Abstract
For label-free expression profiling of tissue proteomes, efficient protein extraction, thorough and quantitative sample cleanup and digestion procedures, as well as sufficient and reproducible chromatographic separation, are highly desirable but remain challenging. However, optimal methodology has remained elusive, especially for proteomes that are rich in membrane proteins, such as the mitochondria. Here we describe a straightforward and reproducible sample preparation procedure, coupled with a highly selective and sensitive nano-LC/Orbitrap analysis, which enables reliable and comprehensive expression profiling of tissue mitochondria. The mitochondrial proteome of swine heart was selected as a test system. Efficient protein extraction was accomplished using a strong buffer containing both ionic and non-ionic detergents. Overnight precipitation was used for cleanup of the extract, and the sample was subjected to an optimized 2-step, on-pellet digestion approach. In the first step, the protein pellet was dissolved via a 4 h tryptic digestion under vigorous agitation, which nano-LC/LTQ/ETD showed to produce large and incompletely cleaved tryptic peptides. The mixture was then reduced, alkylated, and digested into its full complement of tryptic peptides with additional trypsin. This solvent precipitation/on-pellet digestion procedure achieved significantly higher and more reproducible peptide recovery of the mitochondrial preparation, than observed using a prevalent alternative procedure for label-free expression profiling, SDS-PAGE/in-gel digestion (87% vs. 54%). Furthermore, uneven peptide losses were lower than observed with SDS-PAGE/in-gel digestion. The resulting peptides were sufficiently resolved by a 5 h gradient using a nano-LC configuration that features a low-void-volume, high chromatographic reproducibility, and an LTQ/Orbitrap analyzer for protein identification and quantification. The developed method was employed for label-free comparison of the mitochondrial proteomes of myocardium from healthy animals vs. those with hibernating myocardium. Each experimental group consisted of a relatively large number of animals (n=10), and samples were analyzed in random order to minimize quantitative false-positives. Using this approach, 904 proteins were identified and quantified with high confidence, and those mitochondrial proteins that were altered significantly between groups were compared with the results of a parallel 2D-DIGE analysis. The sample preparation and analytical strategy developed here represents an advancement that can be adapted to analyze other tissue proteomes.
Introduction
The capability to compare and contrast proteomes using techniques of protein expression profiling (1, 2) can add immensely to our understanding of the integration of biological processes, as well as the identification of biomarkers of disease progression or drug action. The overall objective of expression profiling is to determine comprehensively the identity, abundance, and modification state of proteins in the sets of proteomic samples to be compared. However, accurate and reliable expression profiling remains challenging on the proteomic scale. The approaches generally employed include two-dimensional (2D) gel electrophoresis (3) or LC/MS-based methods, such as isotope labeling by metabolic incorporation (e.g. SILAC) (4, 5) and chemical/enzymatic labeling (e.g. ICAT, iTRAQ and 18O-incorporation) (6–8), and more recently, label-free protein expression profiling approaches (2, 9–14).
Label-free methods employ a “shotgun” approach that is particularly effective for large-scale protein analysis (15): samples are digested enzymatically into a large number of small peptide fragments and then subjected to LC/MS analysis without labeling. Because there exists a linear correlation between MS signal intensities and the relative quantity of peptides (16), the relative quantification of proteins in these samples is carried out by direct comparison of the peak areas of proteolytic peptides among LC/MS runs. A recent evaluation by the Association of Biomolecular Resource Facilities (www.abrf.org/prg) suggested that label-free approaches provided the best accuracy for relative protein quantification in protein mixtures, compared to other expression profiling methods. In addition to the potential for providing higher quantitative accuracy, the label-free approach holds advantages of (i) applicability to a wide range of proteomic samples, such as those derived from tissues, (ii) the ability to quantify and compare multiple samples, and (iii) simplicity and less expensive sample preparation.
Because the label-free proteomic analysis approach often does not employ internal standards, quantitative and reproducible sample preparation is particularly important for obtaining reliable results (17). However, processing samples for label-free expression profiling in a quantitative manner, particularly for analyzing tissue proteomes, represents a fundamental challenge for several reasons (18). First, although tissue samples are believed to provide a highly focused pool of biomarkers (19), efficient extraction of these marker proteins can be difficult because of the necessity to disrupt tissue structural interactions, as well as the low solubility of some proteins, such as membrane proteins (20). Therefore, a ‘strong’ solvent that contains significant amounts of detergents often is necessary to dissolve membranes, disrupt protein-lipid interactions, and achieve protein solubilization (20). However, it is both necessary and a challenge to remove these detergents and other buffer components without incurring uneven loss of proteins, which can severely compromise quantitative LC/MS analysis. Second, endogenous non-protein compounds are co-extracted from tissue samples, and these can co-elute with the proteolytic peptides during LC/MS analysis, causing undesirable effects such as compromise of chromatographic reproducibility, ion suppression, and/or interference. Therefore, elimination of such compounds before analysis is essential to ensure reproducible label-free quantification. Finally, protease inhibitors that were added during protein extraction also must be eliminated.
To address these challenges, numerous sample preparation strategies have been employed for label-free expression profiling of tissue samples, including 1-dimensional SDS/PAGE electrophoresis (1-DE) followed by in-gel digestion (21–23), strong cation exchange (SCX) chromatography (18) and the use of non-detergent extraction buffers (24). Among these approaches, the 1-DE/in-gel digestion approach is the most prevalent. This method not only fractionates the sample proteins, but also effectively reduces or eliminates detergents, protease inhibitors, and non-protein components, so that the samples are suitable for enzymatic digestion and LC/MS analysis (20, 25–27). Nevertheless, 1-DE/in-gel digestion has several limitations. First, the in-gel digestion procedure is susceptible to serious and non-uniform peptide losses, mainly arising from loss of proteins during destaining, and from incomplete peptide extraction from the polyacrylamide gel after proteolysis (28). Figeys and co-workers reported peptide losses ranging from 15 to 50%, depending on peptide properties and concentrations (29). Non-uniform peptide losses may adversely affect both the sensitivity and quantitative accuracy of label-free profiling. Efforts to increase the efficiency of peptide extraction from gels generally have achieved only marginal gains in peptide yields (e.g. 5%) (28). A second problem with 1-DE/in-gel digestion is that many proteins span the borders of gel slices, and therefore will be present in multiple gel fractions. Because the separation of proteins by the 1-DE method is not highly reproducible for parallel samples (20), artificial quantitative differences in “boundary proteins” thus may be created among proteomic samples. As a result, additional normalizations based on informatics approaches may be necessary to reduce this confounding factor (23).
A robust and traditional approach to protein sample cleanup is protein precipitation employing denaturing organic solvents such as acetone, which has long been regarded as an effective means for the removal of detergents, lipids, small molecular-mass nucleic acids, and other contaminant species, while providing high protein recoveries (30–33). A significant disadvantage of protein precipitation is that the proteins are denatured and aggregated, rendering the pellet difficult to re-solubilize unless detergent-containing buffers are used. Difficulties in re-dissolution are especially problematic for samples rich in membrane proteins (33). Therefore, the protein precipitation approach is usually reserved for those applications in which components that aid in re-solubilizing the protein pellet can be tolerated, such as SDS-PAGE or 2-D electrophoresis (33).
Here we report the development of a facile, efficient, and reproducible “precipitation/on-pellet digestion” strategy for label-free expression profiling of tissue proteomes. In this approach, a strong, detergent-containing buffer is employed to extract tissue proteins efficiently. The proteins in the extract are then precipitated using acetone to remove detergents, protease inhibitors, and non-protein matrix components. A 2-step enzymatic digestion procedure subsequently brings the precipitated proteins back into solution as soluble, completely-cleaved peptides, without introducing detergents. This method is simple to follow, highly quantitative in protein recovery, and provides a clean peptide mixture suitable for nano-LC/MS analysis. Combined with efficient chromatographic separation and sensitive MS detection, as developed in this study, this strategy enables relatively comprehensive label-free expression profiling.
As a proof of concept, this strategy was applied for large-scale comparative expression profiling of the mitochondrial proteome of myocardium from healthy swine and those with chronic hibernating myocardium. Hibernating myocardium develops as an intrinsic adaptive response to repetitive myocardial ischemia, and is characterized by reduced regional contraction, resting blood flow, and regional oxygen consumption, which is correlated with reductions in multiple mitochondrial proteins involved in oxidative metabolism (34, 35). Identification of changes in the abundance of specific proteins in hibernating myocardium can shed light upon the mechanisms responsible for compensatory physiological changes, and identify novel cellular adaptations to chronic ischemia (35). A major challenge for label-free profiling of the mitochondrial proteome of myocardium is the presence of a high percentage of membrane proteins; approx. 80% of the inner mitochondrial membrane mass is comprised of membrane proteins (36, 37). The approach developed here addresses the need for efficient extraction of membrane proteins from tissue sub-fractions, adequate sample cleanup, and a sufficient chromatographic separation to resolve peptides of this highly complex sample, thus permitting label-free, comparative quantification.
Experimental
Reagents
HPLC grade methanol, acetonitrile, and water were from B&J (Muskegon, MI). Formic acid was from Fluka (Buchs, Switzerland). For Obitrap calibration, caffeine, MAFA peptide, and Ultramark were from Thermo Scientific (San Jose, CA). HPLC grade acetone, Tris(2-carboxyethyl) phosphine (TCEP), Tris, iodoacetamide (IAA), NP-40, sodium chloride, sodium deoxycholate, sodium dodecyl sulfate (SDS) and phosphate-buffered saline (PBS, tablets) were obtained from Sigma-Aldrich (St. Louis, MO). Sequencing grade TPCK-treated trypsin and ProteasMax® cleavable detergent (sodium 3-((1-(furan-2-yl)undecyloxy)carbonylamino)propane-1-sulfonate) were from Promega (Madison WI). Protease, phosphatase, and kinase inhibitor cocktail tablets were from Roche (Basel, Switzerland). Protein and peptide concentrations were determined using bicinchoninic acid (BCA) protein assay reagents (Pierce, Rockford, IL).
