Abstract
Objectives
The p1 region of HIV-1 gag contains the frameshift stem-loop, gag–pol transframe and a protease cleavage site that are crucial for viral assembly, replication and infectivity. Identifying and characterizing CD8+ epitopes that are under host immune selection in this region will help in designing effective vaccines for HIV-1.
Design
An approach combining bioinformatical analysis and interferon gamma enzyme-linked immunosorbent spot (ELISPOT) assays is used to identify and characterize the epitopes. Potential human leukocyte antigen (HLA)-restricted epitopes were identified by correlating the positively selected mutations with host HLA alleles.
Methods
ELISPOT analysis with overlapping peptides was used to confirm and characterize the epitopes.
Results
Four positively selected residues were significantly associated with HLA class I alleles, including HLA B*1302 (K4R, P=0.0008 and I5L, P=0.0108), A*7401 (S9N, P=0.0002) and A*30 genotypes (P7S, P=0.009), suggesting epitopes restricted by these alleles are present in this region. ELISPOT analysis with patient peripheral blood mononuclear cells identified 7 novel epitopes restricted by the 3 alleles. Two types of epitopes were observed in this region based on the ELISPOT responses, Type I: the positively selected variation does not affect CD8+ T-cell responses; and Type II: the CD8+ T-cell responses are determined by the epitope variants.
Conclusion
We identified and characterized seven novel CD8+ epitopes in the p1 spacer protein region. Classifying the effects of positively selected variants on CD8+ T-cell responses will help in designing effective vaccines for HIV-1.
Keywords: epitope, Gag, HIV-1, human leukocyte antigens, p1
Introduction
The error-prone reverse transcriptase leads to substantial sequence variations, enabling the rapid mutation of HIV-1. Because of viral variability, an HIV vaccine must elicit immune responses to a wide array of viral variants within and between clades [1]. Human leukocyte antigen (HLA)-restricted cytotoxic T lymphocyte (CTL) responses are critical for immune control of HIV infection and recent data suggest that CTL targeting Gag contributes to this control [1,2]. HLA-restricted CTL escape mutations may compromise the ability of the CTL to suppress HIV replication and influence viral evolution in a given host and at the population level [1,3]. Escape can result from changes that impact processing, HLA binding and T-cell receptor (TCR) binding [1,3]. The sequence variants escaping host immune responses have been defined as positively selected mutations. The effect of positively selected mutations are not equal, some positively selected mutations allow the virus to survive but at a fitness cost. Identification and study of these positively selected mutations can not only identify conserved regions of the viral genome that could potentially be included in vaccine candidates but also the regions that contain HLA-restricted epitopes [3–6]. Testing the regions containing positively selected amino acids with overlapping peptides using enzyme-linked immunosorbent spot (ELISPOT) assays can facilitate epitope identification and overcome the disadvantages of using traditional methods.
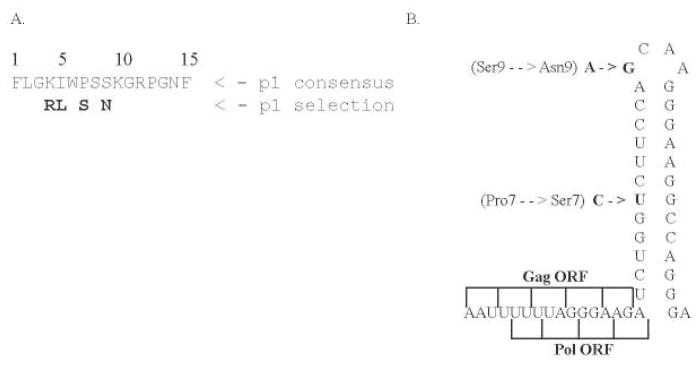
Our previous study [4] of HIV-1 gag in a large HIV-1-infected cohort (n=431) identified four positively selected amino acids within the p1 spacer polyprotein (Fig. 1a Fig. 1). The percentage of positively selected residues among the six gag proteins ranges from zero (p2) to 48% (p6), and these positively selected sites overlap with many confirmed CTL epitopes and protease cleavage sites [4]. Among these gag proteins, a small spacer protein, p1, has a high percentage of positively selected residues compared with other small gag proteins, with five out of 16 amino acids (31%) being positively selected sites. The high proportion of positively selected amino acids in this protein implies that the region contains multiple HLA epitopes as suggested by the significant correlations of K4R with B*1302 (P=0.0008) and S9N with A*7401 (P=0.0002) [4].
Fig. 1.
p1 protein sequence and stemloop structure. gr1
(a) Identified positively selected amino acid residues in the p1 spacer protein (in bold).
