Abstract
Deubiquitinating enzymes (DUBs) are proteases that process ubiquitin or ubiquitin-like gene products, reverse the modification of proteins by a single ubiquitin (or ubiquitin-like protein), and remodel polyubiquitin (or ubiquitin-like) chains on target proteins. The human genome encodes nearly 100 DUBs with specificity for ubiquitin in five families: the UCH, USP, OTU, Josephin, and JAMM families. Four families are cysteine proteases, while the later is a family of metalloproteases. Most DUB activity is cryptic and active site rearrangements often occur during the binding of ubiquitin and/or scaffold proteins. DUBs with specificity for ubiquitin contain multiple domains with insertions and extensions modulating DUB substrate specificity, protein-protein interactions, and cellular localization. Binding partners and multi-protein complexes with which DUBs associate modulate DUB activity and substrate specificity. Quantitative studies of activity and protein-protein interactions, together with genetic studies and the advent of RNAi, have lead to new insights into the function of yeast and human DUBs. This review will discuss ubiquitin-specific DUBs, some of the generalizations emerging from recent studies of the regulation of DUB activity, and their roles in various cellular processes. Specific examples are drawn from studies of protein degradation, DNA repair, chromatin remodeling, cell cycle regulation, endocytosis, and modulation of signaling kinases.
Keywords: Ubiquitin, Proteasome, Histone, Cell cycle, Endocytosis, DNA Damage, Signal Transduction
Introduction
Ubiquitination
Ubiquitination, the covalent attachment of ubiquitin to a target protein, is a posttranslational modification that regulates the stability, function, and/or localization of the modified protein (1-4). Thus, ubiquitin acts as a signal that can be used to target proteins to specific location in the cell. Ubiquitination is catalyzed by the sequential action of three enzymes, a ubiquitin activating enzyme, E1, a ubiquitin conjugating enzyme, E2, and a ubiquitin ligase, E3. The ubiquitin-activating enzyme activates ubiquitin through an ATP dependent step forming a thiol ester bond between the C terminus of ubiquitin and the active cysteine of the E1. The activated ubiquitin is subsequently transferred to the active site cysteine of a ubiquitin conjugating enzyme which together with a ubiquitin ligase, transfers the ubiquitin to a lysine residue in the target protein. This later step is highly regulated and occurs in a substrate specific manner (3-6).
Deubiquitination
Deubiquitinating enzymes (DUBs) are proteases that cleave ubiquitin or ubiquitin-like proteins from pro-proteins or target proteins. They play several roles in the ubiquitin pathway (7). First, DUBs carry out activation of the ubiquitin pro-proteins, probably co-translationally. Ubiquitin is always expressed as a pro-protein fused to either ribosomal proteins or as linear polyubiquitin consisting of multiple copies of mono ubiquitin that must be processed to yield the mature ubiquitin monomer (8-10). The polyubiquitin gene product also contains an additional residue at the C-terminus that must be removed in order to activate ubiquitin. Secondly, DUBs recycle ubiquitin that may have been adventitiously trapped by the reaction of small cellular nucleophiles with the thiol ester intermediates involved in the ubiquitination of proteins (11). Thirdly, DUBs reverse the ubiquitination or ubiquitin-like modification of target proteins (7, 12). In this role DUBs antagonize the ubiquitination of proteins, playing a role analogous to that of the phosphatases in a kinase/phosphatase regulatory pathway. Finally, deubiquitinating enzymes are also responsible for the regeneration of monoubiquitin from unanchored polyubiquitin, i.e. free polyubiquitin that is synthesized de novo by the conjugating machinery or that has been released from target proteins by other deubiquitinating enzymes (13, 14).
Several earlier reviews specifically discuss DUBs from pathogens (15, 16) and those with specificity for ubiquitin-like proteins (17, 18). This review will be limited to discussing only those DUBs with specificity for ubiquitin. Nearly 100 putative DUBs are encoded by the human genome and they belong to five different families (12). Four families, the ubiquitin C-terminal hydrolases (UCH), the ubiquitin specific protease (USP/UBP), the ovarian tumor (OTU), the Josephin domain are papain-like cysteine proteases. The fifth family belongs to the JAB1/MPN/Mov34 metalloenzyme (JAMM) domain zinc-dependent metalloprotease family. One other small family of DUBs is known but their activity on, and specificity for, ubiquitin is low. The Adenain family of cysteine proteases include: the ubiquitin-like proteases (ULP, also known as SENPs in humans) that are specific for the ubiquitin-like proteins SUMO (small ubiquitin-like modifier) or Nedd8 (neural precursor cell expressed, developmentally down-regulated 8); and DUBs resembling the adenovirus protease that some bacteria and viruses have acquired and that probably play a role in infectivity by cleaving Ub and ISG15 (interferon stimulated gene 15) conjugates (17, 19-22). The large number of gene families and individual members suggests that they exhibit a significant degree of substrate specificity.
Like ubiquitination, deubiquitination is a highly regulated process that has been implicated in numerous cellular functions, including cell cycle regulation (23), proteasome-and lysosome-dependent protein degradation (24-26), gene expression (27), DNA repair (28), kinase activation (25, 29), microbial pathogenesis (15, 16), and more (5, 12). A number of pathogenic microorganisms have acquired genes encoding DUBs suggesting that disruption of ubiquitination in the host cell may confer a selective advantage for these bacteria (16, 30-35) and viruses (15, 36-43). Furthermore, mutations in several deubiquitinating enzymes have been linked to disease ranging from cancer to neurological disorders (44-46). Although a few substrates have been identified for a handful of DUBs, the substrates and physiological role of most DUBs is poorly defined.
General properties of DUBs
In spite of the paucity of knowledge about the regulation and roles of many DUBs, several generalizations have emerged in recent years. Each will be discussed in more detail below.
Most DUB activity is cryptic. That is, the energy of associating with the substrate or a scaffolding protein is required to achieve the catalytically competent conformation. Thus, like most other proteases, their activity is carefully controlled to prevent adventitious cleavage of inappropriate substrates (47). Other DUBs are covalently modified by phosphorylation, ubiquitination or sumoylation, all modifications that are likely to affect activity, localization or half-life.
DUBs are modular, containing not only catalytic domains but also additional ubiquitin binding domains and various protein-protein interaction domains. These modules contribute to the binding and recognition of different chain linkages (48) and direct the assembly of multi-protein complexes that localize DUBs and assist in substrate selection. DUBs require these localization and substrate specificity determinants in order to function physiologically. The association of DUBs with substrate adapters, scaffolds, and inhibitors are regulatory interactions driving specificity. A recurring theme has also emerged in which DUBs associate with complexes containing E3 ligases, thus negatively regulating ubiquitin conjugation (49).
A more detailed understanding of these protein-protein interactions and substrate selectivity will require development of quantitative assays of activity and binding. Only by comparing the absolute activities on related substrates can we define substrate specificity. For example, simply observing that an enzyme preparation can completely cleave both K48- and K63-linked chains does not give much information about their relative preferences. This is particularly important when a single DUB can cleave multiple substrates, albeit with vastly different efficiencies (19, 50).
Many excellent reviews on DUBs have appeared in recent years (15-17, 23-25, 29, 44, 49, 51-53) and we will try to minimize overlap with those. This review will expound upon the above generalizations and provide an overview of the current body of knowledge regarding cellular functions and regulation of ubiquitin specific DUBs. Because of space limitations this review cannot be fully comprehensive. Rather, we will use selected examples to illustrate these generalizations (Table 1).
Table 1.
List of DUBs discussed in this review and the cellular processes in which they are involved. References are given in the text.
| Cellular process | DUB (yeast/human) | Substrate or interacting partners | Notes |
|---|---|---|---|
| Ubiquitin processing | Isopeptidase T | Polyubiquitin | Processes ubiquitin chains to regenerate free ubiquitin |
| Rpn11/POH1 | Polyubiquitin | Associates with proteasome to process ubiquitin chains from proximal end before substrate degradation | |
| UCH37 | Polyubiquitin | Associates with proteasome to process ubiquitin chains before substrate degradation | |
| Ubp6/Usp14 | Polyubiquitin | Regulates chain length of proteasome substrates | |
| Histone deubiquitination | Ubp8/Usp22 | Histone H2A and H2B | Component of SAGA complex regulating transcription and mRNA export |
| Ubp10 | Histone H2A | Functions in silencing through its association with the Sir4 protein | |
| Usp16/Ubp-M | Histone H2A | Chromatin remodeling | |
| Usp21 | Histone H2A | Chromatin remodeling | |
| 2A-DUB | Histone H2A | Chromatin remodeling | |
| Cell cycle regulation and DNA repair | Usp28 | FBW7 | Involved in cell cycle regulation and DNA damage repair pathway |
| Usp44 | APC | Negatively regulates activation of the anaphase promoting complex | |
| Usp1 | FANCD2 | Regulates DNA repair, defective in Fanconi anemia. | |
| Usp11 | BRCA2 (indirect) | DNA damage repair functions | |
| Usp3 | Histone H2A and H2B | Chromatin remodeling | |
| Kinase Signaling | A20 | RIP1, NEMO | Downregulation of NF-κB pathway by DUB and ligase activities |
| CYLD | TRAF2/6, RIP1, NEMO | Downregulation of NF-κB pathway by DUB activity | |
| Usp15 | IκB | Downregulates NF-κB pathway by stabilizing IκB | |
| Usp9Y | MARK4, NUAK1 | Regulates AMPK-related Kinases | |
| Endocytosis | Doa4/Usp8 | Polyubiquitin | Recycles Ub at the late endosome |
| AMSH | EGFR (indirect) | Accelerates EGFR down-regulation | |
| Fat facets (Drosophila), Usp9X | Liquid facets (Lqf) | Adaptor molecules involved in the initial steps of endocytosis |
Families of Ubiquitin Specific DUBs
The ubiquitin C-terminal hydrolase (UCH) domain
The UCH catalytic core consists of a 230 amino acid domain as exemplified by the human UCH-L1 and -L3 isozymes. Humans have four UCH domain deubiquitinating enzymes, while S. cerevisiae only has one. Like all thiol protease DUBs, UCHs adopts a core fold and a catalytic triad geometry closely resembling papain (54). UCH DUBs preferentially process small leaving groups or substrates with disordered sequences linking the leaving group to the C-terminus of ubiquitin (7, 54-56). Thus, UCH-L1 and -L3 deubiquitinating enzymes are thought to be involved in processing pro-ubiquitin proteins and salvaging adventitiously trapped ubiquitin (7, 55, 57). This may not be the only role, as the UCH domain deubiquitinating enzyme from Drosophila can cleave polyubiquitin from polyubiquitinated proteins (58) and deletion of the single UCH in yeast does not impair pro-ubiquitin processing. Two other mammalian UCHs have additional C-terminal extensions: UCH37 contains about 100 additional amino acids at the C-terminus that direct it to the proteasome where it is involved in trimming polyubiquitin from proteins as they are degraded (24, 59); and BAP1 contains an additional 500 amino acids containing a nuclear localization signal and the binding site for interaction with the N-terminal ring finger of BRCA1, a ubiquitin ligase (60, 61).
The ubiquitin specific protease (USP) domain
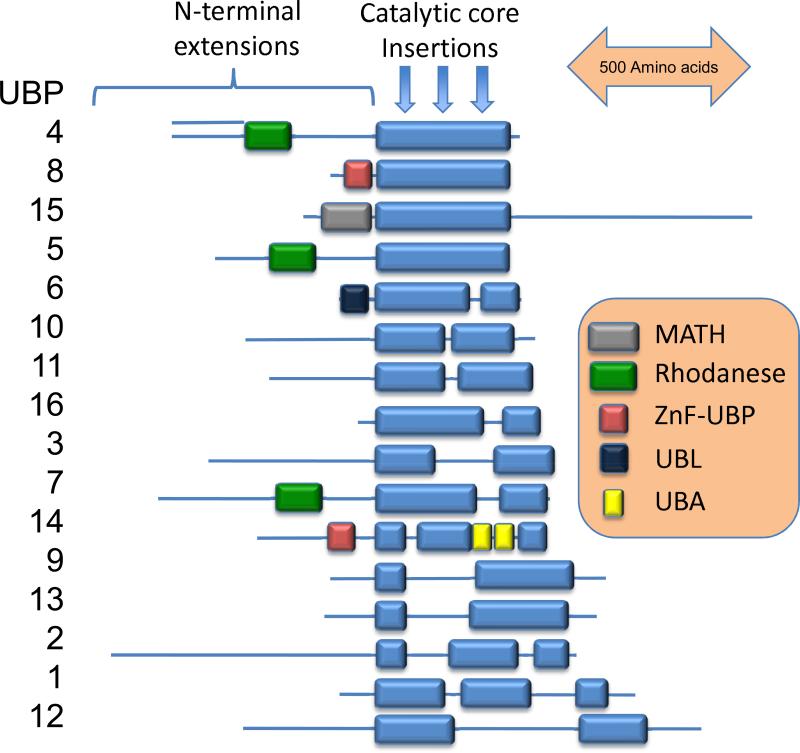
USP (UBP in yeast) family members usually process larger leaving groups. Yeast have 16 USP family DUBS (Figure 1) while humans are predicted to have over 50 USP family deubiquitinating enzymes, making this class of DUBs the largest (12). Structures of six USP domain deubiquitinating enzymes indicate that the USP domain fold is highly conserved despite low sequence similarity (62-66). The USP fold consists of three sub-domains: the finger, palm and thumb, which together resemble a right hand. Only CYLD, a deubiquitinating enzyme implicated in the human benign tumor syndrome cylindromatosis, lacks the finger sub-domain (66). Structures of USP domains bound to ubiquitin or a ubiquitin based inhibitor show that the C-terminus of ubiquitin sits in a cleft located between the thumb and the palm sub-domains, while the globular portion of ubiquitin interacts with the finger (62, 63, 65). Most USPs contain a core catalytic domain with insertions and terminal extensions bearing additional protein interactions domains (illustrated for yeast UBPs in Figure 1).
Figure 1.
Domain structure of yeast UBP deubiquitinating enzymes. The catalytic core is indicated by the blue boxes. Other common domains are indicated with differently colored boxes: MATH, meparin and TRAF homology; Rhodanese, rhodanese-like; ZnF-UBP, zinc finger common in UBP DUBs, UBL, ubiquitin-like, and UBA, ubiquitin-associated. Three sites of common insertions within the catalytic core are indicated by arrows. These insertions and the N- and C-terminal extensions are thought to provide specific sequences that define substrate specificity and provide interaction surfaces for binding to adapters and scaffolds.
The ovarian tumor (OTU) domain
The OTU domain family of deubiquitinating enzymes was identified based on their homology to the ovarian tumor gene involved in the development of the ovaries of fruit flies (67-69). Yeast encode two OTU domain DUBs and humans encode fourteen. Subsequent studies demonstrated that several OTU domain proteins have deubiquitinating activity (70-76), although not all have been shown to have DUB activity. For instance, the Drosophila ovarian tumor protein contains a serine at the active site and may not be active. Recent structural studies indicate that the OTU core domain is composed of five β-strands sandwiched between helical domains that vary in size among OTU family members (73, 74, 77, 78).
The Josephin domain (MJD)
The Machado-Joseph disease protein Ataxin-3, a protein implicated in the neurodegenerative disorder spinocerebellar ataxia type 3, is the best-studied member of the four human Josephin family proteins (79-84). The structure of the Josephin domain of Ataxin-3 was solved by NMR and resembles that of UCH domain deubiquitinating enzymes. The active site residues of Ataxin-3 are positioned in a catalytically competent conformation and it has been reported to edit K63-linked chains specifically (85).
The JAB1/MPN/Mov34 metalloenzyme (JAMM) domain
Al least four different JAMM domain DUBs exists (86-89). Three of them are known to process ubiquitinated substrates (87-90), and one acts on proteins modified with the ubiquitin-like modification Nedd8 (86). Recently the structure of the AMSH-like protein (associated molecule with the SH3 domain of STAM), was solved bound to K63-linked diubiquitin (91). AMSH-LP is a deubiquitinating enzyme that functions in vesicle trafficking and specifically cleaves K63-linked polyubiquitin. This structure represents the first structure of a deubiquitinating enzyme bound to polyubiquitin and explains the linkage selectivity displayed by AMSH DUBs. AMSH-LP binds two zinc ions, one of which is involved in catalysis and another that is part of an AMSH-specific insert forming a motif responsible for the recognition of the proximal ubiquitin in the K63 diubiquitin. A second AMSH-specific insert interacts with the distal ubiquitin. The linkage selectivity results from the interaction of the JAMM domain of AMSH-LP with both the distal ubiquitin and the tri-peptide sequence Gln62-Lys62-Glu64 of the proximal ubiquitin, a unique surface that is only found in K63-linked polyubiquitin. Other JAMM domain proteases that lack specificity for polyubiquitin lack the AMSH-specific inserts that determine this specificity,
DUB activity is regulated and often cryptic
Since DUBs are proteases, it is likely important to regulate their enzymatic activity to avoid the inadvertent cleavage of non-substrate proteins. Further regulation may be anticipated as a mechanism to control the timing and localization of substrate cleavage.
