Abstract
Spliced leader (SL) trans-splicing is a common mRNA processing mechanism in dinoflagellates, in which a 22-nt sequence is transferred from the 5′-end of a small noncoding RNA, the SL RNA, to the 5′-end of mRNA molecules. Although the SL RNA gene was shown initially to be organized as tandem repeats with transcripts of 50–60 nt, shorter than most of their counterparts in other organisms, other gene organizations and transcript lengths were reported subsequently. To address the evolutionary gradient of gene organization complexity, we thoroughly examined transcript and gene organization of the SL RNA in a phylogenetically and ecologically diverse group of dinoflagellates representing four Orders. All these dinoflagellates possessed SL RNA transcripts of 50–60 nt, although in one species additional transcripts of up to 92 nt were also detected. At the genomic level, various combinations of SL RNA and 5S rRNA tandem gene arrays, including SL RNA–only, 5S rRNA–only, and mixed SL RNA–5S rRNA (SL–5S) clusters, were amplified by polymerase chain reaction for six dinoflagellates, containing intergenic spacers ranging from 88 bp to over 1.2 kb. Of these species, no SL–5S cluster was detected in Prorocentrum minimum, and only Karenia brevis showed the U6 small nuclear RNA gene associated with these mixed arrays. The 5S rRNA–only array was also found in three dinoflagellates, along with two SL–5S-adjacent arrangements found in two other species that could represent junctions. Two species contained multimeric SL exon repeats with no associated intron. These results suggest that 1) both the SL RNA tandem repeat and the SL–5S cluster genomic organizations are an “ancient” and widespread feature within the phylum of dinoflagellates and 2) rampant genomic duplication and recombination are ongoing independently in each dinoflagellate lineage, giving rise to the highly complex and diversified genomic arrangements of the SL RNA gene, while conserving the length and structure of the functional SL RNA.
Keywords: Dinoflagellate, SL RNA, complex genomic arrangement
Introduction
Spliced leader (SL) trans-splicing has been found in a phylogenetically disjointed group of eukaryotes (Hastings 2005), in which a short RNA fragment (i.e., SL, ∼15–50 nt) from a small noncoding RNA (SL RNA) is spliced at the splice acceptor site in the 5′-untranslated region of an independently transcribed pre-mRNA. Mature mRNAs are formed with the SL sequence occupying their 5′ ends (for reviews see Blumenthal 2005; Hastings 2005; Mayer and Floeter-Winter 2005). This process can have a multitude of functions: 1) generating translatable monocistronic mRNAs from polycistronic precursor transcripts; 2) sanitizing the 5′ end of mRNAs; 3) stabilizing mRNAs; and 4) regulating gene translation. Among the organisms examined, SL trans-splicing is found in Euglenozoa, nematodes, platyhelminthes, cnidarians, rotifers, ascidians, and appendicularia. The SL RNA contains two functional domains: an exon (i.e., SL) that is transferred to a pre-mRNA and an intron that contains a consensus U-rich binding site (Sm-binding motif) for the assembly of small nuclear ribonucleoprotein particles (snRNPs). The SL RNA bears low sequence similarity across phyla; however, a three-stem-loop secondary structure is conserved in most lineages (Bruzik et al. 1988; Mayer and Floeter-Winter 2005). The SL 5′-cap structure in different organisms is not always the same. In trypanosomes, the SL carries a hypermethylated structure, consisting of an inverted 7-methylguanosine (m7G) followed by 4 nt (nucleotides) with 2′-O-ribose and three base methylations (termed cap 4). In worms, the SL carries an inverted 2,2,7-trimethylguanosine 5′-cap. In both groups, the heptameric Sm-protein complex, a structure formed on several U-rich snRNPs and involved in both cis- and trans-splicing, interacts with the SL RNAs through the Sm-binding motif.
The presence of SL trans-splicing was described recently in dinoflagellates (Zhang, Hou, et al. 2007), a group of unicellular eukaryotes belonging to the Alveolata lineage that contribute significantly to marine primary production, growth of coral reefs, and harmful algal blooms. Through the analysis of hundreds of full-length cDNAs from 15 representative species of dinoflagellates, we demonstrated that nuclear-encoded mRNAs in all species, from ancestral to derived lineages, are trans-spliced with the addition of the 22-nt conserved SL, DCCGUAGCCAUUUUGGCUCAAG (D = U, A, or G). In dinoflagellates, the primary structure of SL RNA appears to be different from most of its counterparts in other organisms: 1) the SL RNA transcripts are unusually short at 50–64 nt, with a conserved Sm-binding motif (AUUUUGG) located in the SL (exon) rather than the intron, as in other organisms; and 2) the 5′-terminal position is predominantly U or A, a feature that may affect capping and subsequent translation and stability of the recipient mRNA. Because the association of the Sm complex with U-rich small nuclear RNAs (snRNAs) in vertebrates signals nuclear import, its presence in the dinoflagellate SL creates the paradox as to how the Sm-binding motif could remain on mature mRNAs without impeding cytosolic localization or translation of the mRNAs.
From another dinoflagellate, Karenia brevis (Wilson isolate), an SL RNA gene (KbrSL) was found locating in a 5S ribosomal RNA (5S rRNA) gene cluster (Lidie and van Dolah 2007), an organization detected in the kinetoplastids (Santana et al. 2001), cnidarian (Stover and Steele 2001), and nematodes (Blaxter and Liu 1996). The transcript of KbrSL was estimated to be ∼150 nt based on RNA blot analysis although a ∼60-nt RNA band was also apparent (however was not discussed). A predicted 148-nt KbrSL transcript was modeled into a typical three-stem-loop structure with a potential SL splice site at nucleotide 32 and a potential Sm-binding motif in the intron. The same gene, however, was also predicted to be 125 bp with the SL splice site at nucleotide 27 in the same study. These results prompted us to ask if organization of SL RNA and its gene represented an evolutionary trend within the dinoflagellate phylum. To address this issue, we systematically examined SL RNA size and genomic structure for K. brevis strain CCMP2228, along with a selection of phylogenetically diverse dinoflagellates (table 1). The species chosen for this study includes representatives of five dinoflagellate Orders including Gymnodiniales, Peridiniales, Prorocentrales, and Suessiales that are distributed throughout the phylogenetic spectrum (Saldarriaga et al. 2001; Zhang, Bhattacharya, and Lin 2007; Zhang, Hou, et al. 2007). The species represent isolates with distinct autotrophic, heterotrophic, and mixotrophic nutritional requirements and polar and subtropical ecological niches. We found that although the size of dinoflagellate SL RNA transcripts can vary from 42 to 92 nt, the major ones are 56–59 nt; there was no evidence for an SL RNA transcript of 150 nt in any of these dinoflagellates. Both the length and sequence of SL RNA are conserved in all dinoflagellates, including K. brevis, and the SL RNA gene is organized both in single gene tandem repeats and in mixed SL RNA–5S rRNA (SL–5S) arrangements, with numerous variations. Evolution of the SL RNA gene organization among phylogenetically diverse dinoflagellate lineages and of gene arrangement within a species are demonstrated. The diverse SL genomic structure appears to be a result of rampant genomic duplication and chromosomal recombination; however, the complexity of SL gene structure does not mirror the proposed evolutionary tree.
Table 1.
Taxonomic and Ecotypic Distribution of Dinoflagellates Studied and Their SL RNA Genomic Structures
| Kbr | Kve | Pmi | Ppi | Pgl | Har | |
| Straina | CCMP2228 | CCMP1975 | CCMP696 | CCMP1831 | CCMP2088 | CCMP445 |
| Ecotype | Toxic, autotrophic, subtropical | Toxic, mixotrophic, temperate | Autotrophic, global | Heterotrophic, wide geographic range | Autotrophic, polar regions | Autotrophic, Arctic |
| SL RNA repeat type | SL; SL-U6-5S | SL; SL–5S | SL | SL; SL–5S | SL; SL–5S | SL; SL–5S |
Kbr, Karenia brevis; Kve, Karlodinium veneficum; Pmi, Prorocentrum minimum; Ppi, Pfiesteria piscicida; Pgl, Polarella glacialis; and Har, Heterocapsa arctica.
From Provasoli–Guillard National Center for Culture of Marine Phytoplankton, West Boothbay Harbor, Maine.
Materials and Methods
Selection of Dinoflagellate Species and Cultures
A new strain of K. brevis and other dinoflagellate species ranging from more basal lineages (Polarella, Heterocapsa) to more derived (Prorocentrum, Pfiesteria) as well as a phylogenetically related species (Karlodinium veneficum; Zhang, Bhattacharya, and Lin 2007) were selected for analysis in this study. The phylogenetic affiliations of these lineages were examined using multi-gene analysis except Polarella, whose basal position was presumed based on its close relationship with Symbiodinium (Montresor et al. 1999). Karenia brevis (CCMP2228) and the two arctic dinoflagellates, Polarella glacialis (CCMP2088) and Heterocapsa arctica (CCMP445), were grown in f/2 seawater medium at 25 °C (K. brevis) and 4 °C (P. glacialis and H. arctica), respectively, at a 12 h:12 h light:dark photocycle with a photon flux of approximately 50 μE·m−2 s−1. When the cultures were in the exponential growth phase, cells were harvested by centrifugation at 3,000 × g at 25 °C (for K. brevis) or 4 °C (for P. glacialis and H. arctica), and the cell pellet for each species was resuspended thoroughly in TRIzol (Invitrogen) for RNA extraction or in DNA buffer for DNA extraction (Zhang, Hou, et al. 2007). Cultures of other dinoflagellate species have been reported previously (Zhang, Hou, et al. 2007).