Animal model and mitochondria preparation
The housing and experimental use of animals adhered to the National Institutes of Health guidelines and procedures were approved by the Institutional Animal Care and Use Committee. Briefly, farm-bred juvenile pigs were chronically instrumented with a 1.5- to 2.0-mm fixed diameter stenosis placed on the proximal left anterior descending (LAD) coronary artery. This model progresses from chronically stunned- to hibernating myocardium after 3 months, with physiological features persisting unchanged for up to 5 months after instrumentation. Sham (control) animals were either untreated or underwent thoracotomy and dissection of the LAD without placement of an occlusive stenosis. Hibernating myocardium animals (n=10) were evaluated physiologically 3 months after instrumentation in the closed-chest sedated state (with Telazol/xylazine IM and propofol 2 to 5 mg/kg/min IV). Resting and vasodilated coronary flows were assessed using fluorescent microspheres as previously described (35)and left ventricle (LV) function was assessed with M-mode echocardiography. Animals were allowed to recover before tissue harvesting to circumvent potential transient effects of pharmacological stimulation secondary to agents used in the physiological study. After at least 72 h, a surgical plane of anesthesia was induced and pigs were rapidly euthanized with an injection of KCl. The hearts were immediately excised for protein sampling and microsphere flow analysis. The LV was weighed and sectioned into 1-cm rings parallel to the AV groove from apex to base. Subendocardial samples (inner third of the wall) immediately adjacent to the LAD and posterior descending arteries were flash frozen in liquid nitrogen and stored at −80 °C.
Protein extraction
An enriched mitochondrial protein fraction was prepared from 0.5–1g pieces of the collected myocardium using a mitochondrial isolation kit (MITO-ISO1) from Sigma-Aldrich (St. Louis, MO). In brief, the tissue was minced and washed in an extraction buffer (10 mM HEPES, pH 7.5, 0.2 M mannitol, 70 mM sucrose and 1 mM EGTA). Samples were incubated on ice with 0.25 mg/ml trypsin in extraction buffer for a total of 25 minutes. Albumin was added to a final concentration of 10 mg/ml to quench the proteolytic activity.
Following homogenization in extraction buffer, samples were centrifuged twice at low speed (800 g for 8 minutes, 700 g for 7 minutes) and then at 11,000 g for 12 minutes. The supernatant and white flocculant material was discarded. The brown mitochondrial pellet was resuspended in an extraction buffer (50 mM Tris, pH 8, 150 mM NaCl, 2% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS). Mitochondria were lysed with 3 freeze/thaw cycles;for each cycle, the sample was frozen in liquid nitrogen for 5 min and then thawed under 37 °C for 10 min, followed by a 1-min vortex. The resultant solution was clarified by ultracentrifugation (140,000 g, 40 minutes, 4 °C) and then the protein concentration was determined with the BCA protein assay.
Precipitation/on-pellet digestion procedure
For protein precipitation, the total concentration of the protein solution was adjusted to 4 µg/µL and 25 µL was precipitated by stepwise addition of 3 aliquots of cold acetone (50 µL each) while vortexing. Stepwise addition of the acetone caused the protein solution to become cloudy with no large aggregates of sediment appearing. The mixture was incubated overnight at −20 °C, and then centrifuged at 12,000 g for 20 min at 4°C. The supernatant was then removed and the pellet was allowed to air dry for 3 min.
The on-pellet-digestion procedure consisted of two steps. In Step 1, a volume (50 µL) of Tris buffer (50 mM, pH 8.5) that was twice the volume of the original protein solution was added, which contained trypsin at an enzyme:substrate ratio of 1:30 (w/w). After a brief vortexing (30 s), the solution was incubated at 37°C and 120 rpm for 4 h in a New Brunswick shaking incubator (Edison, NJ), in order to dissolve the pellet. An alternative approach also researched in this study was to include 0.05% of cleavable detergent (ProteasMAX™) in the solution to expedite the digestion and improve the completeness of digestion. Step 1 resulted in large and incompletely cleaved peptides (Fig. 1). In Step 2, the peptide sample was reduced with 1 mM TCEP at 95 °C for 5 min, and then alkylated with 100 mM iodoacetamide at 37°C for 30 min in the dark. A second aliquot of trypsin was added at an enzyme:substrate ratio of 1:25 (w/w). The mixture was incubated at 37 °C overnight to achieve complete digestion. The final volume of the digestion mixture was approximately 100 µL.
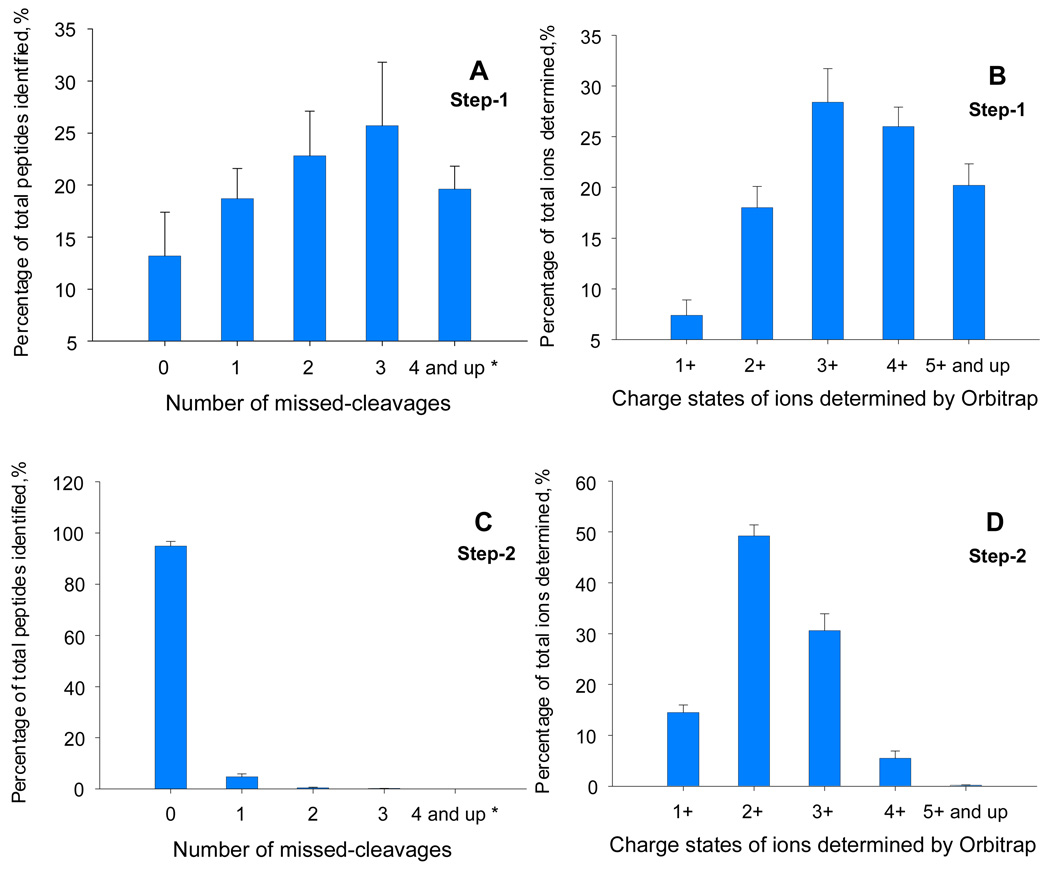
Figure 1.
Completeness of digestion for each step of the two-step digestion procedure (n=3 replicates). The 5 h nano-LC gradient (Experimental section) was employed for all analyses. (A) The distribution of the number of missed-cleavages in tryptic peptides identified by LTQ/ETD after the first digestion step (4 hr). The average number of identified peptides was 4215. (B) The distribution of charge states of the precursor ions determined by Orbitrap after the first digestion step. The average number of total precursors was 14195. (C) The distribution of numbers of missed-cleavages for tryptic peptides identified by LTQ/ETD after the second digestion step (10 hr). The average number of identified peptides was 9653. (D) The distribution of charge states of the precursor ions detected by Orbitrap after the second digestion step. The average number of total precursors was 27,316. The mapping of charge state distribution was carried out by using Rawmeat® program (Thermo) to analyze Orbitrap data.
Modified BCA method for evaluation of peptide recoveries from different sample preparation methods
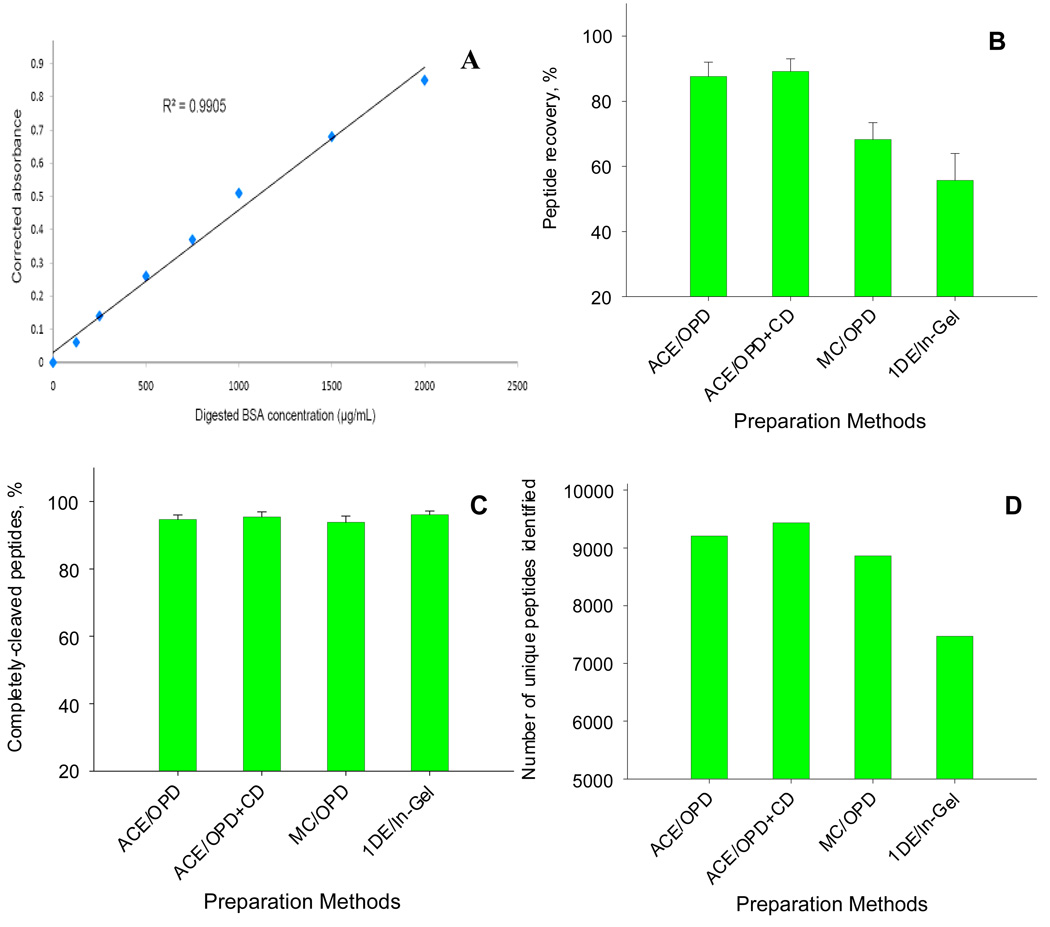
A modified BCA method was devised to permit comparison of the peptide recoveries for different sample preparation techniques (cf. Fig 2). Bovine serum albumin (BSA) standard solutions (125–2000 µ/mL) were prepared in 50 mM Tris buffer (pH=8.5) and digested in-solution overnight with 5 µg trypsin. A solution of 50 mM Tris buffer containing 5 µg trypsin but lacking BSA was incubated along with the standards as a blank sample. The absorption of the solutions was measured at a wavelength of 562 nm, and then the calibration curve was constructed based on the blank-corrected absorption (i.e. the sample absorption following subtraction of the absorption of the blank sample). Linearity, accuracy and variability were investigated (cf. Fig. 2 and SI table 1).