Substrate induced conformational changes
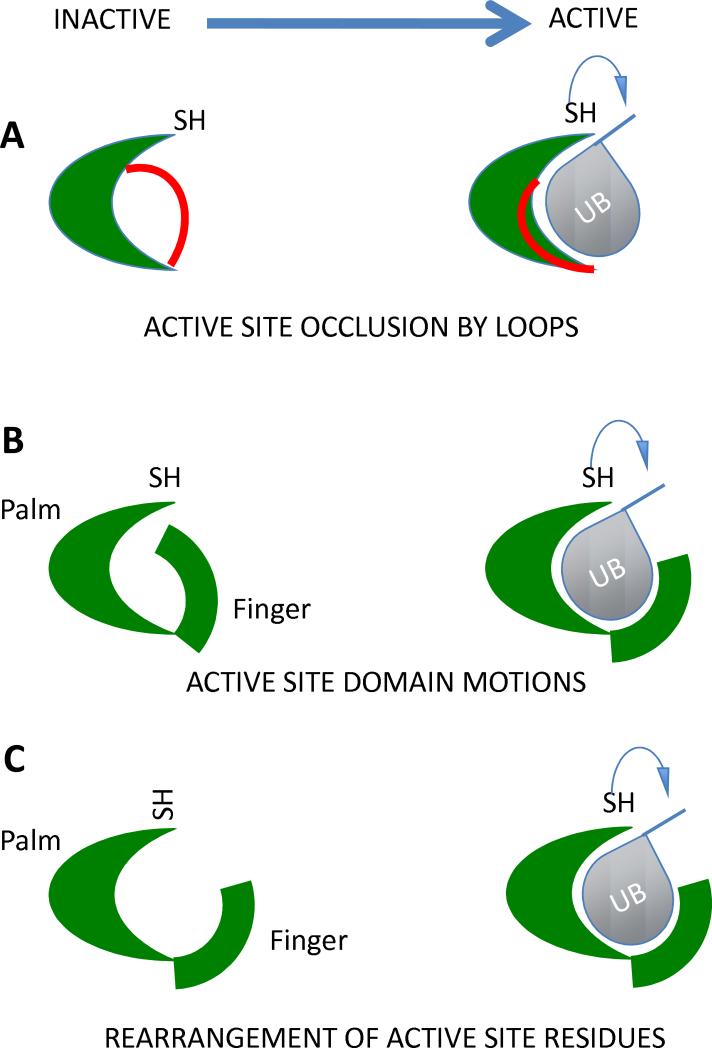
Structures of at least one member of each DUB family are currently available, and many have been solved in the free form or bound to ubiquitin or ubiquitin-based inhibitors (54, 56, 62, 64-66, 73, 74, 78, 82, 91-93). These structures reveal that active site rearrangements occur upon binding to ubiquitin and these changes must occur to productively bind and catalyze hydrolysis. This requirement prevents spurious cleavages at Gly-Gly sequences and enhances specificity. These conformational changes are shown schematically in Figure 2.
Figure 2.
Substrate induced conformational changes upon ubiquitin binding to DUBs. A) a loop occluding the active must be displaced to allow substrate binding and access to the active site. B) A larger domain occluding the active site must be displaced upon substrate binding or association with a scaffold protein. C) The binding of the substrate induces a conformational change in the catalytic triad allowing it to achieve a catalytically competent geometry.
Crystal structures of human UCH-L1 and UCH-L3 and the S. cerevisiae YUH1 reveal that these proteins undergo conformational changes that are required for catalysis (54, 56, 92, 94). In UCH-L3, an active site loop is disordered but appears to cross over the active site (54). In contrast, when UCH-L3 is bound to an ubiquitin-based inhibitor, ubiquitin vinyl methyl ester, the crossover loop becomes stabilized in an α-helix followed by an S shaped loop forming contacts with the C terminus of ubiquitin (92). This conformation was also seen in the structure of YUH1 bound to a different active site inhibitor (56), ubiquitin aldehyde, indicating that the crossover loop conformation is conserved among UCH domain deubiquitinating enzymes (Figure 2A). This loop may generally prevent UCH domains from accommodating tightly folded domains C-terminal to ubiquitin since folded domains cannot be inserted through the loop. Alternatively, this loop may be repositioned between the C-terminal leaving group and the body of the UCH domain forming an interaction surface that imparts specificity as to the types of substrates that may be hydrolyzed. Such a model could explain why the Drosophila UCH can cleave ubiquitin attached to larger proteins (58).
The structure of yeast OTU1 bound to a ubiquitin based inhibitor has been solved and comparison with the free structure of human Otubain-2 suggests that the OTU domain also may exhibit active site rearrangements upon binding to ubiquitin (77). The active site residues in Otubain-2 adopt a catalytically competent conformation but an active site loop is partially disordered and appears to adopt a position that interferes with ubiquitin binding. In the substrate bound OTU1 structure, this loop becomes ordered into a β-strand allowing the binding to ubiquitin. Thus, until repositioned this loop would interfere with ubiquitin binding.
Like the UCH family, USP domain deubiquitinating enzymes also undergo conformational changes upon binding to ubiquitin. These conformational changes include: alignment of the catalytic triad in a productive conformation in USP7 (Figure 2C) (63), displacement of active site loops that would otherwise block the binding of the C-terminal tail of ubiquitin in USP14 (62), or movement of the finger domain outwards to accommodate the globular body of ubiquitin in USP8 (Figure 2B) (64). USP5 (more commonly known as isopeptidase T or IsoT) presents a final dramatic example that is less well defined structurally, but which illustrates the importance of these types of conformational changes in enforcing substrate specificity. USP5 hydrolyzes unanchored polyubiquitin chains in vitro and in vivo (13, 95-97). A free ubiquitin carboxyl terminus is required at the proximal end of the chains (the end where the chain would be attached to a target protein) to achieve efficient catalysis. USP5 will also hydrolyze a substrate analog, Ubiquitin-7-amido-4-methylcoumarin (Ub-AMC) slowly. However, addition of ubiquitin greatly increases the rate of hydrolysis by occupying the S1’ (leaving group) site of USP5 mimicking the proximal ubiquitin in a physiological chain (98, 99). This activation only occurs when the stimulatory ubiquitin bears a free C-terminus. The conformational change induced by occupancy of the S1’ site by ubiquitin (or the proximal ubiquitin in a polyubiquitin chain) is necessary for physiological activity. A polyubiquitin chain still attached to a peptide or lacking the C-terminal glycine on the proximal ubiquitin is not hydrolyzed efficiently (13, 99). Thus, USP5 proofreads the C-terminus of polyubiquitin chains and will not cleave them if they are still attached to a protein or peptide. In this way USP5 can remove the polyubiquitin end products of proteasomal degradation but cannot prematurely deubiquitinate a polyubiquitinated protein.
Activity can also be induced by scaffold or adapter binding
Maintaining DUBs in an inactive state until they are properly localized enforces another level of control. A clear example of this is provided by the three proteasome associated DUBs discussed in more detail below. USP14, UCH37, and POH1 are largely inactive until they are associated with the proteasome (100-103). In the case of the first two, the activation associated with binding to the proteasome may be important in limiting their activity as these proteins both exist in free and proteasome-bound states (101, 104).
Another challenge faced by DUBs is achieving substrate specificity. The core catalytic domains themselves only recognize ubiquitin and in order to select the proper ubiquitinated protein substrate additional interactions with the target protein are required. A number of DUBs have been shown to have good catalytic capability but little affinity for ubiquitin, and must therefore depend on other interactions to acquire their substrates. Interactions with scaffolding proteins can assure proper localization of the DUB to the same locale as the substrate. Interactions with substrate adapters can bridge the gap between the DUB/scaffold and the target protein by binding to both. In this way, the proper substrate is delivered to the DUB in the right place and time for its catalytic action. A recent review catalogs these and other DUB-protein interactions (49).
Transcriptional regulation of DUB expression
Specificity of DUB action can also be achieved by regulating the time when a particular DUB is accumulated. Perhaps the best-studied examples are the cytokine inducible DUBs of murine lymphocytes (105-108). Stimulation of lymphocytes with different cytokines results in the induction of one of a family of 60 kDa DUBs most closely related to human USP17. Thus, DUB-1 is induced by interleukins 3 and 5 and GM-CSF (granulocyte-macrophage colony stimulating factor). DUB-2 is stimulated by interleukin 2. These DUBs are immediate early genes whose transcription is rapidly induced under the control of cytokine responsive elements in the DUB genes (106). They are also rapidly degraded as the cytokine response is down regulated, probably by becoming polyubiquitinated and delivered to the proteasome (109, 110). This temporal regulation suggests that DUB-1 and -2 may play a role in down regulating the cytokine response, perhaps by virtue of its DUB activity (111). Other DUBs are probably regulated in a similar manner. For instance, the transcription of CYLD is induced by activation of the NF-κB and MKK3/6-p38 pathways (112)
Post-translational covalent modifications
We have come to expect that key regulatory enzymes will be modulated by a host of signal transduction pathways and DUBs are no exception. Most regulated proteins are subject to phosphorylation by one or more kinases. The majority of DUBs are phosphorylated on serine, threonine and tyrosine residues in vivo (http://www.phosphosite.org), although with few exceptions the consequences are unknown. Two recent proteomic analyses found that a large number of components of the ubiquitin proteasome system, including USP15, USP19, USP28 and USP34, are regulated by phosphorylation by the Ataxia Telangiasia-mutated (ATM)/ATRRad3-related (ATR) kinase in response to DNA damage (113, 114). Several of these components regulate DNA damage checkpoints, with USP34 being involved in regulating the rapid G1 arrest in response to ionizing radiation mediated by ATM/ATR kinases. USP28 is also phosphorylated by ATR/ATM and is involved in the response to DNA damage by virtue of its ability to deubiquitinate and stabilize FBW7 (see below), the ubiquitin ligase involved in the degradation of the myc transcription factor (115). Finally, USP15 is phosphorylated by ATR/ATM and is known to cleave ubiquitin from IκB, thus dampening the NF-κB response (see below). Another DUB, A20, also regulates the NF-κB pathway by first removing K63-linked chains from the RIP1 (receptor interacting protein 1), TRAF2 (tumor-necrosis factor receptor (TNFR)-associated factor 2), TRAF6, and NEMO (NF-κB essential modulator) components of this pathway. Subsequently, the ubiquitin ligase domain of A20 facilitates K48-ubiquitination of at least some of these, thereby increasing their degradation and down regulating the NF-κB pathway (76). Phosphorylation of A20 on S381 by IκB kinase β increases the ability of A20 to inhibit signaling through this pathway (116). It is not clear whether the DUB activity is increased or the ligase activity is decreased by phosphorylation of A20, but the net result is a feedback inhibition of NF-κB signaling.
Ubiquitination itself is another regulatory modification and a small number of DUBs have been shown to be ubiquitinated, probably as a signal for their degradation. In addition to the murine cytokine induced DUBs mentioned above, USP4 is also polyubiquitinated. USP4 is responsible for deubiquitinating Rho52, an oncoprotein that is an E3 ligase. Rho52 in turn can ubiquitinate USP4 (117). In the USP4-Rho52 complex USP4 is inhibited, suggesting a model whereby the heterodimeric complex acts as a ligase in the presence of substrates. When substrate is not present RHO52 may auto-ubiquitinate itself and USP4 reverses this ubiquitination. However, if Rho52 ubiquitinates USP4 the DUB may be degraded. Thus, USP4 and Rho52 transregulate each other (117). Another oncoprotein, TRE17 (a fusion protein between a Rab GTPase activating protein homology domain at the N-terminus and USP6 at the C-terminus) is monoubiquitinated in a calcium/calmodulin dependent pathway (118). This too suggests regulation modulated by cellular signaling pathways. USP7 (119), USP36 (120) and DUB-1 (109, 110) are also ubiquitinated, although the importance of these modifications is unknown. Finally, it has been shown that USP25 is sumoylated and that this modification decreases USP25 binding and hydrolysis of polyubiquitin (121).
DUBs are modular
In addition to their active site core domains, most deubiquitinating enzymes contain insertions and N and C-terminal extensions that participate in substrate recognition, regulate the deubiquitinating enzyme cellular localization, catalytic activity and/or protein-protein interactions (Figure 1). These extensions include: protein-protein interaction domains that interact with substrates, scaffolds or substrate adapters; and ubiquitin-binding domains that in some instances participate in ubiquitin and/or polyubiquitin recognition. We will discuss two examples to highlight this principle.
Multiple ubiquitin binding domains
The observation that different polyubiquitin chain linkages direct proteins to different cellular fates suggests that receptors and enzymes operating on polyubiquitin are capable of distinguishing among these chains. One model for this recognition suggests that the receptor/enzyme will recognize several ubiquitins in a chain. A number of proteins of the ubiquitin system have multiple ubiquitin binding domains that in principle can accomplish this type of interaction (85, 121, 122).
USP5, also referred to as IsoT, was one of the first DUBs to be characterized (13, 98, 123, 124). IsoT has orthologs in yeast, slime mold, plants, flies, and other eukaryotes. Functional analyses of IsoT orthologs in three different model organisms demonstrate that IsoT is responsible for most of unanchored polyubiquitin disassembly in vivo (95-97). The levels of unanchored polyubiquitin are regulated by the deubiquitinating activity of yeast Isopeptidase T (13, 95). Accumulation of free polyubiquitin is probably detrimental as it acts as a competitive inhibitor of substrate binding to the proteasome and other ubiquitin receptors (14). Unanchored polyubiquitin can be either generated de novo by the ubiquitin conjugation machinery and/or by the release of ubiquitin chains from target proteins by deubiquitinating enzymes associated with the proteasome (13, 95).
Deletion of UBP14, the IsoT ortholog in S. cerevisiae, leads to defects similar to those seen in other mutants of the ubiquitin-proteasome system (95), i.e., a strong sporulation defect, hypersensitivity to canavanine, and a decrease in the rate of the degradation of two well-characterized artificial substrates of the ubiquitin-proteasome system. However, unlike other ubiquitin–proteasome mutants, deletion of UBP14 leads to an accumulation of unanchored polyubiquitin chains as well as ubiquitin conjugates. Although Ubp14 is not essential in yeast its ortholog is required for development in multicellular organisms. Dictyostelium discoideum cells with a mutant IsoT ortholog gene, ubpA accumulate unanchored polyubiquitin and ubiquitin conjugates (97). UbpA is not necessary for growth in D. discoideum, however it is crucial for aggregation, chemotaxis, and cell adhesion. These defects are thought to result from inhibition of the proteasome by excess unanchored polyubiquitin (125). The A. thaliana gene is essential and mutation of the AtUbp14 gene causes embryonic lethality (96).
IsoT recognizes multiple ubiquitin molecules in the polyubiquitin chain (13) and is capable of cleaving polyubiquitin linked by K29, K48 and K63 linkages (126). Analysis of the mechanism of polyubiquitin processing showed that IsoT sequentially cleaves the ubiquitin from the proximal end of the chain, thereby acting as an exo-isoamidase (13). IsoT has at least four ubiquitin binding sites for both linear and K48-linked polyubiquitin, making IsoT a bona fide polyubiquitin binding protein (13). These sites are formed by four ubiquitin binding domains-an N-terminal ZnF UBP domain, a USP/UBP domain, and two UBA domains (99, 122). By definition proximal ubiquitin in the chain contacts the S1’ leaving group pocket, which is formed by the ZnF UBP domain (99). This domain is important for inducing a conformational change leading to activation when the proximal ubiquitin contains a free C terminus. The ZnF UBP domain interacts intimately with the C-terminus of the proximal ubiquitin and binding of the proximal ubiquitin is greatly diminished by truncation or elongation of this C-terminal tail. This mandatory substrate-induced conformational change prevents IsoT from disassembling polyubiquitin chains before they have been released from the target protein by the action of other DUBs. The second ubiquitin binds to the S1 pocket, which corresponds to the active site of the enzyme and is formed by the USP/UBP domains. The third and fourth ubiquitins in polyubiquitin bind the S2 and S3 sites respectively (13). The S2 and S3 sites of IsoT are formed by two UBA domains, UBA2 and UBA1 respectively (122), which are located in insertions between the Cys and His boxes of the core USP domain.
Although polyubiquitin isoforms adopt different three-dimensional structures (127-130), IsoT uses the same set of domains to interact with the corresponding ubiquitin subunit irrespective of the chain linkage (122). Thus, either IsoT must be flexible enough to allow significant motion of the binding domains to accommodate the different chain geometries or the ubiquitins in different polyubiquitin chains must be able to adopt similar three-dimensional orientations. In reality, both mechanisms may be involved.
Direct recognition of substrates
In addition to recognizing the specific chain linkages of a polyubiquitin chain some DUBs also exhibit direct affinity for the ubiquitinated protein itself. In one case there is structural data describing these interactions. USP7, also known as Herpes associated USP (HAUSP), deubiquitinates p53 and Mdm2 and is inhibited by the Epstein-Barr nuclear antigen 1 (EBNA1) protein of Epstein-Barr virus (EBV). The N-terminal domain of USP7 binds two closely spaced 4-residue sites in both p53 and MDM2 (p53 residues 359−367 and MDM2 residues 147−159) (131). The crystal structure of the p53-binding domain of USP7 bound to an EBNA1 peptide has been described (132). NMR studies of USP7 bound by EBNA1 and p53 indicated that p53 and EBNA bind the same pocket but p53 but makes less extensive contacts with USP7. Functional studies indicated that EBNA1 binding to USP7 inhibits its deubiquitination of p53 and protects cells from apoptotic challenge by lowering p53 levels (132).