RNA Blot Analyses of SL RNA and 5S rRNA in Dinoflagellates
Total RNA from 106 cells of K. brevis, P. glacialis, K. veneficum (formerly Karlodinium micrum; CCMP1975), Pfiesteria piscicida (CCMP1831), and P. minimum (CCMP696) were loaded in an 8% acrylamide/8 M urea gel, subjected to electrophoresis and transferred to nylon membranes (Sturm et al. 1999). This medium resolution gel is optimal for small RNAs below 350 nt, but does not reveal the SL-containing mRNA bands for kinetoplastids or dinoflagellates (Sturm et al. 1999; Zhang, Hou, et al. 2007). Oligonucleotide probes used for hybridization included dinoSLa/s for detection of the general dinoflagellate SL RNA transcripts, and dino5Sa/s for dinoflagellate 5S rRNA (all oligonucleotides used in this study are listed in table 2). Total RNA from Leishmania tarentolae cells was included to provide known size markers. Oligonucleotide probes were labeled with 32P-γATP for hybridization (Sturm et al. 1999).
Table 2.
Oligonucleotides Used in This Study
| Primer Name | Sequence (5′–3′) | Application; Referencea |
| dinoSLg-F | cgagagtatcAGCCATTTTGGCTCAAGb | PCR of genomic SL RNA tandem repeats; Zhang, Hou, et al. (2007) |
| dinoSLg-R | acagaacaAGCCAAAATGGCTACGGb | PCR of genomic SL RNA tandem repeats; Zhang, Hou, et al. (2007) |
| dino5SF1 | GCCATACCGTGTCGAATGC | 5S rRNA forward primer |
| dino5SF2 | CGACCTCCGAAGTTAAGCG | 5S rRNA forward primer |
| dino5SR1 | ACAGCACCCTAAGGWCTTCC | 5S rRNA reverse primer |
| KbrSL1-F1 | AGCCATTTTGGCTCAAGGTACAAG | KbrSL-1 forward primer |
| KbrSL3-F1 | CGTAGCCATTTTGGCTCAAGGC | KbrSL-3 forward primer |
| KbrSL4-F1 | AGCCATTTTGGCTCAAGGTCTAC | KbrSL-4 forward primer |
| KmiSL-F3 | GCTCAAGGTACAAGTTGGGCTG | KbrSL-1 forward primer; Zhang, Hou, et al. (2007) |
| KbrSL3-F2 | AGCCATTTTGGCTCAAGGCACC | KbrSL-3 forward primer |
| KbrSL4-F2 | ATTTTGGCTCAAGGTCTACATCTG | KbrSL-4 forward primer |
| dinoSLa/s | TGTACCTTGAGCCAAAATG | General dinoflagellate SL RNA detection; Zhang, Hou, et al. (2007) |
| dinoSL | NCCGTAGCCATTTTGGCTCAAG | PCR amplifying dinoflagellate full-length cDNAs as well as SL RNA genomic DNA |
| dino5Sa/s | GGACTTCCCGGGCGGTC | General 5S rRNA detection |
| KbrSL-4a/s | AGCCCAGATGTAGACCT | Karenia brevis SL RNA type SL-4 detection |
Oligonucleotides from this study show no reference.
Lowercase letters represent random nucleotides.
Isolation of Nucleic Acids from Dinoflagellates
Genomic DNA from K. brevis, P. glacialis, and H. arctica was extracted following a protocol using Cetyltrimethylammonium bromide (Zhang and Lin 2005). DNA from K. veneficum, P. piscicida, and Prorocentrum minimum was prepared as reported (Zhang, Hou, et al. 2007). Total RNA from K. brevis and P. glacialis was extracted following Lin et al. (2002). Total RNA isolation from P. piscicida, P. minimum, and K. veneficum has been reported previously (Zhang, Hou, et al. 2007).
Analyses of SL RNA Genomic Arrangements in Six Dinoflagellates
The SL RNA gene encompassing at least two tandem loci (Zhang, Hou, et al. 2007) was amplified by using primers dinoSLg-F and dinoSLg-R (table 2). DNA extracted from 104 and 103 dinoflagellate cells was used as the template in polymerase chain reaction (PCR) under a touchdown PCR program: 5 cycles of 95 °C for 20 s, and 72 °C for 1.5 min; 5 cycles of 95 °C for 20 s, 65 °C for 30 s, and 72 °C for 1 min; 5 cycles of 95 °C for 20 s, 60 °C for 30 s, and 72 °C for 1 min; and 15 cycles of 95 °C for 20 s, 58 °C for 30 s, and 72 °C 1 min. This PCR program had been shown to be efficient to amplify up to eight SL RNA gene repeats in the three dinoflagellates (Zhang, Hou, et al. 2007). PCR products were purified and cloned into a T-vector, and the resulting colonies were picked randomly and sequenced as reported (Zhang, Hou, et al. 2007).
Two forward primers (dino5SF1 and dino5SF2) and one reverse primer (dino5SR1) were designed in the conserved region of the reported 5S rRNA gene in Crypthecodinium cohnii (GenBank accession number M25115) and K. brevis (Lidie and van Dolah 2007). PCR was run under conditions that favored the generation of larger products following Zhang and Lin (2003) with some modification: TAKARA LA Taq Polymerase (Takara Mirus Bio) was used, and 35 cycles were run with 20 s of denaturation at 98 °C, 30 s of annealing at 60 °C, and 3 min of elongation at 72 °C. Using this Taq polymerase and a similar PCR program, we have amplified successfully tandem repeats of Rubisco gene (1.7–4.7 kb) from P. minimum (Zhang and Lin 2003). Combinations of the forward primers with dinoSL were also used. Two rounds of PCR were carried out for K. brevis, K. veneficum, P. piscicida, and P. minimum, as well as the two arctic species P. glacialis and H. arctica. The first round of PCR was performed using primer set dino5SF1–dino5SR1, dino5SF2–5SR1, dino5SF1–dinoSL, and dino5SF2–dinoSL, whereas the second round (nested) PCR was conducted using a 100-fold dilution of the first-round products as template with primer sets dino5SF2–dino5SR1, dino5SF1–dinoSL, and dino5SF2–dinoSL (for dino5SF1–dino5SR1 products), dino5SF1–dinoSL, and dino5SF2–dinoSL (for dino5SF1–dino5SR1 products), or dino5SF2–dinoSL (for dino5SF2–dinoSL products). For comparison, PCR with TAKARA EX Taq Polymerase (Takara Mirus Bio) and the above-mentioned primer combinations was also run using the following program: 35 cycles of 94 °C for 30 s, 58 °C for 40 s, 72 °C for 1 min followed by 1 cycle of 10 min at 72 °C.
Rapid Amplification of cDNA 3′ End (3′-RACE) of K. brevis SL RNA
Poly(A) mRNA was depleted from K. brevis total RNA, and a poly(A) tail was added to the remaining population using Escherichia coli Poly(A) Polymerase (Takara Mirus Bio) as reported (Zhang, Hou, et al. 2007). First-strand cDNA synthesized using GeneRacer Oligo dT primer (Invitrogen) was used as PCR template. Two rounds of touchdown PCR were carried out as described above with the extension time at 72 °C for 5 s. The first round of PCR was performed using KbrSL1F1, KbrSL2F1, or KbrSL3F1 paired with GeneRacer3 as the primers. The second round PCR used a 100-fold dilution of the first-round PCR products as the template with KbrSL1F2, KbrSL2F2, or KbrSL3F2 paired with GeneRacer3 as the nested primers (table 2).
Modeling of RNA Structure in K. brevis, P. piscicida, P. glacialis, and H. arctica
Based on the cDNA sequences obtained, we modeled SL RNA structures for K. brevis using MFOLD: prediction of RNA secondary structure modeling program (http://bioweb.pasteur.fr/seqanal/interfaces/mfold-simple.html). Modeling was also done for SL RNA predicted based on genomic sequences for the P. piscicida SL–5S form, P. glacialis, and H. arctica, by identifying conserved regions in the alignment of SL RNA genes with all the mapped dinoflagellate SL RNA transcripts (see fig. 3). Folding was performed at 25 °C for K. brevis, 20 °C for P. piscicida, and 4 °C for P. glacialis and H. arctica, temperatures at which these algae were cultured. The default setting was used because the constraints used in previous SL RNA models (Bruzik et al. 1988) stipulating that the splice-donor dinucleotide [“gu(c)” in “GCUCAAGgu(c)”] be double stranded and the putative Sm-binding site (AUUUUGG) be single stranded would lead to unstable structures for all but P. glacialis tandem repeat type SL RNA. Folding was also performed at 10 and 30 °C or K. brevis and P. piscicida, and 20 °C for P. glacialis and H. arctica to examine theoretical effects of temperature.
FIG. 3.—
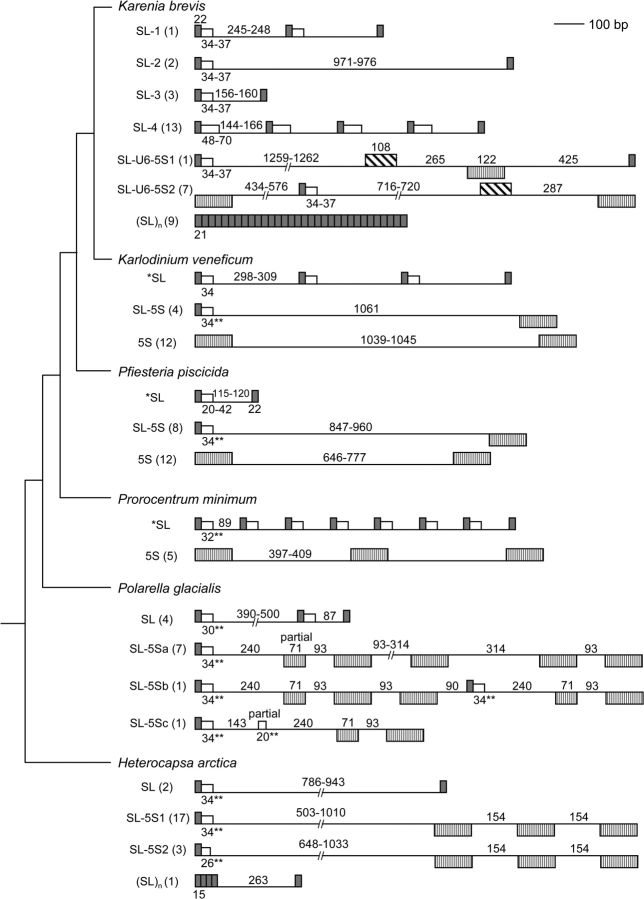
The complex genomic organizations of SL RNA genes in six dinoflagellates. Genes are oriented relative to the direction of the SL RNA gene when in tandem, and gene boxes below the line are transcribed from the opposite strand. Gray box, SL exon; open box, SL intron; vertically hatched box, 5S rRNA gene; diagonally hatched box, U6 snRNA gene; thin line, intergenic region; numbers near the lines or boxes depict length of sequence segments; unnumbered boxes are of the same length as immediately prior counterpart. *SL, structures reported previously (Zhang, Hou, et al. 2007); **, predicted intron sizes. Numbers in parentheses indicate the clones obtained for that type of SL RNA gene. Species are arranged so that basal taxa are on the bottom and later diverging taxa on the top, and their phylogenetic positions based on Zhang, Bhattacharya, and Lin (2007) are indicated in the clustering pattern shown on the left.