Figure 2.
A comparison of peptide recoveries, digestion completeness and numbers of unique peptides identified by different sample preparation methods for a pooled mitochondrial extract. The protein concentration was 5.8 mg/mL (n=3). (A) The calibration curve of the revised BCA method for peptide concentration measurement; the absorbance was corrected using a blank sample containing trypsin. (B) peptide recoveries of the mitochondria preparation by different sample preparation methods; the method for calculation of recovery is described in the Experimental section. (C) Digestion completeness, as expressed as the percent of completely-digested peptides out of the total peptides identified, for the different sample preparation methods. (D) numbers of unique peptides identified by the different sample preparation methods; a stringent set of criteria was used to filter the identification (cf. Experimental section) and the number of peptide IDs for each preparation method was obtained by combining the data from triplicate, 5 h nano-LC/MS runs. Abbreviation for sample preparation methods: ACE/OPD: acetone precipitation with on-pellet-digestion; ACE/OPD+CD: acetone precipitation with on-pellet-digestion with cleavable detergent; MC/OPD: methanol/chloroform precipitation with on-pellet-digestion; 1DE/In-Gel: 1D SDS-PAGE electrophoresis with in-Gel digestion.
To determine the efficiency of peptide recovery for various different sample preparation methods, all samples were taken from the same pooled mitochondrial extract. The tryptic digests obtained from each sample preparation method, which were 5-fold diluted relative to the original extract, were further diluted 3-fold with Tris buffer, centrifuged, and the supernatant was quantified by the BCA method described above (treated sample). Tris buffer containing trypsin was used as the blank. As a reference, the original protein extract was diluted 15-fold with Tris buffer, digested with trypsin overnight, and then quantified for protein concentration (non-treated sample), using 15-fold diluted extraction buffer containing trypsin as the blank. The tryptic peptide recovery was calculated as the ratio of the corrected absorption of a treated sample to that of the non-treated sample. All analyses were performed in triplicate.
SDS-PAGE and in-gel digestion
For comparison with the methods developed here, the pooled mitochondrial preparation was also cleaned up using 1D SDS-PAGE. Precast gradient minigels (Invitrogen; Carlsbad, CA) were used as described previously (22). The gel was cut into 4 slices, destained, and subjected to a standard overnight in-gel digestion procedure (22). The tryptic peptides were then extracted and the solutions obtained from different slices were combined. The final solution was dried under vacuum, reconstituted with 50 mM Tris buffer, and then analyzed for peptide concentration in triplicate using the modified BCA method described above.
Nano-LC/MS
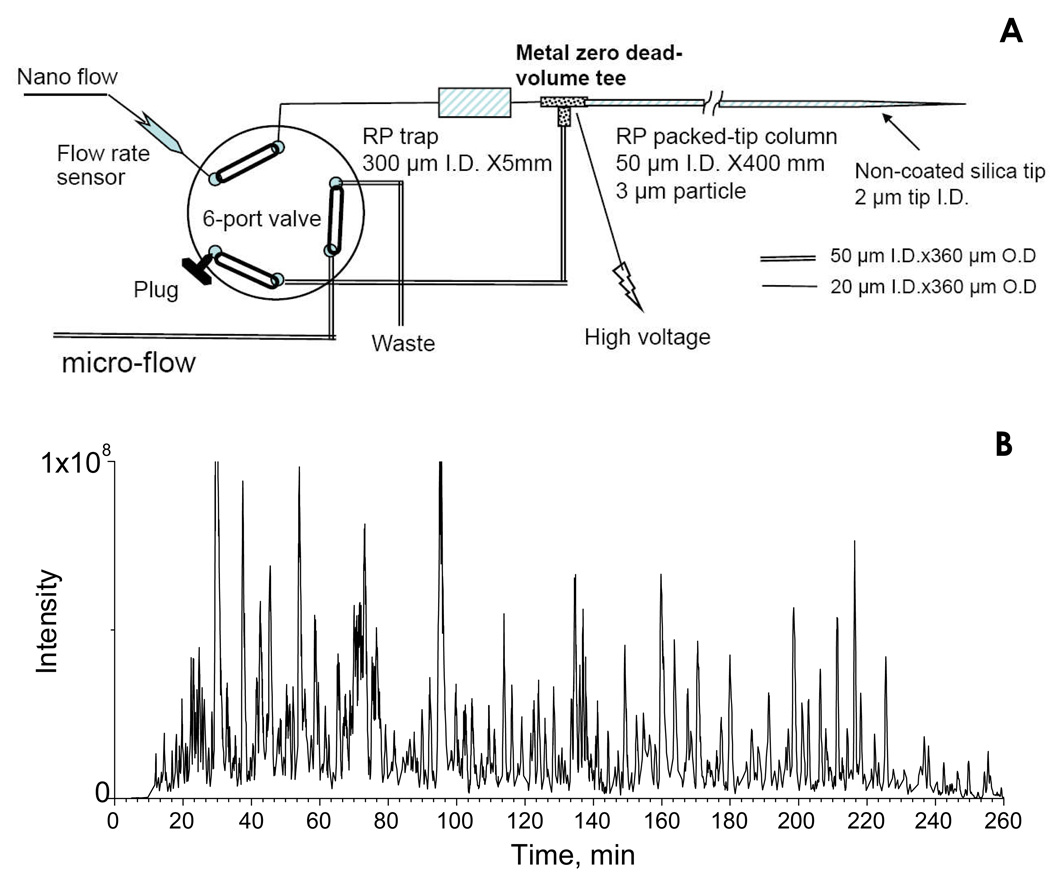
A nano-LC system consisting of a Spark Endurance autosampler (Emmen, Holland) and four Eksigent direct-flow capillary/nano-LC pumps (Dublin, CA) that were powered by pressurized nitrogen (100 p.s.i) were used for all analyses. In order to achieve a comprehensive separation of the complex peptide mixture, a nano-LC/nanospray setup, which features low void volume and high chromatographic reproducibility (Fig. 3A), was developed. The trap and the nano column were connected back-to-back on a Valco (Houston, TX) metal zero-dead-volume (ZDV) tee, and a waste line was connected to the 90° arm. Between the trap and the tee, a ZDV conductivity sensor (GE, Fairfield, CT) was optionally connected (omitted in Fig. 3A) to monitor the gradient change and trap washing efficiency. The high voltage (1.7–2.5 kV) was applied to the metal tee for nanospray. At the loading position (valve position opposite to the position shown in Fig. 3A), the sample was loaded onto the trap with 3% B at a flow rate of 5 µL/min, and the trap was washed for 3 min. The valve was then switched to the analysis position (as shown on Fig 3A), and the spray voltage was applied on the tee. Because the waste line was connected to a dead end at this position, the system pressure built up quickly and the sample peptides were conveyed to the nano column. A series of nano flow gradients was used; mobile phase A consisted of 0.1% formic acid in 2% acetonitrile and mobile phase B was 0.1% formic acid in 84% acetonitrile. The flow rate was 200 nL/min and the gradient profile was (i) a linear increase from 3% to 10% B over 5 min; (ii) an increase from 10 to 24% B over 115 min; (iii) an increase from 24 to 38% B over 70 min; (iv) an increase from 38 to 60% B over 50 min; (v) an increase from 60 to 97% B in 35 min, and finally (vi) isocratic at 97% B for 25 min.
Figure 3.
Nano-LC strategy for the analysis of the highly complex swine heart mitochondria proteome. (A) Schematic of the low void volume and highly reproducible nano-LC flow path employed in this study; (B) Base peak chromatogram for the separation of the tryptic peptides derived from 12 µg of mitochondrial proteins, using a long, shallow gradient. The Orbitrap was used as the detector.
An LTQ/Orbitrap hybrid mass spectrometer (Thermo Fisher Scientific, San Jose, CA) was used for label-free quantification, and an LTQ/ETD (Thermo Fisher Scientific) was employed to evaluate the completeness of the digestion of the tryptic peptides. Both mass spectrometers were connected to the same nano-LC/Nanospray setup as described above. For LTQ/Orbitrap analysis, one scan cycle included an MS1 scan (m/z 300–2000) at a resolution of 60,000 followed by seven MS2 scans by LTQ, to fragment the seven most abundant precursors found in the MS1 spectrum. The target value for MS1 by Orbitrap was 3×106. For LTQ/ETD, the MS was working under data-dependent mode; one scan cycle was comprised of an MS1 scan (m/z range from 300–2000) followed by six sequential dependent MS2 scans (the maximum injection time was 250 ms). The first, third, and fifth MS2 scans were CID fragmentations of the first, second, and third most-abundant precursors found in the MS1 spectrum, respectively. The second, fourth, and sixth MS2 scans were ETD fragmentations corresponding to the same group of precursors. For CID, the activation time was 30 ms, the isolation width was 1.5 amu, the normalized activation energy was 35%, and the activation q was 0.25. For ETD, a mixture of ultrapure helium and nitrogen (25% helium and 75% nitrogen, purity >99.995%) was used as the reaction gas. The ETD reaction time was set at 120 ms and the isolation width was 2 amu; supplemental activation, which uses a short CID activation process to dissociate the charge-stripped precursors, was employed to enhance the fragmentation efficiency for doubly-charged precursors. For both LTQ/ETD and LTQ/Orbitrap experiments, dynamic exclusion was used with one repeat count, 35s repeat duration, and 40s exclusion duration. All samples were analyzed in random order, in order to eliminate quantitative false-positives arising from analytical artifacts such as possible drift in nano-LC or MS performance.