Protein-Protein interaction domains
Another common feature of DUBs is that they contain other protein-protein interaction domains (Figure 1). As a consequence, many DUBs are found associated with substrates, adaptors and scaffolds. We will use the example of proteasome associated DUBs to highlight this aspect of DUBs structure and function. A recent review catalogs more of these interactions and points out that many DUBs form macromolecular complexes with ubiquitin ligases (49). This suggests a coupled regulation of ubiquitination and deubiquitination that may be an important part of the functions of ubiquitination.
The proteasome is a large multi-protein complex catalyzing ubiquitin dependent protein degradation (4, 26, 133). The 26S proteasome contains more than 30 proteins that are found in two main subcomplexes: the 20S core particle and the 19S regulatory particle (134, 135). Four stacked heptameric rings form the 20S core particle (Figure 3). The two inner rings are formed by β-subunits, which contain the active site of the protease catalyzing protein degradation. The outer rings are formed by seven α-subunits, which together create a narrow gated pore through which unfolded polypeptides must pass in route the proteolytic chamber. Because the polyubiquitin chain attached to the protein would present a steric block to translocation of the substrate into the lumen of the proteasome the chains must be removed for optimal proteolysis.
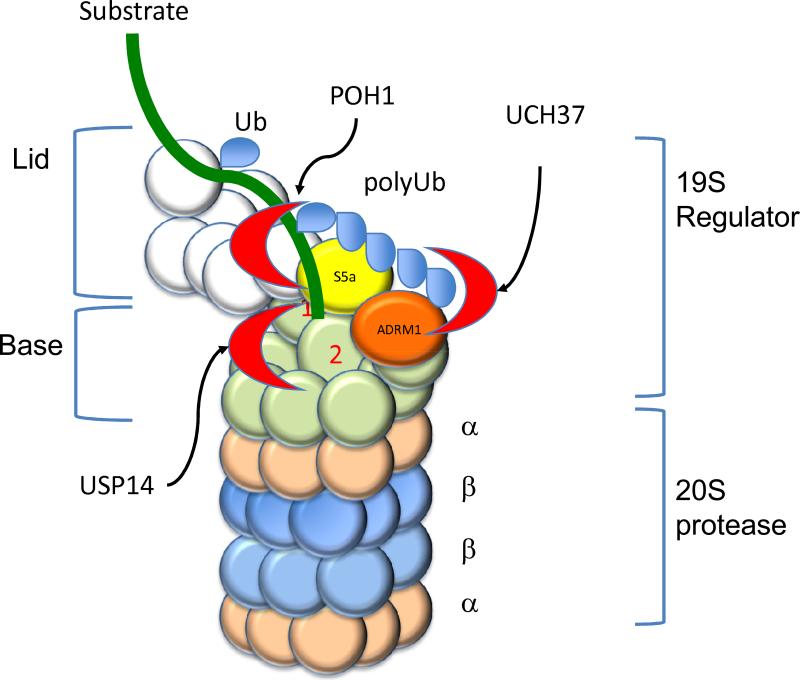
Figure 3.
Proteasome bound deubiquitinating enzymes. Deubiquitinating enzymes are indicated by red crescents, the substrate as a green line, and ubiquitin as blue ovals. POH1 catalyzes the release of a polyubiquitin chain “en bloc” as the substrate is engaged and translocated through the gated pore of the 20S protease. RPN10 (yellow) binds the polyubiquitin chain and the distal end of the chain can be removed by the action of UCH37 bound to ADRM1 (orange). USP14 is bound to the proteasome via interactions with RPN1 (purple) and probably removes mono ubiquitin attached to the substrate.
Three DUBs (USP14, UCH37 and POH1) belonging to different DUB families are associated with the proteasome in higher eukaryotes (59, 87, 90, 100, 102, 103, 136-142). S. cerevisiae lacks UCH37, although most other eukaryotes contain an ortholog of this enzyme (59, 138, 143). Together, these DUBs remove polyubiquitin chains from substrates during proteasomal degradation. All three require association with the proteasome and subsequent conformational changes to exhibit significant catalytic activity. Two of them, POH1 and UCH37 are integral subunits of the 19S regulatory particle (59, 103, 136, 144). UCH37 contains a C-terminal extension that interacts with the Admr1 subunit of the 19S regulatory particle (103). Analysis of various truncation mutants suggest that the N-terminal JAMM domain of the yeast homolog of POH1 (RPN11) is required for association with other lid subunits but it is not clear if all the truncated proteins are properly folded. Thus, other protein-protein interactions may also be involved. The third deubiquitinating enzyme, Usp14, reversibly associates with the Rpn1 (PSMD2) subunit of the 19S base subcomplex of the proteasome through an N-terminal ubiquitin-like domain (102, 145).
Cellular roles of DUBs often involve multi-protein complexes
There is now much direct evidence for the generalization that many DUBs must associate with multi-protein complexes to exert their physiological functions. Not only does this help co-localize the substrates and the DUB, but it also allows a sequential ubiquitination and deubiquitination cycle to give directionality to the pathways controlled by ubiquitination and deubiquitination.
Processing of polyubiquitinated proteins by proteasome-associated DUBs
The three proteasome-associated DUBs of the proteasome are associated with the 19S regulatory particle of the proteasome. The 19S regulatory particle unfolds and deubiquitinates polyubiquitinated proteins (26, 134, 135). The 19S regulatory particle can be further divided into two subcomplexes, the base and the lid (Figure 3). The lid (Figure 3, white, yellow and orange spheres) includes multiple polyubiquitin receptors. Two of these receptors, S5a (Figure 3, yellow sphere) and Adrm1 (Figure 3, orange sphere), are integral components of the 19S regulatory particle while the remaining polyubiquitin receptors reversibly associate with the proteasome (144, 146-148). The later type of receptors associate with the Rpn1 subunit (PSMD2 in humans) of the 19S proteasome through ubiquitin like (UBL) domains and contain ubiquitin binding domains belonging to the ubiquitin associated domains (UBA) family (148, 149). They act as adapters linking polyubiquitinated substrates to the proteasome. The base subcomplex (Figure 3, white spheres) contains six AAA ATPases that are thought to assist in unfolding and translocating the substrate into the lumen of the proteasome. Thus, the 19S regulatory particle binds polyubiquitin while the base subcomplex probably unfolds and translocates the target protein through the gated pore of the 20S protease.
Functional studies suggest that UCH-37 inhibits the degradation of ubiquitinated proteins by the proteasome by removing ubiquitin from the distal end of the polyubiquitin chain (59). Depletion of UCH37 leads to accelerated hydrolysis of model proteasome substrates with a concomitant decrease in the levels of cellular polyubiquitinated proteins (59). The C-terminal domain of UCH37 associates with C-terminus of the lid subunit Adrm1 (103, 136, 144). Association of UCH37 with Adrm1 increases the efficiency of hydrolysis of an artificial substrate, Ub-AMC, by decreasing the KM for the substrate without modifying the Vmax (103). The C-terminus of free UCH37 inhibits its own catalytic activity and binding of Adrm1 relieves this autoinhibition (103). In addition to interacting with and activating UCH37 hydrolysis of Ub-AMC, Adrm1 also interacts with Rnp2/PSM1D and polyubiquitin through different surfaces of its N-terminal domain (137, 146). Since the N-terminal ubiquitin-binding domain of Adrm1 interacts with the juxtadistal subunit of polyubiquitin, it serves to bind both the enzyme and one end of the substrate. The catalytic center of UCH37 then binds the distal subunit of polyubiquitin and hydrolyzes it from the chain (103, 136, 144). However activation of the hydrolysis of diubiquitin only occurs upon the association of UCH37 with the whole 19S regulatory particle, suggesting that other proteins participate in the activation of UCH37 and/or that there is an additional mechanism for activation.
Like UCH37, Usp14 is activated upon association with the 19S regulatory particle. Depletion of Usp14 in human cells or knockout of UBP6, its ortholog in S. cerevisiae, leads to accelerated protein degradation (59, 145). Surprisingly, the inhibition of protein degradation mediated by Ubp6 is independent of its deubiquitinating activity but depends on its association with the base of the 19S regulatory particle through its UBL domain (102). Ubp6 acts catalytically in cooperation with a proteasomal associated E3 ligase, Hul5, to regulate the length polyubiquitin chains attached to target proteins (101). Consistent with a role in polyubiquitin remodeling at the proteasome, Usp14 disassembles polyubiquitin chains from the distal end (62). This dynamic elongation and shortening of polyubiquitin chains may represent a balance between keeping the substrate associated with the proteasome and the necessity to remove polyubiquitin from the substrate as it is translocated into the proteasome. In addition to its role in regulating chain length at the proteasome, Ubp6 also functions to regulate ubiquitin levels by recycling ubiquitin at the proteasome (100). In agreement with this data, deletion of the mouse ortholog USP14 causes a depletion of free ubiquitin and ataxic mice (axJ) that contain a mutant form of the Usp14 gene display a reduction in free monoubiquitin levels in the brain (141, 150).
A third deubiquitinating enzyme, Rpn11/POH1 is an integral part of the 19S regulatory particle (87, 142). Rpn11 belongs to the JAMM domain family of metalloproteases. Rpn11 is essential for viability is yeast, and depletion of its human ortholog, POH1, by RNAi inhibits cellular growth (59). Knockdown of POH1 leads to an increase in polyubiquitinated proteins, defective cellular protein degradation, and compromised proteasomal activity due to a defect in 26S proteasome maturation or stability. Thus, unlike the other two proteasome associated DUBs POH1 is required for proteasome integrity (59). Several groups have reported that mutations in the active site of Rpn11 lead to a loss of viability in Drosophila (143), human cells (59) and yeast (87, 90). However, Guterman and Glickman have shown that a D121A active site mutation in yeast Rpn11 causes a defect in protein degradation, but not in cell viability (142). Association of Rpn11 with the lid of the 19S regulatory particle and hydrolysis of ATP are required for Rpn11 dependent deubiquitination, suggesting that deubiquitination may be coupled to protein unfolding by the ATPases of the base subcomplex. Unlike Usp14 and UCH37, Rpn11 cleaves polyubiquitin from substrate proteins at or near the proximal end of the chains thereby releasing polyubiquitin “en bloc” (103).
Histone deubiquitination
In higher eukaryotes, monoubiquitinated histones H2A (uH2A) and H2B (uH2B) are critical in the regulation of multiple nuclear processes including mitosis, transcriptional initiation and elongation, mRNA export and silencing (151-154). In contrast, histone H2A is not ubiquitinated in Saccharomyces cerevisiae (152). Deubiquitinating enzymes have been recently identified that can reverse ubiquitination of both H2A and H2B (151, 155-157), or are specific for only H2B (93, 154, 158-160) or only H2A (89, 153, 161, 162). As discussed for proteasome-associated DUBs, many of the histone deubiquitinating enzymes also work only in the context of multi-protein complexes (Figure 3) (151, 155, 156, 159, 162-164). While it is tempting to think of this as an example of several different DUBs acting on the same substrate we must keep in mind that it is both the identity of the histone and its specific location within chromatin that seems to be important in determining which population of histones is deubiquitinated. Thus, ubiquitinated histones in active chromatin may not be equivalent to those in silenced chromatin because of other associated proteins or epigenetic marks.
Sequential ubiquitination/deubiquitination of H2B regulates transcription and silencing
A specific E2 and E3 catalyze monoubiquitination of H2B at K123 in yeast and K120 in mammals. In yeast, Rad6 and Bre1 catalyze H2b ubiquitination while in humans the corresponding enzymes are UbcH6 and RNF20. (Figure 4A). This modification is enriched at promoters and the 5’end of many genes during transcriptional activation. H2B ubiquitination is a pre-requisite for di- and tri-methylation of Lys4 and Lys79 of histone H3 (H3K4me and H3K79me), a modification that correlates with transcriptionally active chromatin and helps recruit the SAGA co-activator complex and additional chromatin modification activities (27, 158, 165).
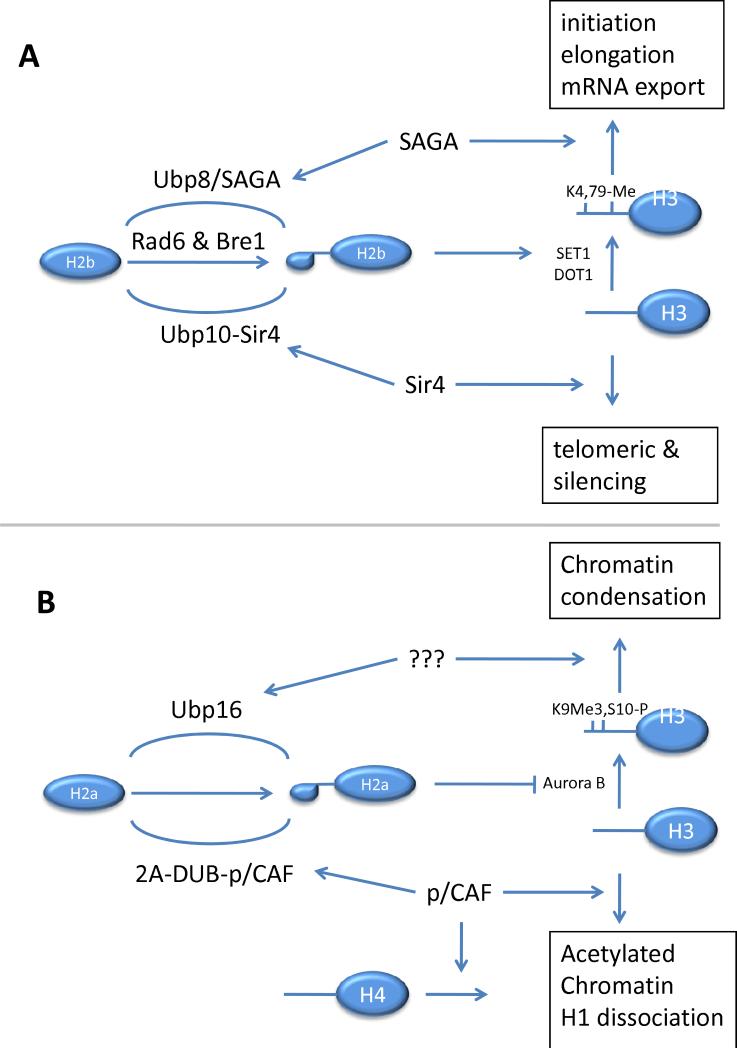
Figure 4.
Deubiquitination of histones H2A and H2B regulate chromatin structure and activity. Panel A details the dynamic ubiquitination/deubiquitination of H2B in yeast. These DUBs are localized to sites of action by interaction with other chromatin bound proteins that act as scaffolds and activate the DUB activity. Panel B shows the ubiquitination/deubiquitination of H2A in higher eukaryotes. A putative binding partner for UBP16 with a role in chromatin condensation is shown with question marks.
The removal of ubiquitin from histone H2B at promoter and coding regions of a small number of genes has also been shown to be required for their optimal transcription (158). S. cerevisiae Ubp8 and its human ortholog, Usp22, are part of the Spt-Ada-Gcn5-acetyltransferase (SAGA) complex (151, 155, 156, 159, 164). SAGA is a co-activator complex that regulates transcriptional activation, elongation and mRNA export of multiple genes, in part by modulating the state of histone post-translational modifications such as histone acetylation and H2B ubiquitination (uH2B) (27, 165, 166). Thus, the recruitment of the SAGA complex after methylation of Lys4 and 79 of histone H3 results in the deubiquitination of uH2B during elongation and facilitates the phosphorylation of Ser2 in the RNA polymerase II C-terminal domain. Mechanistically, the ubiquitination of H2B blocks the recruitment of the RNA polymerase II (RNAPII) kinase Ctk1, thereby preventing the phosphorylation of Ser2 of the C-terminal domain (CTD) of RNAPII (154). Deubiquitination of H2B by Ubp8/USP22 results in the recruitment of Ctk1 leading to Ser2 phosphorylation, a modification associated with elongation.
Ubp8 has also been linked to mRNA export by virtue of its association with a protein module within SAGA that contains three additional proteins Sgf73, the yeast ortholog of Ataxin-7, Sus1, a member of the TREX-2 mRNA export complex, and Sgf11 (163, 164, 167). This module mediates the binding of SAGA to additional members of the TREX-2 mRNA export complex factors, Sac3 and Thp1, that interact with the nuclear pore complex thereby physically coupling transcription with mRNA export (163). Consistent with a role of SAGA in mRNA export, several genes regulated by SAGA, including GAL1, are localized to the nuclear periphery during transcription, and this localization is dependant on Sus1 (167). Furthermore, loss of Sgf73, which activates Ubp8 deubiquitinating activity, results in a defect in the export of mRNA of GAL1, supporting a genetic link between SAGA, histone H2B deubiquitination and mRNA export.