Results
Major Dinoflagellate SL RNA Transcripts Are 50–60 nt
In the previous study on the SL RNA transcripts for the Wilson isolate of K. brevis (Lidie and van Dolah 2007), two RNA bands were visualized by RNA blot analysis, with the larger being about 150 nt, and the smaller migrating in the size range predicted for other dinoflagellate SL RNA substrate transcripts (Zhang, Hou, et al. 2007). A 125- to 148-nt SL RNA transcript including an intronic Sm-binding site would alter the direction of future experiments in dinoflagellate trans-splicing. To confirm the larger SL RNA transcript size in this and possibly other dinoflagellate lineages, K. brevis strain CCMP2228 was examined along with a diverse cohort of dinoflagellates for both transcript size and genomic organization.
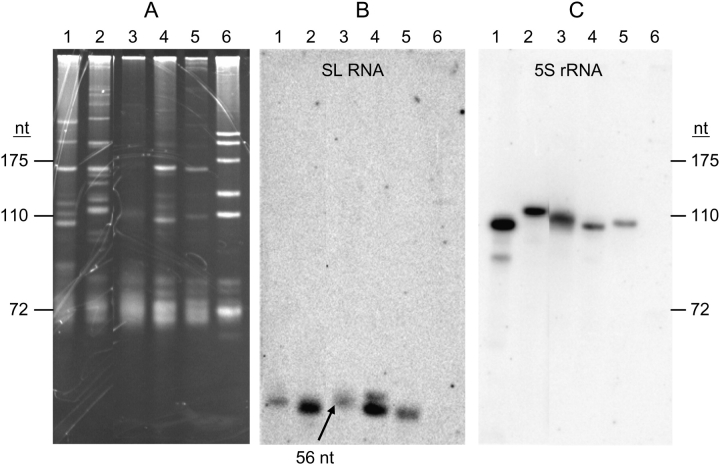
The SL RNA transcripts from K. brevis, three dinoflagellates studied previously (K. veneficum, P. piscicida, and P. minimum; Zhang, Hou, et al. 2007) and the unstudied P. glacialis were examined by RNA blot. Specific oligonucleotide probe dinoSLa/s was designed to recognize 14 nt of the exon and the first 5 nt of the intron, sequences that are conserved throughout the dinoflagellate SL RNAs we examined previously (Zhang, Hou, et al. 2007). Ethidium bromide staining of a medium resolution polyacrylamide gel showed that the small RNA molecules were not identical in different dinoflagellate species (fig. 1A). In contrast to L. tarentolae RNA, which contained five major small rRNA bands and a spectrum of smaller tRNA bands, the dinoflagellates contained two major bands and a number of fainter bands, depending on the species. Eight to 10 bands were visible in the K. brevis RNA sample. Hybridization of the subsequent RNA blot revealed that the SL RNA transcripts detected by probe dinoSLa/s migrated faster than 72 nt in all genera of dinoflagellates (fig. 1B). Probe specificity was indicated by absence of hybridization bands in L. tarentolae. Pfiesteria piscicida had two SL RNA size classes, of which the smaller was more abundant, consistent with our previous result (Zhang, Hou, et al. 2007). The size of the K. brevis SL RNA transcripts (KbrSL) appeared to be identical with its counterpart in K. veneficum (56 nt) and similar to those in P. piscicida and P. glacialis. In the present study, to avoid the potential signal decrease when the same blot was reused to detect the sizes of 5S rRNA for dinoflagellates (see the next section), we did not use the 32P-labeled oligonucleotide S-255 to hybridize with the heterogeneous cytochrome oxidase subunit III guide RNA of L. tarentolae (55–60 nt) as the size marker for dinoflagellate SL RNA, as we have done in the previous study (Zhang, Hou, et al. 2007). However, because SL RNA transcripts in K. veneficum, P. piscicida, and P. minimum have been clearly shown to be in the 50–60 nt size range (Zhang, Hou, et al. 2007), and the hybridized bands for the two newly examined taxa (K. brevis and P. glacialis) fell also into the same range (fig. 1B), despite the lack of a hybridized marker lane in RNA blot, it is clear that for all the dinoflagellates examined so far, the size of SL RNA transcripts ranged 50–60 nt. This result was further supported by 3′-RACE of SL RNA (see the section “3′-End Analysis for K. brevis SL RNA Transcripts”). The total RNA amount of K. brevis used in the RNA blot was similar to those of P. glacialis and P. piscicida as revealed by ethidium bromide staining (fig. 1A); however, the intensity of KbrSL hybridized band was weaker, suggesting a lower expression level of SL RNA in K. brevis than other dinoflagellates. The size of this band is consistent with the “minor” band reported for K. brevis (Lidie and van Dolah 2007), whereas the “major” ∼150-nt species detected in that study did not appear in our analysis (fig. 1B), and is likely to have been an artifact of the probe used.
FIG. 1.—
Dinoflagellate SL RNAs are 50–60 nt. Polyacrylamide gel electrophoresis of total RNA (A), and blot hybridization of probes for SL RNA using probe DinoSLa/s (B), and 5S rRNA using probe Dino5S (C). Lane 1, Karenia brevis; 2, Polarella glacialis; 3, Karlodinium veneficum; 4, Pfiesteria piscicida; 5, Prorocentrum minimum; and 6, Leishmania tarentolae. Size standards (marked on the left and the right) are L. tarentolae 5.8S rRNA (175 nt), 5S rRNA (110 nt) and tRNAGly (72 nt), and the RNA blots (B,C) were aligned with the gel so that the size standards apply for the RNA blots. The single band of K. veneficum SL RNA transcript (marked by an arrow) has been proven to be 56 nt in length by RACE-based cloning and sequencing (Zhang, Hou, et al. 2007).
Dinoflagellate 5S RNA Transcripts Are 100–120 nt
Based on the alignment of 5S rRNA genes obtained with the reported sequences (see below for details), the predicted sizes of 5S rRNA for the six dinoflagellates were all 122 nt (fig. 2). To validate the identity of the 5S rRNA band from the small size RNA mixture around 120 nt (fig. 1A), the RNA blot was rehybridized with radiolabeled oligonucleotide dino5Sa/s (fig. 1C), whose sequence was based on conservation of the K. brevis 5S rRNA with that from C. cohnii (Hinnebusch et al. 1981). One major band of hybridization was observed at approximately 100–120 nt for K. brevis, P. glacialis, K. veneficum, P. piscicida, and P. minimum, although the band for K. brevis, K. veneficum, and P. minimum appeared to be a little larger (110 nt) than that of P. piscicida (100 nt), and P. glacialis 5S rRNA the largest of all (120 nt). Differences in relative hybridization strength (fig. 1C) were consistent with the different intensities of the corresponding RNA bands visualized by ethidium bromide staining (fig. 1A) and the sequence variation in the region covered by the oligonucleotide probe (positions 89–105; fig. 2).
FIG. 2.—
Alignment of dinoflagellate 5S ribosomal RNA gene using ClustalX. Promoter elements are indicated in Box A, I, and Box C. The region of dino5S probe binding is indicated by an arrow; positions where nucleotides differ from the probe are marked with shading. Har, Heterocapsa arctica; Kbr, Karenia brevis; Kve-1, Karlodinium veneficum, 5S in tandem repeats; Kve-2, K. veneficum, 5S clustered with SL RNA; Pgl, Polarella glacialis; Pmi, Prorocentrum minimum; Ppi-1, Pfiesteria piscicida, 5S in tandem repeats; and Ppi-2, P. piscicida, 5S clustered with SL RNA. “–” indicates missing nucleotides. Cco, Crypthecodinium cohnii (M25115); Pat (Perkinsus atlanticus, AF509333) are shown for reference.
Karenia brevis SL RNA Genes: SL–SL Amplification
To address the genomic organization of the SL RNA gene in K. brevis, we first amplified KbrSL from K. brevis strain CCMP2228 using the opposing SL primer set dinoSLg-F/dinoSLg-R. Agarose gel electrophoresis of the PCR products revealed various bands of ∼0.3 to >2 kb. Twenty-seven clones obtained from these PCR products revealed a total of six different types of SL RNA gene repeats containing either one to five full units of exon and intron with an intergenic region, or up to 24 units of 21-bp exon repeats (SL)n. Although the possible existence of additional forms cannot be excluded, the arrangements of the SL RNAs found in K. brevis and the other dinoflagellates examined in this and previous studies are already highly diverse, as shown in schematic diagrams (fig. 3) and alignments of the SL RNA (fig. 4).