Protein identification and quantification
Peptide/protein identification was first performed with BioWorks3.3.1 embedded with Sequest (Thermo Scientific), against a non-redundant, pig, human and cattle protein sequence database derived in-house from SWISS-PROT as of November 2008. The precursor mass tolerances were 10 ppm and 1.5 mass units, respectively, for Orbitrap and LTQ; the mass tolerance for the fragments of both CID and ETD was 1.0 unit. A stringent set of score filters was employed. Correlation score (Xcorr) criteria were as follows: ≥4 for quadruply-charged (4+) and higher charge states, ≥3 for 3+ ions, ≥2.2 for 2+ ions, and ≥1.7 for 1+ ions. The CID results were further analyzed using Scaffold 2 proteome software (Portland, OR) which integrates both Protein Prophet and Peptide Prophet: additional criteria were that two unique peptides must be identified independently for each protein, the peptide probability must be 95% or higher, and the protein probability must be 99% or higher. For ETD spectra, a final score (Sf) of 0.85 was required for each identification.
A commercial label-free quantification package, Sieve (Fiona build, v. 1.2, Thermo Scientific), was used for comparing relative abundance of peptides and proteins between the control and experimental groups. Briefly, the chromatographic peaks detected by Orbitrap were aligned and the peptides peaks were detected with a minimum signal intensity of 2×105; peptide extracted ion current (XIC) peaks were matched by their retention time (± 1 min) and mass (± 0.025 unit) among sample runs. Each subset of matched peaks was termed a “frame”. The area under the curve (AUC) of each matched peptide within a frame was calculated and compared to the corresponding peak in the control sample. Fisher’s combined probability test was performed to determine whether there was any significant difference in peptide abundances between the two experimental groups. Relative abundance of an individual protein was calculated as the mean AUC ratio for all peptides derived from that protein. All proteins differing significantly between the two groups (quantitative difference <0.8 for down-regulation or >1.25 for up-regulation, and a p-value <0.05) were confirmed by a stringent manual inspection of the fragmentation spectra and the XIC of the ions within a 5-min elution window.
Results and discussion
1. Development of the precipitation/on-pellet-digestion approach
Successful label-free expression profiling of tissue samples requires a method for quantitative sample preparation that includes efficient protein extraction from tissues, removal of components from the sample matrix and extraction buffer that may compromise subsequent digestion of the protein extract or nano-LC/MS analysis, and maintains a high efficiency of peptide recovery. To achieve these goals, we devised a strategy that employs an overnight cold acetone precipitation for effective and quantitative sample cleanup, followed by a 2-step enzymatic digestion: the first step brings the protein pellet into solution by digesting the precipitated proteins into large, soluble tryptic fragments (digestion step 1), and the second step involves reduction, alkylation, and completion of enzymatic digestion (digestion step 2), rendering a sample that is suitable for nano-LC/MS analysis. To develop and optimize this procedure, the swine heart mitochondrial complex was used as a test system.
1.1 Cleanup of mitochondrial protein extract by acetone precipitation
To weaken repulsive electrostatic forces in protein extracts and induce precipitation, a variety of reagents have been used, including organic solvents (e.g. ethanol and acetone), acids (e.g. trichloroacetic acid), and salts (e.g. ammonium sulfate) (33). In pilot studies, we evaluated these and other precipitation strategies in terms of protein recovery from mitochondrial extracts. We observed that acetone precipitation provided superior recovery (cf. Fig. 2B) and did not result in noticeable interference in the subsequent digestion and nano-LC/MS steps. Moreover, others have suggested that acetone precipitation can efficiently remove detergents, protease inhibitors, lipids, and other buffer- or cellular components without introducing salts or strong acids to the sample (25, 38). Based on these considerations, acetone was selected as the precipitant in this study. One notable drawback of acetone precipitation is that it is not compatible with high concentrations of urea (Table 1). For cases in which the extracting buffer contains a high concentration of urea, it may be possible to substitute the methanol/chloroform precipitation procedure developed by Wessel and co-workers (39), which was also evaluated in the present studies.
Table 1.
The Compatibility of Some Commonly Used Protein Extraction Buffer Components with Cold Acetone Precipitation a
| Buffer components | Maximum concentration |
|---|---|
| NP-40 | >5% b |
| SDS | 4% |
| CHAPS | 3% |
| Sodium deoxycholate | 4% |
| Triton X-100 | 5% |
| MOPS | 2% |
| TW-80 | 4% |
| Tris | >300 mM b |
| EDTA | <5 mM |
| KCl | <1 M |
| NaCl | <1M |
| Urea | <4M |
| EGTA | Not compatible |
Each buffer component was prepared in water and then subjected to overnight acetone precipitation at −20 °C with a 6-fold excess of acetone. The maximum concentration was defined as 80% of the concentration at which a visible precipitate formed.
Maximum soluble concentration was not determined.
Because a large portion of the mitochondrial proteome is comprised of hydrophobic membrane proteins (36, 37), it is challenging to disrupt the membrane complex and extract these proteins efficiently (25). A recent report suggested that ‘strong’ buffers containing a combination of relatively high concentrations of both ionic and non-ionic detergents were necessary for an efficient extraction (38). However, it is essential that the extracting detergents and buffer components remain soluble in the subsequent precipitation step. Therefore, commonly used buffer components for membrane protein extraction, including detergents, salts, and protease inhibitors, were tested for compatibility with the acetone precipitation procedure (Table 1). After balancing the considerations of extraction efficiency and compatibility with subsequent sample preparation steps, a buffer containing 2% NP-40, 0.5% sodium deoxycholate and 0.1% SDS was selected, and is described in detail in the Experimental section. Although the procedure was optimized here for membrane protein extraction from swine heart mitochondria, this extraction strategy has been demonstrated in our lab to be applicable to a variety of other complex samples, such as human liver or cultured cells, and reproducible extractions with high yields have been observed for proteins of a wide range of polarities (data not shown).
Conditions for cold acetone precipitation were optimized for mitochondrial extracts having protein concentrations in the range of 5–7 mg/mL. From a survey of many combinations of conditions, optimal protein recovery was achieved using a 6-fold (v/v) excess of acetone for precipitation, and ≥10 hr for incubation at −20°C (SI Fig 1). In the course of this survey, it was discovered that if large clumps or sediment formed at the time of cold acetone addition, detergents would be detected in the final digestion mixture. This is highly undesirable for label-free profiling and probably resulted from the inclusion of extraction buffer components within the rapidly-forming precipitate. Two approaches were found to circumvent this problem: (i) dropwise addition of acetone with continuous vortexing, or (ii) addition of the acetone in multiple small steps. This optimized acetone precipitation procedure provided efficient cleanup and high peptide recovery from mitochondrial extracts, and eliminated detergents and non-protein matrix components that could severely compromise chromatographic separation and ionization (cf. Fig 2b and Table 3).
Table 3.
Evaluation of the Reproducibility of Retention Times and Peak Areas (AUCs) of Representative Peptides from the Mitochondrial Preparation using a 5-hr Nano-LC Gradient (n=20)
| Peptide | Xcorr/charge | Peptide probability a |
Protein | Retention timeb min (RSD%) |
AUCb (RSD%) |
|---|---|---|---|---|---|
| ILDDPSPPQPGEEK | 2.95/2 | 99% | Q5US13 | 33.4 (0.65) | 2.7E7 (9.2) |
| AAYFGVYDTAK | 2.67/2 | 99% | Q9XS69 | 58.5 (0.45) | 2.4E8 (4.8) |
| AFDQGADAIYEHINQGK | 3.15/2 | 99% | Q2EN79 | 79.2 (0.32) | 2.6E7 (5.1) |
| FTQAGSEVSALLGR | 3.57/2 | 99% | Q0QEM6 | 99.7 (0.51) | 1.9E9 (10.7) |
| YLESVQHLLDDEEYSR | 3.46/2 | 98% | Q5US13 | 122.4 (0.39) | 9.6E6 (11.3) |
| LALFNPDVCWDR | 2.45/2 | 99% | A1XQS3 | 146.8 (0.24) | 9.2E7 (3.5) |
| YFPTQALNFAFK | 2.63/2 | 98% | A5PI51 | 167.1 (0.57) | 1.9E8(8.9) |
| LGDGLFLQCCEEVAELYPK | 3.51/2 | 99% | Q1G1K7 | 189.6 (0.28) | 2.6 E6(8.1) |
| DVVDYLVFGTVIQEVK | 2.98/2 | 98% | Q9TUF6 | 209.3 (0.21) | 8.2E7 (12.4) |
| SEVELVQIVIDGVNYLVDCEK | 4.00/3 | 99% | Q2HYU1 | 231.4 (0.17) | 2.8E8 (8.6)) |
Determined by the Peptide Prophet program.
The reproducibility data were obtained by twenty repetitive analysis of the same sample over a 5-day period
1.2 Development of on-pellet-digestion procedure
For conventional in-solution digestion procedures, the sample must be fully dissolved for optimal and reproducible disulfide reduction, thiol alkylation, and enzymatic digestion (38). However, precipitated pellets of mitochondrial proteins, which contained highly aggregated, denatured membrane proteins, are difficult to reconstitute. For example, acetone-precipitated pellets were not soluble even in a relatively strong buffer such as those containing 0.2% SDS and 6 M urea. Only high concentrations of detergents, similar to those in the extraction buffer, were able to solubilize the pellets, but these were not compatible with subsequent tryptic digestion and nano-LC/MS analysis procedures.
The alternative approach developed here is based on the observation that tryptic digestion for several hours under vigorous agitation is an effective means to dissolve the acetone-precipitated protein pellet. It is likely that trypsin degrades the integrity of the pellet by continuously cleaving proteins into more soluble peptides. With time, the aggregated proteins in the pellet are sufficiently cleaved so as to dissolve completely. However, analysis (described below) showed that many peptides produced were incompletely cleaved, and thus a 2-step on-pellet digestion procedure evolved: in the first step, digestion buffer and trypsin were added to the pellet, followed by incubation under agitation until the pellet was completely dissolved; in the second step, the resulting tryptic fragments were reduced, alkylated, and then cleaved to completion by incubation with freshly added trypsin.