In addition to its role in transcriptional initiation, elongation, and mRNA export, H2B deubiquitination is required for gene silencing (160, 168). Deletion of Ubp8 or Ubp10 leads to an increase in uH2B; however while Ubp8 is involved in gene activation (see above), Ubp10 functions in gene silencing by deubiquitinating uH2B in silenced regions (Figure 4A) (160, 168). Consistent with a role in silencing, Ubp10 localizes to two silenced domains, telomeres and rDNA (168) and associates with the silencing protein Sir4, a member of the Sir complex composed of two additional proteins, Sir3 and the histone deacetylase Sir2 (169). Deletion of Ubp10 affects telomeric silencing by increasing the levels of uH2B and subsequently H3K4me and H3K79me, which in turn inhibits the association of the Sir proteins to silent loci (160, 168). Overexpression of Ubp10 also leads to a loss of silencing at telomeres by globally decreasing uH2B, H3K4me and H3K79me, and the spreading of the Sir complex to non-silenced regions. This delocalization of the SIR complex is thought to be due to the binding of Sir3 to unmethylated Lys4 of histone H3 in normally non-silenced regions, thus preventing its binding to silenced regions (160, 168, 170) In addition to a role in telomere silencing, Ubp10 appears to have roles in other regions of the genome since the majority of the genes with altered expression in ubp10Δ are not located at telomeres (160). In support of a role outside telomeric loci, a mutant form of Ubp10 that specifically causes a defect in the silencing of telomeres does not lead to a global increase in H2B ubiquitination or H3K4me. Furthermore, ubp8Δ ubp10Δ cells display a synergistic increase in H2B ubiquitination levels and the expression of non-telomeric genes, indicating that Ubp10 and Ubp8 may act at overlapping regions of the genome (160).
Ubiquitination/deubiquitination of H2A in higher eukaryotes
H2A ubiquitination is linked to silencing of developmental genes, X-chromosome inactivation cell cycle progression, and transcriptional initiation and elongation in higher eukaryotes (152, 162, 171, 172). There is considerable confusion and uncertainty as to the precise role (s) of uH2A in transcription, but in some genes it seems to inhibit initiation by preventing the K4 methylation of H3 (opposite to the effects of uH2B). In other genes elongation appears to be decreased by the presence of uH2A. It should be mentioned that the importance of different epigenetic histone modifications varies depending on the gene examined and so the effects of ubiquitinated histones on these modifications will also vary in different genes.
Three deubiquitinating enzymes, Usp16 (Ubp-M), Usp21, and 2A-DUB have been shown to specifically deubiquitinate uH2A (Figure 4B) (89, 153, 162). One H2A deubiquitinating enzyme, 2A-DUB, belongs to the JAMM domain family of isopeptidases (89). Recent work has shown that 2A-DUB acts as a co-activator of the androgen receptor (AR), modulating full activation of AR-dependent gene expression through deubiquitination of H2A (89). 2A-DUB associates with the histone acetyltransferase p300/CBP-associated factor (p/CAF), a complex that mediates acetylation of histones H3 and H4 in transcriptionally active chromatin (Figure 4B). 2A-DUB preferentially deubiquitinates H2A in hyperacetylated nucleosomes, suggesting that histone acetylation facilitates H2A deubiquitination (89). However, uH2A deubiquitination does not appear to affect the pCAF acetyltransferase activity, hinting that these epigenetic modifications may occur through a series of sequential steps. The increase in uH2A that results from knockdown of 2A-DUB can be partially antagonized by knockdown of p/CAF, in agreement with a role of histone acetylation in the control of H2A deubiquitination. The deubiquitinating activity of 2A-DUB also results in the destabilization of the linker histone H1, an event that also requires histone acetylation and that is generally associated with gene activation.
Usp16 functions both in the regulation of gene expression and cell cycle progression (153, 161, 173). Early studies showed wild type Usp16 localizes to the cytoplasm; however an active site mutant of the enzyme was shown to associate with chromosomes during mitosis and to remain in the nucleus after mitosis, suggesting a chromatin associated function (161). Consistent with a role in chromatin functions, Usp16 was shown to deubiquitinate H2A in vitro and overexpression of Usp16 lead to a decrease in the levels of uH2A (161, 173). More recently, Joo et al. (153) demonstrated that Usp16 removes ubiquitin from H2A within nucleosomes, and that immunodepletion or knockdown of Usp16 results in increased uH2A levels. Normally uH2A levels decrease upon entering mitosis, and subsequently increase as the cells exit mitosis. Knockdown of Usp16 leads to an increase in uH2A and a decrease in the number of cells undergoing mitosis (153). Ubiquitination of H2A is inversely correlated with phosphorylation of H3 Ser10 by Aurora B kinase (153). Ubiquitination of H2A inhibits Aurora B kinase binding to nucleosomes where it is required for chromatin condensation, chromosome segregation, and progression through mitosis (Figure 4B). The physiological consequences of Usp16 depletion appear to be broader than simply interfering with mitosis and chromatin condensation. In Xenopus laevis Usp16 also functions to regulate the expression of the Hox gene required for anterior-posterior patterning (153). Knockdown of Usp16 causes a decrease in Hox expression and the rescue from this defect requires the deubiquitinating activity of Usp16. Given that previous studies indicate that H2A ubiquitination regulates Hox gene silencing, it is possible that Usp16 may repress Hox gene expression by modulating the levels of uH2A at the promoter and 5’ end regulatory region of the Hox gene (83, 153, 174, 175)
Finally, there is recent evidence that, in addition to a role in silencing, uH2A may also control transcriptional initiation. Usp21 is a third deubiquitinating enzyme shown to deubiquitinate H2A in humans (162). Usp21 was shown to regulate transcription in the liver during hepatocyte regeneration. This enzyme functions to relieve uH2A mediated repression of di- and tri-methylation of H3K4, thereby activating transcription initiation.
A unifying model
Figure 3 presents a unifying model for histone DUB activity. In general, ubiquitination of H2A or H2B is an epigenetic mark that controls the binding of other histone modifying enzymes, thus altering additional covalent modifications of other histones in the same or nearby nucleosomes. Often, the other modifying enzymes are macromolecular complexes containing the deubiquitinating activity and uH2A or uH2B are subsequently deubiquitinated. This allows sequential ubiquitination, factor recruitment and deubiquitination giving the reaction a directionality that enforces a defined sequence of events. One prediction of this model arises from an inspection of figure 4. For instance, this postulates that USP16 will be localized to mitotic chromatin by interacting with a protein that binds to S10 phosphorylated histone H3 or a subsequent downstream modification. The identity of such a putative binding protein is unknown.
RNAi reveals roles for DUBs in cell cycle regulation and DNA repair
Cell cycle regulation requires the timed degradation of numerous checkpoint and signaling molecules to allow an orderly progression through replication, growth and mitosis. Often these checkpoints involve surveillance of genome integrity and many mutations that affect cell cycle regulation and DNA repair are incompatible with the establishment of stable cell lines (23, 113, 153, 176). Thus, a more transient and acute “knockdown” is needed and the technique of RNAi has proven productive in revealing the role of DUBs in these processes (12, 59, 177, 178). A few illustrative examples are discussed below.
A search for DUBs that act on uH2A and uH2b implicated Usp3 (157). Wild type Usp3 localizes to the nucleus, where it interacts with chromatin. Mutation of the active cysteine results in the stabilization of the interaction between chromatin and Usp3, presumably by acting as a substrate trap. In addition to its catalytic core domain, Usp3 harbors a ZnF Ubp domain in its N-terminus. This putative ubiquitin binding domain is required to mediate binding to uH2A (157). Usp3 can deubiquitinate both H2A and H2B in humans (157). Overexpression of Usp3 in HeLa cells causes the reduction in both uH2A and uH2B without altering the bulk pool of ubiquitinated proteins (157). Depletion of Usp3 by RNAi lead to a increase in the levels of uH2A and to a lesser extent uH2B demonstrating that it regulates the level of histone ubiquitination in vivo (157). The functional consequences of depleting Usp3 by RNAi include: a delay in S phase progression and decreased incorporation of BrdU suggesting a defect in replication; and the accumulation of DNA breaks, which leads to an induction of the Ataxia Telangiectasia mutated (ATM) and ATM/Rad3 related (ATR) checkpoint kinases that regulate DNA damage response pathways. Together these observations suggest that knockdown of Usp3 causes defects in replication that cause activation of the ATM/ATR DNA damage response and delays progression though the S-phase (157).
In a study of DNA damage induced apoptosis, Usp28 was shown to be involved in the checkpoint kinase 2 (Chk2)-p53-PUMA pathway, a major regulator of DNA-damage-induced apoptosis, in response to double-strand breaks in vivo (179). Usp28 was originally found to physically interact with the checkpoint mediators 53BP1, Claspin and Mdc1 (179). These scaffolding proteins mediate DNA-damage responses that result from ionizing radiation (IR). Knockdown of Usp28 leads to a decrease in the levels of all three interacting proteins as well as the checkpoint kinase Chk2, suggesting that USP28 functions to stabilize these proteins. Both Claspin and Mdc1 appear to be substrates but there is no evidence that 53BP1 is ubiquitinated, bringing up the possibility that association with the 53BP1 scaffolding protein is important for USP28 function. The absence of Usp28 or of its catalytic activity causes attenuation of DNA damage signals and inactivation of p53 mediated apoptosis. Knockdown of Usp28 leads to resistance in IR induced apoptosis similarly to what had been observed with p53, chk2 and PUMA null mice, again consistent with a role in p53 dependent apoptosis. Usp28 also regulates the G2 DNA-damage-response checkpoint (180). In DNA-damaged G2 cells, Claspin is ubiquitinated by the anaphase promoting complex and targeted for proteasomal degradation. Usp28 counteracts this process by deubiquitination of Claspin allowing it to activate Chk1 in response to DNA damage (180).
Usp28 also controls the cellular levels of the transcription factor MYC (115). MYC is a proto-oncogene that regulates cell growth, apoptosis and cell proliferation. MYC is degraded in a ubiquitin dependent manner by the E3 ligase FBW7 (F-box and WD repeat-domain containing 7) that is a component of a SCF (complex of SKP1, CUL1 and F-box protein)-type of ubiquitin ligases. MYC is ubiquitinated by the FBW7γ isoform in the nucleolus and by the FBW7α isoform in the nucleoplasm. Together they control the growth promoting activity of MYC. MYC ubiquitination catalyzed by the FBW7α isoform in the nucleoplasm, is antagonized by Usp28 DUB activity. Knockdown of Usp28 results in a decrease of c-MYC while overexpression of the enzyme and leads to stabilization of the transcription factor. The effect is limited to FBW7α because Usp28 specifically interacts with FBW7α and this interaction mediates complex formation between MYC and Usp28. Since FBW7γ lacks the USP28 interaction motif of FBW7α, Usp28 selectively antagonize nucleoplasmic MYC degradation while not affecting nucleolar degradation. Given that the catalytic activity of Usp28 is required for MYC stabilization (179), inhibition of this DUB may be of pharmacological interest.
A directed RNAi screen of all USP family DUBs revealed that Usp1 is involved in DNA repair processes through its deubiquitination of the Fanconi anemia protein FANCD2 (178, 181) and PCNA (182). Usp1 and FANCD2 were found to interact in vivo and co-localize on chromatin after DNA damage (178). Knockdown of Usp1 by small interfering RNA lead to hyper-accumulation of monoubiquitinated FANCD2 (178). Usp1 is proposed to deubiquitinate FANCD2 when cells exit S phase or recommence cycling after DNA damage and may play a critical role in the FA pathway by recycling FANCD2 (49, 178). USP1 is found in a macromolecular complex with the 80kDa WD40 repeat containing protein USP1-associated factor 1 (UAF1) (183). The formation of this heterodimeric complex greatly activates Usp1, suggesting a role for UAF1 as a scaffold. Transcription of USP1 is terminated in response to DNA damage and the protein disappears rapidly. Interestingly, USP1 is subject to autocatalytic cleavage at an internal Gly-Gly sequence and it is possible that this leads to a dissociation of the complex and consequent degradation of the Usp1 fragments (182).
Finally, Usp11 has been described as a DUB that exhibits pro-survival functions as part of the cellular response to DNA damage (184). In response to Mitomycin C (MMC)-induced DNA damage, Usp11 participates in DNA damage repair functions within the breast cancer 2 (BRCA2) pathway, but does so independently of BRCA2 deubiquitination (184). The ubiquitination and degradation of BRCA2 has been implicated in prostate cancer cell proliferation. This down-regulation of BRCA2 levels requires phosphatidylinositol (PI) 3-kinase signaling and up-regulation of Skp2, a subunit of the Skp1-Cul1-F-box protein ubiquitin complex (185).
DUBs modulate kinase cascades of signal transduction pathways
Ubiquitination modulates signal transduction pathways by at least two mechanisms. First, K63-linked polyubiquitin chains serve as recognition signals that recruit adapters and signaling kinases to propagate the signals, and secondly ubiquitination contributes to regulation of these pathways by controlling the half-life of signaling complexes and components. The best-studied example is the nuclear factor kappa B (NF-κB) pathway responsible for propagating signals from tumor necrosis factor (TNF), interleukin-1/TLR (TOLL-like receptors), and T- and B- cell antigen receptors to the nucleus to control immune, inflammatory and anti-microbial responses (29, 186). Deubiquitination downregulates these responses and defects in at least two DUBs result in sustained signaling. We will briefly discuss this pathway as an example of the regulation of signaling cascades.
Ubiquitination and NF-κB signaling
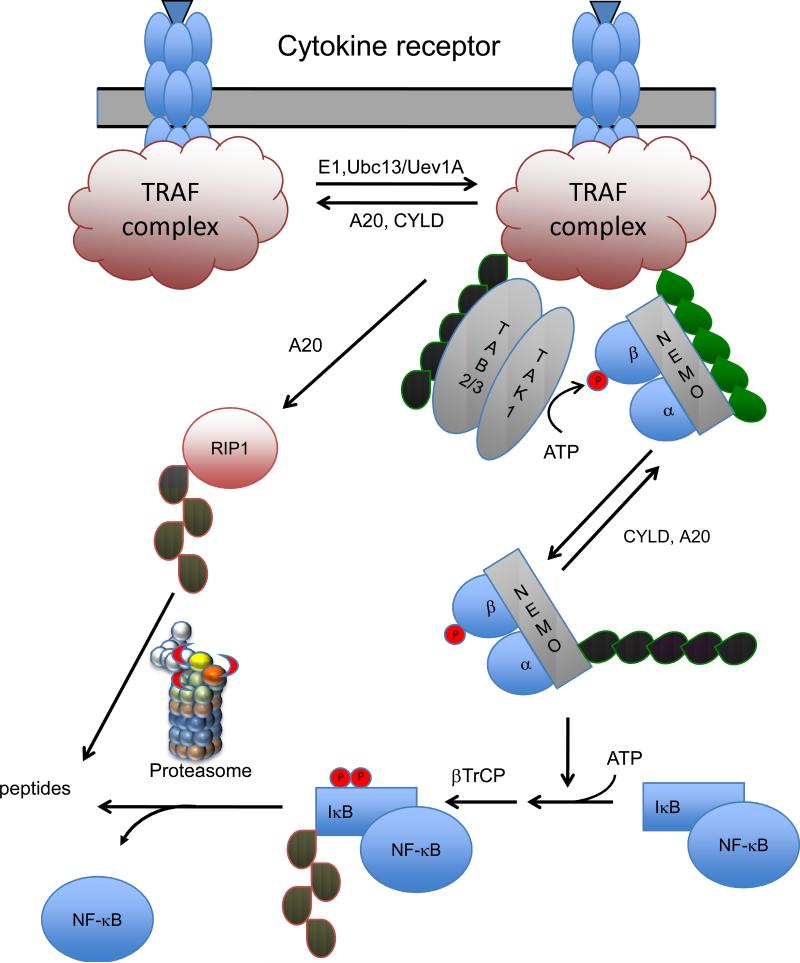
The role of regulatory ubiquitination in the NF-κB pathway was first suggested by the finding that enzymes catalyzing K63-linked polyubiquitination were necessary for efficient signaling through the NF-κB pathway (187). These chains do not target proteins for degradation by the proteasome and so another function was suggested. However, K48-linked ubiquitination is involved as it is necessary to trigger the degradation of IκB, an inhibitor of NF-κB that retains the transcription factor in the cytoplasm, and to target the degradation of the TNF receptor interacting protein 1 (RIP1) during downregulation of the signaling pathway. Finally, the discovery two other inhibitors of signaling, A20 (76)and CYLD (188, 189), are deubiquitinating enzymes has emphasized the importance of these modifications. A general model of activation of the NF-κB pathway is given in Figure 5. The details of this regulation vary depending on the particular receptor triggering the signaling events (29, 186, 190, 191). In general however, ubiquitin dependent regulation of the NF-κB pathway consists of several steps: 1) receptor engagement activates members of the TRAF (TNF receptor-associated factor) family of E3 ligases to assemble a K63-linked polyubiquitin chain on itself (autoubiquitination) or an associated protein such as RIP1; 2) the K63-linked polyubiquitin then recruits the TAK1/TAB2/3 protein kinase and its substrate IκB Kinase (IKK); 3) This assembled complex catalyzes the phosphorylation of the β subunit of IKK by the TAK1/TAB2/3 protein kinase and the K-63 polyubiquitination of NEMO (NF-κB essential modulator, the regulatory γ subunit of IKK) by the TRAF ligases; 4) the phosphorylated and ubiquitinated IKK is now active to phosphorylate IκB resulting in its K48-polyubiquitination by β-TrCP (beta-transducin repeat-containing protein) to trigger its degradation by the proteasome. 5) Finally, the removal of the K63-linked polyubiquitination by the DUB A20 downregulates the signaling response and the subsequent K48-linked ubiquitination of RIP1 by A20 triggers its degradation by the proteasome (Figure 5).