FIG. 4.—
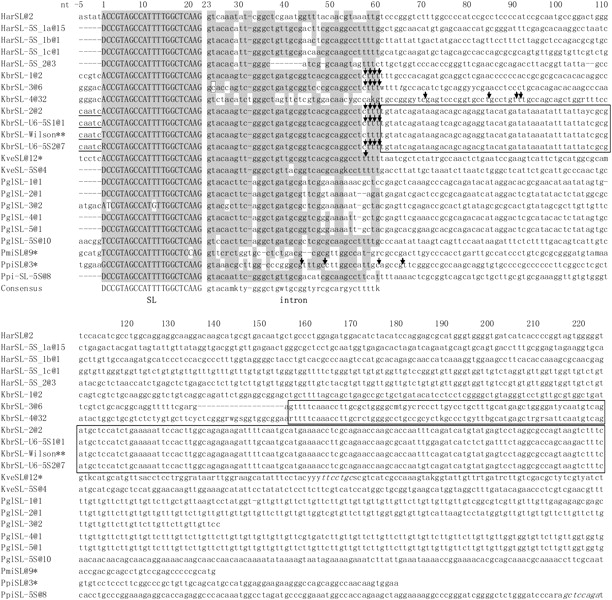
Conservation of the ∼60-bp dinoflagellate SL RNA coding region (SL + intron). In the alignment are representative sequences for each type; the number of identical clones retrieved for each type is indicated by “@number” following the species abbreviation and type number. Har, Heterocapsa arctica; Kbr, Karenia brevis; Kve, Karlodinium veneficum; Ppi, Pfiesteria piscicida; Pgl, Polarella glacialis; and Pmi, Prorocentrum minimum. SL refers to SL RNA sequences obtained from SL-only repeats; SL–5S indicates SL RNA sequences from genes associated with 5S rRNA genes. *: Sequence from Zhang, Hou, et al. (2007); **: sequence from Lidie and van Dolah (2007). SL RNAs mapped by 3′-RACE analyses are denoted by arrows to indicate the terminal positions. The 22-nt SL (exon) was shown in uppercase letters, whereas the intron and the downstream region were in lowercase letters. Shaded are conserved positions defined as identical in over six sequences in at least three species. Where available, 5nt upstream of the 22-bp SL are shown for evaluation of a potential initiator element; sequences consistent with the initiator motif are underlined. Nucleotides existing in some clones but absent in others were italicized. A noncanonical C in the splice-donor site of KbrSL-3 is boxed. The conserved intergenic spacer region between different types of KbrSL are also boxed. Gaps introduced in the sequence alignment are shown as –.
Among these six different types of SL RNA, KbrSL-1 contained two full SL RNA repeats (GenBank accession number FJ434700). The length of a full repeat unit for this type was 304 bp, falling between that of the K. veneficum (354–365 bp) and of P. piscicida (179 bp) or P. minimum (143 bp) SL RNA genes (Zhang, Hou, et al. 2007) (fig. 3). KbrSL-1 shared the greatest similarity (95%) to K. veneficum SL RNA in the first 60 bp, the region corresponding to the predicted dinoflagellate SL RNA (Zhang, Hou, et al. 2007), and similarity diminished downstream (fig. 4).
Two clones of the KbrSL-2 type contained two SL repeats with one full repeat unit (FJ434701–2); the unit lengths were 1,030 and 1,032 bp, respectively. Sequences of these two clones were very similar (98%), with two indels and a few nt substitutions. The KbrSL-2 sequences were identical to KbrSL-1 for the first 60 bp and then diverged. Despite their large size, no additional genes were found in the intergenic spacers (figs. 3 and 4).
Six clones contained one full unit of KbrSL-3 (215–216 bp for one full SL unit; FJ434692, FJ434703–7) with some nt substitutions and a deletion in one of the clones. This type was similar to KbrSL-1, KbrSL-2, and the KbrSL-U6-5S types (see below) for the first 59 bp, except for one “T” to “C” transition at the splice-donor dinucleotide (“GT” to “GC”) and two “A” to “C” transversions in the intron but shared little similarity downstream (figs. 3 and 4).
Thirteen clones were found to contain one to four full repeats designated as KbrSL-4 (FJ434692–94, FJ434704–13), including five clones that contained one unit of KbrSL-3 in the upstream region (FJ434692, FJ434704–7). The length of each KbrSL-4 unit was identical (236 bp) in all clones, with a number of nt substitutions found in different units (fig. 4). Intron sequence of this type was distinct compared with the other types of KbrSL as well as other dinoflagellate SL RNAs. Notably, the poly T tracts that exist in the other types of KbrSL and most of the dinoflagellate SL RNA in the 55–60-nt region were absent in this type. In RNA blot analysis using DinoSLa/s as the probe, only one band of 55–60 nt was detected (fig. 1B). A probe derived from the intron sequence of KbrSL-4 (table 2) did not yield any signal (data not shown), suggesting that its expression level may be too low to be detected by RNA blot, or that the signal may be masked by the abundant tRNA population, as we have observed before in other organisms (Sturm S and Campbell D, unpublished data); however, expression of KbrSL-4 was confirmed by the presence of KbrSL-4 sequences in 3′ RACE (see fig. 4). The last 81 bp of the intergenic region of KbrSL-4 shared 90–97% identity to that of KbrSL-3, suggesting these two types of KbrSL might have similar origin.
A bizarre arrangement of SL RNA was found in the Kbr(SL)n type, which consisted of 10–24 exon-only repeats, mostly 21 bp with the first nucleotide missing (fig. 3; FJ434695, FJ434714–21). In some clones, insertions/deletions (indels) and nucleotide substitutions were observed.
One clone belonged to the KbrSL-U6-5S1 type (one full unit to be 2,238 bp; FJ434722). The gene structure and sequence consisted of an SL RNA, a 5S rRNA gene, a U6 snRNA gene, then another SL RNA (fig. 3). Structurally, KbrSL-U6-5S1 was identical to KbrSL-U6-5S2 (see below), although there were numerous indels in the intergenic regions between these two types of SL RNA gene. KbrSL-U6-5S1 was identical to KbrSL-1, KbrSL-2, and KbrSL-U6-5S2 (see below) in the first 60 bp, suggesting that these four types of KbrSL have identical transcripts (fig. 4).
Karenia brevis SL RNA Genes: SL–5S Amplification
PCR using K. brevis genomic DNA as the template with primer sets dino5SF1-R1 and dino5SF2-R1 amplified a strong band of about 1.9 kb and some weaker bands with sizes larger than 3.5 kb. The 1.9-kb band by primer set dino5SF1-R1 was cut from the gel, DNA purified and cloned into a T-vector. Seven of the resultant plasmid clones with the size of 1,836–1,979 bp were sequenced (FJ434723–29). Sequencing results indicated that these clones belonged to a new type of KbrSL designated as KbrSL-U6-5S2. This type of KbrSL had the SL RNA gene located in the 5S rDNA tandem repeats as reported by Lidie and van Dolah (2007) for K. brevis Wilson isolate (figs. 3 and 4). The sequences of these clones were similar to one another and to that of the K. brevis Wilson isolate (97–99%). There was microsatellite variance in the “ATG” triple nucleotide repeats in the 5S–SL intergenic spacer region (six iterations in the Wilson isolate, 17 to 47 repeats in strain CCMP2228). The 22-bp SL initiated with a “T” in the Wilson isolate, however either A (four clones) or G (three clones) was found in strain CCMP2228. The first 529 bp starting from the 22-bp SL of KbrSL-2, KbrSL-U6-5S1, and KbrSL-U6-5S2 were similar (see fig. 3 for part of the alignment), indicating that these types of KbrSL have a similar origin. When all dinoflagellate SL sequences are aligned together, only the first 59–60 bp were conserved, whereas the sequence immediately downstream diverged largely between these different types, within or between species (fig. 4).
SL RNA Genes in K. veneficum, P. piscicida, and P. minimum: 5S–SL Amplification
In a previous study, we detected SL RNA tandem repeat genomic arrangements in K. veneficum, P. piscicida, and P. minimum with primer set dinoSLg-F/dinoSLg-R (Zhang, Hou, et al. 2007). In this study, we reanalyzed these species with the primer sets that amplified KbrSL-U6-5S2 from K. brevis. Primer sets (dino5SF1–R1 and dino5SF2–R1) that efficiently amplified the tandem repeats of 5S rRNA gene cluster for K. brevis yielded a faint band of about 1.3 kb for K. veneficum under both PCR conditions used. Cloning and sequencing of the amplicon revealed tandem repeats of 5S rRNA gene with unique intergenic sequence, in which neither SL RNA nor U6 sequences were present (Kve5S, figs. 2 and 3; FJ434789–800). With primer sets dino5SF1–dinoSL and dino5SF2–dinoSL, PCR produced a ∼1.2-kb band that contained SL RNA and 5S rRNA genes (KveSL-5S; figs. 3 and 4; GenBank accession nos FJ434777–780), as well as several other bands from 0.4 to 3 kb in length, which proved to be nonspecific amplicons (GenBank accession nos FJ434845–92). In KveSL-5S, a unique intergenic region was found between the SL and 5S rRNA genes, without the presence of the U6 snRNA gene, in contrast to the SL-U6-5S structure found in K. brevis (fig. 3). The KveSL-5S sequence was identical in the first 60 nt (SL RNA region) to K. veneficum SL RNA tandem repeats reported previously (Zhang, Hou, et al. 2007) with only one A to G substitution in the intron, but similarity diminished beyond, a feature similar to that of KbrSL-1 versus KbrSL-U6-5S types. The 5S rRNA gene sequence in the K. veneficum SL–5S gene cluster had nucleotide substitution at three cites compared with the 5S rRNA gene tandem repeats, and the sequences flanking the 5S rRNA gene showed no similarity.