(b) Amino acid positions 7 and 9 and their corresponding single nucleotide changes in the p1 stem loop structure. The Gag and GagProPol polyprotein domains are also shown as overlapping regions and frameshift coding regions.
The p1 spacer protein contains the critical RNA structure of the frameshift, stem-loop and gag–pol transframe open reading frames (Fig. 1b) [2,7]. Mutations in this region, particularly the proline residues at positions 7 and 13, negatively affect protein processing, RNA dimer stability and abolish viral infectivity in multiple strains of HIV-1 [7]. Although the role of p1 in viral replication is not well understood, it has been suggested that mutations in p1 impact nucleocapsid function, which is important in the early stages of reverse transcription and integration [7]. The cleavage of p1 may therefore be important in the regulation of various nucleocapsid functions. The ability of CTL targeting Gag is important in controlling HIV viremia; therefore, identification and characterization of HLA epitopes in this region will provide critical insight about vulnerable regions of the virus and help to develop control strategies [8]. In this study, we identified seven novel epitopes and characterized the CD8+ epitopes targeting this region by combining bioinformatical analysis, correlation of host HLA types with positively selected amino acids and ELISPOT assays with patient peripheral blood mononuclear cells (PBMCs) and overlapping peptides.
Patients and methods
Study cohort
Patients involved in this study were HIV-1 positive, antiretroviral therapy-naive women (at the time of sampling) followed longitudinally in a sex worker cohort established in 1985 in the Pumwani District of Nairobi, Kenya [9]. All patients gave informed consent to participate, and the study has been approved by the Institutional Review Boards at the Universities of Manitoba and Nairobi.
Human leukocyte antigen class I typing
Genomic DNA was isolated from all study participants. PCR amplification of HLA-A, B and C genes was carried out using gene-specific primers. Products were sequenced using the ABI 3100 Genetic Analyzer (Applied Biosystems, Foster City, California, USA). Sequence outputs were genotyped with CodonExpress software developed based on a taxonomy-based sequence analysis (TBSA) [10].
P1 spacer protein sequencing and analysis
The HIV-1 gag gene was amplified from genomic DNA of all study patients. When multiple dominant quasispecies were present, the PCR products were cloned and analyzed. P1 sequences were analyzed with Sequencher 4.5 and aligned with Mega 3.0 [11]. Quasi, a selection-mapping algorithm, was used to identify positively selected amino acids [12]. SPSS 11.0 (SPSS Inc., Chicago, Illinois, USA) was used to correlate the positively selected amino acids with HLA data. In cases in which multiple sequences were obtained, all sequences were analyzed for the presence or absence of a positively selected residue, ensuring that each patient was counted only once [4]. In a 2×2 cross-tabulation in which all expected counts are above 10, Pearson’s chi-square test was used to determine associations between the presence of a specific HLA allele and positively selected at a given site [4]. For any cell that contains an expected count below 10, the Fisher’s exact test was used [4] and false discovery rate was used to control for multiple comparisons [4,13].
Peptides
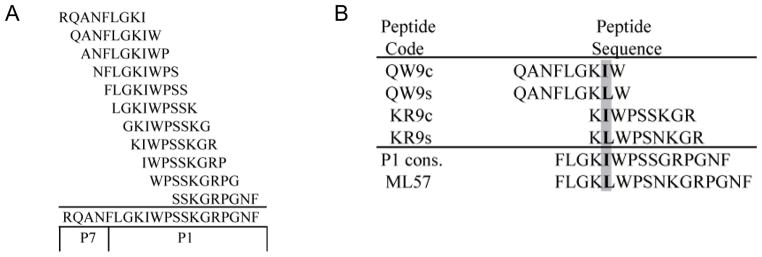
Overlapping peptides (Sigma Genosys, Oakville, Ontario, Canada) were designed in sequences of nine amino acid residues overlapping by eight, to span part of the p7 region and the entire p1 region (20-amino acid residues in length, Fig. 2a Fig. 2). The library contained peptides with the consensus residue and a positively selected residue at various positions. An example is shown in Fig. 2b for the positively selected residue in position 5 of p1 (Ile→Leu). The peptides were also selected on the basis of the p1 sequence(s) found in each patient to ensure both the consensus and positively selected, autologous residues were tested (Fig. 2b).
Fig. 2.
Peptide alignment. gr2
(a) Consensus peptides designed to overlap by eight amino acid residues spanning the p1 region and part of the p7 region. (b) Alignment of consensus and selection peptides tested by ELISPOT to determine whether positively selected amino acid at position 5 of p1 could effect epitope recognition at position 2 or 8 of the peptide. ELISPOT, enzyme-linked immunosorbent spot.