Figure 5.
A generalized model for signaling through the NF-κB pathway. Upon ligand binding to receptors for Tumor Necrosis Factor, Interleukins, Toll ligand, or T- and B-cell antigens a signaling complex is assembled. This complex contains TRAFs (TNF receptor associated factors) that catalyze the formation of a K63-linked polyubiquitin chain (green) on themselves and/or other proteins in the complex. The K63-linked polyubiquitin then recruits the TAB2/3-TAK1 kinase and its substrate IκB kinase (consisting of kinases IKKα, IKKβ, and the regulatory subunit NEMO). Subsequently the phosphorylation of IKKβ and the K63 polyubiquitination of NEMO activates the IKK activity. IKK phosphorylates IκB triggering its K48 polyubiquitination (red) by βTRCP and its degradation by the proteasome. The liberated NF-κB then enters the nucleus and acts as a transcription factor activating genes involved in inflammation and immune responses. The signaling is downregulated by two K63 specific DUBs, A20 and CYLD and defects in either lead to prolonged signaling. RIP1, a component of the TRAF complex assembled upon TNFR1 signaling is one of the targets for K63 polyubiquitination. K63 polyubiquitinated RIP1 is deubiquitinated by A20. This DUB is also a ubiquitin ligase that then assembles a K48 polyubiquitin chain on RIP1 leading to its degradation by the proteasome.
K63-specific DUBs inhibit and downregulate NF-κB signaling
When overexpressed, the DUB A20 is an inhibitor of TNF mediated cell death as well as the NF-κB pathway (191, 192). A20 expression is induced by TNF and required for the downregulation of signaling. Its involvement in ubiquitin-dependent regulation was suggested by the presence of both an OTU DUB domain and a ring finger ubiquitin ligase domain. A20 deficient mice are prone to inflammation and fail to downregulate the NF-κB response due to a constitutively active IKK. A20 binds to NEMO, RIP1, and TRAFs and its DUB activity results in the deubiquitination of these proteins, inhibiting the NF-κB response by preventing the TRAF-mediated ubiquitination of NEMO. The presence of a ring finger E3 ligase domain in A20 is reminiscent of the tendency of DUBs and E3s to associate non-covalently. The combined DUB and E3 ligase activity of A20 results in the remodeling of the polyubiquitin chains on its substrates (191). The DUB activity first removes the activating K63 polyubiquitin chain and the ligase subsequently assembles a K48-linked chain that targets these proteins for degradation by the proteasome. Thus, the absence of A20 results in hyper-accumulation of K63 polyubiquitin on these signaling components and a failure to degrade them as part of the normal downregulation of the response.
Another DUB, CYLD, was identified as an inhibitor of NF-κB signaling in an siRNA screen searching for DUB functions and as a protein whose mutation causes a benign tumor syndrome called cylindromatosis (188, 189). CYLD binds to and deuibuquitinates K63 polyubiquitinated TRAF2 and TRAF6. CYLD interacts directly with TRAF2, an adaptor molecule involved in signaling by members of the family of TNF/nerve growth factor receptors (188, 189). CYLD also negatively regulates TRAF7, a recently identified TRAF family member, likely via a deubiquitination-dependent mechanism (112). Like A20, CYLD inhibits signaling by disassembling the K63 polyubiquitin recruiting signal. CYLD also deubiquitinates IκB kinase gamma (IKKγ, also known as NEMO), the regulatory subunit of IKK (188, 189) thus inhibiting the activation of IKK. The two proteins interact via the third cytoskeleton-associated protein-glycine conserved (CAP-Gly) domain of CYLD and a proline-rich sequence on IKKγ (193). CYLD also negatively regulates the IKK-related kinases, IKKε and TBK1, by the same mechanisms (194). Because CYLD deficiency causes constitutive activation of IKKε and TBK1, this DUB has been suggested to play an essential role in preventing the aberrant activation of these kinases during the induction of type1 interferon that occurs during viral infection (194).
Key to these regulatory pathways is the specificity of the DUBs for K63-linked polyubiquitin. Recently, the crystal structure of CYLD has been solved and it reveals two features that contribute to the specificity of CYLD (66). First, the finger domain of the USP core is truncated allowing facile interactions with polyubiquitin. Secondly, compared to other USPs, a CYLD-specific insertion in a loop generates a surface that is important for activity on with K63 polyubiquitin. The structure of A20 has been solved by two groups (73, 78), both of whom use model building to highlight common surfaces that might be involved in the specific binding of K63-linked chains. In vitro studies showed that A20 cleaves both free K48- and K63-linked chains, while when chains are attached to a protein only K63 is cleaved rapidly. Intact K63 chains are released via an endo-cleavage (chain amputation) whereas K48-liked chains attached to a protein are cleaved from the distal end (chain trimming). It is suggested that this preference arises from the additional interactions between substrate and enzyme that position the DUB for efficient cleavage near the attachment site for polyubiquitin (195),
Variations on the theme
The IKK complexes contain at least two kinases, IKKα and IKKβ, which are separately regulated. IKKα for instance is involved in the regulation of responses to a subset of TNF family members and in controlling expression of p53. Additional IKK-related complexes also couple other signaling pathways to NF-κB (196). The expression of the alpha subunit of IKK (IKKα) is regulated by the DUB USP11. Inhibition of USP11 expression reduces p53 levels and IKKα expression, as well as increasing the response of the NF-κB pathway to TNFα (197). It is not known whether this regulation involves K63, K48, or other forms of polyubiquitin.
Similar combinations of the canonical K48 polyubiquitin degradation signal and/or other non-canonical forms may regulate other signaling pathways. Activation of AMPK (AMP-activated protein kinase)-related kinases by the master kinase LKB1 is inhibited by ubiquitination of the AMPK-related kinases, apparently via unusual K29/K33 polyubiquitin chains, rather than the more common K48/K63 linkages (198, 199). Many AMPK-related kinases contain UBA domains (that in other proteins function as ubiquitin binding modules) and the binding of AMPK-related kinases to LKB1 requires the UBA domain. Thus, the UBA domain of AMPK-related kinases may associate intramolecularly with the polyubiquitin chain preventing AMPK-related kinases from interacting with the LKB1 kinase. Deubiquitination would free the UBA domain for participation in LKB1 kinase binding. USP9 associates with and stimulates the LKB1-mediated phosphorylation of two AMPK kinases: MARK4 (microtubule-affinity-regulating kinase 4) and NUAK1 (AMPK-related kinase 5) through its deubiquitinating activity (198).
Endocytosis
The involvement of the ubiquitin system in endocytosis has long been recognized (25, 51, 200, 201). Monoubiquitination (primarily in yeast), and the attachment of K63-linked Ub oligomers in mammalian proteins, play an important role in endocytosis of receptors and sorting of endocytosed proteins (200, 201). DUBs are implicated in the endocytic pathway at multiple levels and also play important roles in other types of intracellular traffic. For example, in addition to regulating the kinase activity of several NF-κB-related kinases, the DUB CYLD has also been shown to control other seemingly disparate cellular processes involving K63-linked polyubiquitination, including endocytosis of the NGF receptor, TrkA (190).
In yeast, the DUB Doa4 acts to recycle Ub at the late endosome (202). Doa4 is recruited to the late endosome after assembly of the ESCRT-III (endosomal sorting complex required for transport III). The N-terminus of Doa4 interacts with, and it is activated by, the class E Vps protein Bro1 on endosomal membranes. The recruitment of Doa4 to endosomes requires Bro1 and inactivation of Bro1 or Doa4 interferes with sorting of integral membrane proteins to the multivesicular body (203). Other Ub-related processes are also inhibited since interfering with DOA4 function results in the internalization of ubiquitinated receptors by the vacuole and depletion of free Ub. Many of the defects observed on Doa4 mutant cells are restored upon expression of additional Ub (202). Usp8 (also known as UBPY) is the closest human ortholog of Doa4 and is regulated by phosphorylation and 14−3−3 binding (204). Usp8 regulation of endocytic traffic requires its interaction with the ESCRT machinery and it inhibits EGF receptor (EGFR) endocytosis (205-207).
AMSH, is a JAMM domain DUB that also deubiquitinates cargo at the MVB, However, as described above, AMSH appears to be specific for K63-linked chains. Inhibition of AMSH accelerates EGFR down-regulation suggesting a role in antagonizing ubiquitin-dependent sorting of cargo to the lysosome (88). Growth factor-activated receptor tyrosine kinases (RTKs) such as EGFR are ubiquitinated in response to ligand engagement and undergo rapid endocytosis and degradation in lysosomes (25). On the endosomal membrane, ubiquitinated RTKs are sorted by the coordinated actions of the class E vacuolar protein sorting (Vps) proteins. Like Doa4 in yeast, mammalian Usp8 and AMSH associate with class E Vps proteins on endosomes. Thus, RTK downregulation appears to be controlled not only by ubiquitination but also by deubiquitination of RTKs and other endosomal proteins (25). AMSH and USP8 exert opposite effects on the rate of epidermal growth factor receptor downregulation. This may be due to their distinct specificities for different types of polyubiquitin chain linkage. It has been suggested that AMSH might rescue ubiquitinated cargo from lysosomal degradation through disassembly of K63-linked polyubiquitin chains. USP8 function is essential for effective downregulation but is likely to be multifaceted, encompassing activity against both K63-linked and K48-linked polyubiquitin chains (51).
In Drosophila, Fat facets (Faf; the ortholog of human Usp9X), deubiquitinates Liquid facets (Lqf), resulting in enhanced Lqf activity and levels (208). The human homologs of Lqf are known as epsins, adaptor proteins involved in the initial steps of endocytosis (12). Lqf and Faf play a role in Drosophila eye development by enhancing the internalization of the Delta receptor implicated in cell patterning (208).
Quantitative assays of activity and binding
The above examples point out that the effects of a given DUB will be determined by its substrate specificity. One of the biggest hurdles to understanding specificity of DUBs and the significance of their protein interactions is the qualitative nature of most available assays. Protein interaction assays have been often validated by co-overexpressing a DUB and an interaction partner in cells and showing that there is some degree of association in pull-down assays. These can be criticized as simply using the cell as a “test tube”, and in general do not address the question of whether the interactions occur at physiological concentrations and localization. This problem may be amplified when the DUB contains an interaction module that normally recognizes a domain in its substrate that may also be present in other proteins. A module that recognizes a single ring finger in a specific E3 partner may bind several other ring fingers from other ligases when they are co-expressed at high concentrations. Another limitation to keep in mind is that, by their nature, pull down assays do not measure simple affinity; the rates of dissociation must be slow in order for the complex to survive the pull down and wash conditions. DUB substrate interactions may be considerably less stable than DUB scaffold interactions as the former are expected to be rather transient because the enzyme must be able to release its products.
Substrates
Early assays for activity involved long incubations with unknown amounts of enzyme preparations or co-expression of the substrate and enzyme in cells. Thus, the observation that an enzyme preparation can cleave a particular substrate is difficult to interpret if a rate cannot be determined. Many DUBs have been shown to act on ubiquitinated proteins or ubiquitin AMC substrates but at rates that are probably too slow to be of real significance. Both UCH37 and Usp14 will hydrolyze Ub-AMC, albeit very slowly. When bound to the proteasome however, the rate of hydrolysis is increased hundreds of fold (103, 145). A simple “end point” assay that only determined whether a substrate could or could not be cleaved would not have revealed this physiologically important activation. Further, several DUBs have been shown to hydrolyze more than one ubiquitin-like proteins. IsoT can hydrolyze ubiquitin, Nedd8 and ISG15 derivatives ((209), Gan-Erdene and Wilkinson, unpublished), SENP8 hydrolyzes ubiquitin and Nedd8 derivatives (19), and many SENPs act on both SUMO1 and SUMO2/3 (210, 211). Understanding their cellular capabilities requires quantitative assays with specific substrates. Fluorescent derivatives of all the ubiquitin-like (UBL) proteins with aminomethyl coumarin or rhodamine leaving groups can be conveniently produced using the ubiquitin-intein technology (212). Assays using these substrates can be easily quantitated and conventional steady-state kinetic approaches can be applied (98, 213). A variety of UBL protein fusions have also been used, although most require SDS-PAGE to visualize cleavage and quantitation is difficult (46, 214, 215). Various fluorescence energy transfer approaches could be used but are highly specialized and difficult to use. Finally, an enzyme-linked assay has been described where a fusion protein between ubiquitin and an enzyme is used as the substrate (216). The fusion protein has minimal enzymatic activity and is significantly activated upon the cleavage of the ubiquitin-enzyme junction by DUBs. This assay is also challenging to quantitate because it is a coupled assay, but it will probably be useful in high-throughput screens for inhibitors and activators of DUB action.
Active site-directed irreversible inhibitors as probes of catalytic capability
One of the more useful reagents that can be applied to determine the specificity of DUBs, both purified and in crude extracts, is a family of UBL-X derivatives where X is an electrophile. Ubiquitin vinylsulfone (Ub-VS) was first synthesized to specifically label ubiquitin specific DUBs (140), This reagent reacts specifically with the active site of DUBs where the catalytic thiol can react with the electrophile to covalently label the DUB. If the UBL also contains an epitope tag the labeling can be detected by SDS-PAGE and immunoblotting and is thus useful to inventory the complement of DUBs present in lysates or crude extracts (22). These reagents show little or no reactivity with other proteins since the electrophiles used are not very reactive and require a suitable nucleophile nearby before crosslinking can occur. Thus, incubation with low levels of UBL-X for a few minutes is sufficient to label many DUBs, These reagents must be used with caution however, since incubations with large amounts of the UBL-X reagent or longer reaction times can result in spurious labeling of DUBs that actually prefer another UBL. Both the identity of the UBL domain and the nature of the electrophile determine the specificity of this reaction (22), demonstrating that DUBs have significant specificity for the UBL domain (Ub vs. Nedd8 vs. ISG15, etc.) and that differences in the active site environment can be exploited to develop DUB specific reagents. The latter observation suggests that it may be possible to develop specific small molecule inhibitors of different DUBs.
Use of substrate-based reagents to define the specificity of a DUB
One example of the use of these substrates and active site directed inhibitors is instructive. DEN1 (Deneddylating enzyme 1) is a member of the ULP-family of DUBs that was originally thought to be SUMO-specific (19). Using the substrates and inhibitors described above we showed that it was a Nedd8-specific DUB with weak activity as a Ub-specific DUB. In a search for Nedd8-specific DUBs we reacted cell lysates with FLAG-Nedd8-VS. Analysis by SDS-PAGE showed that two proteins were rapidly labeled. Using the FLAG epitope we immunopurified the covalently labeled forms of both. The major protein was UCH-L3 (see above) and the minor labeled protein proved to be identical to a member of the ULP DUB family, SENP8. UCH-L3 was labeled more strongly because it is present at much higher concentrations than was DEN1. A steady state kinetic analysis showed that DEN1 bound Nedd8 much tighter than it bound Ub and that the catalytic capability (kcat/Km) of DEN1 was five orders of magnitude greater for Nedd8 substrates than for Ub substrates. Conversely, UCH-L3 greatly preferred to bind and catalyze the hydrolysis of Ub over that of Nedd8. Without these quantitative comparisons it would have been easy to conclude that UCH-L3 was a very good deneddylating enzyme when in fact, labeling was largely due to the fact that its abundance overwhelmed the signal from the more specific but less abundant DEN1. (19).
Small molecule inhibitors
While RNAi and genetic studies have been invaluable in defining the physiological roles of DUBs it would prove useful to have small molecule inhibitors that could be used to achieve rapid and specific inhibition of selected DUBs. This is because RNAi methods require considerable time to achieve ablation of activity and because cells lacking specific DUBs may not be available or viable. Further, some of these inhibitors could prove to be useful drugs in treating various pathological conditions. The development of such small molecules is just beginning and there is still some question about whether DUBs are “druggable”. Inhibitors of UCH-family DUBs have been reported with modest selectivity and affinity (177, 217), and J-series prostaglandins have been reported to inhibit IsoT (218). There has been more progress in developing good inhibitors of the SARS coronavirus DUB PLpro and a family of inhibitors with sub-micromolar IC50 values have been reported and shown to interfere with viral propagation (40). These preliminary efforts are encouraging and suggest the possibility of developing small molecule inhibitors with useful selectivity profiles.