Similar to the case in K. veneficum, PCR with dino5SF1-R1 and dino5SF2-5SR1 primer sets for P. minimum and P. piscicida led only to isolation of tandem repeats of the 5S rRNA gene (two to four repeats), with unique intergenic regions lacking the SL RNA or U6 snRNA genes (figs. 2 and 3; FJ434801–5, FJ434806–17). Several bands of 0.4–1.8 kb were obtained with primer set dino5SF1–dinoSL. When these PCR products were diluted 100-fold and used as the template with primer set dino5SF2–dinoSL in the second round of nested PCR, bands of about 1 and 1.6 kb were detected for P. piscicida and P. minimum, respectively. Cloning and sequencing of the amplicons indicated that the PCR product for P. minimum was nonspecific amplification (FJ434910–13), whereas that for P. piscicida contained the targeted amplicon (figs. 2– 4; FJ434781–88).
The eight sequenced clones from the P. piscicida amplicon (986–1,099 bp) showed an identical SL–5S structure and sequence, with some indels occurring approximately 210 bp downstream of the SL RNA gene. This P. piscicida SL RNA was a new type (PpiSL-5S), similar to KbrSL-1 for the first 60 bp, but different from the tandemly arrayed SL RNA previously reported for P. piscicida (Zhang, Hou, et al. 2007) in both intron and intergenic spacer regions. Similar to KveSL-5S, the PpiSL-5S cluster also consisted of one unit of SL RNA followed by a unique intergenic region and a 5S rRNA gene. No U6 snRNA was detected in this gene cluster.
SL RNA Genes in P. glacialis and H. arctica
To investigate whether multiple genomic arrangements of the SL RNA gene found in the above-mentioned dinoflagellates also occur in basal lineages, we tested various primer sets for P. glacialis and H. arctica. When genomic DNA of P. glacialis and H. arctica were used as the template in PCR with the opposing SL primer set dinoSLg-F/dinoSLg-R, multiple bands of ∼0.3 to 2.2 kb were obtained. PCR products were cloned into a T-vector, and clones with a variety of insert sizes were sequenced. Most of the clones proved to be nonrelated sequences (FJ434818–41, FJ434893–909), whereas four clones for P. glacialis contained inserts of 464–713 bp, and two clones for H. arctica with inserts of 864–1,012 bp contained two to three units of SL RNA with moderate similarity at the first 57 bp to that of the other dinoflagellates (figs. 3 and 4; FJ434764–67, FJ434741–42). In contrast to the SL RNA tandem repeats discovered so far in most dinoflagellates that were similar in length and sequence for each unit, different adjacent units of the P. glacialis and H. arctica SL RNA genes were obtained. Two units of P. glacialis SL RNA had two substitutions in the SL (C → T at position 2 and T → G at position 12) with much shorter (87 bp) intergenic region compared with the other four units (390–500 bp), in which various lengths and combinations of T, G, and C repeats were found with a number of indels (figs. 3 and 4). For the two H. arctica SL RNA clones, SL RNA and the intergenic region was identical for the first 350 bp, then the sequence similarity decreased by various indels in the rest of the intergenic region. For H. arctica, one clone contained three partial SL repeats (15 bp, CCATTTTGGCTCAAG), followed by a unique intergenic spacer sequence, then a single complete SL sequence, without the splice-donor dinucleotide “GT” immediately after the SL exon (FJ434743).
PCR with primer sets dino5SF1-5SR1, dino5SF2-5SR1, and dino5SF1–dinoSL primer sets for P. glacialis and H. arctica did not amplify any visible bands. When using 100-fold diluted dino5SF1–dinoSL PCR products as the template with dino5SF2–dinoSL as the primer set, several bands of 0.3–1.5 kb for P. glacialis and 0.7–2.2 kb for H. arctica were generated. For P. glacialis, nine clones were sequenced (328–1,430 bp). Two of these contained a full unit of SL RNA, an intergenic spacer and an incomplete 5S rRNA gene, whereas five contained a full unit of SL RNA, an intergenic spacer, an incomplete 5S rRNA gene, followed by two to five units of the 5S rRNA gene (P. glacialis SL-5Sa in fig. 3; FJ434768–74). One clone contained one unit of SL RNA, an intergenic spacer, and an incomplete 5S rRNA gene, followed by two units of 5S rRNA gene, an intergenic spacer, a second unit of SL RNA, an incomplete 5S rRNA gene, an intergenic spacer, and another unit of 5S rRNA gene (P. glacialis SL-5Sc in fig. 3; FJ434775). The remaining clone contained one unit of SL RNA, an intergenic spacer, and an incomplete intron of SL RNA, followed by an intergenic spacer, an incomplete 5S rRNA gene, an intergenic spacer, and another unit of 5S rRNA gene (FJ434776). The sequences of these clones were very similar in the corresponding regions. No U6 snRNA genes were identified. The SL RNA gene in this SL–5S genomic arrangement was different from the SL RNA tandem repeats mentioned above in the same species, and the first 60 bp were similar to KbrSL-1 (figs. 2–4).
For H. arctica, the sequences from the 20 clones were analyzed (642–1,715 bp) and clustered into two types (HacSL-5S1, FJ434744–60; HacSL-5S2, FJ434761–63; figs. 3 and 4). Both had a similar gene structure, containing one unit of SL RNA, an intergenic spacer, and one to three units of 5S rRNA gene but without U6 snRNA gene in the cluster. They differed in the length of the intron and the intergenic spacer and combinations of T–G repeats. KbrSL-2 SL RNA had a 7-bp deletion in the middle of the predicted intron region, reducing intron size to 24 bp for a total predicted SL RNA of 46 bp, the shortest of the known SL RNA and possibly representing a pseudogene. Both types of SL RNA had different sequences and structure from the SL tandem repeat organization mentioned above for the same species, but the first 60 bp of the SL–5S types were more similar to SL RNAs in other dinoflagellates than the SL tandem repeat in the same species. Heterocapsa arctica was not included in the direct RNA analysis due to the slow growth of this species, which did not produce enough biomass for the RNA blot.
Absence of an Initiator Element in dinoSL RNA
The KbrSL of K. brevis Wilson isolate was proposed to contain four additional nucleotides, AATC, at the 5′ end of the 22-nt DinoSL, and together with the “C” immediately upstream to form a potential initiator element CA+1ATCTC (Lidie and van Dolah 2007), a loose consensus defined in metazoans as YA+1NT/AYYY. Thus, we looked for this consensus sequence in our KbrSL genomic clones, as well as the SL RNA genomic clones from other dinoflagellates, with both initiation site predictions in mind. We obtained the upstream sequences beyond the 5′ end of SL in cases where the DNA clones contained at least two tandem repeats of the SL gene. We examined the 5 nt immediately upstream of the 22-bp SL and found that “CAATC” existed only in KbrSL-2 and both KbrSL-U6-5S variants, the three types with similar sequences in the first 529 bp starting from the 22-bp SL. The corresponding upstream region differed between other types of KbrSL as well as SL RNAs of other dinoflagellates (fig. 4). Thus, the “CAATC” sequence does not seem to act as a universal transcription initiator element for dinoflagellate SL RNA.
3′-End Analysis for K. brevis SL RNA Transcripts
To map precisely the size of SL RNA determined by RNA blotting, we designed specific primers and performed 3′-RACE for various types of K. brevis SL RNA transcripts (figs. 3 and 4; FJ434696–97, FJ434730–40) using the reported method (Zhang, Hou, et al. 2007). Because KbrSL-1, KbrSL-2, KbrSL-U6-5S1, and KbrSL-U6-5S2 were identical for the first 60 bp, and the cDNAs obtained ended at positions 56–59, it was not possible to distinguish the origin of these cDNAs, and the cDNAs are referred to as KbrSLx. Sixteen cDNA clones were obtained for this group with slightly different ends: three cDNAs ended at position 59, four at position 58, three at position 57, and six at position 56. Another five clones with one C to U substitution at nt 55 in the intron also ended at position 56; these five clones might represent another type of KbrSL, or could be a PCR/cloning error. Seven cDNAs clones were obtained for KbrSL-3, which had similar ends to KbrSLx (one clone ended at positions 59, 58, and 57, respectively, four clones ended at position 56) with one C to A substitution in all seven cDNA clones compared with their genomic counterpart at nt 46; the substitution might represent a PCR error or a new type of KbrSL similar to KbrSL-3. In total, 54% of the cDNAs obtained ended at position 56, whereas the rest ended at positions 57–59 for KbrSLx and KbrSL-3.
cDNAs from the KbrSL-4 locus were longer and contained identifying sequences. Ten clones were obtained for this distinct KbrSL variant, one ending at position 92, one at position 91, four at position 84, and four at position 70 (figs. 3 and 4).
Predicted SL RNA Structures and Sm-Binding Site Locale
Modeling analysis (fig. 5) was performed using MFOLD online service (http://mobyle.pasteur.fr/cgi-bin/MobylePortal/portal.py?form=mfold). Similar to K. veneficum SL (Zhang, Hou, et al. 2007), under the constraint that the splice-donor dinucleotide [“gu” in “Ggua(u)c(a)”] is double stranded and the putative Sm-binding site (AUUUUGG) single stranded, the resultant structures were unstable thermodynamically for most of the dinoflagellate SL RNAs obtained in this study; therefore, the default settings of MFOLD were used. For the transcripts of KbrSLx and KbrSL-3, as well as the predicted transcripts of H. arctica, P. piscicida, K. veneficum, and one type of P. glacialis SL RNA (Pgl SL-5S), modeling consistently yielded a two-stem-loop structure, with the 22-nt SL forming a stem, the splice-donor dinucleotide “gu” double-stranded, and 3–6 nt of the Sm-binding motif located in the double-stranded region; for a different type of P. glacialis SL RNA (Pgl SL-r), a two stem-loop structure with the Sm-binding motif remaining single stranded was obtained (fig. 5B). For KbrSL-4, several different structures consisting of two to four stem loops were formed for different transcripts, and the most thermodynamically stable structures are shown in figure 5. The same models were produced when folds were computed at 10 and 30 °C for K. brevis and P. piscicida, temperatures at which these two algae are found in the natural marine environment. For P. glacialis and H. arctica, in addition to 4 °C, the temperature under which these cultures were normally grown, folds were also computed at 20 °C, the temperature at which these polar algae can survive but do not grow (Lin S et al., unpublished results) and the same models were obtained.