All key conditions, such as the enzyme/substrate ratio and digestion time, were optimized by monitoring the completeness of digestion of tryptic peptides generated in both digestion steps by nano-LC/LTQ/ETD and nano-LC/LTQ/Orbitrap. The digestion buffer was 50 mM Tris (pH=8.4), in 5-fold excess with respect to the original sample volume. For digestion step 1, an enzyme:substrate ratio of 1:30 (w/w), incubation at 37°C, and 4 hrs agitation resulted in complete dissolution of the protein pellet. Examination of the first digestion mixture by nano-LC/LTQ, using alternating CID and ETD fragmentation, showed that more than 65% of the tryptic fragments identified carried 2 or more internal lysine or arginine (Fig 1a). Most of these missed cleavages were detectable by ETD but not by CID, likely because of the high charge states of the peptides. Because sequencing of very long, mis-cleaved peptides is difficult even with ETD, these peptides could escape identification and thus made it difficult to obtain a comprehensive evaluation of digestion completeness. As a complementary approach, the Orbitrap MS analyzer was employed to determine the charge state distribution of the ions in the digestion mixture. Because the charge states of long, mis-cleaved peptides, which contain multiple internal basic amino acids (such as K and R), are high under ESI (40) (41), the mapping of peptide charge state by Orbitrap provides additional information for evaluating the completeness of digestion without the need for peptide identification.
Following the first digestion step, more than 73% of the ions detected exhibited charge states ≥3+ (Fig. 1b), suggesting that most contained multiple internal basic amino acid residues. Overall, the analysis suggested that while tryptic digestion in step 1 was sufficient to solubilize proteins, cleavage was incomplete, which would severely compromise protein identification and quantitative accuracy during label-free profiling.
Interestingly, though prolonged incubation improved digestion completeness, it was not able to achieve a complete digestion (SI Fig 2; therefore a second digestion step was incorporated. The mixture was reduced and alkylated, and trypsin was added at an enzyme:substrate ratio of 1:25 (w/w). Incubation at 37°C was continued for 10 h. By this means, complete tryptic digestion was achieved, as indicated by the fact that more than 94% of the identified peptides were completely cleaved (Fig. 1c) and the charge state distributions were dominated by double charges, i.e., the predominant charge state for completely cleaved tryptic peptides (Fig 1D).
An alternative approach for digestion step 1 is to employ a cleavable surfactant to expedite the digestion of the protein pellet, and the effective use of such reagents in a one-step tryptic digestion procedure has been described recently (42). To investigate the potential benefits of a degradable detergent, the acid-cleavable surfactant ProteasMax® was added in digestion step 1 at a concentration of 0.02%. Although the mitochondrial protein pellet initially did not dissolve, the cleavable surfactant provided two benefits: (i) dissolution in the presence of trypsin was accelerated, and typically required <2 h; (ii) the completeness of digestion after 4 h was improved markedly (data not shown). One disadvantage, however, is that the cleaved product of the degradable detergent appeared to diminish the reproducibility of chromatographic separation and the ion intensities when a shallow gradient and long column were employed on nano-LC/MS analysis (below; cf. Fig 3).
1.3 Evaluation of peptide recoveries for the precipitation/on-pellet-digestion procedure
Because label-free quantification utilizes peptide ion currents among parallel runs for quantification, without internal standards, a sample preparation procedure of highly reproducible efficiency is essential for achieving high quantitative precision and accuracy (17). We observed previously that sample preparation and digestion procedures could result in perplexing preferences for certain peptides (7). Therefore, firstly we examined whether the precipitation/on-pellet digestion procedure causes uneven losses for individual peptides by selecting a single protein and comparing the results with those obtained by 1DE/in-gel digestion. BSA at a concentration of 1 mg/mL was used as a substrate, and the relative quantities of individual tryptic peptides produced by the acetone precipitation/on-pellet-digestion approach were compared with those produced by 1DE/in-gel digestion using the Sieve™ program. Digests of the original BSA solution, digested overnight, was used as the control group for Sieve analysis. Quantification was performed in triplicate, and the results are shown in Table 2. It is evident that the precipitation/on-pellet-digestion approach resulted in significantly higher recoveries for most tryptic peptides than did 1DE/in-gel digestion, especially for longer peptides. The difficulty of extracting longer peptides from the gel likely contributed to the lower recovery. Furthermore, at least one peptide detected in the precipitation/on-pellet-digestion group was not observed for the 1DE/in-gel digestion procedure. Thus in-gel digestion appeared to result in lower recovery and more uneven peptide loss than the precipitation/on-pellet-digestion procedure for BSA.
Table 2.
Comparison of the Recoveries of Individual Tryptic Peptides from BSA by the Precipitation/ on-pellet-digestion and 1DE/in-gel Digestion Procedures a, b
| Recovery (RSD%) | ||
|---|---|---|
| Peptide | On-pellet- digestion |
In-gel- digestion |
| ATEEQLK | 0.93 (14) | 0.95 (8) |
| AEFVEVTK | 0.98 (7) | 0.91 (4) |
| YLYEIAR | 1.02 (5) | 1.11 (12) |
| DLGEEHFK | 0.96 (9) | 0.85 (5) |
| LVVSTQTALA | 0.95 (7) | 0.69 (11) |
| QTALVELLK | 1.04 (11) | 0.88 (4) |
| LVNELTEFAK | 0.98 (9) | 0.84 (10) |
| HPEYAVSVLLR | 1.21 (10) | 1.17 (28) |
| HLVDEPQNLIK | 1.04 (10) | 0.78 (14) |
| SLHTLFGDELCK | 1.14 (7) | 0.73 (9) |
| LGEYGFQNALIVR | 0.87 (9) | 0.61 (3) |
| VPQVSTPTLVEVSR | 1.13 (11) | 0.54 (14) |
| DAFLGSFLYEYSR | 1.22 (8) | 0.39 (5) |
| HPYFYAPELLYYANK | 0.94 (7) | ND |
| DAIPENLPPLTADFAEDK | 0.89 (12) | 0.23 (19) |
| WVTFISLLLLFSSAYSR | 1.06 (13) | 0.18 (26) |
| Average recovery | 1.02 (9) | 0.72 (11) |
BSA digests obtained from overnight, direct, in-solution digestion were used as the control group
To minimize quantitative artifacts, only completely digested peptides were selected; also peptides containing amino acids amendable to modification( M or C), were not included.
Peptide recoveries from the more complex mitochondrial protein extracts were also assessed for different sample preparation methods. An assay of peptide concentration was required that could be applied to the samples produced by the different preparation procedures compared, and therefore a modified BCA method was developed. Digested BSA solutions were used to construct the calibration curve, as described in the Experimental section, rather than intact BSA protein. This approach provides good linearity over the protein concentration range investigated (Fig 2a) and was validated using BSA quality control solutions (SI Table 2). It is important to emphasize this method provides a reproducible and standardized means for the calculation of relative peptide recovery, but absolute concentrations are not necessarily determined, because the tryptic pattern of the BSA standard may not reflect precisely the tryptic pattern of the total mitochondrial proteins.
Using this modified BCA method, mitochondrial peptide recoveries were compared for the precipitation/on-pellet-digestion and 1DE/in-gel digestion procedures. For precipitation/on-pellet-digestion, the effect of a cleavable detergent and alternative precipitants upon peptide recovery was also evaluated (Fig 2b). All analyses were carried out in triplicate and with identical amounts of proteins from the same pooled mitochondrial extract. Recoveries were calculated relative to a digest of the original extract without any preparation step, diluted appropriately to contain the same amount of total proteins (detailed in Experimental section).
The highest peptide recoveries were observed for the acetone precipitation, with or without degradable detergent; differences in peptide recovery between the two procedures were not statistically significant. Recovery using ethanol/chloroform as an alternative precipitation approach was significantly lower (p<0.05), and the lowest recovery observed was for 1DE/in-gel digestion (p<0.05). This lower peptide recovery likely arises from the lower efficiency of in-gel digestion and extraction, and the fact that membrane proteins in the mitochondrial preparation may fail to migrate into the gel (28). The reproducibility of recovery for 1DE/in-gel digestion also was poorer than for the precipitation/on-pellet digestion procedures, which could compromise the quantitative precision of label-free profiling.
We also evaluated the completeness of digestion and the number of unique proteins/peptides identified for the four sample preparation methods (Fig 2c–2d). For all, nearly complete digestion was achieved; > 94% of the observed tryptic peptides were completely digested (Fig 2c). For acetone precipitation/on-pellet-digestion, the number of proteins and peptides identified with and without the cleavable detergent were similar. Fewer peptides were identified with chloroform/methanol precipitation/on-pellet-digestion and 1DE/in-gel digestion (Fig 2d).
2. Optimization of the nano-LC/Orbitrap approach
2.1 Nano-LC/Orbitrap system
Because the enriched mitochondrial preparations are highly complex (35, 36, 43), a large number of tryptic peptides are retrieved by the precipitation/on-pellet digestion procedure. As a result, sufficient chromatographic separation is required to achieve the most comprehensive identification/quantification of the proteome, especially for lower abundance peptides. Furthermore, for reliable label-free expression profiling, high run-to-run reproducibility of retention times and MS signal intensities is essential (2). To address these requirements, we devised a nano-LC/nanospray configuration that features a low void volume, high separation efficiency, and high reproducibility (Fig 3a), and has several advantageous features over more conventional arrangements. First, the trap and column were connected back-to-back on a metal zero-dead-volume tee, eliminating the in-valve void volume between trap and column that is typical of a more conventional configuration (44). As a result, the chromatographic peak shapes and widths for tryptic peptides were improved significantly (detailed data not shown). Second, a relatively long (40 cm) reversed-phase nano-column was used for separation; such a column, in conjunction with a long, shallow elution gradient, can enhance proteomic coverage and enable analysis of low abundance peptides, as suggested by our pilot experiments and by others (45). Here, a 5 h gradient was optimized to resolve the mitochondrial samples, and a typical base peak chromatogram is shown in Fig 3b. An extended peptide elution window of more than 240 min was achieved, and this high level of chromatographic separation enabled extensive identification and profiling of the proteome (cf. SI. Table 2).