Concluding remarks
It is evident that the reversal of ubiquitination by deubiquitinating enzymes plays an important role in regulating diverse cellular processes including 1) the maintenance of steady state levels of monoubiquitin, 2) regulation of substrate degradation at the proteasome, 3) chromatin remodeling through histone deubiquitination and the subsequent effects on other histone modifications, 4) cell cycle regulation, 5) DNA damage repair processes 6) activation of kinases and other enzymes and 7) endocytosis. DUBs are highly specific and subject to multiple modes of regulation, including transcriptional control, substrate or scaffold induced activation, post-translational modifications, and rapid degradation. DUBs are often found as part of large multi-protein complexes that function to regulate their localization, substrate availability and activity in cellular processes. As a result, DUBs have emerged as important pharmacologic targets in the in the quest to develop more specific agents to treat various diseases. It is becoming evident that DUBs are involved in regulating almost every cellular process and as the body of knowledge grows, DUBs may prove to be important targets of therapeutic drug action.
Article Components
Terms/Definitions list:
Ubiquitination. The covalent attachment of ubiquitin, a 76 amino acid protein, to a target protein via formation of an isopeptide bond between the C-terminal Gly of ubiquitin and the e-amino group lysine in the target protein or another ubiquitin.
Deubiquitination. The hydrolysis of the isopeptide bond between ubiquitin and itself or a target protein.
Deubiquitinating enzyme. Enzymes that catalyze deubiquitination.
Polyubiquitin. Polymer of ubiquitin linked by isopeptide bonds to itself. All seven lysines of ubiquitin may participate.
Proubiquitin. Primary gene products where the C-terminus of one ubiquitin is attached to the N-terminus of itself or another protein in a peptide bond.
Distal subunit of polyubiquitin. The subunit attached to the target protein or containing a free C-terminus.
Proximal subunit of polyubiquitin. The subunit most distant from the proximal subunit.
Acronyms list:
DUB. Deubiquitinating Enzyme
UCH. A DUB family with the Ubiquitin C-terminal Hydrolase domain (Pfam PF01088, EC:3.4.19.12)
USP. A DUB family with the Ubiquitin specific protease domain (Pfam PF00443, EC 3.1.2.15)
OTU. A DUB family with the Ovarian Tumor Domain (Pfam PF02338, EC 3.1.2.-)
MJD. A DUB family with the Machado-Joseph Disease protein Domain (Pfam PF02099, EC 3.4.22.-)
JAMM. A DUB family with the Jab1/MPN metalloenzyme domain (Pfam PF01398, EC 3.1.2.15)
E1. Ubiquitin activating enzyme responsible for adenylation and subsequent thiolesterification of the C-terminus of ubiquitin.
E2. Ubiquitin conjugating enzyme that acts as an acyl-carrier protein by forming a thiol ester bond with the C-terminus of ubiquitin. Some E2s can catalyze transfer to substrates directly while others require the participation of an E3 ligase.
E3. Ubiquitin ligase that mediates the ubiquitination of target proteins by facilitating the attack of a lysine side chain on the E2∼Ubiquitin thiol ester.
Summary Points list:
The large number of DUBs suggests considerable specialization and specificity.
Activity is controlled conformational changes induced by DUB binding to substrate or to other binding partners.
Localization of DUBs is often controlled by binding to adapters or scaffolds.
Co-localization with substrates by binding to adapters is an important part of limiting specificity.
Some substrates can be acted upon by several DUBs.
Because of the high specificity of DUBs they are attractive targets for drug development.
Future Issues list:
What is the nature of conformational changes that activate DUBs?
Using siRNA, genetics and interaction data can we define the role of individual DUBs in cellular regulation?
What is the role of microbial DUBs in infectivity and pathology?
Is it possible to develop “specific” inhibitors of DUBs that might be useful probes of function or drugs to treat disease?
References
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Hicke L. Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 3.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 4.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–6. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–33. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 6.Fang S, Weissman AM. A field guide to ubiquitylation. Cell Mol Life Sci. 2004;61:1546–61. doi: 10.1007/s00018-004-4129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilkinson KD. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. Faseb J. 1997;11:1245–56. doi: 10.1096/fasebj.11.14.9409543. [DOI] [PubMed] [Google Scholar]
- 8.Baker RT, Board PG. The human ubiquitin gene family: structure of a gene and pseudogenes from the Ub B subfamily. Nucleic Acids Res. 1987;15:443–63. doi: 10.1093/nar/15.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozkaynak E, Finley D, Solomon MJ, Varshavsky A. The yeast ubiquitin genes: a family of natural gene fusions. EMBO J. 1987;6:1429–39. doi: 10.1002/j.1460-2075.1987.tb02384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiborg O, Pedersen MS, Wind A, Berglund LE, Marcker KA, Vuust J. The human ubiquitin multigene family: some genes contain multiple directly repeated ubiquitin coding sequences. EMBO J. 1985;4:755–9. doi: 10.1002/j.1460-2075.1985.tb03693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pickart CM, Rose IA. Ubiquitin carboxyl-terminal hydrolase acts on ubiquitin carboxyl-terminal amides. J Biol Chem. 1985;260:7903–10. [PubMed] [Google Scholar]
- 12.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–86. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Wilkinson KD, Tashayev VL, O'Connor LB, Larsen CN, Kasperek E, Pickart CM. Metabolism of the polyubiquitin degradation signal: structure, mechanism, and role of isopeptidase T. Biochemistry. 1995;34:14535–46. doi: 10.1021/bi00044a032. [DOI] [PubMed] [Google Scholar]
- 14.Piotrowski J, Beal R, Hoffman L, Wilkinson KD, Cohen RE, Pickart CM. Inhibition of the 26 S proteasome by polyubiquitin chains synthesized to have defined lengths. J Biol Chem. 1997;272:23712–21. doi: 10.1074/jbc.272.38.23712. [DOI] [PubMed] [Google Scholar]
- 15.Lindner HA. Deubiquitination in virus infection. Virology. 2007;362:245–56. doi: 10.1016/j.virol.2006.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rytkonen A, Holden DW. Bacterial interference of ubiquitination and deubiquitination. Cell Host Microbe. 2007;1:13–22. doi: 10.1016/j.chom.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Ha BH, Kim EE. Structures of proteases for ubiqutin and ubiquitin-like modifiers. BMB Rep. 2008;41:435–43. doi: 10.5483/bmbrep.2008.41.6.435. [DOI] [PubMed] [Google Scholar]
- 19.Gan-Erdene T, Nagamalleswari K, Yin L, Wu K, Pan ZQ, Wilkinson KD. Identification and characterization of DEN1, a deneddylase of the ULP family. J Biol Chem. 2003;278:28892–900. doi: 10.1074/jbc.M302890200. [DOI] [PubMed] [Google Scholar]
- 20.Li SJ, Hochstrasser M. A new protease required for cell-cycle progression in yeast. Nature. 1999;398:246–51. doi: 10.1038/18457. [DOI] [PubMed] [Google Scholar]
- 21.Li SJ, Hochstrasser M. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol Cell Biol. 2000;20:2367–77. doi: 10.1128/mcb.20.7.2367-2377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Love KR, Catic A, Schlieker C, Ploegh HL. Mechanisms, biology and inhibitors of deubiquitinating enzymes. Nat Chem Biol. 2007;3:697–705. doi: 10.1038/nchembio.2007.43. [DOI] [PubMed] [Google Scholar]
- 23.Song L, Rape M. Reverse the curse--the role of deubiquitination in cell cycle control. Curr Opin Cell Biol. 2008;20:156–63. doi: 10.1016/j.ceb.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guterman A, Glickman MH. Deubiquitinating enzymes are IN/ (trinsic to proteasome function). Curr Protein Pept Sci. 2004;5:201–11. doi: 10.2174/1389203043379756. [DOI] [PubMed] [Google Scholar]
- 25.Komada M. Controlling receptor downregulation by ubiquitination and deubiquitination. Curr Drug Discov Technol. 2008;5:78–84. doi: 10.2174/157016308783769469. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt M, Hanna J, Elsasser S, Finley D. Proteasome-associated proteins: regulation of a proteolytic machine. Biol Chem. 2005;386:725–37. doi: 10.1515/BC.2005.085. [DOI] [PubMed] [Google Scholar]
- 27.Daniel JA, Grant PA. Multi-tasking on chromatin with the SAGA coactivator complexes. Mutat Res. 2007;618:135–48. doi: 10.1016/j.mrfmmm.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kennedy RD, D'Andrea AD. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev. 2005;19:2925–40. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- 29.Adhikari A, Xu M, Chen ZJ. Ubiquitin-mediated activation of TAK1 and IKK. Oncogene. 2007;26:3214–26. doi: 10.1038/sj.onc.1210413. [DOI] [PubMed] [Google Scholar]
- 30.Catic A, Misaghi S, Korbel GA, Ploegh HL. ElaD, a Deubiquitinating protease expressed by E. coli. PLoS ONE. 2007;2:e381. doi: 10.1371/journal.pone.0000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Negrate G, Faustin B, Welsh K, Loeffler M, Krajewska M, et al. Salmonella secreted factor L deubiquitinase of Salmonella typhimurium inhibits NF-kappaB, suppresses IkappaBalpha ubiquitination and modulates innate immune responses. J Immunol. 2008;180:5045–56. doi: 10.4049/jimmunol.180.7.5045. [DOI] [PubMed] [Google Scholar]
- 32.Le Negrate G, Krieg A, Faustin B, Loeffler M, Godzik A, et al. ChlaDub1 of Chlamydia trachomatis suppresses NF-kappaB activation and inhibits IkappaBalpha ubiquitination and degradation. Cell Microbiol. 2008;10:1879–92. doi: 10.1111/j.1462-5822.2008.01178.x. [DOI] [PubMed] [Google Scholar]
- 33.Misaghi S, Balsara ZR, Catic A, Spooner E, Ploegh HL, Starnbach MN. Chlamydia trachomatis-derived deubiquitinating enzymes in mammalian cells during infection. Mol Microbiol. 2006;61:142–50. doi: 10.1111/j.1365-2958.2006.05199.x. [DOI] [PubMed] [Google Scholar]
- 34.Rytkonen A, Poh J, Garmendia J, Boyle C, Thompson A, et al. SseL, a Salmonella deubiquitinase required for macrophage killing and virulence. Proc Natl Acad Sci U S A. 2007;104:3502–7. doi: 10.1073/pnas.0610095104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye Z, Petrof EO, Boone D, Claud EC, Sun J. Salmonella effector AvrA regulation of colonic epithelial cell inflammation by deubiquitination. Am J Pathol. 2007;171:882–92. doi: 10.2353/ajpath.2007.070220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arguello MD, Hiscott J. Ub surprised: viral ovarian tumor domain proteases remove ubiquitin and ISG15 conjugates. Cell Host Microbe. 2007;2:367–9. doi: 10.1016/j.chom.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 37.Barretto N, Jukneliene D, Ratia K, Chen Z, Mesecar AD, Baker SC. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J Virol. 2005;79:15189–98. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iyer LM, Koonin EV, Aravind L. Novel predicted peptidases with a potential role in the ubiquitin signaling pathway. Cell Cycle. 2004;3:1440–50. doi: 10.4161/cc.3.11.1206. [DOI] [PubMed] [Google Scholar]
- 39.Kattenhorn LM, Korbel GA, Kessler BM, Spooner E, Ploegh HL. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol Cell. 2005;19:547–57. doi: 10.1016/j.molcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 40.Ratia K, Pegan S, Takayama J, Sleeman K, Coughlin M, et al. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc Natl Acad Sci U S A. 2008;105:16119–24. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schlieker C, Korbel GA, Kattenhorn LM, Ploegh HL. A deubiquitinating activity is conserved in the large tegument protein of the herpesviridae. J Virol. 2005;79:15582–5. doi: 10.1128/JVI.79.24.15582-15585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sompallae R, Gastaldello S, Hildebrand S, Zinin N, Hassink G, et al. Epstein-barr virus encodes three bona fide ubiquitin-specific proteases. J Virol. 2008;82:10477–86. doi: 10.1128/JVI.01113-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sulea T, Lindner HA, Purisima EO, Menard R. Deubiquitination, a new function of the severe acute respiratory syndrome coronavirus papain-like protease? J Virol. 2005;79:4550–1. doi: 10.1128/JVI.79.7.4550-4551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fischer JA. Deubiquitinating enzymes: their roles in development, differentiation, and disease. Int Rev Cytol. 2003;229:43–72. doi: 10.1016/s0074-7696(03)29002-1. [DOI] [PubMed] [Google Scholar]
- 45.Jiang YH, Beaudet AL. Human disorders of ubiquitination and proteasomal degradation. Curr Opin Pediatr. 2004;16:419–26. doi: 10.1097/01.mop.0000133634.79661.cd. [DOI] [PubMed] [Google Scholar]
- 46.Shanmugham A, Ovaa H. DUBs and disease: activity assays for inhibitor development. Curr Opin Drug Discov Devel. 2008;11:688–96. [PubMed] [Google Scholar]
- 47.Liz MA, Sousa MM. Deciphering cryptic proteases. Cell Mol Life Sci. 2005;62:989–1002. doi: 10.1007/s00018-005-4544-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reyes-Turcu FE, Wilkinson KD. Polyubiquitin binding and disassembly by deubiquitinating enzymes. Chemical Reviews. 2009 doi: 10.1021/cr800470j. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ventii KH, Wilkinson KD. Protein partners of deubiquitinating enzymes. Biochem J. 2008;414:161–75. doi: 10.1042/BJ20080798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu K, Yamoah K, Dolios G, Gan-Erdene T, Tan P, et al. DEN1 is a dual function protease capable of processing the C terminus of Nedd8 and deconjugating hyper-neddylated CUL1. J Biol Chem. 2003;278:28882–91. doi: 10.1074/jbc.M302888200. [DOI] [PubMed] [Google Scholar]
- 51.Clague MJ, Urbe S. Endocytosis: the DUB version. Trends Cell Biol. 2006;16:551–9. doi: 10.1016/j.tcb.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 52.Soboleva TA, Baker RT. Deubiquitinating enzymes: their functions and substrate specificity. Curr Protein Pept Sci. 2004;5:191–200. doi: 10.2174/1389203043379765. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y. Transcriptional regulation by histone ubiquitination and deubiquitination. Genes Dev. 2003;17:2733–40. doi: 10.1101/gad.1156403. [DOI] [PubMed] [Google Scholar]
- 54.Johnston SC, Larsen CN, Cook WJ, Wilkinson KD, Hill CP. Crystal structure of a deubiquitinating enzyme (human UCH-L3) at 1.8 A resolution. EMBO J. 1997;16:3787–96. doi: 10.1093/emboj/16.13.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Larsen CN, Krantz BA, Wilkinson KD. Substrate specificity of deubiquitinating enzymes: ubiquitin C-terminal hydrolases. Biochemistry. 1998;37:3358–68. doi: 10.1021/bi972274d. [DOI] [PubMed] [Google Scholar]
- 56.Johnston SC, Riddle SM, Cohen RE, Hill CP. Structural basis for the specificity of ubiquitin C-terminal hydrolases. EMBO J. 1999;18:3877–87. doi: 10.1093/emboj/18.14.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilkinson KD, Laleli-Sahin E, Urbauer J, Larsen CN, Shih GH, et al. The binding site for UCH-L3 on ubiquitin: mutagenesis and NMR studies on the complex between ubiquitin and UCH-L3. J Mol Biol. 1999;291:1067–77. doi: 10.1006/jmbi.1999.3038. [DOI] [PubMed] [Google Scholar]
- 58.Zhang N, Wilkinson K, Bownes M. Cloning and analysis of expression of a ubiquitin carboxyl terminal hydrolase expressed during oogenesis in Drosophila melanogaster. Dev Biol. 1993;157:214–23. doi: 10.1006/dbio.1993.1125. [DOI] [PubMed] [Google Scholar]
- 59.Koulich E, Li X, DeMartino GN. Relative structural and functional roles of multiple deubiquitylating proteins associated with mammalian 26S proteasome. Mol Biol Cell. 2008;19:1072–82. doi: 10.1091/mbc.E07-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ventii KH, Devi NS, Friedrich KL, Chernova TA, Tighiouart M, et al. BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer Res. 2008;68:6953–62. doi: 10.1158/0008-5472.CAN-08-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jensen DE, Proctor M, Marquis ST, Gardner HP, Ha SI, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097–112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- 62.Hu M, Li P, Song L, Jeffrey PD, Chenova TA, et al. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 2005;24:3747–56. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu M, Li P, Li M, Li W, Yao T, et al. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–54. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- 64.Avvakumov GV, Walker JR, Xue S, Finerty PJ, Jr., Mackenzie F, et al. Amino-terminal dimerization, NRDP1-rhodanese interaction, and inhibited catalytic domain conformation of the ubiquitin-specific protease 8 (USP8). J Biol Chem. 2006;281:38061–70. doi: 10.1074/jbc.M606704200. [DOI] [PubMed] [Google Scholar]
