Abstract
Male reproductive fitness is strongly affected by seminal fluid. In addition to interacting with the female environment, seminal fluid mediates important physiological characteristics of sperm, including capacitation and motility. In mammals, the male reproductive tract shows a striking degree of compartmentalization, with at least six distinct tissue types contributing material that is combined with sperm in an ejaculate. Although studies of whole ejaculates have been undertaken in some species, we lack a comprehensive picture of the specific proteins produced by different accessory tissues. Here, we perform proteomic investigations of six regions of the male reproductive tract in mice—seminal vesicles, anterior prostate, dorsolateral prostate, ventral prostate, bulbourethral gland, and bulbourethral diverticulum. We identify 766 proteins that could be mapped to 506 unique genes and compare them with a high-quality human seminal fluid data set. We find that Gene Ontology functions of seminal proteins are largely conserved between mice and humans. By placing these data in an evolutionary framework, we show that seminal vesicle proteins have experienced a significantly higher rate of nonsynonymous substitution compared with the genome, which could be the result of adaptive evolution. In contrast, proteins from the other five tissues showed significantly lower nonsynonymous substitution, revealing a previously unappreciated level of evolutionary constraint acting on the majority of male reproductive proteins.
Keywords: Mus, reproduction, proteomics
Introduction
For sexually reproducing organisms, the process of fertilization involves complex interactions between male- and female-derived proteins. These interactions extend beyond the fusion of gametes and can influence many important events leading up to fertilization. When internally fertilizing species mate, males contribute not only sperm but also a complex mixture of seminal fluid derived from a suite of accessory tissues.
Seminal fluid has many functions during reproduction, including regulation of sperm motility (Peitz and Olds-Clarke 1986; Agrawal and Vanha-Perttula 1987; Peitz 1988), mediation of sperm capacitation (Huang et al. 2000; Kawano and Yoshida 2007), suppression of the female immune system (Peitz and Bennett 1981; Anderson and Tarter 1982; Thaler 1989), and interaction with sperm from other males (Prout and Clark 2000). In mammals, experimental removal of accessory glands results in reduced pregnancy rate, a reduction in the number of young produced, and/or delays in embryonic development (Pang et al. 1979; Queen et al. 1981; Peitz and Olds-Clarke 1986; O et al. 1988; Henault et al. 1995; Carballada and Esponda 1999). Many seminal fluid proteins bind directly to sperm and comigrate with sperm through the female reproductive tract (Irwin et al. 1983; Robinson et al. 1987; Carballada and Esponda 1997, 1998).
Seminal fluid proteins might also contribute to reproductive isolation between closely related species. For example, Dean and Nachman (2009) showed that fertilization rate was significantly faster in conspecific versus heterospecific matings involving Mus domesticus and Mus musculus, and seminal fluid may mediate these interactions.
Despite the central role of seminal fluid in reproductive processes, relatively little is known about the proteins produced by specific accessory glands in mammals. Previous work usually focused on a few highly abundant proteins derived from the seminal vesicles and the prostate (Platz and Wolfe 1969; Mann and Lutwak-Mann 1981; Shivaji et al. 1990; Carballada and Esponda 1991), whereas more comprehensive expression studies focused on a single region of the reproductive tract, such as the prostate or epididymis (Abbott et al. 2003; Berquin et al. 2005; Johnston et al. 2005; Fujimoto et al. 2006; Jelinsky et al. 2007).
One striking feature of the male reproductive tract of mammals is its complex compartmentalization. Upon ejaculation, sperm is mixed with fluids from at least four distinct male accessory organs—the seminal vesicles, the prostate, the bulbourethral gland, and the bulbourethral diverticulum. Anatomical, histological, and gene expression data suggest that the mouse prostate can be further subdivided into at least three distinct biological regions: the anterior, dorsolateral, and ventral lobes (Hayashi et al. 1991; Abbott et al. 2003; Berquin et al. 2005). Although one of the central findings of molecular evolution has been that genes underlying male reproduction evolve rapidly (reviewed by Clark et al. 2006), it is unknown whether this pattern applies to genes expressed throughout all compartments of the male reproductive tract or whether it is localized to a few tissues that produce a specific suite of proteins.