A third advantage of this configuration is that it provides exceptionally high reproducibility in terms of both retention times and the AUC of peptide ion currents. This was achieved by the use of a large-I.D. (300 µm I.D.) trap, which was 6-fold larger than that of the column, and a bi-direction flow path in the nano-LC analysis cycle. Our initial results for nano-LC optimization indicated that reproducible chromatographic separations were difficult to obtain for 5 h gradient runs when a standard 100-µm-I.D. trap (i.e., 2 fold larger than the nano-column I.D.) was used. By connecting a zero-dead-volume onductivity sensor between the trap and the tee (not shown in Fig 3a), it was observed that the poor reproducibility arose from (i) incomplete mixing of the two mobile phases at nanoliter/min flow rates, and (ii) inaccuracy/variation in mobile phase delivery by the two nano-pumps (i.e. pump noise) during the long, shallow gradient. We found that a large-I.D. trap promoted complete gradient mixing, dampened pump noise, and provided reproducible gradient delivery to the downstream nano-column. In addition, by employing a low-organic loading solvent (3% B) and a bidirectional flow path, which permitted peptide loading using a retrograde flow, peptides were focused on the column side of the trap (Fig. 3a), and then eluted to the nano-column for separation by a series of nano flow gradients. By this strategy, the deterioration of chromatographic performance that could result from using a large-I.D. trap, such as peak broadening and tailing, was avoided. The cost of using a large-I.D. trap here was that an additional 4–5 min void time was observed.
An additional benefit of this configuration is that the large-I.D. trap provides a larger loading capacity for peptides when a long gradient is used. The rationale is that with a long gradient elution, the loading capacity is determined by the capacity of the trap, rather than that of the nano-column; it would be expected that the amount of peptides delivered from the trap to the column at any given time is relatively small when a shallow gradient is employed, and therefore does not exceed column capacity easily. By this mechanism, the large loading capacity thus increased the sensitivity for peptide identification/quantification. Here, tryptic peptides derived from 12 µg mitochondrial proteins were loaded for each run.
The Orbitrap was chosen as the MS analyzer because its high mass resolution/accuracy enables high specificity for quantification. For example, at the resolution used here (HWFM=60,000), a peptide ion current can be extracted using a narrow m/z window (0.03 mass units) without losing sensitivity. As a result of this high specificity, the resulting ion currents exhibit very low chemical noise. Optimal MS conditions, such as the position of the nanospray tip, ionization voltages, and dynamic exclusion conditions were experimentally identified. It is important to note that a high target ion value (4×106) was used for the Orbitrap, which improved signal-to-noise ratios for peptides by at least 4 fold over the default value (0.5 ×106) without compromising the accuracy for m/z measurement (data not shown).
2.2 Reproducibility of the developed analytical system
To investigate the performance of the developed analytical system for a relatively large-scale expression profiling of mitochondrial proteome, reproducibility was evaluated with 20 repeated injections of the same pooled mitochondrial sample over a 5-day period. The reproducibility of chromatographic separation and signal intensities for the twenty 5-h runs was excellent, as assessed from data for selected tryptic peptides identified in the mitochondrial preparation. Representative data are shown in Table 3. Variations in retention time for the selected peptides were in the range of 0.17–0.65%, and variations for precursor ion current AUCs were in the range of 3.5–12.4% over the 5 day period. This high level of reproducibility can be attributed to two factors: (i) the highly reproducible chromatographic configuration described above, and (ii) the efficient precipitation/on-pellet-digestion procedure that removed of detergents and other undesirable compounds, which could otherwise compromise the reproducibility of the nano-LC/MS analysis, particularly given the long gradient employed.
For samples in which the cleavable detergent was included in the first step of the on-pellet digestion procedure, variations of 0.9–4.5% were observed in retention time, and variations of 5–47% were observed in the AUC of the ion current for the select peptides monitored (data not shown). This variability, which was not acceptable for label-free expression profiling, likely results from presence of detergent degradation products in the sample, which could alter the chromatographic behavior of the trap/column for a shallow, long gradient, as well as the ionization efficiency. The impact of this variability was concluded to outweigh the advantage of more rapid solubilization of the protein pellet when the cleavable detergent was included.
3. Label-free expression profiling of the mitochondrial proteome in myocardium of swine with hibernating myocardium
The approaches developed here for sample preparation and analysis were applied to label-free expression profiling of enriched mitochondrial samples from swine heart, for the purpose of identifying novel proteins differentially regulated in hibernating myocardium (35), which is an adaptive response to repetitive myocardial ischemia. Chronically altered myocytes in viable but dysfunctional myocardium can intrinsically down-regulate their metabolic needs in terms of resting blood flow, function, and oxygen consumption in order to achieve a balance between metabolic demand at reduced regional workload and supply. Previously we used 2D-DIGE to investigate changes in the level of regulatory proteins in whole myocardial tissue, and it was observed that many of the proteins altered in hibernating myocardium are associated with oxidative metabolism and electron transport (35). In order to gain a more comprehensive understanding of the effects and processes involved in myocardial hibernation, label-free expression profiling was performed in this study.
Mitochondrial preparations derived from healthy animals (control group) and those with hibernating myocardium (experimental group) were compared. In the design of these experiments, it was recognized that a larger number of biological replicates is desirable to increase the quantitative reliability and statistical power of the results (2), and to improve the chance to identify low abundance proteins, because more fragmentation data will be collected. However, a larger number of biological replicates not only increase the number of animals and samples, but also, the protracted analysis time may increase the variability in separation/ionization performance, which is highly undesirable for label-free profiling. After balancing these factors, a sample size of 10 animals per group was selected. After tissue acquisition, the enriched mitochondrial preparations were prepared in parallel, cleaned up and digested using the precipitation/on-pellet digestion approach, and then subjected to nano-LC/Orbitrap analysis as described above. To eliminate quantitative false-positives arising from time-dependent factors such as drift in nano-LC or ionization performance, and possible instability of certain tryptic peptides, samples from the 20 animals were analyzed in a random order. Peak alignment, peptide ion matches, relative quantification, result validation, and statistics were carried out using the Sieve software package.
Prior to comparison of samples from the two groups, the false-positive rate in relative quantification was evaluated. From the 20 repetitive analyses of a pooled mitochondrial quality control sample (above), 10 runs were randomly assigned as the control group, and the remaining 10 were designated as the experimental group. Expression profiles between the two groups were then compared. In total, 14,237 ion-current frames were matched among the two groups of samples using Sieve. The observed distribution of peptide ratios (experimental:control) concentrated narrowly around 1.0, with 93% of ion-current frames in the range of 0.9–1.1. Approx. 1% of ions differed by more than 15% of the 1.0. Only 1 peptide was identified as significantly changed between the two groups at p< 0.05. Such a low false-positive rate and high quantitative precision supported the suitability of this method for profiling of the mitochondrial proteome using the replicate number (n=10) selected.
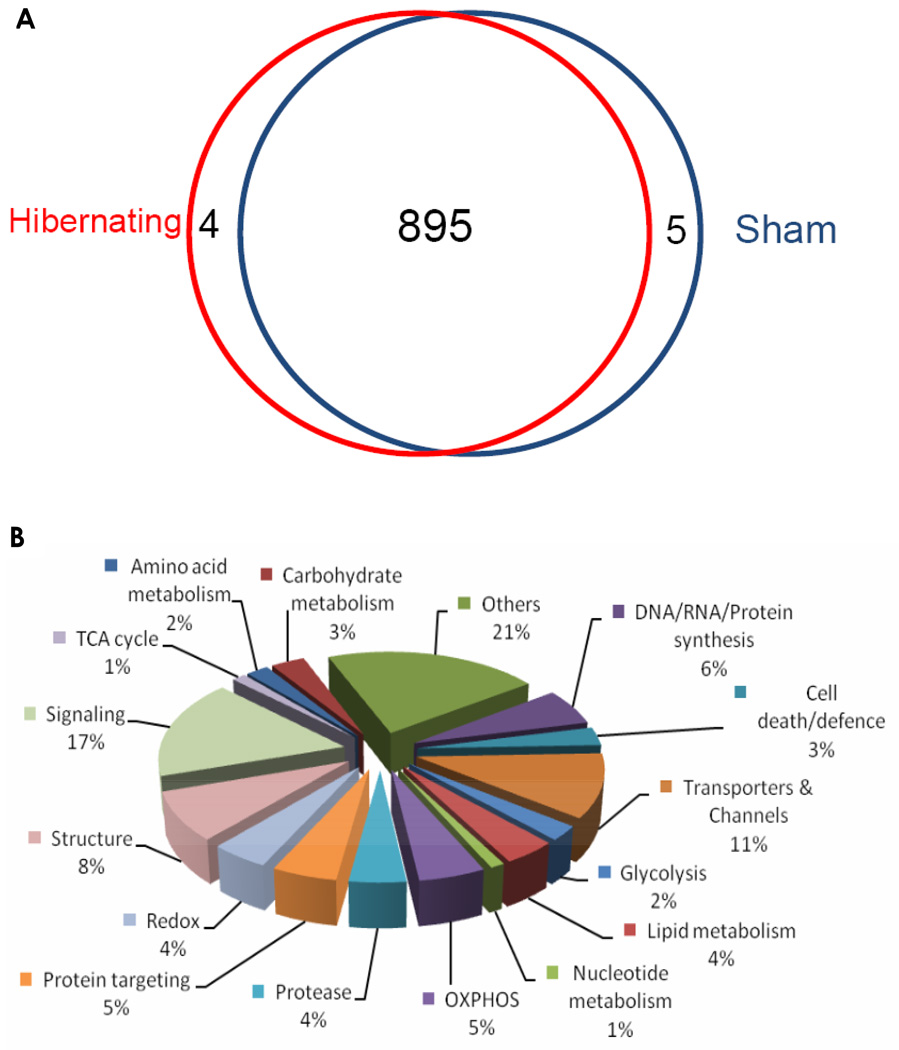
Analysis of normal and hibernating myocardium mitochondrial proteomes identified 904 proteins after application of a set of strict criteria, which included a stringent threshold for Xcorr, 10 ppm for precursor m/z tolerance, a peptide probability >95%, a protein probability >99%, and the requirement that two unique peptides must be identified for each protein (Experimental section). The complete list of identified proteins is shown in SI Table 2. Almost all proteins (895 out of 904) were identified as present in both normal and hibernating myocardium mitochondrial samples (Fig 4a). Analysis of the data using Gene Ontology database revealed that 502 proteins possessed a mitochondrial and/or membrane annotation. The distribution of functional classifications of the identified proteins is shown in Fig. 4b. A number of low abundance proteins, such as mitochondrial ribosomal proteins, uncoupling proteins, pyruvate dehydrogenase complex, and heart fatty acid-binding protein, were identified with high confidence, which suggested that the sample preparation and nano-LC/MS strategy provided a comprehensive proteomic analysis. Significant numbers of non-mitochondrial proteins were also observed. Possible explanations include contamination of the mitochondria-enriched preparation or the possibility that some proteins may be present in multiple cellular locations. Support for the latter hypothesis is provided in a previous study (36).
