to the ribosome during translocation.
4. Concluding remarks and future challenges
The structural dynamics of a small RNA three-way junction were measured for the first time by single-molecule fluorescence only a decade ago [35]. Since that initial ground breaking study, a wide variety of RNA systems have been investigated using single-molecule fluorescence, and it is now possible to observe even the ratcheting motion of single ribonucleoprotein complexes as large as the ribosome. These studies have exposed the rich dynamic properties of RNA molecules, and from the hairpin ribozyme to the ribosome, almost every RNA enzyme studied thus far exhibits a close link between folding dynamics and efficient catalysis. It is possible that this innate ability of RNAs to be dynamic may serve to compensate for their lack of chemical variety compared to that of proteins.
While the progress up to now has been extraordinary, there are still many challenges and limitations yet to overcome during the next decade. We expect future technical improvements will help advance single-molecule RNA science. For example, more sensitive CCD cameras to enable the monitoring of RNA folding events with sub-millisecond time resolution. New fluorescent probes with improved photophysical properties (quantum yield, blinking and photobleaching) will enhance future single molecule florescence studies and will offer the possibility of monitoring local conformational dynamics in parallel with the global dynamics already possible with FRET.
Single-molecule RNA studies will soon expand their repertoire to include other large ribonucleoprotein complexes, such as the spliceosome. Despite a large number of biochemical and genetic studies, this highly dynamic, multi-megadalton complex still remains one of the least understood molecular machines in the cell. The dynamic nature of the spliceosome makes it a particularly challenging system to characterize with ensemble-averaged methods, and single-molecule fluorescence is an ideal approach to provide new insights. Further improvements in fluorophore labeling strategies of large RNAs and novel surface modifications to prevent non-specific binding are currently being developed to generate a complete understanding of spliceosome dynamics [96].
Another interesting area that is likely to develop steadily over the next decade is the study of RNA folding and catalysis in live cells. The environmental differences between the test tube and the cell may impose unforeseeable constraints to the folding potential energy surfaces and to catalysis. Only in vivo studies will serve to fully elucidate the function of RNA molecules in their natural environment. An issue of particular significance is that RNA molecules are edited and fold co-transcriptionally: the 5′ end of a long RNA may fold long before the 3′ end is even transcribed. How the processes of transcription and RNA editing are coupled with RNA folding remains poorly understood. Some progress in this direction has been demonstrated by a recent study from the Block lab in which co-transcriptional folding in vitro is monitored using single-molecule force spectroscopy [46].
Acknowledgements
This work was supported by the National Science Foundation (NSF CAREER award 0747285) and the National Institutes of Health (R01GM085116).
Biographies
 Krishanthi Karunatilaka received her B.Sc. degree in Biochemistry and Molecular Biology from the University of Colombo, Sri Lanka in 2002. She is a Ph.D. student in the Department of Chemistry, Wayne State University, Detroit, Michigan. She is currently studying the mechanisms of group II intron and nuclear pre-mRNA splicing by single-molecule fluorescence under the supervision of Dr. David Rueda.
Krishanthi Karunatilaka received her B.Sc. degree in Biochemistry and Molecular Biology from the University of Colombo, Sri Lanka in 2002. She is a Ph.D. student in the Department of Chemistry, Wayne State University, Detroit, Michigan. She is currently studying the mechanisms of group II intron and nuclear pre-mRNA splicing by single-molecule fluorescence under the supervision of Dr. David Rueda.
 David Rueda is currently an Assistant Professor in the Department of Chemistry at Wayne State University, Detroit, Michigan. He graduated in 1997 with a degree in chemical engineering from the Swiss Federal Institute of Technology in Lausanne, Switzerland, specialized in Physical Chemistry. For his Diploma Thesis research, he studied the non-linear intensity dependence in the infrared multiphoton excitation and dissociation of methanol pre-excited to different energies in the laboratory of Prof. Martin Quack at the Swiss Federal Institute of Technology in Zürich, Switzerland. In 2001, he obtained his Ph.D. under the supervision of Prof. Thomas R. Rizzo back in Lausanne, where he characterized the vibrational dependence of the torsional potential of jet-cooled methanol. Since then, he has been studying the correlation between the structure, function and dynamics in RNA and ribonucleoprotein complexes using single-molecule fluorescence. His current research interests are to develop and apply single-molecule fluorescence approaches to study the structural dynamics and function of RNA and RNA-protein complexes. Dr. Rueda has been awarded the Ciba Specialty Chemical Award for the best Diploma thesis (1997), a Postdoctoral Research Fellowship from the Swiss National Funds (2001) and a CAREER award from the National Science Foundation (2008).