We performed tandem mass spectrometry (MS) analyses on proteins isolated from extracellular regions of six distinct male reproductive tissues in Mus—the seminal vesicles, the anterior prostate (also referred to as the coagulating gland), the dorsolateral prostate, the ventral prostate, the bulbourethral gland (also referred to as the Cowper's gland), and the bulbourethral diverticulum. Other regions of the male reproductive tract, such as the ampullary glands, Rete testis, and epididymis, may contribute seminal fluid but were not included because their small size precluded adequate extraction of proteins. We compared our data with a high-quality human seminal fluid data set (Pilch and Mann 2006) and report four key findings: 1) different regions of the male reproductive tract produce different suites of proteins, 2) some male reproductive genes occur in currently unannotated regions of the reference genome, 3) seminal proteins of mouse and human have similar Gene Ontology (GO) functions, and 4) there is heterogeneity in evolutionary rate among proteins from different regions of the male reproductive tract. Although proteins expressed in the seminal vesicles evolve significantly more rapidly than the genome average, proteins from the other five regions show an opposite pattern of significant evolutionary constraint. Our study provides an important future resource for understanding variance in male reproductive fitness.
Materials and Methods
Samples
We avoided wild caught mice in this experiment because wild genotypes cannot be replicated. Classical inbred strains of mice carry replicable genotypes by virtue of their homozygosity, but their genomes include material from three distinct species (M. domesticus, M. musculus, and Mus castaneus, also referred to as subspecies in the literature) (Silver 1995; Wade et al. 2002; Frazer et al. 2007; Yang et al. 2007), so their relevance to natural variation is uncertain. Therefore, we focused on the wild-derived inbred strains LEWES/EiJ (LEWES) and WSB/Eij (WSB). These strains were both derived from natural populations of M. domesticus outside of the European hybrid zone and have the standard karyotype (2n = 40). They were initially obtained from the Jackson Laboratory (Bar Harbor, ME) and maintained in our laboratory for more than 10 generations.
By crossing a female LEWES to a male WSB, we removed potentially confounding effects of inbreeding depression in their progeny, while maintaining the benefits of a replicable genotype. Progeny from this cross are expected to be similar genetically to wild mice. A single F1(LEWES × WSB) male was used. Testing the effects of different social conditions on seminal protein composition, and quantifying variation in protein composition among individuals, is outside the scope of the current study.
The parents of this male were paired for 1 week and then separated so that the dam gave birth in isolation. At 21 days postpartum, the male was weaned individually to avoid dominance interactions among brothers. Grouped males have reduced fertility compared with singly caged males (Snyder 1967), probably because social hierarchy reduces reproduction of subordinate males. Although fertility declines significantly more rapidly in isolated males vs. males caged with females, this effect is only seen after approximately 22 months of age (Schimidt et al. 2009), much older than the mouse used here. All mice were maintained at the University of Arizona Central Animal Facility in accordance with IACUC regulations.
Protein Isolation
Artificial ejaculation techniques produce abnormal and inconsistent ejaculates in mice (Snyder 1966; Tecirlioglu et al. 2002). Therefore, we identified proteins directly from dissected tissues. At 60 days of age, the F1(LEWES × WSB) male was sacrificed, and the seminal vesicles, anterior prostate, ventral prostate, dorsolateral prostate, bulbourethral gland, and the bulbourethral diverticulum were quickly dissected into phosphate buffered saline (PBS). Testis weight, testis histology, sperm count, and fertility data all show that F1(LEWES × WSB) males are sexually mature by 60 days of age (Good et al. 2007). Internal fluids were manually pressed out using sterile 28G needles. The anterior, ventral, and dorsolateral prostates are small and tubular, so they had to be sliced to release adequate amounts of extracellular fluids. Cell lysis solutions were intentionally avoided in order to target extracellular proteins likely to be secreted and ejaculated. Isolated proteins in PBS were stored at −80 °C until MS analysis.
MS and Protein Identification
MS was performed as described in Findlay et al. (2008), with several modifications. Briefly, proteins from each of the six male reproductive tissues were digested with trypsin and prepared for MS as previously described (Aagaard et al. 2006). For each sample, peptides were then loaded onto a 75-μm internal diameter capillary column that had been packed with 40 cm of Jupiter C12 reverse phase material. Two technical replicates, each of 5 μg, were then analyzed by injecting the sample directly on to a column that had been placed online with an LTQ ion-trap mass spectrometer (ThermoElectron). Peptides were eluted off the column over a 4-h gradient, and mass spectra were acquired using data-dependent acquisition. Data files from each sample were analyzed using the CHARGE-CZAR, SEQUEST, and DTASELECT programs (Eng et al. 1994; Tabb et al. 2002; Klammer et al. 2005). MS2 files were searched against all annotated proteins in the mouse genome (NCBI build 37, Ensembl version 48). Only proteins detected with at least two tryptic peptides were included. The false discovery rate (FDR) was estimated by including a set of “decoy” proteins in the database when performing the search. Each decoy was formed by randomly rearranging the amino acids in an annotated protein. The FDR was calculated by dividing the number of decoy proteins identified by the number of annotated proteins identified; in no case were any decoy proteins detected under our stringent requirement of at least two unique tryptic peptides.