Enzyme-linked immunosorbent spot assays
Interferon gamma (IFNγ) ELISPOT assays using patient PBMCs were used to identify and confirm the potential epitopes overlapping the region containing positively selected amino acids. Ninety-six well nitrocellulose plates were coated with anti-IFNγ monoclonal antibody (mAb; Mabtech, Nacka Strand, Sweden) followed by blocking with R-10 media [14]. Peptide stocks were used at a final concentration of 10μg/ml in Royal Park Memorial Institute (RPMI) media (HyClone, Utah, USA). PBMCs were suspended in commercially available RPMI media and 105 cells were stimulated in duplicate overnight at 37°C (with CO2) with each peptide individually without pooling, 1μg/ml phytohemagglutinin (PHA, as positive control) or media (background) [14]. After incubation, the cells were discarded, plates were washed and incubated with a biotinylated anti-IFNγ mAb (Mabtech) followed by streptavidin-conjugated alkaline phosphatase (Mabtech) [15]. Plates were developed using an alkaline phosphatase-conjugate substrate kit (Bio-Rad Laboratories, Ontario, Canada) and the spot-forming units (SFUs) were counted using an automated ELISPOT reader (Autoimmun Diagnostika GmbH, Strassberg, Germany) [15]. Responses were considered positive if there were at least 50 SFU/million PBMC after background subtraction and the positive control was successful [14].
Results
Correlation of positively selected amino acids in p1 with patient human leukocyte antigen alleles suggested potential epitopes for human leukocyte antigen B*1302, A*7401 and A*30
Our previous study [4] identified four positively selected amino acid residues in the p1 spacer protein by Quasi analysis and two of the positively selected residues, K4R and S9N, were significantly correlated with B*1302 (Lys→Arg, P=0.0008) and A*7401 (Ser→Asp, P=0.0002), respectively. Further analysis of the positively selected amino acids at positions 5 and 7 of p1 showed that I5L was also significantly correlated with B*1302 (Ile→Leu, P=0.0108), and P7S was significantly associated with A*30 (Pro→Ser; P=0.009), suggesting that this region contains epitopes of multiple HLA alleles. Thus, a targeted ELISPOT analysis using peptides overlapping this region with PBMCs of patients with defined HLA genotypes was used to identify, confirm and characterize epitopes.
Enzyme-linked immunosorbent spot analysis identified and confirmed multiple epitopes and epitope variants for human leukocyte antigen B*1302, A*7401 and A*30
In a preliminary ELISPOT analysis, we tested all the peptides in the library and were only able to detect IFNγ responses to the peptides with positively selected residues at anchor positions (P2 and P8). Thus, in subsequent assays, peptides were selected with positively selected residues in anchor positions only [16]. HLA data for all ELISPOT patients tested can be found in Table 1.
Table 1.
Human leukocyte antigen data of all patients tested by enzyme-linked immunosorbent spot assay.
| Patient # | [0,3–4]HLA-A | [0,5–6]HLA-B | [0,7–8]HLA-C | ||||
|---|---|---|---|---|---|---|---|
| [6,0]B*1302 | 415 | 3002 | 3402 | 1302 | 5301 | 0401 | 0804 |
| 915 | 0103 | 2301 | 1302 | 5801 | 0602 | 0701 | |
| 1160 | 3402 | 3402 | 1302 | 4403 | 0401 | 0804 | |
| 1720 | 2901 | 3010 | 1302 | 0705 | 0602 | 1505 | |
| 1778 | 0202 | 6802 | 1302 | 5703 | 0602 | 0701 | |
| 1787 | 3001 | 6802 | 1302 | 4201 | 0602 | 1701 | |
| 1937 | 2902 | 3402 | 1302 | 1503 | 0202 | 0602 | |
| [13,0]A*7401 | 602 | 7401 | 7401 | 3501 | 3501 | 0401 | 0401 |
| 672 | 3201 | 7401 | 0705 | 5802 | 0602 | 0804 | |