FIG. 5.—
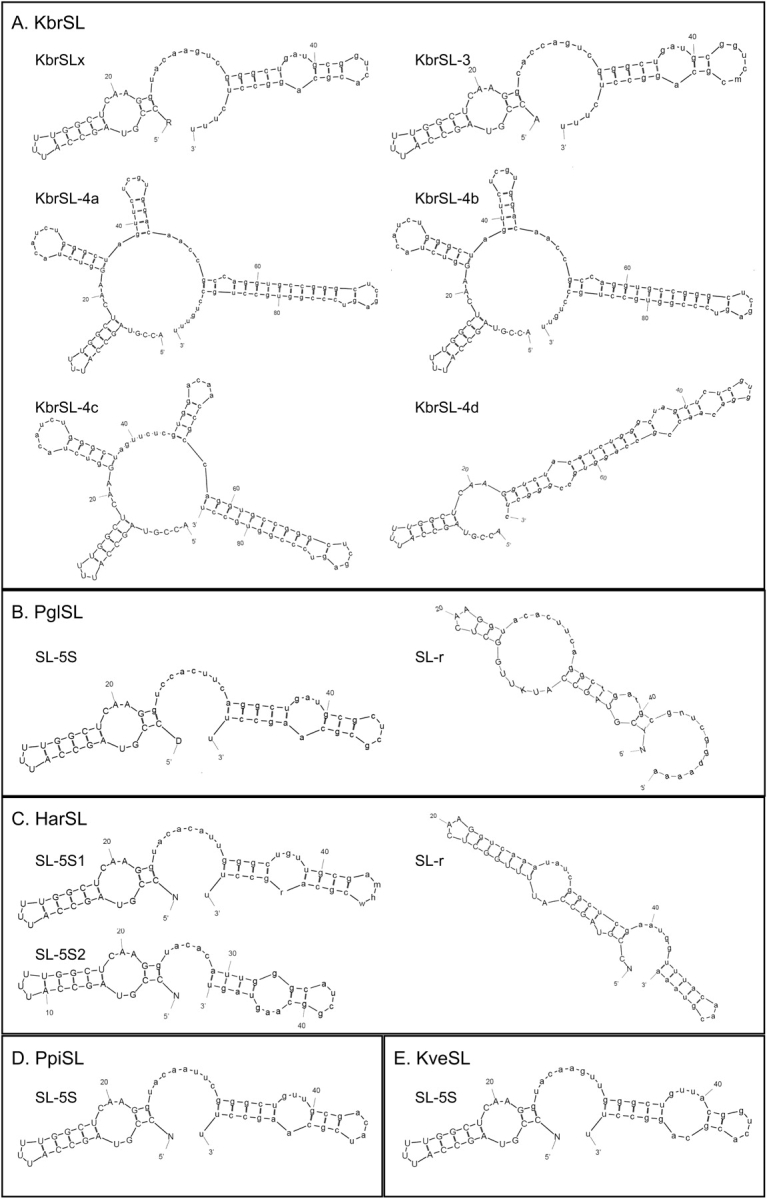
Predicted structures of SL RNA for Karenia brevis (A), Polarella glacialis (B), Heterocapsa arctica (C), and Pfiesteria piscicida (D). Model simulation was run using MFOLD: Prediction of RNA secondary structure modeling program (http://bioweb.pasteur.fr/seqanal/interfaces/mfold-simple.html) under default setting. Simulation for different types of K. brevis SL RNA (KbSL) was based on isolated cDNA and that for P. glacialis (PglSL), H. arctica (HarSL), and the P. piscicida SL–5S form (PpiSL) was based on genomic sequences, by identifying conserved regions in the alignment of SL RNA genes with all the mapped dinoflagellate SL RNA transcripts. SL–5S denotes SL RNA transcribed from the SL–5S genomic cluster; SL-r denotes SL RNA transcribed from genomic tandem repeats of SL RNA gene. For more information, see text.
Discussion
The SL sequence is conserved in dinoflagellates, but distinct compared with counterparts in other organisms in which SL trans-splicing is operative. The only variable positions of the 22-nt SL in dinoflagellates include 1) the first nucleotide position, which is predominantly A or U, with a few G but no C examples, and 2) one or two internal positions in some type of SL RNA in P. minimum and P. glacialis, respectively. However, the genomic arrangement of the SL RNA gene is markedly more complex than suspected initially. The mechanism by which the SL RNA genomic arrangement arose is still unclear. In this study, we have analyzed the genomic structure of SL RNA genes in six species of dinoflagellates representing four Orders with contrasting phylotypes and ecotypes, as discussed below, and provide evidence on how these structures might have evolved.
Molecular Size and Sequence of Dinoflagellate SL RNA Transcripts Are Conserved
Our RNA blot and cDNA 3′-RACE analyses indicate that both the sequence and the size of the SL RNA transcripts are similar in all the genera examined, including dinoflagellates that span a wide phylogenetic and ecological range. Pfiesteria piscicida (heterotrophic) and H. arctica (autotrophic) belong to the Order Peridiniales (a polyphyletic lineage), but Heterocapsa is a basal lineage of dinoflagellates based on a multi-gene phylogeny as well as degree of mitochondrial gene mRNA editing (Zhang, Bhattacharya, and Lin 2007; Zhang and Lin 2008). Polarella glacialis (autotrophic) is closely related to Symbiodinium, from the Order Suessiales (Montresor et al. 1999), a lineage occupying a relatively basal position in dinoflagellate phylogeny (Zhang, Bhattacharya, and Lin 2007; Lin et al. 2008; Zhang and Lin 2008). Following H. arctica and P. glacialis, most likely would be P. minimum (autotrophic) in the Order Prorocentrales, and the later diverging were Pfiesteria, and K. brevis (autotrophic)/K. veneficum (mixotrophic) in the Order Gymnodiniales, both producing ichthyotoxic compounds (fig. 3), although the resolution for Prorocentrales, Gymnodiniales, and Peridiniales is limited (Saldarriaga et al. 2001; Zhang, Bhattacharya, and Lin 2007). Geographically, K. brevis is best known in Gulf of Mexico; the tropical region. Pfiesteria piscicida causes blooms in North Carolina and Chesapeake Bay area (subtropical–temperate). Karlodinium veneficum is found more commonly in temperate waters on both sides of the Atlantic Ocean. Polarella glacialis and H. arctica are only in polar regions. The SL RNA in all these dinoflagellates is essentially the same. In addition, the consistency among the multiple 3′-RACE cDNA clones and the RNA blot demonstrates that the SL sequence is evolutionarily conserved in both length and sequence.
An earlier report on SL RNA from the K. brevis Wilson isolate (Lidie and van Dolah 2007) postulated additional sequence length for both the 5′ and 3′ ends of the transcript for this SL RNA. Four extra nucleotides were proposed for the 5′ end of the exon based on a presumptive initiator element identified in the primary sequence of a single gene. The presence of this element was not supported by our characterization of multiple genomic SL RNA genes in this and other dinoflagellate species. It is possible that the 5′-terminal sequence in cDNAs, especially in the case of modified 5′-cap structures, was missed experimentally. Indeed the inability of reverse transcriptase to read through the hypermethylated cap-4 structure led to early conclusions that the kinetoplastid SL was 35 nt in length (Boothroyd and Cross 1982), rather than the actual 39 nt (Perry et al. 1987). However, the lack of primary sequence conservation in the proposed 5′ extension within K. brevis or among other dinoflagellates throws doubt on the possibility of a 26-nt SL. The 56- to 59-nt size, confirmed by precise 3′-end mapping, indicates that K. brevis does not deviate from the dinoflagellate norm. Coupled with the lack of genomic conservation beyond the 60-bp mark, discussion of an intronic Sm-binding site and extended structural characterization are rendered moot. Although no commonly used molecular data such as 18S rRNA, ITS, mitochondrial gene sequences, nor ultrastructural morphology data are available for us to reconfirm the identity of the K. brevis strain (CCMP2228) in this study to the Wilson strain, the conserved gene structure as well as nucleotide similarity of the SL RNA-U6snRNA-5S rRNA array between these two strains, and the significant difference between K. brevis and the closely related K. veneficum (Zhang, Bhattacharya, and Lin 2007) indicate that Wilson isolate and CCMP2228 indeed are the same species: K. brevis. The larger hybridizing band seen in the K. brevis Wilson isolate RNA blot analysis is most likely an artifact.