We note that MS cannot identify all proteins exhaustively. Some proteins, such as those encoded by “Androgen-binding protein” genes, are resistant to standard trypsin digestion (Karn and Laukaitis 2003). Accordingly, we did not detect this protein even though both the transcript and protein are abundant in mouse prostate (Dlouhy et al. 1986; Laukaitis et al. 2005). Because not all proteins are equally detectable, absolute or relative abundance cannot be reliably compared across proteins.
All spectra were searched against the C57BL/6J reference genome (NCBI build 37, Ensembl version 48). Genetic divergence between the F1(LEWES × WSB) mouse used here and the C57BL/6J reference genome is unlikely to affect protein identification. More than 90% of the C57BL/6J genome is thought to be derived from M. domesticus (Yang et al. 2007). Average pairwise divergence (π, Nei and Li 1979) in the introns of several genes sampled from M. domesticus is <0.003 (Baines and Harr 2007; Geraldes et al. 2008). Average pairwise divergence that results in nonsynonymous substitution is expected to be even lower. Furthermore, because proteins were identified by a median of four peptides (see Results), most proteins would be “hidden” only if amino acid substitutions occurred at multiple peptides.
Six Frame Search
Previous work in Drosophila has shown that genes encoding reproductive proteins are often not detected by gene prediction algorithms (Findlay et al. 2008, 2009). To identify unannotated reproductive genes in mouse tissues, we searched all mass spectra from each tissue against a six reading frame translation of the C57BL/6J reference genome (NCBI build 37, Ensembl version 48). The DNA sequence of each chromosome was translated in all six reading frames. Open-reading frames were discarded if they translated to fewer than 11 amino acids, consisted solely of a single amino acid repeat, or lacked the K or R residues predicted for tryptic peptides (Findlay et al. 2008). Translating and filtering produced ∼137 million possible open-reading frames. We also included annotated proteins that were identified from the searches above, so that six-frame peptides matching them could be easily removed. Because many false positives might be expected from searching against such a large database, we required that six-frame peptides be matched by at least two spectra in order for them to be considered a true positive. We used a combination of BLAST (Altschul et al. 1990) and BLAT (Kent 2002) to map these peptides back to the mouse genome.
Comparison to Human Seminal Fluid Data
We compared our data with a high-quality data set identifying human seminal fluid proteins. Specifically, 923 unique proteins (of which 806 could be unambiguously linked to single Ensembl gene names) were identified from purified human seminal fluid (Pilch and Mann 2006).
Molecular Evolution
Estimating Rates of Nonsynonymous Change (dN/dS)
Orthology was determined using the phylogenetic clustering analyses of Ensembl version 48 (www.ensembl.org) (NCBI mouse build 37). Protein sequences were aligned using CLUSTALW version 1.83 (Thompson et al. 1994), then associated with their coding DNA sequences using REVTRANS version 1.5 (Wernersson and Pedersen 2003). From these alignments, we calculated dN/dS, the number of nonsynonymous substitutions per nonsynonymous site normalized by the number of synonymous substitutions per synonymous site, using the method of Goldman and Yang (1994) as implemented in PAML version 3.15 (Yang 1997). For genes with multiple transcripts, we estimated dN/dS for all possible pairwise comparisons between mouse and rat, then chose the pair with the lowest estimated dS as an indication of the best alignment (following Dean et al. 2008). As quality control, we excluded any genes with fewer than 100 codons (dN/dS is difficult to estimate accurately with small genes), an estimated dN > 1 (probably an analytical artifact indicating more than one nonsynonymous substitution per nonsynonymous site), or an estimated dS ≥ 0.381 (twice the median dS value across the 15,950 genes, which may indicate poor alignment), resulting in 14,963 analyzed genes. We constructed 95% confidence intervals (CIs) of the estimated median by sampling 10,000 bootstrap replicates with R (www.r-project.org).