| 1032 | 2301 | 7401 | 3501 | 8101 | 0401 | 1801 | |
| 1072 | 6802 | 7401 | 1510 | 4901 | 0304 | 0701 | |
| 1076 | 6801 | 7401 | 1517 | 3501 | 0401 | 0701 | |
| 1349 | 6601 | 7401 | 3910 | 5802 | 0602 | 1701 | |
| 1419 | 1010 | 7401 | 4201 | 5702 | 1701 | 1801 | |
| 1481 | 0202 | 7401 | 1406 | 5001 | 0602 | 0701 | |
| 1614 | 0109 | 7401 | 4415 | 1503 | 0202 | 0407 | |
| 1850 | 6802 | 7401 | 4201 | 1801 | 0501 | 1701 | |
| 1881 | 0301 | 7401 | 4901 | 5802 | 0602 | 0701 | |
| 1901 | 0202 | 7401 | 5802 | 5802 | 0602 | 0602 | |
| 1974 | 0202 | 7401 | 5802 | 5301 | 0401 | 0602 | |
| 2026 | 6802 | 7401 | 1503 | 5301 | 0202 | 0415 | |
| [21,0]A*30 | 79 | 3002 | 6802 | 4901 | 5802 | 0602 | 0701 |
| 199 | 3001 | 0201 | 4201 | 5703 | 0701 | 1701 | |
| 273 | 3002 | 6601 | 0801 | 1405 | 0701 | 0802 | |
| 415 | 3002 | 3402 | 1302 | 5301 | 0401 | 0804 | |
| 752 | 3002 | 6601 | 5703 | 5802 | 0602 | 1801 | |
| 810 | 3001 | 0201 | 1503 | 4201 | 0202 | 1701 | |
| 890 | 3004 | 6802 | 5802 | 4503 | 0602 | 0802 | |
| 960 | 3001 | 6601 | 1303 | 4415 | 0401 | 0602 | |
| 1324 | 3001 | 0201 | 3501 | 5301 | 0701 | 1203 | |
| 1410 | 3001 | 2301 | 1801 | 4201 | 0202 | 1701 | |
| 1497 | 3002 | 0205 | 0801 | 1503 | 0401 | 0701 | |
| 1530 | 3009 | 6802 | 1510 | 8101 | 0304 | 0401 | |
| 1594 | 3001 | 0101 | 0801 | 4201 | 0704 | 1701 | |
| 1694 | 3001 | 0201 | 5802 | 1503 | 0202 | 0602 | |
| 1802 | 3002 | 0101 | 5801 | 5801 | 0302 | 0302 | |
| 1843 | 3001 | 0101 | 3910 | 5702 | 1203 | 1801 | |
| 1894 | 3001 | 0202 | 1503 | 5703 | 0202 | 1701 | |
| 1956 | 3001 | 3002 | 4501 | 1510 | 0304 | 1601 | |
| 2033 | 3002 | 3303 | 0702 | 5801 | 0302 | 0701 | |
| 2052 | 3001 | 0103 | 4201 | 5802 | 0602 | 1701 | |
| 2067 | 3002 | 6802 | 1510 | 3910 | 0304 | 1203 | |
| 2144 | 3002 | 3402 | 1503 | 4501 | 0202 | 1601 | |
HLA, human leukocyte antigen.
We observed two types of epitopes through ELISPOT analysis with overlapping peptides. In type I epitopes, positively selected mutations do not affect the CD8+ T-cell responses, whereas in the type II epitopes, positively selected variants determine downstream CD8+ T-cell responses. The two types of epitopes can be seen among three B*1302 epitopes or six epitope variants overlapping the region with the positively selected residues K4R and I5L (Table 2). Peptides RQANFLGKI (RI9c) and RQANFLGRI (RI9s) contain K4R at anchor residue 8. These two peptides were recognized by PBMCs from all seven patients (Tables 2 and 3a Table 3) indicating that these peptides are variants of one epitope of B*1302. The K to R variation does not appear to affect HLA allele recognition or down stream CD8+ T-cell responses. Therefore, the peptide variants RQANFLGKI (RI9c) and RQANFLGRI (RI9s) belong to type I epitope, whereas peptides GKIWPSSKG (GG9c) and GRIWPSNKG (GG9s) with K4R at anchor position 2 represent the type II epitope. ELISPOT assays (Table 3a) showed that four out of seven patient samples (57.1%) had IFNγ responses against one of the two peptides, whereas no patients responded to both. It appears that the difference in CD8+ T-cell response is not due to the differential HLA binding to the peptide variants as patients with the same HLA allele can recognize both peptide variants. The CD8+ T-cell responses appear to be affected by the downstream events after HLA binding, possibly differences in the TCR repertoire. Some epitopes combine the characteristics of types I and II epitopes. For example, four out of five samples tested recognized KIWPSSKGR (KR9c) or KLWPSNKGR (KR9s) containing I5L at anchor position 2, three of which responded to both peptides (Table 3a).
Table 2.