- 65.Renatus M, Parrado SG, D'Arcy A, Eidhoff U, Gerhartz B, et al. Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure. 2006;14:1293–302. doi: 10.1016/j.str.2006.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Komander D, Lord CJ, Scheel H, Swift S, Hofmann K, et al. The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol Cell. 2008;29:451–64. doi: 10.1016/j.molcel.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 67.Goodrich JS, Clouse KN, Schupbach T. Hrb27C, Sqd and Otu cooperatively regulate gurken RNA localization and mediate nurse cell chromosome dispersion in Drosophila oogenesis. Development. 2004;131:1949–58. doi: 10.1242/dev.01078. [DOI] [PubMed] [Google Scholar]
- 68.Steinhauer WR, Walsh RC, Kalfayan LJ. Sequence and structure of the Drosophila melanogaster ovarian tumor gene and generation of an antibody specific for the ovarian tumor protein. Mol Cell Biol. 1989;9:5726–32. doi: 10.1128/mcb.9.12.5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Makarova KS, Aravind L, Koonin EV. A novel superfamily of predicted cysteine proteases from eukaryotes, viruses and Chlamydia pneumoniae. Trends Biochem Sci. 2000;25:50–2. doi: 10.1016/s0968-0004(99)01530-3. [DOI] [PubMed] [Google Scholar]
- 70.Balakirev MY, Tcherniuk SO, Jaquinod M, Chroboczek J. Otubains: a new family of cysteine proteases in the ubiquitin pathway. EMBO Rep. 2003;4:517–22. doi: 10.1038/sj.embor.embor824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Borodovsky A, Ovaa H, Kolli N, Gan-Erdene T, Wilkinson KD, et al. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem Biol. 2002;9:1149–59. doi: 10.1016/s1074-5521(02)00248-x. [DOI] [PubMed] [Google Scholar]
- 72.Evans PC, Smith TS, Lai MJ, Williams MG, Burke DF, et al. A novel type of deubiquitinating enzyme. J Biol Chem. 2003;278:23180–6. doi: 10.1074/jbc.M301863200. [DOI] [PubMed] [Google Scholar]
- 73.Komander D, Barford D. Structure of the A20 OTU domain and mechanistic insights into deubiquitination. Biochem J. 2008;409:77–85. doi: 10.1042/BJ20071399. [DOI] [PubMed] [Google Scholar]
- 74.Messick TE, Russell NS, Iwata AJ, Sarachan KL, Shiekhattar R, et al. Structural basis for ubiquitin recognition by the Otu1 ovarian tumor domain protein. J Biol Chem. 2008;283:11038–49. doi: 10.1074/jbc.M704398200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rumpf S, Jentsch S. Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone. Mol Cell. 2006;21:261–9. doi: 10.1016/j.molcel.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 76.Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–9. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 77.Nanao MH, Tcherniuk SO, Chroboczek J, Dideberg O, Dessen A, Balakirev MY. Crystal structure of human otubain 2. EMBO Rep. 2004;5:783–8. doi: 10.1038/sj.embor.7400201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lin SC, Chung JY, Lamothe B, Rajashankar K, Lu M, et al. Molecular basis for the unique deubiquitinating activity of the NF-kappaB inhibitor A20. J Mol Biol. 2008;376:526–40. doi: 10.1016/j.jmb.2007.11.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Albrecht M, Golatta M, Wullner U, Lengauer T. Structural and functional analysis of ataxin-2 and ataxin-3. Eur J Biochem. 2004;271:3155–70. doi: 10.1111/j.1432-1033.2004.04245.x. [DOI] [PubMed] [Google Scholar]
- 80.Chai Y, Berke SS, Cohen RE, Paulson HL. Poly-ubiquitin binding by the polyglutamine disease protein ataxin-3 links its normal function to protein surveillance pathways. J Biol Chem. 2004;279:3605–11. doi: 10.1074/jbc.M310939200. [DOI] [PubMed] [Google Scholar]
- 81.Mao Y, Senic-Matuglia F, Di Fiore PP, Polo S, Hodsdon ME, De Camilli P. Deubiquitinating function of ataxin-3: insights from the solution structure of the Josephin domain. Proc Natl Acad Sci U S A. 2005;102:12700–5. doi: 10.1073/pnas.0506344102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nicastro G, Menon RP, Masino L, Knowles PP, McDonald NQ, Pastore A. The solution structure of the Josephin domain of ataxin-3: structural determinants for molecular recognition. Proc Natl Acad Sci U S A. 2005;102:10493–8. doi: 10.1073/pnas.0501732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang Q, Li L, Ye Y. Regulation of retrotranslocation by p97-associated deubiquitinating enzyme ataxin-3. J Cell Biol. 2006;174:963–71. doi: 10.1083/jcb.200605100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhong X, Pittman RN. Ataxin-3 binds VCP/p97 and regulates retrotranslocation of ERAD substrates. Hum Mol Genet. 2006;15:2409–20. doi: 10.1093/hmg/ddl164. [DOI] [PubMed] [Google Scholar]
- 85.Winborn BJ, Travis SM, Todi SV, Scaglione KM, Xu P, et al. The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits Lys63 linkages in mixed linkage ubiquitin chains. J Biol Chem. 2008;283:26436–43. doi: 10.1074/jbc.M803692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, et al. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–11. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- 87.Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–5. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 88.McCullough J, Clague MJ, Urbe S. AMSH is an endosome-associated ubiquitin isopeptidase. J Cell Biol. 2004;166:487–92. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, et al. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol Cell. 2007;27:609–21. doi: 10.1016/j.molcel.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–7. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 91.Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, et al. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 2008;455:358–62. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- 92.Misaghi S, Galardy PJ, Meester WJ, Ovaa H, Ploegh HL, Gaudet R. Structure of the ubiquitin hydrolase UCH-L3 complexed with a suicide substrate. J Biol Chem. 2005;280:1512–20. doi: 10.1074/jbc.M410770200. [DOI] [PubMed] [Google Scholar]
- 93.Emre NC, Berger SL. Histone H2B ubiquitylation and deubiquitylation in genomic regulation. Cold Spring Harb Symp Quant Biol. 2004;69:289–99. doi: 10.1101/sqb.2004.69.289. [DOI] [PubMed] [Google Scholar]
- 94.Das C, Hoang QQ, Kreinbring CA, Luchansky SJ, Meray RK, et al. Structural basis for conformational plasticity of the Parkinson's disease-associated ubiquitin hydrolase UCH-L1. Proc Natl Acad Sci U S A. 2006;103:4675–80. doi: 10.1073/pnas.0510403103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Amerik A, Swaminathan S, Krantz BA, Wilkinson KD, Hochstrasser M. In vivo disassembly of free polyubiquitin chains by yeast Ubp14 modulates rates of protein degradation by the proteasome. EMBO J. 1997;16:4826–38. doi: 10.1093/emboj/16.16.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Doelling JH, Yan N, Kurepa J, Walker J, Vierstra RD. The ubiquitin-specific protease UBP14 is essential for early embryo development in Arabidopsis thaliana. Plant J. 2001;27:393–405. doi: 10.1046/j.1365-313x.2001.01106.x. [DOI] [PubMed] [Google Scholar]
- 97.Lindsey DF, Amerik A, Deery WJ, Bishop JD, Hochstrasser M, Gomer RH. A deubiquitinating enzyme that disassembles free polyubiquitin chains is required for development but not growth in Dictyostelium. J Biol Chem. 1998;273:29178–87. doi: 10.1074/jbc.273.44.29178. [DOI] [PubMed] [Google Scholar]
- 98.Dang LC, Melandri FD, Stein RL. Kinetic and mechanistic studies on the hydrolysis of ubiquitin C-terminal 7-amido-4-methylcoumarin by deubiquitinating enzymes. Biochemistry. 1998;37:1868–79. doi: 10.1021/bi9723360. [DOI] [PubMed] [Google Scholar]
- 99.Reyes-Turcu FE, Horton JR, Mullally JE, Heroux A, Cheng X, Wilkinson KD. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell. 2006;124:1197–208. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- 100.Chernova TA, Allen KD, Wesoloski LM, Shanks JR, Chernoff YO, Wilkinson KD. Pleiotropic effects of Ubp6 loss on drug sensitivities and yeast prion are due to depletion of the free ubiquitin pool. J Biol Chem. 2003;278:52102–15. doi: 10.1074/jbc.M310283200. [DOI] [PubMed] [Google Scholar]
- 101.Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–13. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- 102.Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, et al. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 103.Yao T, Song L, Xu W, DeMartino GN, Florens L, et al. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat Cell Biol. 2006;8:994–1002. doi: 10.1038/ncb1460. [DOI] [PubMed] [Google Scholar]
- 104.Yao T, Song L, Jin J, Cai Y, Takahashi H, et al. Distinct modes of regulation of the Uch37 deubiquitinating enzyme in the proteasome and in the Ino80 chromatin-remodeling complex. Mol Cell. 2008;31:909–17. doi: 10.1016/j.molcel.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Baek KH, Mondoux MA, Jaster R, Fire-Levin E, D'Andrea AD. DUB-2A, a new member of the DUB subfamily of hematopoietic deubiquitinating enzymes. Blood. 2001;98:636–42. doi: 10.1182/blood.v98.3.636. [DOI] [PubMed] [Google Scholar]
- 106.Jaster R, Baek KH, D'Andrea AD. Analysis of cis-acting sequences and trans-acting factors regulating the interleukin-3 response element of the DUB-1 gene. Biochim Biophys Acta. 1999;1446:308–16. doi: 10.1016/s0167-4781(99)00095-0. [DOI] [PubMed] [Google Scholar]
- 107.Zhu Y, Lambert K, Corless C, Copeland NG, Gilbert DJ, et al. DUB-2 is a member of a novel family of cytokine-inducible deubiquitinating enzymes. J Biol Chem. 1997;272:51–7. doi: 10.1074/jbc.272.1.51. [DOI] [PubMed] [Google Scholar]
- 108.Zhu Y, Pless M, Inhorn R, Mathey-Prevot B, D'Andrea AD. The murine DUB-1 gene is specifically induced by the betac subunit of interleukin-3 receptor. Mol Cell Biol. 1996;16:4808–17. doi: 10.1128/mcb.16.9.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Baek KH, Kim MS, Kim YS, Shin JM, Choi HK. DUB-1A, a novel deubiquitinating enzyme subfamily member, is polyubiquitinated and cytokine-inducible in B-lymphocytes. J Biol Chem. 2004;279:2368–76. doi: 10.1074/jbc.M304774200. [DOI] [PubMed] [Google Scholar]
- 110.Lee MY, Ajjappala BS, Kim MS, Oh YK, Baek KH. DUB-1, a fate determinant of dynein heavy chain in B-lymphocytes, is regulated by the ubiquitin-proteasome pathway. J Cell Biochem. 2008 doi: 10.1002/jcb.21961. [DOI] [PubMed] [Google Scholar]
- 111.Baek KH. Cytokine-regulated protein degradation by the ubiquitination system. Curr Protein Pept Sci. 2006;7:171–7. doi: 10.2174/138920306776359740. [DOI] [PubMed] [Google Scholar]
- 112.Yoshida H, Jono H, Kai H, Li JD. The tumor suppressor cylindromatosis (CYLD) acts as a negative regulator for toll-like receptor 2 signaling via negative cross-talk with TRAF6 AND TRAF7. J Biol Chem. 2005;280:41111–21. doi: 10.1074/jbc.M509526200. [DOI] [PubMed] [Google Scholar]
- 113.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–6. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 114.Mu JJ, Wang Y, Luo H, Leng M, Zhang J, et al. A proteomic analysis of ataxia telangiectasia-mutated (ATM)/ATM-Rad3-related (ATR) substrates identifies the ubiquitinproteasome system as a regulator for DNA damage checkpoints. J Biol Chem. 2007;282:17330–4. doi: 10.1074/jbc.C700079200. [DOI] [PubMed] [Google Scholar]
- 115.Popov N, Herold S, Llamazares M, Schulein C, Eilers M. Fbw7 and Usp28 regulate myc protein stability in response to DNA damage. Cell Cycle. 2007;6:2327–31. doi: 10.4161/cc.6.19.4804. [DOI] [PubMed] [Google Scholar]
- 116.Hutti JE, Turk BE, Asara JM, Ma A, Cantley LC, Abbott DW. IkappaB kinase beta phosphorylates the K63 deubiquitinase A20 to cause feedback inhibition of the NF-kappaB pathway. Mol Cell Biol. 2007;27:7451–61. doi: 10.1128/MCB.01101-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wada K, Kamitani T. UnpEL/Usp4 is ubiquitinated by Ro52 and deubiquitinated by itself. Biochem Biophys Res Commun. 2006;342:253–8. doi: 10.1016/j.bbrc.2006.01.144. [DOI] [PubMed] [Google Scholar]
- 118.Shen C, Ye Y, Robertson SE, Lau AW, Mak DO, Chou MM. Calcium/calmodulin regulates ubiquitination of the ubiquitin-specific protease TRE17/USP6. J Biol Chem. 2005;280:35967–73. doi: 10.1074/jbc.M505220200. [DOI] [PubMed] [Google Scholar]
- 119.Lee HJ, Kim MS, Kim YK, Oh YK, Baek KH. HAUSP, a deubiquitinating enzyme for p53, is polyubiquitinated, polyneddylated, and dimerized. FEBS Lett. 2005;579:4867–72. doi: 10.1016/j.febslet.2005.07.048. [DOI] [PubMed] [Google Scholar]
- 120.Kim MS, Kim YK, Kim YS, Seong M, Choi JK, Baek KH. Deubiquitinating enzyme USP36 contains the PEST motif and is polyubiquitinated. Biochem Biophys Res Commun. 2005;330:797–804. doi: 10.1016/j.bbrc.2005.03.051. [DOI] [PubMed] [Google Scholar]
- 121.Meulmeester E, Kunze M, Hsiao HH, Urlaub H, Melchior F. Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol Cell. 2008;30:610–9. doi: 10.1016/j.molcel.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 122.Reyes-Turcu FE, Shanks JR, Komander D, Wilkinson KD. Recognition of polyubiquitin isoforms by the multiple ubiquitin binding modules of isopeptidase T. J Biol Chem. 2008 doi: 10.1074/jbc.M800947200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Melandri F, Grenier L, Plamondon L, Huskey WP, Stein RL. Kinetic studies on the inhibition of isopeptidase T by ubiquitin aldehyde. Biochemistry. 1996;35:12893–900. doi: 10.1021/bi9612935. [DOI] [PubMed] [Google Scholar]
- 124.Stein RL, Chen Z, Melandri F. Kinetic studies of isopeptidase T: modulation of peptidase activity by ubiquitin. Biochemistry. 1995;34:12616–23. doi: 10.1021/bi00039a017. [DOI] [PubMed] [Google Scholar]
- 125.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Raasi S, Varadan R, Fushman D, Pickart CM. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol. 2005;12:708–14. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- 127.Cook WJ, Jeffrey LC, Kasperek E, Pickart CM. Structure of tetraubiquitin shows how multiubiquitin chains can be formed. J Mol Biol. 1994;236:601–9. doi: 10.1006/jmbi.1994.1169. [DOI] [PubMed] [Google Scholar]
- 128.Eddins MJ, Varadan R, Fushman D, Pickart CM, Wolberger C. Crystal structure and solution NMR studies of Lys48-linked tetraubiquitin at neutral pH. J Mol Biol. 2007;367:204–11. doi: 10.1016/j.jmb.2006.12.065. [DOI] [PubMed] [Google Scholar]
- 129.Phillips CL, Thrower J, Pickart CM, Hill CP. Structure of a new crystal form of tetraubiquitin. Acta Crystallogr D Biol Crystallogr. 2001;57:341–4. doi: 10.1107/s090744490001800x. [DOI] [PubMed] [Google Scholar]
- 130.Tenno T, Fujiwara K, Tochio H, Iwai K, Morita EH, et al. Structural basis for distinct roles of Lys63- and Lys48-linked polyubiquitin chains. Genes Cells. 2004;9:865–75. doi: 10.1111/j.1365-2443.2004.00780.x. [DOI] [PubMed] [Google Scholar]
- 131.Sheng Y, Saridakis V, Sarkari F, Duan S, Wu T, et al. Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat Struct Mol Biol. 2006;13:285–91. doi: 10.1038/nsmb1067. [DOI] [PubMed] [Google Scholar]
- 132.Saridakis V, Sheng Y, Sarkari F, Holowaty MN, Shire K, et al. Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization. Mol Cell. 2005;18:25–36. doi: 10.1016/j.molcel.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 133.Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol. 2004;5:177–87. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- 134.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 135.Glickman MH, Maytal V. Regulating the 26S proteasome. Curr Top Microbiol Immunol. 2002;268:43–72. doi: 10.1007/978-3-642-59414-4_3. [DOI] [PubMed] [Google Scholar]
- 136.Qiu XB, Ouyang SY, Li CJ, Miao S, Wang L, Goldberg AL. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 2006;25:5742–53. doi: 10.1038/sj.emboj.7601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, et al. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature. 2008;453:548–52. doi: 10.1038/nature06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Stone M, Hartmann-Petersen R, Seeger M, Bech-Otschir D, Wallace M, Gordon C. Uch2/Uch37 is the major deubiquitinating enzyme associated with the 26S proteasome in fission yeast. J Mol Biol. 2004;344:697–706. doi: 10.1016/j.jmb.2004.09.057. [DOI] [PubMed] [Google Scholar]
- 139.Anderson C, Crimmins S, Wilson JA, Korbel GA, Ploegh HL, Wilson SM. Loss of Usp14 results in reduced levels of ubiquitin in ataxia mice. J Neurochem. 2005;95:724–31. doi: 10.1111/j.1471-4159.2005.03409.x. [DOI] [PubMed] [Google Scholar]