David Rueda is currently an Assistant Professor in the Department of Chemistry at Wayne State University, Detroit, Michigan. He graduated in 1997 with a degree in chemical engineering from the Swiss Federal Institute of Technology in Lausanne, Switzerland, specialized in Physical Chemistry. For his Diploma Thesis research, he studied the non-linear intensity dependence in the infrared multiphoton excitation and dissociation of methanol pre-excited to different energies in the laboratory of Prof. Martin Quack at the Swiss Federal Institute of Technology in Zürich, Switzerland. In 2001, he obtained his Ph.D. under the supervision of Prof. Thomas R. Rizzo back in Lausanne, where he characterized the vibrational dependence of the torsional potential of jet-cooled methanol. Since then, he has been studying the correlation between the structure, function and dynamics in RNA and ribonucleoprotein complexes using single-molecule fluorescence. His current research interests are to develop and apply single-molecule fluorescence approaches to study the structural dynamics and function of RNA and RNA-protein complexes. Dr. Rueda has been awarded the Ciba Specialty Chemical Award for the best Diploma thesis (1997), a Postdoctoral Research Fellowship from the Swiss National Funds (2001) and a CAREER award from the National Science Foundation (2008).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].The RNA World . CSHL Press; 2006. [Google Scholar]
- [2].Famulok M, Verma S. Trends Biotechnol. 2002;20:462–6. doi: 10.1016/s0167-7799(02)02063-2. [DOI] [PubMed] [Google Scholar]
- [3].Sullenger BA, Gilboa E. Nature. 2002;418:252–8. doi: 10.1038/418252a. [DOI] [PubMed] [Google Scholar]
- [4].Wood M, Yin H, McClorey G. PLoS Genet. 2007;3:e109. doi: 10.1371/journal.pgen.0030109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bonetta L. Cell. 2009;136:581–4. doi: 10.1016/j.cell.2009.02.010. [DOI] [PubMed] [Google Scholar]
- [6].Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Cell. 1982;31:147–57. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- [7].Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. Cell. 1983;35:849–57. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- [8].Valadkhan S. Biol Chem. 2007;388:693–7. doi: 10.1515/BC.2007.080. [DOI] [PubMed] [Google Scholar]
- [9].Abelson J. Nat Struct Mol Biol. 2008;15:1235–7. doi: 10.1038/nsmb1208-1235. [DOI] [PubMed] [Google Scholar]
- [10].Noller HF, Hoffarth V, Zimniak L. Science. 1992;256:1416–9. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- [11].Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. Science. 2000;289:905–20. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- [12].Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. Science. 2000;289:920–30. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- [13].Bartel DP. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- [14].Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Genes Dev. 2006;20:515–24. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- [15].Carthew RW, Sontheimer EJ. Cell. 2009;136:642–55. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Winkler WC, Breaker RR. Annu Rev Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- [17].Zarrinkar PP, Williamson JR. Science. 1994;265:918–24. doi: 10.1126/science.8052848. [DOI] [PubMed] [Google Scholar]
- [18].Zarrinkar PP, Wang J, Williamson JR. RNA. 1996;2:564–73. [PMC free article] [PubMed] [Google Scholar]
- [19].Pan J, Thirumalai D, Woodson SA. J Mol Biol. 1997;273:7–13. doi: 10.1006/jmbi.1997.1311. [DOI] [PubMed] [Google Scholar]