Dinoflagellate SL RNA Secondary Structure Is Conserved
With few exceptions (Rajkovic et al. 1990; Vandenberghe et al. 2001), the known SL RNAs in other organisms share conserved structures: The exon and the beginning of the intron form a stem loop that contains the splice-donor dinucleotide “GU”, followed by two additional stem loops that flank a single-stranded region in which a binding site for the Sm-protein complex is located (Bruzik et al. 1988). However, the secondary structures of SL RNAs for all dinoflagellates examined to date using MFOLD are essentially the same (two stem loops with the potential Sm-binding site in exon), with the exception of KbrSL–4 SL RNAs (see below); no evolutionary trend has been found in SL RNA structure within the phylum of dinoflagellates. This result suggests that most dinoflagellate SL RNAs share a common secondary structure that is different drastically from other eukaryotic SL RNAs. The sequence of the Sm-binding motif is conserved, with RAU4–6GR in the kinetoplastids, freshwater planarians and Caenorhabditis, RAUUUUCGG in Hydra, AGCUUUGG in Ciona, AGCUUUUCUUUGG in Schistosoma, and AAYUYUGA in Rotifera (Bruzik et al. 1988; Rajkovic et al. 1990; Pellé and Murphy 1993; Stover and Steele 2001; Vandenberghe et al. 2001; Zeiner et al. 2004; Pouchkina-Stantcheva and Tunnacliffe 2005; Zayas et al. 2005). No Sm-binding motifs are found in the introns of the dinoflagellate SL RNAs mapped. Instead, the common variant of the Sm-binding motif AUUUUGG (AUGUUGG in one type of P. glacialis SL RNA) is present in the exon. The exonic Sm-binding motif occurs as a conserved stem-loop in almost all the predicted dinoflagellate SL RNA structures (fig. 5 and Zhang, Hou, et al. 2007), and may act as the Sm-complex binding site. Immunological evidence indicates that dinoflagellates have conserved Sm proteins (Reddy et al. 1983). We have obtained Sm-protein genes from several dinoflagellates (Zhang, Hou, et al. 2007), so Sm-complex formation can be demonstrated in the future. “Abnormal” Sm-binding motifs occur in the SL of two other organisms in which the SL RNA lacks the typical three-stem-loop structure. Although not pointed out by the authors in the original studies, we notice that the two SL RNAs in Hydra have potential Sm-binding motifs in both the exon and the intron (Stover and Steele 2001), as does the Oikopleura SL RNA (Ganot et al. 2004). In the latter case, the potential Sm-binding motif in the SL (exon) has a better fit with the consensus motif than the two candidate Sm-binding motifs in the intron, where the poly-U tracts are interrupted by multiple C residues. Consistent with our proposal that Sm proteins may complex with the SL, we also notice that immunoprecipitation experiments in Oikopleura demonstrate that both full-length SL RNA and a good amount of free SL, or possibly mRNA-derived SL, are captured by antibody raised against Sm proteins (Ganot et al. 2004).
The K. brevis SL RNA primary gene sequences are not conserved absolutely. Type KbrSL-3 exhibited two position changes from the other types of KbrSL. One A to C transversion in the intron appeared to have no profound effect on structure. In contrast, the T to C transition at nt 24 that changed the consensus “GU” splice-donor site to “GC” in KbrSL-3 RNA may render the transcripts nonfunctional. The cDNA amplification of this form indicates that it is transcribed, but trans-splicing remains to be determined. A variant class of AT–AC introns has been characterized in mammals, where splicing is mediated by a U12 snRNA-dependent mechanism (Wu and Krainer 1996; Will and Lührmann 2005). A variant class of GC–AG intron boundaries has also been found in other dinoflagellate genes (Bachvaroff and Place 2008), which may represent a minor class of splicing in dinoflagellates. The mechanisms of trans-splicing and function of GC–AG bounded introns in dinoflagellates remain to be investigated.
A distinct SL RNA class, KbrSL-4, was found in K. brevis. The 70–92-nt transcripts of this SL RNA were not detected in the general dinoflagellate SL RNA blot; however, the cDNAs were obtained by 3′-RACE analysis, indicating that the transcription level of this SL RNA may be relatively low. More stem-loop structures were predicted in this case than for the common dinoflagellate SL RNA, but similar to the other types of dinoflagellate SL RNA, the only potential Sm-binding site was in exon. This class was not the longer form of SL RNA detected in the study for K. brevis Wilson isolate (Lidie and van Dolah 2007).
Genomic Organization of the SL RNA in Dinoflagellates Is Complex but with no Clear Phylogenetic Trend
The occurrence of many different types and variants of SL RNA gene in each dinoflagellate was not unexpected. Southern analyses of K. veneficum, P. piscicida, and P. minimum indicated a complex SL RNA gene arrangement that could not be readily explained (Zhang, Hou, et al. 2007). It is of interest to investigate whether the complexity exhibits a phylogenetic trend. Among the six dinoflagellates examined, the SL RNA gene in the more derived K. brevis seems to have the most complicated genomic organization. In this species, SL RNA genes exist in at least six different types, giving rise to at least three distinct types of transcripts. Five types have almost identical sequences within the 56–59 nt SL RNA transcripts. The bizarre Kbr(SL)n arrangement contains tandem repeats of the 21-bp DinoSL, lacking the first nt but containing no intron sequence. However, although K. veneficum is phylogenetically closer to K. brevis than to P. piscicida (Zhang, Bhattacharya, and Lin 2007), the genomic organization of K. veneficum SL RNA is more similar to that of P. piscicida than to K. brevis (fig. 3). Only two types of SL RNA genomic organizations were found for K. veneficum and P. piscicida, one as SL RNA tandem repeats, whereas the other as SL–5S clusters. Furthermore, although SL RNA transcripts in these two types were almost identical in K. veneficum, they were very different in P. piscicida. In P. minimum, in contrast, SL RNA genes was only found in tandem repeats but not in SL–5S clusters. This might be due to PCR failure, but it is also possible that the SL–5S genomic structure has been lost in this lineage. Complicated genomic organizations of SL RNA genes were also found in the basal dinoflagellates. In both H. arctica and P. glacialis, SL RNA genes are tandemly repeated as well as clustered with 5S RNA gene. The (SL)n arrangement found in K. brevis was also detected in H. arctica with some variation (fig. 3). All these results indicate the absence of a clear phylogenetic trend in complexity of dinoflagellate SL genomic organization.
The multiple types and many variants of SL RNA genomic structures existing in most of the dinoflagellate species analyzed show that the SL RNA genes can reside in a variety of genomic contexts (tandem repeats on same chromosome and varying types likely located on different chromosomes). This may be evidence of the evolutionary history of dinoflagellate genomes in which extensive gene duplication through chromosome crossover and recombination have occurred repeatedly. Dinoflagellates in general have enormous genomes and most of the genes studied so far have many copies (Zhang et al. 2006 and the references therein). Clearly the genomic structure of SL RNA genes in K. brevis and other dinoflagellates is much more complex than previously thought (Lidie and van Dolah 2007; Zhang, Hou, et al. 2007). As these highly diverse genomic structures of SL RNA occur in dinoflagellates of wide phylogenetic and ecologic spectrum, it is apparent that such complex structures are common in the phylum of dinoflagellates (i.e., arose early in dinoflagellate evolution). The varying degrees of complexity (e.g., apparent absence of SL–5S cluster in P. minimum and the presence of SL-U6-5S cluster in K. brevis), as well as the high degree of sequence similarity between tandem repeat type and 5S rRNA-associated type SL RNA genes in some lineages (K. brevis and K. veneficum) but relatively higher variation in the others (H. arctica, P. piscicida, and P. glacialis) suggest that ongoing evolution may be occurring independently in each dinoflagellate lineage.
Extensive Duplication and Recombination of SL RNA Gene in Dinoflagellates as a Potential Evolutionary Mechanism to Diversify SL RNA Gene Structure
The association, or lack thereof, of 5S rRNA genes with SL RNA genes and other small RNA genes has been documented in a variety of organisms (i.e., Drouin et al. 1992; Drouin and de Sá 1995), but the consequences of these associations are unknown. In some dinoflagellate species (e.g., Alexandrium fundyense, Heterocapsa triquetra, and P. minimum), complete or partial SL sequence exist at the 5′ end of some protein coding genes (Zhang, Hou, et al. 2007; Slamovits and Keeling 2008; Zhang and Lin 2008). One possibility is that the dinoflagellate nuclear genome may recycle between trans-spliced mRNA and genomic DNA (Slamovits and Keeling 2008). Although plausible, especially for the case where full or partial SL exists at the 5′ end of a protein-coding gene, our data do not support this hypothesis. The SL RNA gene structure is much more complex than currently understood (Slamovits and Keeling 2008). Different types of SL repeats contain complete or partial SL sequences, with or without intergenic regions, as well as SL–5S tandem arrays with or without the U6 snRNA gene. Within each of these two major classes, numerous variants exist. Furthermore, complete or partial SL sequences are distributed in various parts of the genome, as we have detected in a number of dinoflagellates (FJ434698–99, FJ434818–942). The SL gene in dinoflagellates is more likely to have been duplicated rampantly and propagated in the genome, either through chromosomal recombination, or the SL RNA gene itself behaving as a transposon and inserts itself into various locations in the genome. Consistent with the chromosomal recombination hypothesis, we have observed that many types of KbrSL share similar sequences downstream of the SL RNA gene, although the SL RNA gene itself can be quite divergent (fig. 4). Our hypothesis is further supported by retrieval of non-SL RNA–related clones using primer sets DinoSLg-F-R, DinoSL–Dino5SF1, or DinoSL–Dino5SF2 (FJ434818–FJ434942), which had partial or nearly complete DinoSL sequence (22 bp) yet lacked the splice-donor dinucleotide (“GT”) and did not show similarity to the conserved intron of dinoflagellate SL RNA genes in the downstream region.
Further insights into the evolutionary history of dinoflagellate SL RNA genes as hypothesized above could have been provided by estimation of copy numbers for each form of SL RNA gene detected in this study. Unfortunately, this cannot be achieved by simple Southern hybridization because rampant tandem repeats of dinoflagellate genes would normally cloud the result. It is well recognized that genes in dinoflagellates usually have multiple copies and often array as tandem repeats (i.e., Zhang et al. 2006 and the references therein). SL RNA genes in dinoflagellates are no exception. In our previous study (Zhang, Hou, et al. 2007), Southern hybridization for three dinoflagellates indicated numerous bands with different hybridization strength, rendering it very difficult to identify which band represented which copy and estimate copy number of each type. Furthermore, as revealed by the sequencing of the clones obtained, there are numerous partial or whole copies of the 22-nt DinoSL throughout the genome for all species examined. Therefore, we believe that Southern hybridization will not reveal precisely how many of the SL RNA genes exist in the genome, as also had been demonstrated for Rubisco gene (Rowan et al. 1996). The exhaustive catalogue of SL RNA gene arrangements remains to be obtained through more extensive, ideally whole genome, analyses. A thorough understanding of the evolutionary history of dinoflagellate SL RNA gene, as well as its genome, remains to emerge.
New Sequences
The sequences obtained in this study have been deposited in GenBank, with accession numbers FJ434692–FJ434942.
Acknowledgments
We thank Yunyun Zhung and Ding Wang for technical assistance. This study is supported by the NSF grants EF-0626678 (to S.L./H.Z.) and NIH grant AI056034 (to D.A.C./N.R.S.).