Summary of enzyme-linked immunosorbent spot assay peptide responses to B*1302, A*7401 and A*30 patient samples.
| [0,3-4]Peptide | [0,5-6]Patient samples | [0,7-8]% Responded | ||||||
|---|---|---|---|---|---|---|---|---|
| Allele | Positively selected residue | Codea | Sequence | Testedb | Responded | Peptide | Both | Mean SFU/million |
| [7,0]B*1302 | [3,0]K4R | RI9c | RQANFLGKI | [1,0]7 | 7 | 100.0 | [1,0]100.0 | 365.7 |
| RI9s | RQANFLGRI | 7 | 100.0 | 406.4 | ||||
| GG9c | ------------GKIWPSSKG | [1,0]7 | 2 | 28.6 | [1,0]0.0 | 80.0 | ||
| GG9s | ------------GRIWPSNKG | 2 | 28.6 | 85.0 | ||||
| [3,0]I5L | QW9c | QANFLGKIW | [1,0]5 | 0 | 0.0 | [1,0]0.0 | 0.0 | |
| QW9s | QANFLGKLW | 0 | 0.0 | 0.0 | ||||
| KR9c | ------------KIWPSSKGR | [1,0]5 | 4 | 80.0 | [1,0]60.0 | 73.8 | ||
| KR9s | ------------KLWPSNKGR | 3 | 60.0 | 85.0 | ||||
| [3,0]A*7401 | [3,0]S9N | LK9c | LGKIWPSSK | [1,0]14 | 5 | 35.7 | [1,0]6.7 | 72.5 |
| LK9s | LGKIWSSNK | 2 | 14.3 | 60.0 | ||||
| SF9c | ------------SSKGRPGNF | [1,0]14 | 3 | 21.4 | [1,0]6.7 | 191.7 | ||
| SF9s | ------------SNKGRPGNF | 7 | 50.0 | 105.7 | ||||
| [2,0]A*30 | [2,0]P7S | NS9s | NFLGKIWSS | 22 | 5 | 22.7 | NA | 132.0 |
| WG9c | -----------WPSSKGRPG-- | [1,0]21 | 6 | 28.6 | [1,0]14.3 | 126.7 | ||
| WG9s | -----------WSSNKGRPG-- | 4 | 19.0 | 163.8 | ||||
SFU, spot-forming unit.
c, consensus peptide; s, selection peptide.
Patient samples were split and tested for both consensus and selection peptides.
Table 3.
Detailed enzyme-linked immunosorbent spot data for each peptide tested in each patient for all three alleles.
| [0,3–8]Peptide |
|||||||
|---|---|---|---|---|---|---|---|
| Allele | Patient # | GG9c | GG9s | KR9c | KR9s | RI9c | RI9s |
| [0,1–8]a: B*1302 patient data | |||||||
| [6,0]B*1302 | 415 | nra | 75b | 50 | nr | 60 | 75 |
| 915 | 55 | nr | 115 | 65 | 225 | 85 | |
| 1160 | nr | nr | nr | nt | 710 | 490 | |
| 1720 | 110 | nr | 55 | 135 | 280 | 505 | |
| 1778 | nr | 75 | 75 | 50 | 1055 | 1180 | |
| 1787 | nr | nr | ntc | nt | 145 | 65 | |
| 1937 | nr | nr | nt | nt | 85 | 445 | |
| [0,3–6]Peptide |
|||||
|---|---|---|---|---|---|
| Allele | Patient # | LK9c | LK9s | SF9c | SF9s |
| [0,1–6]b: A*7401 patient data | |||||
| [13,0]A*7401 | 602 | nr | nr | nr | nr |
| 672 | nr | nr | 2652 | 120 | |
| 1032 | nr | 60 | 110 | nr | |
| 1072 | 55 | 60 | nr | 75 | |
| 1076 | nr | nr | nr | nr | |
| 1349 | nr | nr | nr | 78 | |
| 1419 | 80 | nr | nr | 252 | |
| 1481 | nr | nr | nr | 90 | |
| 1614 | 70 | nr | nr | nr | |
| 1850 | 60 | nr | nr | 70 | |
| 1881 | nr | nr | nr | 55 | |
| 1901 | nr | nr | nr | nr | |
| 1974 | 98 | nr | 220 | nr | |
| 2026 | nr | nr | nr | nr | |
| [0,3–5]Peptide |
||||
|---|---|---|---|---|
| Allele | Patient # | NS9s | WG9c | WG9s |
| [0,1–5]c: A*30 patient data | ||||
| [21,0]A*30 | 79 | nr | nr | nr |
| 199 | nr | nr | nr | |
| 273 | nr | nr | nr | |
| 415 | 702 | 165 | 215 | |
| 752 | nr | nr | nr | |
| 810 | nr | nr | nr | |
| 890 | nr | nr | nr | |
| 960 | nr | nr | nr | |
| 1324 | 200 | nr | nr | |
| 1410 | nr | nr | nr | |
| 1497 | 285 | 125 | 323 | |
| 1530 | nr | nr | nr | |
| 1594 | nr | nr | nr | |
| 1694 | nr | 55 | nr | |
| 1802 | nr | 55 | nr | |
| 1843 | nr | nr | 125 | |
| 1894 | nr | nr | nr | |
| 1956 | nr | 75 | nr | |
| 2033 | 55 | nr | nr | |
| 2052 | nr | 60 | 95 | |
| 2067 | nr | nr | nr | |
| 2144 | 50 | 165 | 100 | |
No response.