- 140.Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, Ploegh HL. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 2001;20:5187–96. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wilson SM, Bhattacharyya B, Rachel RA, Coppola V, Tessarollo L, et al. Synaptic defects in ataxia mice result from a mutation in Usp14, encoding a ubiquitin-specific protease. Nat Genet. 2002;32:420–5. doi: 10.1038/ng1006. [DOI] [PubMed] [Google Scholar]
- 142.Guterman A, Glickman MH. Complementary roles for Rpn11 and Ubp6 in deubiquitination and proteolysis by the proteasome. J Biol Chem. 2004;279:1729–38. doi: 10.1074/jbc.M307050200. [DOI] [PubMed] [Google Scholar]
- 143.Lundgren J, Masson P, Mirzaei Z, Young P. Identification and characterization of a Drosophila proteasome regulatory network. Mol Cell Biol. 2005;25:4662–75. doi: 10.1128/MCB.25.11.4662-4675.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hamazaki J, Iemura S, Natsume T, Yashiroda H, Tanaka K, Murata S. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 2006;25:4524–36. doi: 10.1038/sj.emboj.7601338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, et al. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- 146.Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, et al. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–8. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Elsasser S, Chandler-Militello D, Muller B, Hanna J, Finley D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J Biol Chem. 2004;279:26817–22. doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- 148.Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 149.Madura K. Rad23 and Rpn10: perennial wallflowers join the melee. Trends Biochem Sci. 2004;29:637–40. doi: 10.1016/j.tibs.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 150.Crimmins S, Jin Y, Wheeler C, Huffman AK, Chapman C, et al. Transgenic rescue of ataxia mice with neuronal-specific expression of ubiquitin-specific protease 14. J Neurosci. 2006;26:11423–31. doi: 10.1523/JNEUROSCI.3600-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 152.Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–63. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 153.Joo HY, Zhai L, Yang C, Nie S, Erdjument-Bromage H, et al. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature. 2007;449:1068–72. doi: 10.1038/nature06256. [DOI] [PubMed] [Google Scholar]
- 154.Wyce A, Henry KW, Berger SL. H2B ubiquitylation and de-ubiquitylation in gene activation. Novartis Found Symp. 2004;259:63–73. discussion −7, 163−9. [PubMed] [Google Scholar]
- 155.Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, et al. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell. 2008;29:102–11. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhang XY, Pfeiffer HK, Thorne AW, McMahon SB. USP22, an hSAGA subunit and potential cancer stem cell marker, reverses the polycomb-catalyzed ubiquitylation of histone H2A. Cell Cycle. 2008;7:1522–4. doi: 10.4161/cc.7.11.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nicassio F, Corrado N, Vissers JH, Areces LB, Bergink S, et al. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr Biol. 2007;17:1972–7. doi: 10.1016/j.cub.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 158.Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, et al. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–63. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Daniel JA, Torok MS, Sun ZW, Schieltz D, Allis CD, et al. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J Biol Chem. 2004;279:1867–71. doi: 10.1074/jbc.C300494200. [DOI] [PubMed] [Google Scholar]
- 160.Gardner RG, Nelson ZW, Gottschling DE. Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: distinct roles in telomeric silencing and general chromatin. Mol Cell Biol. 2005;25:6123–39. doi: 10.1128/MCB.25.14.6123-6139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Cai SY, Babbitt RW, Marchesi VT. A mutant deubiquitinating enzyme (Ubp-M) associates with mitotic chromosomes and blocks cell division. Proc Natl Acad Sci U S A. 1999;96:2828–33. doi: 10.1073/pnas.96.6.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Nakagawa T, Kajitani T, Togo S, Masuko N, Ohdan H, et al. Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation. Genes Dev. 2008;22:37–49. doi: 10.1101/gad.1609708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kohler A, Schneider M, Cabal GG, Nehrbass U, Hurt E. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat Cell Biol. 2008;10:707–15. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- 164.Ingvarsdottir K, Krogan NJ, Emre NC, Wyce A, Thompson NJ, et al. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol Cell Biol. 2005;25:1162–72. doi: 10.1128/MCB.25.3.1162-1172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Baker SP, Grant PA. The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene. 2007;26:5329–40. doi: 10.1038/sj.onc.1210603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Pijnappel WW, Timmers HT. Dubbing SAGA unveils new epigenetic crosstalk. Mol Cell. 2008;29:152–4. doi: 10.1016/j.molcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 167.Kohler A, Pascual-Garcia P, Llopis A, Zapater M, Posas F, et al. The mRNA export factor Sus1 is involved in Spt/Ada/Gcn5 acetyltransferase-mediated H2B deubiquitinylation through its interaction with Ubp8 and Sgf11. Mol Biol Cell. 2006;17:4228–36. doi: 10.1091/mbc.E06-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Emre NC, Ingvarsdottir K, Wyce A, Wood A, Krogan NJ, et al. Maintenance of low histone ubiquitylation by Ubp10 correlates with telomere-proximal Sir2 association and gene silencing. Mol Cell. 2005;17:585–94. doi: 10.1016/j.molcel.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 169.Kahana A, Gottschling DE. DOT4 links silencing and cell growth in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6608–20. doi: 10.1128/mcb.19.10.6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Santos-Rosa H, Bannister AJ, Dehe PM, Geli V, Kouzarides T. Methylation of H3 lysine 4 at euchromatin promotes Sir3p association with heterochromatin. J Biol Chem. 2004;279:47506–12. doi: 10.1074/jbc.M407949200. [DOI] [PubMed] [Google Scholar]
- 171.Clague MJ, Coulson JM, Urbe S. Deciphering histone 2A deubiquitination. Genome Biol. 2008;9:202. doi: 10.1186/gb-2008-9-1-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Vissers JH, Nicassio F, van Lohuizen M, Di Fiore PP, Citterio E. The many faces of ubiquitinated histone H2A: insights from the DUBs. Cell Div. 2008;3:8. doi: 10.1186/1747-1028-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Mimnaugh EG, Kayastha G, McGovern NB, Hwang SG, Marcu MG, et al. Caspase-dependent deubiquitination of monoubiquitinated nucleosomal histone H2A induced by diverse apoptogenic stimuli. Cell Death Differ. 2001;8:1182–96. doi: 10.1038/sj.cdd.4400924. [DOI] [PubMed] [Google Scholar]
- 174.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–54. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 175.Wei J, Zhai L, Xu J, Wang H. Role of Bmi1 in H2A ubiquitylation and Hox gene silencing. J Biol Chem. 2006;281:22537–44. doi: 10.1074/jbc.M600826200. [DOI] [PubMed] [Google Scholar]
- 176.Stegmeier F, Rape M, Draviam VM, Nalepa G, Sowa ME, et al. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature. 2007;446:876–81. doi: 10.1038/nature05694. [DOI] [PubMed] [Google Scholar]
- 177.Liu Y, Lashuel HA, Choi S, Xing X, Case A, et al. Discovery of inhibitors that elucidate the role of UCH-L1 activity in the H1299 lung cancer cell line. Chem Biol. 2003;10:837–46. doi: 10.1016/j.chembiol.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 178.Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, et al. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell. 2005;17:331–9. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 179.Zhang D, Zaugg K, Mak TW, Elledge SJ. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell. 2006;126:529–42. doi: 10.1016/j.cell.2006.06.039. [DOI] [PubMed] [Google Scholar]
- 180.Bassermann F, Frescas D, Guardavaccaro D, Busino L, Peschiaroli A, Pagano M. The Cdc14B-Cdh1-Plk1 axis controls the G2 DNA-damage-response checkpoint. Cell. 2008;134:256–67. doi: 10.1016/j.cell.2008.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Fujiwara T, Saito A, Suzuki M, Shinomiya H, Suzuki T, et al. Identification and chromosomal assignment of USP1, a novel gene encoding a human ubiquitin-specific protease. Genomics. 1998;54:155–8. doi: 10.1006/geno.1998.5554. [DOI] [PubMed] [Google Scholar]
- 182.Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–47. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 183.Cohn MA, Kowal P, Yang K, Haas W, Huang TT, et al. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol Cell. 2007;28:786–97. doi: 10.1016/j.molcel.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 184.Schoenfeld AR, Apgar S, Dolios G, Wang R, Aaronson SA. BRCA2 is ubiquitinated in vivo and interacts with USP11, a deubiquitinating enzyme that exhibits prosurvival function in the cellular response to DNA damage. Mol Cell Biol. 2004;24:7444–55. doi: 10.1128/MCB.24.17.7444-7455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Moro L, Arbini AA, Marra E, Greco M. Up-regulation of Skp2 after prostate cancer cell adhesion to basement membranes results in BRCA2 degradation and cell proliferation. J Biol Chem. 2006;281:22100–7. doi: 10.1074/jbc.M604636200. [DOI] [PubMed] [Google Scholar]
- 186.Li H, Lin X. Positive and negative signaling components involved in TNFalpha-induced NF-kappaB activation. Cytokine. 2008;41:1–8. doi: 10.1016/j.cyto.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 187.Deng L, Wang C, Spencer E, Yang L, Braun A, et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–61. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 188.Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature. 2003;424:801–5. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- 189.Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature. 2003;424:793–6. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- 190.Courtois G. Tumor suppressor CYLD: negative regulation of NF-kappaB signaling and more. Cell Mol Life Sci. 2008;65:1123–32. doi: 10.1007/s00018-007-7465-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Heyninck K, Beyaert R. A20 inhibits NF-kappaB activation by dual ubiquitin-editing functions. Trends Biochem Sci. 2005;30:1–4. doi: 10.1016/j.tibs.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 192.Cooper JT, Stroka DM, Brostjan C, Palmetshofer A, Bach FH, Ferran C. A20 blocks endothelial cell activation through a NF-kappaB-dependent mechanism. J Biol Chem. 1996;271:18068–73. doi: 10.1074/jbc.271.30.18068. [DOI] [PubMed] [Google Scholar]
- 193.Saito K, Kigawa T, Koshiba S, Sato K, Matsuo Y, et al. The CAP-Gly domain of CYLD associates with the proline-rich sequence in NEMO/IKKgamma. Structure. 2004;12:1719–28. doi: 10.1016/j.str.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 194.Zhang M, Wu X, Lee AJ, Jin W, Chang M, et al. Regulation of IkappaB kinase-related kinases and antiviral responses by tumor suppressor CYLD. J Biol Chem. 2008;283:18621–6. doi: 10.1074/jbc.M801451200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Balakirev MY, Wilkinson KD. OTU takes the chains OUT. Nat Chem Biol. 2008;4:227–8. doi: 10.1038/nchembio0408-227. [DOI] [PubMed] [Google Scholar]
- 196.Hacker H, Karin M. Regulation and function of IKK and IKK-related kinases. Sci STKE. 2006:re13. doi: 10.1126/stke.3572006re13. 2006. [DOI] [PubMed] [Google Scholar]
- 197.Yamaguchi T, Kimura J, Miki Y, Yoshida K. The deubiquitinating enzyme USP11 controls an IkappaB kinase alpha (IKKalpha)-p53 signaling pathway in response to tumor necrosis factor alpha (TNFalpha). J Biol Chem. 2007;282:33943–8. doi: 10.1074/jbc.M706282200. [DOI] [PubMed] [Google Scholar]
- 198.Al-Hakim AK, Goransson O, Deak M, Toth R, Campbell DG, et al. 14−3−3 cooperates with LKB1 to regulate the activity and localization of QSK and SIK. J Cell Sci. 2005;118:5661–73. doi: 10.1242/jcs.02670. [DOI] [PubMed] [Google Scholar]
- 199.Thomson DM, Hansen MD, Winder WW. Regulation of the AMPK-related protein kinases by ubiquitination. Biochem J. 2008;411:e9–10. doi: 10.1042/BJ20080459. [DOI] [PubMed] [Google Scholar]
- 200.Haglund K, Di Fiore PP, Dikic I. Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem Sci. 2003;28:598–603. doi: 10.1016/j.tibs.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 201.Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–72. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 202.Swaminathan S, Amerik AY, Hochstrasser M. The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol Biol Cell. 1999;10:2583–94. doi: 10.1091/mbc.10.8.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Richter C, West M, Odorizzi G. Dual mechanisms specify Doa4-mediated deubiquitination at multivesicular bodies. Embo J. 2007;26:2454–64. doi: 10.1038/sj.emboj.7601692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Mizuno E, Kitamura N, Komada M. 14−3−3-dependent inhibition of the deubiquitinating activity of UBPY and its cancellation in the M phase. Exp Cell Res. 2007;313:3624–34. doi: 10.1016/j.yexcr.2007.07.028. [DOI] [PubMed] [Google Scholar]
- 205.Kato M, Miyazawa K, Kitamura N. A deubiquitinating enzyme UBPY interacts with the Src homology 3 domain of Hrs-binding protein via a novel binding motif PX (V/I) (D/N)RXXKP. J Biol Chem. 2000;275:37481–7. doi: 10.1074/jbc.M007251200. [DOI] [PubMed] [Google Scholar]
- 206.Mizuno E, Iura T, Mukai A, Yoshimori T, Kitamura N, Komada M. Regulation of epidermal growth factor receptor down-regulation by UBPY-mediated deubiquitination at endosomes. Mol Biol Cell. 2005;16:5163–74. doi: 10.1091/mbc.E05-06-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Row PE, Liu H, Hayes S, Welchman R, Charalabous P, et al. The MIT domain of UBPY constitutes a CHMP binding and endosomal localization signal required for efficient epidermal growth factor receptor degradation. J Biol Chem. 2007;282:30929–37. doi: 10.1074/jbc.M704009200. [DOI] [PubMed] [Google Scholar]
- 208.Overstreet E, Fitch E, Fischer JA. Fat facets and Liquid facets promote Delta endocytosis and Delta signaling in the signaling cells. Development. 2004;131:5355–66. doi: 10.1242/dev.01434. [DOI] [PubMed] [Google Scholar]
- 209.Catic A, Fiebiger E, Korbel GA, Blom D, Galardy PJ, Ploegh HL. Screen for ISG15-crossreactive deubiquitinases. PLoS ONE. 2007;2:e679. doi: 10.1371/journal.pone.0000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Mikolajczyk J, Drag M, Bekes M, Cao JT, Ronai Z, Salvesen GS. Small ubiquitin-related modifier (SUMO)-specific proteases: profiling the specificities and activities of human SENPs. J Biol Chem. 2007;282:26217–24. doi: 10.1074/jbc.M702444200. [DOI] [PubMed] [Google Scholar]
- 211.Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem Sci. 2007;32:286–95. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 212.Wilkinson KD, Gan-Erdene T, Kolli N. Derivitization of the C-terminus of ubiquitin and ubiquitin-like proteins using intein chemistry: methods and uses. Methods Enzymol. 2005;399:37–51. doi: 10.1016/S0076-6879(05)99003-4. [DOI] [PubMed] [Google Scholar]
- 213.Hassiepen U, Eidhoff U, Meder G, Bulber JF, Hein A, et al. A sensitive fluorescence intensity assay for deubiquitinating proteases using ubiquitin-rhodamine110-glycine as substrate. Anal Biochem. 2007;371:201–7. doi: 10.1016/j.ab.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 214.Kang SH, Park JJ, Chung SS, Bang OS, Chung CH. Strategies for assaying deubiquitinating enzymes. Methods Enzymol. 2005;398:500–8. doi: 10.1016/S0076-6879(05)98041-5. [DOI] [PubMed] [Google Scholar]
- 215.Russell NS, Wilkinson KD. Deubiquitinating enzyme purification, assay inhibitors, and characterization. Methods Mol Biol. 2005;301:207–19. doi: 10.1385/1-59259-895-1:207. [DOI] [PubMed] [Google Scholar]
- 216.Nicholson B, Leach CA, Goldenberg SJ, Francis DM, Kodrasov MP, et al. Characterization of ubiquitin and ubiquitin-like-protein isopeptidase activities. Protein Sci. 2008;17:1035–43. doi: 10.1110/ps.083450408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Mermerian AH, Case A, Stein RL, Cuny GD. Structure-activity relationship, kinetic mechanism, and selectivity for a new class of ubiquitin C-terminal hydrolase-L1 (UCH-L1) inhibitors. Bioorg Med Chem Lett. 2007;17:3729–32. doi: 10.1016/j.bmcl.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 218.Mullally JE, Moos PJ, Edes K, Fitzpatrick FA. Cyclopentenone prostaglandins of the J series inhibit the ubiquitin isopeptidase activity of the proteasome pathway. J Biol Chem. 2001;276:30366–73. doi: 10.1074/jbc.M102198200. [DOI] [PubMed] [Google Scholar]