- [20].Russell R, Herschlag D. J Mol Biol. 1999;291:1155–67. doi: 10.1006/jmbi.1999.3026. [DOI] [PubMed] [Google Scholar]
- [21].Pan J, Deras ML, Woodson SA. J Mol Biol. 2000;296:133–44. doi: 10.1006/jmbi.1999.3439. [DOI] [PubMed] [Google Scholar]
- [22].Zhuang X, Bartley LE, Babcock HP, Russell R, Ha T, Herschlag D, Chu S. Science. 2000;288:2048–51. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- [23].Russell R, Millett IS, Tate MW, Kwok LW, Nakatani B, Gruner SM, Mochrie SG, Pande V, Doniach S, Herschlag D, Pollack L. Proc Natl Acad Sci U S A. 2002;99:4266–71. doi: 10.1073/pnas.072589599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Russell R, Zhuang X, Babcock HP, Millett IS, Doniach S, Chu S, Herschlag D. Proc Natl Acad Sci U S A. 2002;99:155–60. doi: 10.1073/pnas.221593598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Tan E, Wilson TJ, Nahas MK, Clegg RM, Lilley DM, Ha T. Proc Natl Acad Sci U S A. 2003;100:9308–13. doi: 10.1073/pnas.1233536100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhuang X. Annu Rev Biophys Biomol Struct. 2005;34:399–414. doi: 10.1146/annurev.biophys.34.040204.144641. [DOI] [PubMed] [Google Scholar]
- [27].Ditzler MA, Rueda D, Mo J, Hakansson K, Walter NG. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Woodside MT, Garcia-Garcia C, Block SM. Curr Opin Chem Biol. 2008;12:640–6. doi: 10.1016/j.cbpa.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Aleman EA, Lamichhane R, Rueda D. Curr Opin Chem Biol. 2008 doi: 10.1016/j.cbpa.2008.09.010. [DOI] [PubMed] [Google Scholar]
- [30].Hammann C, Cooper A, Lilley DM. Biochemistry. 2001;40:1423–9. doi: 10.1021/bi002231o. [DOI] [PubMed] [Google Scholar]
- [31].Feig AL. Biopolymers. 2007;87:293–301. doi: 10.1002/bip.20816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Woodson SA. Curr Opin Chem Biol. 2008;12:667–73. doi: 10.1016/j.cbpa.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhuang X, Kim H, Pereira MJ, Babcock HP, Walter NG, Chu S. Science. 2002;296:1473–6. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
- [34].Steiner M, Karunatilaka KS, Sigel RK, Rueda D. Proc Natl Acad Sci U S A. 2008;105:13853–8. doi: 10.1073/pnas.0804034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ha T, Zhuang X, Kim HD, Orr JW, Williamson JR, Chu S. Proc Natl Acad Sci U S A. 1999;96:9077–82. doi: 10.1073/pnas.96.16.9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bokinsky G, Zhuang X. Acc Chem Res. 2005;38:566–73. doi: 10.1021/ar040142o. [DOI] [PubMed] [Google Scholar]
- [37].Mukhopadhyay S, Deniz AA. J Fluoresc. 2007;17:775–83. doi: 10.1007/s10895-007-0214-0. [DOI] [PubMed] [Google Scholar]
- [38].Cornish PV, Ha T. ACS Chem Biol. 2007;2:53–61. doi: 10.1021/cb600342a. [DOI] [PubMed] [Google Scholar]
- [39].Wilson TJ, Nahas M, Araki L, Harusawa S, Ha T, Lilley DM. Blood Cells Mol Dis. 2007;38:8–14. doi: 10.1016/j.bcmd.2006.10.004. [DOI] [PubMed] [Google Scholar]
- [40].Tinoco I, Jr., Li PT, Bustamante C. Q Rev Biophys. 2006;39:325–60. doi: 10.1017/S0033583506004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Onoa B, Tinoco I., Jr. Curr Opin Struct Biol. 2004;14:374–9. doi: 10.1016/j.sbi.2004.04.001. [DOI] [PubMed] [Google Scholar]
- [42].Zhuang X, Rief M. Curr Opin Struct Biol. 2003;13:88–97. doi: 10.1016/s0959-440x(03)00011-3. [DOI] [PubMed] [Google Scholar]
- [43].Mollova ET. Curr Opin Chem Biol. 2002;6:823–8. doi: 10.1016/s1367-5931(02)00394-0. [DOI] [PubMed] [Google Scholar]
- [44].Weiss S. Nat Struct Biol. 2000;7:724–9. doi: 10.1038/78941. [DOI] [PubMed] [Google Scholar]