Figure 4.
The distributions of the 904 proteins identified in the enriched swine heart mitochondrial preparation. (A) Protein distribution in the hibernating and sham groups; clearly, vast majority of proteins were identified in both groups (cf. SI Table 2); (B) Protein distribution by their function classifications.
Proteins significantly up- or down- regulated in hibernating myocardium (compared to normal animals) were defined as those for which abundance was statistically different between the two groups (p<0 .05), and the magnitude of change in abundance ratios was <0.8 (for down-regulation) or >1.25 (for up-regulation). These thresholds reduced the potential impact of variations in mitochondrial enrichment, protein sample preparation, and nano-LC/MS analysis. Under these criteria, less than 5% of the 904 identified proteins were significantly altered in hibernating myocardium. This finding suggests that many biological activities remain intact in hibernating myocardium.
As a preliminary validation of these results, the significantly changed proteins identified were compared to results obtained in a comprehensive 2D-DIGE analysis of total myocardial proteins from the same set of animal samples, in which the objective was to identify all gel spots, regardless of whether they were differentially expressed (35). The altered mitochondrial membrane proteins that are common to the results by both methods, which were identified as changed significantly by at least one of the methods, are summarized in Table 4. Of 7 differentially regulated proteins in common between the two methods, 6 were significant as determined by the label-free profiling method. The ratios for most proteins by the label-free profiling correlated well with the DIGE data. This preliminary comparison with 2D-DIGE, which is an established method for proteomic profiling, provides further support for the quantitative results obtained using the label-free expression profiling approach described here.
Table 4.
Comparison of Protein Expression Ratios Between Hibernating and Sham Swine for Common Myocardium Mitochondrial Membrane Proteins Identified by both 2D-DIGE and Label-free LC/MS Profiling.
| Protein Name | Accession | 2D-DIGE a | LC/MSa |
|---|---|---|---|
| NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial |
P19404 | 0.73* (n=6) |
0.75* (n=10) |
| NADH dehydrogenase [ubiquinone] iron-sulfu protein 3, mitochondrial |
O75489 | 0.84 (n=8) |
0.71* (n=10) |
| NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial |
P28331 | 0.55 (n=6) |
0.68* (n=10) |
| Succinate dehydrogenase flavoprotein subunit, mitochondrial |
P31040 | 0.89 (n=7) |
0.62* (n=10) |
| Cytochrome b-c1 complex subunit 1, mitochondrial |
P31930 | 0.77* (n=8) |
0.72* (n=10) |
| ATP synthase subunit alpha, mitochondrial | P25705 | 0.62* (n=7) |
0.86 (n=10) |
| ATP synthase subunit beta, mitochondrial | Q0QEM6 | 0.56* (n=8) |
0.62* (n=10) |
Ratio of protein abundance for hibernating : sham swine
p<0.05 hibernating vs. sham
In addition to the proteins identified in common, DIGE identified 3 additional mitochondrial proteins that were significantly changed in hibernating animals vs. sham (control) animals (35). Conversely, 24 unique mitochondria-associated proteins were identified as changed significantly by label-free expression profiling but not by 2D-DIGE. Among these, 21 proteins responsible for oxidative phosphorylation, metabolism of lipids and carbohydrates, and transporters and channels were significantly down-regulated. These changes may result from reduced ATP production, metabolism, and oxygen/energy consumption in hibernating myocardial cells. Only 3 proteins were significantly up-regulated in animals with hibernating myocardium, which may reflect the stress of oxygen deprivation. Finally, a total of 20 non-mitochondrial membrane proteins were identified as significantly changed between the two groups. As discussed above, these proteins may appear in the enriched mitochondrial fractions as contamination, or may exist in multiple subcellular locations including mitochondria (36).
Further validation and extension of these unique markers is under way, including more sensitive and accurate quantification using nano-LC/triple-quadrupole MS analysis (7), as well as more extensive functional experiments. The primary objectives are to refine the list of candidate proteins that could have potential utility for clinical detection of the presence of hibernating myocardium, and to understand its physiological significance. The findings obtained will contribute to understanding the biological activities that are up- and down-regulated during myocardial hibernation, and may have significant impact upon cardiac care. From the perspective of the proteomic methodology objectives pursued in this study, the combined advances in sample preparation and analysis permitted the identification of highly informative changes in protein abundance that track with the pathophysiological condition under investigation.
Conclusion
For label-free expression profiling of tissue proteomes, a highly efficient and quantitative sample preparation method that produces a proteolytic mixture containing minimal levels of detergents, lipids and other non-protein matrix components is highly desirable but challenging to achieve, and the difficulties are accentuated for samples rich in membrane proteins, such as mitochondria. In this study, we developed a precipitation/on-pellet-digestion procedure, coupled to a nano-LC/Orbitrap, which combines efficient chromatographic separation with highly sensitive and accurate MS detection, for application to label-free expression profiling of the swine heart mitochondrial proteome. The advances and optimizations developed to meet this challenge were focused upon protein extraction, sample cleanup, and digestion, in order to obtain samples suitable for nano-LC/MS analysis. Peptide recovery, the completeness of digestion, and proteolytic products were analyzed by LTQ/ETD and LTQ/Orbitrap during method development. Under optimized conditions, complete sample protein digestion and high peptide recovery were achieved.
In addition, a nano-LC configuration featuring low void volume, highly reproducible chromatographic separation, and a long, shallow chromatographic gradient was developed to separate the highly complex tryptic peptide mixture derived from mitochondrial preparations. The sensitivity of the Orbitrap analyzer for peptide quantification was enhanced by employing a high target ion value. The overall nano-LC/MS strategy developed achieved comprehensive proteomic coverage; 904 proteins were identified with high confidence, including many low abundance proteins. The retention times and AUCs of ion currents for peptides were highly reproducible among repetitive runs, as demonstrated with a 5 day evaluation consisting of 20 replicate runs. This high level of reproducibility is essential for comprehensive profiling and label-free quantification. As proof of concept, a comparison of mitochondrial samples from healthy animals and those with hibernating myocardium was carried out. A large number of biological replicates (n=10) was employed. The quantitative false-positives were found to be very low in evaluation studies, due to (i) the large number of replicates used for each group; (ii) high and quantitative protein recoveries achieved by the sample preparation strategy, and (iii) an experimental design minimizing potential false positives arising from the analytical procedures. The proteins identified as significantly changed by this method were compared with those identified by 2D-DIGE. Proteins identified in common between the method showed good correlation in terms of magnitude of change. However, the strategy developed here identified 24 additional mitochondrial proteins that were down- or up- regulated with statistic significance, and these are under further investigation as biomarkers of hibernating myocardium.
The precipitation/on-pellet-digestion procedure, coupled to a long-gradient nano-LC/Orbitrap strategy, is robust, sensitive and straightforward, and can be adapted to the label-free expression profiling in other proteomic matrices, such as cell cultures, biofluids, and other biologically-derived samples.
Supplementary Material
Acknowledgement
This work was supported by NIH grants HL061610 and HL055324, the Department of Veterans Affairs, and the University at Buffalo Center of Protein Therapeutics grant. The LTQ/ETD system was obtained by Shared Instrumentation Grant S10-RR021221 from the National Center for Research Resources of NIH. We thank Jennifer N. Sutton from Thermo Scientific for her helpful discussions of the statistics with Sieve software.