References
- Bachvaroff TR, Place AR. From stop to start: tandem gene arrangement, copy number and trans-splicing sites in the Dinoflagellate Amphidinium carterae. PLoS ONE. 2008;3:e2929. doi: 10.1371/journal.pone.0002929. doi:10.1371/journal.pone.0002929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaxter ML, Liu LX. Nematode spliced leaders: ubiquity, evolution and utility. Int J Parasitol. 1996;26:1025–1033. [PubMed] [Google Scholar]
- Blumenthal T. Tran-splicing and operons. In: WormBook, editor. The C. elegans Research Community, WormBook. 2005. doi/10.1895/wormbook.1.5.1 http://www.wormbook.org. [Google Scholar]
- Boothroyd JC, Cross GAM. Transcripts coding for variant surface glycoproteins of Trypanosoma brucei have a short, identical exon at their 5′ end. Gene. 1982;20:281–289. doi: 10.1016/0378-1119(82)90046-4. [DOI] [PubMed] [Google Scholar]
- Bruzik JP, Doren KV, Hirsh D, Steitz JA. Trans splicing involves a novel form of small nuclear ribonucleoprotein particles. Nature. 1988;335:559–562. doi: 10.1038/335559a0. [DOI] [PubMed] [Google Scholar]
- Drouin G, de Sá MM. The concerted evolution of 5S ribosomal genes linked to the repeat units of other multigene families. Mol Biol Evol. 1995;12:481–493. doi: 10.1093/oxfordjournals.molbev.a040223. [DOI] [PubMed] [Google Scholar]
- Drouin G, Sévigny M, Mclaren IA, Hofman JD, Doolittle WF. Variable arrangement of 5S ribosomal genes within the ribosomal DNA repeats of arthropods. Mol Biol Evol. 1992;9:826–835. doi: 10.1093/oxfordjournals.molbev.a040764. [DOI] [PubMed] [Google Scholar]
- Ganot P, Kallesøe T, Reinhardt R, Chourrout D, Thompson EM. Spliced-leader RNA trans splicing in a chordate, Okopleura dioca, with a compact genome. Mol Cell Biol. 2004;24:7795–7805. doi: 10.1128/MCB.24.17.7795-7805.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings KEM. SL trans-splicing: easy come or easy go? Trends Genet. 2005;21:240–247. doi: 10.1016/j.tig.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG, Klotz LC, Blanken RL, Loeblich AR., III An evaluation of the phylogenetic position of the dinoflagellate Crypthecodinium cohnii based on 5S rRNA characterization. J Mol Evol. 1981;17:334–337. doi: 10.1007/BF01734355. [DOI] [PubMed] [Google Scholar]
- Lidie KB, van Dolah FM. Spliced leader RNA-mediated trans-splicing in a dinoflagellate, Karenia brevis. J Eukaryot Microbiol. 2007;54:427–435. doi: 10.1111/j.1550-7408.2007.00282.x. [DOI] [PubMed] [Google Scholar]
- Lin S, Zhang H, Hou Y, Zhuang Y, Miranda L. High-level diversity of dinoflagellates in the natural environment, revealed by assessment of mitochondrial cox1 and cob for dinoflagellate DNA barcoding. Appl Environ Microbiol. 2008;75:1279–1290. doi: 10.1128/AEM.01578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Zhang H, Spencer D, Norman J, Gray MW. Widespread and extensive editing of mitochondrial mRNAs in dinoflagellates. J Mol Biol. 2002;320:727–739. doi: 10.1016/s0022-2836(02)00468-0. [DOI] [PubMed] [Google Scholar]
- Mayer MM, Floeter-Winter LM. Pre-mRNA trans-splicing: from kinetoplastids to mammals, an easy language for life diversity. Mem Inst Oswaldo Cruz, Rio de Janeiro. 2005;100:501–513. doi: 10.1590/s0074-02762005000500010. [DOI] [PubMed] [Google Scholar]
- Montresor M, Procaccini G, Stoecker DK. Polarella glacialis, gen nov., sp. Mov. (Dinophyceae): suessiaceae are still alive! J Phycol. 1999;35:186–197. [Google Scholar]
- Pellé R, Murphy NB. Stage-specific differential polyadenylation of mini-exon derived RNA in African trypanosomes. Mol Biochem Parasitol. 1993;59:277–286. doi: 10.1016/0166-6851(93)90225-m. [DOI] [PubMed] [Google Scholar]
- Perry KL, Watkins KP, Agabian N. Trypanosome mRNAs have unusual “cap 4” structures acquired by addition of a spliced leader. Proc Natl Acad Sci USA. 1987;84:8190–8194. doi: 10.1073/pnas.84.23.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouchkina-Stantcheva NN, Tunnacliffe A. Spliced leader RNA-mediated trans-splicing in Phylum Rotifera. Mol Biol Evol. 2005;22:1482–1489. doi: 10.1093/molbev/msi139. [DOI] [PubMed] [Google Scholar]
- Rajkovic A, Davis R, Simonsen J, Rottman F. A spliced leader present on subsets of mRNAs from the human parasite Shistosoma mansoni. Proc Natl Acad Sci USA. 1990;87:8879–8883. doi: 10.1073/pnas.87.22.8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy R, Spector D, Henning D, Liu MH, Busch H. Isolation and partial characterization of dinoflagellate U1–U6 small RNAs homologous to rat U small nuclear RNAs. J Biol Chem. 1983;258:13965–13969. [PubMed] [Google Scholar]
- Rowan R, Whitney SM, Fowler A, Yellowlees D. Rubisco in marine symbiotic dinoflagellates: form II enzymes in eukaryotic oxygenic phototrophs encoded by a nuclear multigene family. Plant Cell. 1996;8:539–553. doi: 10.1105/tpc.8.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldarriaga JF, Taylor FJR, Keeling PJ, Cavalier-Smith T. Dinoflagellate nuclear SSU rRNA phylogeny suggests multiple plastid losses and replacements. J Mol Evol. 2001;53:204–213. doi: 10.1007/s002390010210. [DOI] [PubMed] [Google Scholar]
- Santana DM, Lukes J, Sturm NR, Campbell DA. Two sequence classes of kinetoplastid 5S ribosomal RNA gene revealed among bodonid spliced leader RNA gene arrays. FEMS Microbiol Lett. 2001;204:233–237. doi: 10.1111/j.1574-6968.2001.tb10890.x. [DOI] [PubMed] [Google Scholar]
- Slamovits CH, Keeling PJ. Widespread recycling of processed cDNAs in dinoflagellates. Curr Biol. 2008;18:R550–R552. doi: 10.1016/j.cub.2008.04.054. [DOI] [PubMed] [Google Scholar]
- Stover NA, Steele RE. Trans-spliced leader addition to mRNAs in a cnidarian. Proc Natl Acad Sci USA. 2001;98:5693–5698. doi: 10.1073/pnas.101049998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm NR, Yu MC, Campbell DA. Transcription termination and 3′-End processing of the spliced leader RNA in kinetoplastids. Mol Cell Biol. 1999;19:1595–1604. doi: 10.1128/mcb.19.2.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberghe AE, Meedel TH, Hastings KE. mRNA 5′-leader trans-splicing in the chordates. Genes Dev. 2001;15:294–303. doi: 10.1101/gad.865401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Lührmann R. Splicing of a rare class of introns by the U12-dependent spliceosome source. Biol Chem. 2005;386:713–724. doi: 10.1515/BC.2005.084. [DOI] [PubMed] [Google Scholar]
- Wu Q, Krainer AR. U1-mediated exon definition interactions between AT–AC and GT–AG introns. Science. 1996;274:1005–1008. doi: 10.1126/science.274.5289.1005. [DOI] [PubMed] [Google Scholar]
- Zayas RM, Bold TD, Newmark PA. Spliced-leader trans-splicing in freshwater Planarians. Mol Biol Evol. 2005;22:2048–2054. doi: 10.1093/molbev/msi200. [DOI] [PubMed] [Google Scholar]
- Zeiner GM, Foldynova S, Sturm NR. SmD1 is required for spliced leader RNA. Biogenesis Euk Cell. 2004;3:241–244. doi: 10.1128/EC.3.1.241-244.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Bhattacharya D, Lin S. A three-gene dinoflagellate phylogeny suggests reconciliation of Exuviaella with Prorocentrum and a basal position for Amphidinium and Heterocapsa. J Mol Evol. 2007;65:463–474. doi: 10.1007/s00239-007-9038-4. [DOI] [PubMed] [Google Scholar]
- Zhang H, Hou Y, Lin S. Isolation and characterization of PCNA from the dinoflagellate Pfiesteria piscicida. J Eukaryot Microbiol. 2006;53:142–150. doi: 10.1111/j.1550-7408.2005.00085.x. [DOI] [PubMed] [Google Scholar]
- Zhang H, Hou Y, Miranda L, Campbell DA, Sturm NR, Gaasterland T, Lin S. Spliced leader RNA trans-splicing in dinoflagellates. Proc Natl Acad Sci USA. 2007;104:4618–4623. doi: 10.1073/pnas.0700258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lin S. Complex gene structure of the form II Rubisco in the dinoflagellate Prorocentrum minimum (Dinophyceae) J Phycol. 2003;39:1160–1171. [Google Scholar]
- Zhang H, Lin S. Development of a cob-18S rDNA real-time PCR assay for quantifying Pfiesteria shumwayae in the natural environment. Appl Environ Microbiol. 2005;71:7053–7063. doi: 10.1128/AEM.71.11.7053-7063.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lin S. Status of mRNA editing and SL RNA trans-splicing groups Oxyrrhis, Noctiluca, Heterocapsa, and Amphidinium as basal lineages of dinoflagellates. J Phycol. 2008;44:703–711. doi: 10.1111/j.1529-8817.2008.00521.x. [DOI] [PubMed] [Google Scholar]