- [45].Onoa B, Dumont S, Liphardt J, Smith SB, Tinoco I, Jr., Bustamante C. Science. 2003;299:1892–5. doi: 10.1126/science.1081338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM. Science. 2008;319:630–3. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Stryer L. Annu Rev Biochem. 1978;47:819–46. doi: 10.1146/annurev.bi.47.070178.004131. [DOI] [PubMed] [Google Scholar]
- [48].Selvin PR. Methods Enzymol. 1995;246:300–34. doi: 10.1016/0076-6879(95)46015-2. [DOI] [PubMed] [Google Scholar]
- [49].Nie S, Zare RN. Annu Rev Biophys Biomol Struct. 1997;26:567–96. doi: 10.1146/annurev.biophys.26.1.567. [DOI] [PubMed] [Google Scholar]
- [50].Ha T, Enderle T, Ogletree DF, Chemla DS, Selvin PR, Weiss S. Proc Natl Acad Sci U S A. 1996;93:6264–8. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zhao R, Rueda D. Methods. 2009 doi: 10.1016/j.ymeth.2009.04.017. In Press. [DOI] [PubMed] [Google Scholar]
- [52].Rueda D, Walter NG. J Nanosci Nanotechnol. 2005;5:1990–2000. doi: 10.1166/jnn.2005.505. [DOI] [PubMed] [Google Scholar]
- [53].Deniz AA, Dahan M, Grunwell JR, Ha T, Faulhaber AE, Chemla DS, Weiss S, Schultz PG. Proc Natl Acad Sci U S A. 1999;96:3670–5. doi: 10.1073/pnas.96.7.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Lu HP, Xun L, Xie XS. Science. 1998;282:1877–82. doi: 10.1126/science.282.5395.1877. [DOI] [PubMed] [Google Scholar]
- [55].Bokinsky G, Rueda D, Misra VK, Rhodes MM, Gordus A, Babcock HP, Walter NG, Zhuang X. Proc Natl Acad Sci U S A. 2003;100:9302–7. doi: 10.1073/pnas.1133280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Rueda D, Bokinsky G, Rhodes MM, Rust MJ, Zhuang X, Walter NG. Proc Natl Acad Sci U S A. 2004;101:10066–71. doi: 10.1073/pnas.0403575101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Xie Z, Srividya N, Sosnick TR, Pan T, Scherer NF. Proc Natl Acad Sci U S A. 2004;101:534–9. doi: 10.1073/pnas.2636333100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Qu X, Smith GJ, Lee KT, Sosnick TR, Pan T, Scherer NF. Proc Natl Acad Sci U S A. 2008;105:6602–7. doi: 10.1073/pnas.0801436105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Smith GJ, Lee KT, Qu X, Xie Z, Pesic J, Sosnick TR, Pan T, Scherer NF. J Mol Biol. 2008;378:943–53. doi: 10.1016/j.jmb.2008.01.078. [DOI] [PubMed] [Google Scholar]
- [60].Pereira MJ, Nikolova EN, Hiley SL, Jaikaran D, Collins RA, Walter NG. J Mol Biol. 2008;382:496–509. doi: 10.1016/j.jmb.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Wilson TJ, Nahas M, Ha T, Lilley DM. Biochem Soc Trans. 2005;33:461–5. doi: 10.1042/BST0330461. [DOI] [PubMed] [Google Scholar]
- [62].Ditzler MA, Aleman EA, Rueda D, Walter NG. Biopolymers. 2007;87:302–16. doi: 10.1002/bip.20819. [DOI] [PubMed] [Google Scholar]
- [63].Walter NG, Burke JM. Curr Opin Chem Biol. 1998;2:24–30. doi: 10.1016/s1367-5931(98)80032-x. [DOI] [PubMed] [Google Scholar]
- [64].Walter NG, Hampel KJ, Brown KM, Burke JM. EMBO J. 1998;17:2378–91. doi: 10.1093/emboj/17.8.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Fiore J, Kraemer B, Koberling F, Erdmann R, Nesbitt D. Biochemistry. 2009 [Google Scholar]
- [66].Fiore JL, Hodak JH, Piestert O, Downey CD, Nesbitt DJ. Biophys J. 2008;95:3892–905. doi: 10.1529/biophysj.108.134346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Downey CD, Fiore JL, Stoddard CD, Hodak JH, Nesbitt DJ, Pardi A. Biochemistry. 2006;45:3664–73. doi: 10.1021/bi0520941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Hodak JH, Downey CD, Fiore JL, Pardi A, Nesbitt DJ. Proc Natl Acad Sci U S A. 2005;102:10505–10. doi: 10.1073/pnas.0408645102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Stahley MR, Strobel SA. Curr Opin Struct Biol. 2006;16:319–26. doi: 10.1016/j.sbi.2006.04.005. [DOI] [PubMed] [Google Scholar]