Spot-forming units per million.
Not tested because of lack of cells.
Only type II epitopes for A*7401 and A*30 were identified in the p1 region. ELISPOT assays for A*7401-positive patient samples confirmed two epitopes overlapping the region containing the positively selected residue S9N (Table 2). Peptides LGKIWPSSK (LK9c) and LGKIWPSNK (LK9s) with the S9N mutation in anchor position 8 were tested with PBMCs from 14 patients. Positive ELISPOT responses were detected in six out of 14 (42.9%) samples to either one of the two peptides and only one sample was ELISPOT positive to both peptides (6.7%) (Tables 2 and 3b). Peptides SSKGRPGNF (SF9c) and SNKGRPGNF (SF9s) containing S9N in anchor position 2 were tested with the same 14 patients. Nine of the patient samples (64.3%) showed positive ELISPOT responses to either one of the two peptides and one sample (6.7%) had responses to both peptides (Tables 2 and 3b).
Two epitopes for the A*30 group were identified by ELISPOT assays for the region containing the P7S mutation. Peptides WPSSKGRPG (WG9c) and WSSNKGRPG (WG9s) contain P7S in anchor position 2. ELISPOT responses were observed in eight out of 21 (38.1%) patient samples to either one of the peptides, four of which (19%) recognized both peptides (Tables 2 and 3c). The NFLGKIWSS (NS9s) peptide contains S9N in anchor position 8. Five out of 22 patient samples responded (22.7%) in ELISPOT assays, and there was no significant difference in peptide recognition between A*3001 and A*3002 patients.
Discussion
It is well accepted that HLA-restricted T-cell responses can influence the evolution of HIV-1 and select mutations at the individual and population level [17]. During HIV infection, there are two opposing selection forces, the one driving immune escape and the one striving to conserve the amino acid sequence and maintain viral fitness [2]. The best scenario for the virus is escape with little fitness cost. However, many mutations under selective pressure from the host immune system or drug treatment are at a considerable fitness cost for the virus [3]. Positively selected mutations reflect viral escape from host immune responses in the context of HLA-restricted CD8+ T-cell responses. Identifying epitopes by correlating positively selected mutations in HIV-1 with specific host HLA alleles has certain advantages. The epitopes identified are immunologically relevant because of the fact that they are under immune pressure. However, the specific epitopes within these regions must be confirmed and characterized by direct biological assays. Characterizing the epitopes that are under host selection can provide critical insight in identifying vulnerable regions of the virus and the targets for candidate vaccines.
The present study identified seven novel HLA epitopes in the p1 spacer protein region of gag that fall into two different categories, those containing positively selected mutations that do not affect the CD8+ T-cell response (type I) and those with positively selected mutations that determine the downstream CD8+ T-cell responses (type II). Epitopes are not equal, as seen in this study; some epitopes can induce T-cell responses in the majority of patients, if not in all, whereas others only induce T-cell responses in a subset of patients. Understanding the characteristics of the epitopes is important in designing effective vaccines.
It is possible that the type I epitopes point to the regions of the virus that require functional conservation where there are few options for mutation or escape. For example, epitope variants RQANFLGKI (RI9c) and RQANFLGRI of B*1302 contain positively selected variants arginine or lysine (K4R) at position 8. Arginine and lysine are positively charged, hydrophilic amino acids and the change from K to R represents a conserved substitution. The K to R mutants have been shown to maintain viral replication and infectivity [2]. Furthermore, the quasispecies detected in the B*1302 patients contained only K or R variants at position 4 of p1, suggesting that arginine or lysine at this location is required to maintain the function of the virus, thus limiting viral escape from B*1302-restricted T-cell responses. Although targeting multiple sites may be required for effective viral control by CD8+ T-cell responses, the type I characteristics of B*1302 epitopes at the p1 location could contribute to its association with slower disease progression in a South African population [2,18].