References
- 1.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 2.Wang G, Wu WW, Zeng W, Chou CL, Shen RF. Label-free protein quantification using LC-coupled ion trap or FT mass spectrometry: Reproducibility, linearity, and application with complex proteomes. J Proteome Res. 2006;5(5):1214–1223. doi: 10.1021/pr050406g. [DOI] [PubMed] [Google Scholar]
- 3.Lilley KS, Razzaq A, Dupree P. Two-dimensional gel electrophoresis: recent advances in sample preparation, detection and quantitation. Curr Opin Chem Biol. 2002;6(1):46–50. doi: 10.1016/s1367-5931(01)00275-7. [DOI] [PubMed] [Google Scholar]
- 4.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1(5):376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 5.Oda Y, Huang K, Cross FR, Cowburn D, Chait BT. Accurate quantitation of protein expression and site-specific phosphorylation. Proc Natl Acad Sci U S A. 1999;96(12):6591–6596. doi: 10.1073/pnas.96.12.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17(10):994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 7.Qu J, Jusko WJ, Straubinger RM. Utility of cleavable isotope-coded affinity-tagged reagents for quantification of low-copy proteins induced by methylprednisolone using liquid chromatography/tandem mass spectrometry. Anal Chem. 2006;78(13):4543–4552. doi: 10.1021/ac0521697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeSouza L, Diehl G, Rodrigues MJ, Guo J, Romaschin AD, Colgan TJ, Siu KW. Search for cancer markers from endometrial tissues using differentially labeled tags iTRAQ and cICAT with multidimensional liquid chromatography and tandem mass spectrometry. J Proteome Res. 2005;4(2):377–386. doi: 10.1021/pr049821j. [DOI] [PubMed] [Google Scholar]
- 9.Li XJ, Pedrioli PG, Eng J, Martin D, Yi EC, Lee H, Aebersold R. A tool to visualize and evaluate data obtained by liquid chromatography-electrospray ionization-mass spectrometry. Anal Chem. 2004;76(13):3856–3860. doi: 10.1021/ac035375s. [DOI] [PubMed] [Google Scholar]
- 10.Chelius D, Zhang T, Wang G, Shen RF. Global protein identification and quantification technology using two-dimensional liquid chromatography nanospray mass spectrometry. Anal Chem. 2003;75(23):6658–6665. doi: 10.1021/ac034607k. [DOI] [PubMed] [Google Scholar]
- 11.Wang W, Zhou H, Lin H, Roy S, Shaler TA, Hill LR, Norton S, Kumar P, Anderle M, Becker CH. Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Anal Chem. 2003;75(18):4818–4826. doi: 10.1021/ac026468x. [DOI] [PubMed] [Google Scholar]
- 12.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics. 2005;4(10):1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Fang R, Elias DA, Monroe ME, Shen Y, McIntosh M, Wang P, Goddard CD, Callister SJ, Moore RJ, Gorby YA, Adkins JN, Fredrickson JK, Lipton MS, Smith RD. Differential label-free quantitative proteomic analysis of Shewanella oneidensis cultured under aerobic and suboxic conditions by accurate mass and time tag approach. Mol Cell Proteomics. 2006;5(4):714–725. doi: 10.1074/mcp.M500301-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Higgs RE, Knierman MD, Gelfanova V, Butler JP, Hale JE. Label-free LC-MS method for the identification of biomarkers. Methods Mol Biol. 2008;428:209–230. doi: 10.1007/978-1-59745-117-8_12. [DOI] [PubMed] [Google Scholar]
- 15.Florens L, Washburn MP, Raine JD, Anthony RM, Grainger M, Haynes JD, Moch JK, Muster N, Sacci JB, Tabb DL, Witney AA, Wolters D, Wu Y, Gardner MJ, Holder AA, Sinden RE, Yates JR, Carucci DJ. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002;419(6906):520–526. doi: 10.1038/nature01107. [DOI] [PubMed] [Google Scholar]
- 16.Silva JC, Denny R, Dorschel CA, Gorenstein M, Kass IJ, Li GZ, McKenna T, Nold MJ, Richardson K, Young P, Geromanos S. Quantitative proteomic analysis by accurate mass retention time pairs. Anal Chem. 2005;77(7):2187–2200. doi: 10.1021/ac048455k. [DOI] [PubMed] [Google Scholar]
- 17.Ono M, Shitashige M, Honda K, Isobe T, Kuwabara H, Matsuzuki H, Hirohashi S, Yamada T. Label-free quantitative proteomics using large peptide data sets generated by nano flow liquid chromatography and mass spectrometry. Mol Cell Proteomics. 2006;5(7):1338–1347. doi: 10.1074/mcp.T500039-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Le Bihan T, Goh T, Stewart II, Salter AM, Bukhman YV, Dharsee M, Ewing R, Wisniewski JR. Differential analysis of membrane proteins in mouse fore- and hindbrain using a label-free approach. J Proteome Res. 2006;5(10):2701–2710. doi: 10.1021/pr060190y. [DOI] [PubMed] [Google Scholar]
- 19.Qian WJ, Jacobs JM, Liu T, Camp DG, 2nd, Smith RD. Advances and challenges in liquid chromatography-mass spectrometry-based proteomics profiling for clinical applications. Mol Cell Proteomics. 2006;5(10):1727–1744. doi: 10.1074/mcp.M600162-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Canas B, Pineiro C, Calvo E, Lopez-Ferrer D, Gallardo JM. Trends in sample preparation for classical and second generation proteomics. J Chromatogr A. 2007;1153(1–2):235–258. doi: 10.1016/j.chroma.2007.01.045. [DOI] [PubMed] [Google Scholar]
- 21.Gu Y, Wu SL, Meyer JL, Hancock WS, Burg LJ, Linder J, Hanlon DW, Karger BL. Proteomic analysis of high-grade dysplastic cervical cells obtained from ThinPrep slides using laser capture microdissection and mass spectrometry. J Proteome Res. 2007;6(11):4256–4268. doi: 10.1021/pr070319j. [DOI] [PubMed] [Google Scholar]
- 22.Alldridge L, Metodieva G, Greenwood C, Al-Janabi K, Thwaites L, Sauven P, Metodiev M. Proteome profiling of breast tumors by gel electrophoresis and nanoscale electrospray ionization mass spectrometry. J Proteome Res. 2008;7(4):1458–1469. doi: 10.1021/pr7007829. [DOI] [PubMed] [Google Scholar]
- 23.Cutillas PR, Vanhaesebroeck B. Quantitative profile of five murine core proteomes using label-free functional proteomics. Mol Cell Proteomics. 2007;6(9):1560–1573. doi: 10.1074/mcp.M700037-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Liu X, Miller BR, Rebec GV, Clemmer DE. Protein expression in the striatum and cortex regions of the brain for a mouse model of Huntington's disease. J Proteome Res. 2007;6(8):3134–3142. doi: 10.1021/pr070092s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bodzon-Kulakowska A, Bierczynska-Krzysik A, Dylag T, Drabik A, Suder P, Noga M, Jarzebinska J, Silberring J. Methods for samples preparation in proteomic research. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;849(1–2):1–31. doi: 10.1016/j.jchromb.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 26.Wang H, Hanash S. Intact-protein based sample preparation strategies for proteome analysis in combination with mass spectrometry. Mass Spectrom Rev. 2005;24(3):413–426. doi: 10.1002/mas.20018. [DOI] [PubMed] [Google Scholar]
- 27.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68(5):850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 28.Granvogl B, Gruber P, Eichacker LA. Standardisation of rapid in-gel digestion by mass spectrometry. Proteomics. 2007;7(5):642–654. doi: 10.1002/pmic.200600607. [DOI] [PubMed] [Google Scholar]
- 29.Stewart II, Thomson T, Figeys D. 18O labeling: a tool for proteomics. Rapid Commun Mass Spectrom. 2001;15(24):2456–2465. doi: 10.1002/rcm.525. [DOI] [PubMed] [Google Scholar]
- 30.Buxton TB, Crockett JK, Moore WL, 3rd, Moore WL, Jr, Rissing JP. Protein precipitation by acetone for the analysis of polyethylene glycol in intestinal perfusion fluid. Gastroenterology. 1979;76(4):820–824. [PubMed] [Google Scholar]
- 31.Chen YY, Lin SY, Yeh YY, Hsiao HH, Wu CY, Chen ST, Wang AH. A modified protein precipitation procedure for efficient removal of albumin from serum. Electrophoresis. 2005;26(11):2117–2127. doi: 10.1002/elps.200410381. [DOI] [PubMed] [Google Scholar]
- 32.Levin B, Oberholzer VG, Whitehead TP. Serum Protein Fractions: A Comparison of Precipitation Methods with Electrophoresis. J Clin Pathol. 1950;3(3):260–265. doi: 10.1136/jcp.3.3.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang L, He L, Fountoulakis M. Comparison of protein precipitation methods for sample preparation prior to proteomic analysis. J Chromatogr A. 2004;1023(2):317–320. doi: 10.1016/j.chroma.2003.10.029. [DOI] [PubMed] [Google Scholar]
- Fallavollita JA, Malm BJ, Canty JM., Jr Hibernating myocardium retains metabolic and contractile reserve despite regional reductions in flow, function, and oxygen consumption at rest. Circ Res. 2003;92(1):48–55. doi: 10.1161/01.res.0000049104.57549.03. [DOI] [PubMed] [Google Scholar]
- 35.Page B, Young R, Iyer V, Suzuki G, Lis M, Korotchkina L, Patel MS, Blumenthal KM, Fallavollita JA, Canty JM., Jr Persistent regional downregulation in mitochondrial enzymes and upregulation of stress proteins in swine with chronic hibernating myocardium. Circ Res. 2008;102(1):103–112. doi: 10.1161/CIRCRESAHA.107.155895. [DOI] [PubMed] [Google Scholar]
- 36.Taylor SW, Fahy E, Zhang B, Glenn GM, Warnock DE, Wiley S, Murphy AN, Gaucher SP, Capaldi RA, Gibson BW, Ghosh SS. Characterization of the human heart mitochondrial proteome. Nat Biotechnol. 2003;21(3):281–286. doi: 10.1038/nbt793. [DOI] [PubMed] [Google Scholar]
- 37.Gaucher SP, Taylor SW, Fahy E, Zhang B, Warnock DE, Ghosh SS, Gibson BW. Expanded coverage of the human heart mitochondrial proteome using multidimensional liquid chromatography coupled with tandem mass spectrometry. J Proteome Res. 2004;3(3):495–505. doi: 10.1021/pr034102a. [DOI] [PubMed] [Google Scholar]
- 38.Kashino Y. Separation methods in the analysis of protein membrane complexes. J Chromatogr B Analyt Technol Biomed Life Sci. 2003;797(1–2):191–216. doi: 10.1016/s1570-0232(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 39.Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Analytical Biochemistry. 1984;138(1):141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 40.Qu J, Straubinger RM. Improved sensitivity for quantification of proteins using triply charged cleavable isotope-coded affinity tag peptides. Rapid Commun Mass Spectrom. 2005;19(19):2857–2864. doi: 10.1002/rcm.2138. [DOI] [PubMed] [Google Scholar]
- 41.Wang H, Straubinger RM, Aletta JM, Cao J, Duan X, Yu H, Qu J. Accurate localization and relative quantification of arginine methylation using nanoflow liquid chromatography coupled to Electron Transfer Dissociation and Orbitrap mass spectrometry. J. Amer. Soc. Mass Spectr. 2008p doi: 10.1016/j.jasms.2008.11.008. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen EI, Cociorva D, Norris JL, Yates JR., 3rd Optimization of mass spectrometry-compatible surfactants for shotgun proteomics. J Proteome Res. 2007;6(7):2529–2538. doi: 10.1021/pr060682a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruiz-Romero C, Calamia V, Mateos J, Carreira V, Martinez-Gomariz M, Fernandez M, Blanco FJ. Mitochondrial dysregulation of osteoarthritic human articular chondrocytes analyzed by proteomics: A decrease in mitochondrial superoxide dismutase points to a redox imbalance. Mol Cell Proteomics. 2008 doi: 10.1074/mcp.M800292-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Premstaller A, Oberacher H, Walcher W, Timperio AM, Zolla L, Chervet JP, Cavusoglu N, van Dorsselaer A, Huber CG. High-performance liquid chromatography-electrospray ionization mass spectrometry using monolithic capillary columns for proteomic studies. Anal Chem. 2001;73(11):2390–2396. doi: 10.1021/ac010046q. [DOI] [PubMed] [Google Scholar]
- 45.Shen Y, Zhang R, Moore RJ, Kim J, Metz TO, Hixson KK, Zhao R, Livesay EA, Udseth HR, Smith RD. Automated 20 kpsi RPLC-MS and MS/MS with chromatographic peak capacities of 1000–1500 and capabilities in proteomics and metabolomics. Anal Chem. 2005;77(10):3090–3100. doi: 10.1021/ac0483062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.