- [70].Russell R, Herschlag D. J Mol Biol. 2001;308:839–51. doi: 10.1006/jmbi.2001.4751. [DOI] [PubMed] [Google Scholar]
- [71].Lee TH, Lapidus LJ, Zhao W, Travers KJ, Herschlag D, Chu S. Biophys J. 2007;92:3275–83. doi: 10.1529/biophysj.106.094623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Sattin BD, Zhao W, Travers K, Chu S, Herschlag D. J Am Chem Soc. 2008;130:6085–7. doi: 10.1021/ja800919q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Bartley LE, Zhuang X, Das R, Chu S, Herschlag D. J Mol Biol. 2003;328:1011–26. doi: 10.1016/s0022-2836(03)00272-9. [DOI] [PubMed] [Google Scholar]
- [74].Block SM. Nature. 1992;360:493–5. doi: 10.1038/360493a0. [DOI] [PubMed] [Google Scholar]
- [75].Chu S, Bjorkholm JE, Ashkin A, Cable A. Phys Rev Lett. 1986;57:314–17. doi: 10.1103/PhysRevLett.57.314. [DOI] [PubMed] [Google Scholar]
- [76].Pace NR, Brown JW. J Bacteriol. 1995;177:1919–28. doi: 10.1128/jb.177.8.1919-1928.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Fang XW, Pan T, Sosnick TR. Nat Struct Biol. 1999;6:1091–5. doi: 10.1038/70016. [DOI] [PubMed] [Google Scholar]
- [78].Fedorova O, Zingler N. Biol Chem. 2007;388:665–78. doi: 10.1515/BC.2007.090. [DOI] [PubMed] [Google Scholar]
- [79].Michel F, Ferat JL. Annu Rev Biochem. 1995;64:435–61. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
- [80].Su LJ, Waldsich C, Pyle AM. Nucleic Acids Res. 2005;33:6674–87. doi: 10.1093/nar/gki973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Su LJ, Brenowitz M, Pyle AM. J Mol Biol. 2003;334:639–52. doi: 10.1016/j.jmb.2003.09.071. [DOI] [PubMed] [Google Scholar]
- [82].Herschlag D. J Biol Chem. 1995;270:20871–4. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- [83].Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, Jantsch MF, Konrat R, Blasi U, Schroeder R. RNA Biol. 2007;4:118–30. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- [84].Stone MD, Mihalusova M, O'Connor C M, Prathapam R, Collins K, Zhuang X. Nature. 2007;446:458–61. doi: 10.1038/nature05600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Witkin KL, Collins K. Genes Dev. 2004;18:1107–18. doi: 10.1101/gad.1201704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Fedor MJ, Williamson JR. Nat Rev Mol Cell Biol. 2005;6:399–412. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- [87].Nahas MK, Wilson TJ, Hohng S, Jarvie K, Lilley DM, Ha T. Nat Struct Mol Biol. 2004;11:1107–13. doi: 10.1038/nsmb842. [DOI] [PubMed] [Google Scholar]
- [88].Ramakrishnan V. Cell. 2002;108:557–72. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- [89].Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. Nat Struct Mol Biol. 2004;11:1008–14. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- [90].Blanchard SC, Kim HD, Gonzalez RL, Jr., Puglisi JD, Chu S. Proc Natl Acad Sci U S A. 2004;101:12893–8. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Green R, Noller HF. Annu Rev Biochem. 1997;66:679–716. doi: 10.1146/annurev.biochem.66.1.679. [DOI] [PubMed] [Google Scholar]
- [92].Munro JB, Altman RB, O'Connor N, Blanchard SC. Mol Cell. 2007;25:505–17. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Cornish PV, Ermolenko DN, Noller HF, Ha T. Mol Cell. 2008;30:578–88. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr. Mol Cell. 2008;30:348–59. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- [95].Cornish PV, Ermolenko DN, Staple DW, Hoang L, Hickerson RP, Noller HF, Ha T. Proc Natl Acad Sci U S A. 2009;106:2571–6. doi: 10.1073/pnas.0813180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Crawford DJ, Hoskins AA, Friedman LJ, Gelles J, Moore MJ. RNA. 2008;14:170–9. doi: 10.1261/rna.794808. [DOI] [PMC free article] [PubMed] [Google Scholar]