The B*1302-restricted epitope that overlaps the I5L region exhibits mixed characteristics of both types I and II epitopes, depending on the patient. Some patients are able to recognize only one variant, whereas others are able to recognize both or neither. Many factors might contribute to the observed T-cell response variation, including differences in TCR repertoire or other host genetic factors. It is important to clarify them in future studies. The mutation at position 5 from isoleucine to leucine is a conserved substitution, suggesting that the functional conservation of this residue is important for the virus. A recent study [2] showed that I5L mutants have reduced infectivity and viral replication.
Two type II A*7401-restricted epitopes were identified in this study, both overlapping the positively selected residue S9N. It appears that more A*7401 patients recognize SF9s (containing the positively selected residue N) than SF9c (containing the consensus residue S). A*7401, a high-frequency allele (30%) in this East African population, is significantly associated with long-term nonprogression in the Pumwani cohort (P=0.008, odds ratio 3.4, 95% confidence interval 1.3–9.4). It is possible that the preferred recognition of SF9s is driving the fixation of consensus SF9c in the population.
Three type II A*30 epitopes overlap the positively selected residue P7S. Overall, responses to these peptides were less frequently detected when compared with the other two alleles (Table 2). The patients expressed several different A*30 alleles, mainly A*3001 and A*3002 (Table 1), and there was no significant difference in responses when grouped by allele. The proline at position 7 of p1 was reported to be critical for the stability of the entire p1 stem loop and the ability of the virus to infect host cells [19]; therefore, selection pressure by the host is likely to drive the mutation to result in a less-fit virus.
In summary, our study showed that identification and characterization of epitopes not only can provide essential information for selecting optimal components for vaccines but also additional knowledge to interpret immune responses after vaccination. The type 1 epitopes are perhaps better to be included in a vaccine because of their location and the T-cell responses with broader population coverage. Identifying epitopes by correlating positively selected mutations in HIV-1 with specific host HLA alleles has advantages in identifying immune relevant epitopes. Characterizing these epitopes can provide critical insight in identifying vulnerable regions of the virus and the targets for candidate vaccines. Identification of regions of HIV that elicit effective CD8+ T-cell responses leading to attenuation of virus due to structural constraints is an important goal of HIV vaccine design.
Acknowledgments
The present study was funded by the NIH (#R01 A1 49383), CIHR (#HOP-43135) and the National Microbiology Laboratory, Public Health Agency of Canada. Thanks to John Rutherford, Mark Mendoza and Rupert Capina of Dr Plummer’s lab group for their technical and editorial assistance. Thanks are extended to the nurses and staff working with the Pumwani Sex Worker Cohort, Jane Njoki, Jane Kamene, Elizabeth Bwibo and Edith Amatiwa. Special thanks to the women enrolled in the Pumwani Sex Worker Cohort for their involvement and contribution to HIV research.
Christina A. Semeniuk wrote and edited the drafts of the manuscript, performed experiments and conducted data analysis. Lyle McKinnon took part in early experiments and editing of the paper. Harold O. Peters conducted data analysis and editing of the paper. Michael Gubbins conducted data analysis of the p1 spacer protein stem loop and editing of the paper. Xiaojuan Mao conducted lab work, including sequencing of the p1 spacer protein. Terry B. Ball provided experimental support and edited the paper. Ma Luo designed the study and helped in writing the manuscript. Francis A. Plummer edited the paper, established the cohort, maintained the cohort and secured funding.
Footnotes
Information Presented at: Public Health Agency of Canada Conference, Winnipeg, Manitoba, Canada, 17–19 March 2008.
Keystone Symposia, Banff, Alberta, Canada, 27 March 1 April 2008.
There are no conflicts of interest.
References
- 1.Gudmundsdotter L, Bernasconi D, Hejdeman B, Sandstrom E, Alaeus A, Lisman K, et al. Cross-clade immune responses to Gag p24 in patients infected with different HIV-1 subtypes and correlation with HLA class I and II alleles. Vaccine. 2008 doi: 10.1016/j.vaccine.2008.03.094. [DOI] [PubMed] [Google Scholar]
- 2.Prado JG, Honeyborne I, Brierley I, Puertas MC, Martinez-Picado J, Goulder PJ. Functional consequences of HIV Escape from an HLA-B*13-restricted CD8+ T-cell epitope in the p1 Gag protein. J Virol. 2008 doi: 10.1128/JVI.01882-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rousseau CM, Daniels MG, Carlson JM, Kadie C, Crawford H, Prendergast A, et al. HLA Class-I Driven Evolution of Human Immunodeficiency Virus Type 1 Subtype C Proteome: Immune Escape and Viral Load. J Virol. 2008 doi: 10.1128/JVI.02455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peters HO, Mendoza MG, Capina RE, Luo M, Mao X, Gubbins M, et al. An integrative bioinformatic approach for studying escape mutations in human immunodeficiency virus type 1 gag in the Pumwani Sex Worker Cohort. J Virol. 2008;82:1980–1992. doi: 10.1128/JVI.02742-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker BD, Goulder PJ. AIDS. Escape from the immune system. Nature. 2000;407:313–314. doi: 10.1038/35030283. [DOI] [PubMed] [Google Scholar]
- 6.Takiguchi M. Role of HLA in HIV-1 infection. Uirusu. 2000;50:47–55. [PubMed] [Google Scholar]
- 7.Hill MK, Bellamy-McIntyre A, Vella LJ, Campbell SM, Marshall JA, Tachedjian G, et al. Alteration of the proline at position 7 of the HIV-1 spacer peptide p1 suppresses viral infectivity in a strain dependent manner. Curr HIV Res. 2007;5:69–78. doi: 10.2174/157016207779316323. [DOI] [PubMed] [Google Scholar]
- 8.Brumme ZL, Tao I, Szeto S, Brumme CJ, Carlson JM, Chan D, et al. Human leukocyte antigen-specific polymorphisms in HIV-1 Gag and their association with viral load in chronic untreated infection. AIDS. 2008;22:1277–1286. doi: 10.1097/QAD.0b013e3283021a8c. [DOI] [PubMed] [Google Scholar]
- 9.Fowke KR, Nagelkerke NJ, Kimani J, Simonsen JN, Anzala AO, Bwayo JJ, et al. Resistance to HIV-1 infection among persistently seronegative prostitutes in Nairobi, Kenya. Lancet. 1996;348:1347–1351. doi: 10.1016/S0140-6736(95)12269-2. [DOI] [PubMed] [Google Scholar]
- 10.Luo M, Blanchard J, Pan Y, Brunham K, Brunham RC. High-resolution sequence typing of HLA-DQA1 and -DQB1 exon 2 DNA with taxonomy-based sequence analysis (TBSA) allele assignment. Tissue Antigens. 1999;54:69–82. doi: 10.1034/j.1399-0039.1999.540108.x. [DOI] [PubMed] [Google Scholar]
- 11.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 12.Stewart JJ, Watts P, Litwin S. An algorithm for mapping positively selected members of quasispecies-type viruses. BMC Bioinformatics. 2001;2:1. doi: 10.1186/1471-2105-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 14.McKinnon LR, Ball TB, Wachihi C, McLaren PJ, Waruk JL, Mao X, et al. Epitope cross-reactivity frequently differs between central and effector memory HIV-specific CD8+ T cells. J Immunol. 2007;178:3750–3756. doi: 10.4049/jimmunol.178.6.3750. [DOI] [PubMed] [Google Scholar]
- 15.Kaul R, Dong T, Plummer FA, Kimani J, Rostron T, Kiama P, et al. CD8(+) lymphocytes respond to different HIV epitopes in seronegative and infected subjects. J Clin Invest. 2001;107:1303–1310. doi: 10.1172/JCI12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korber B, Brander C, Haynes BF, Koup R, Moore JP, Walker BD, et al. HIV Molecular Immunology Database. 2007;2007 [Google Scholar]
- 17.van Opijnen T, de Ronde A, Boerlijst MC, Berkhout B. Adaptation of HIV-1 depends on the host-cell environment. PLoS ONE. 2007;2:e271. doi: 10.1371/journal.pone.0000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honeyborne I, Prendergast A, Pereyra F, Leslie A, Crawford H, Payne R, et al. Control of human immunodeficiency virus type 1 is associated with HLA-B*13 and targeting of multiple gag-specific CD8+ T-cell epitopes. J Virol. 2007;81:3667–3672. doi: 10.1128/JVI.02689-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill MK, Shehu-Xhilaga M, Crowe SM, Mak J. Proline residues within spacer peptide p1 are important for human immunodeficiency virus type 1 infectivity, protein processing, and genomic RNA dimer stability. J Virol. 2002;76:11245–11253. doi: 10.1128/JVI.76.22.11245-11253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]