Testing for Recurrent Positive Selection among One-to-One Orthologs
To test for recurrent positive selection acting on genes, we used a maximum likelihood framework implemented in the CODEML routine of PAML version 3.15 (Yang 1997), as well as a fixed effects likelihood (FEL) implemented in HYPHY version 0.9920070619beta (Kosakovsky Pond et al. 2005). The CODEML framework considers dS to be constant across the gene, whereas FEL allows dS to vary among codons (Kosakovsky Pond and Frost 2005).
Using the same pair of sequences chosen in the above mouse–rat comparisons, we retrieved all one-to-one orthologs in human, cow, and dog. A total of 10,912 genes had one-to-one orthologs across these five species. For genes with multiple transcripts in any of these latter three species, we chose the longest transcript. Alignments were made as described above. In a CODEML framework, we fit the data to three alternative models of molecular evolution (the M7, M8a, and M8 models as described by Yang et al. 2000; Swanson et al. 2003). In essence, M7 and M8a represent different null hypotheses, as neither allows for codons within a sequence to experience recurrent positive selection, whereas model M8 relaxes this constraint.
We considered a gene to have experienced recurrent positive selection if all five of the following criteria were met: 1) M8 fit the data significantly better than M7 at P < 0.01, using a likelihood ratio test, 2) M8 fit the data significantly better than M8a at P < 0.01, 3) the additional class of dN/dS estimated by M8 was greater than 1.1, 4) at least 1% of the codons belonged to this additional class of dN/dS, and 5) FEL analyses revealed significant evidence of positive selection in at least one codon (dN/dS > 1.1 at P < 0.10, the P value recommended by Kosakovsky Pond and Frost 2005). As further quality control, we estimated pairwise dS between mouse and each of the four other species using the runmode = −2 option in CODEML. We excluded any genes whose five species alignment produced fewer than 100 analyzed codons or produced pairwise dS of mouse–rat ≥ 0.38, mouse–human ≥ 1.20, mouse–dog ≥ 1.39, or mouse–cow ≥ 1.43 (each representing greater than twice the median dS estimated from these respective genome pairs, possibly indicating poor alignment). These quality control measures resulted in 9,071 analyzed genes.
Results
Male Reproductive Glands Produce Unique Sets of Proteins
Among the six male reproductive tissues, 43,076 spectra identified 3,056 peptides that mapped to 1,057 known proteins. A median number of 10 spectra from a median of four peptides, covering a median 16% of the protein, were used to identify each protein or protein family. Of the 1,057 proteins identified, 766 could be mapped to a single region in the genome. These 766 proteins annotated to 506 genes (supplementary table 1, Supplementary Material online), indicating a high level of alternative splicing. The other 291 proteins could not be unambiguously assigned to a single region in the genome, usually because they were members of large gene families containing highly similar genes (supplementary table 2, Supplementary Material online). It should be noted that not all proteins identified here are necessarily ejaculated. However, the main conclusions that follow remain unchanged if we consider only the most conservative set of proteins, which are those whose one-to-one ortholog in humans is also found in human ejaculates.
Of the 506 “single region” genes, 228 (45%) were detected in only one of the six tissues (table 1), suggesting that regions of the male reproductive tract often contribute unique proteins. Of the remaining 278 genes, 95, 68, 69, 37, and 9 were detected in two, three, four, five, and six tissues, respectively.
Table 1.
Male Reproductive Proteins Identified Using Mass Spectrometry and Their Characteristics
| Pairwise, Mouse–Rate |
Codon Models, 5 Speciesf |
||||||||
| Tissuea | Proteinsb | Single Region Genesc | Singletonsd | N | Length | dN/dS | N | Length | Pos. sel. |
| SV | 47 | 37 | 7 | 23 | 429 | 0.273 (0.162–0.360) | 12 | 458 | 1 (0.08) |
| AP | 107 | 87 | 5 | 49 | 416 | 0.088 (0.049–0.163) | 26 | 494 | 1 (0.04) |
| DP | 305 | 272 | 52 | 177 | 381 | 0.073 (0.053–0.095) | 101 | 444 | 3 (0.03) |
| VP | 294 | 255 | 42 | 159 | 429 | 0.058 (0.046–0.095) | 92 | 471 | 7 (0.08) |
| BU | 343 | 298 | 95 | 205 | 417 | 0.061 (0.049–0.079) | 133 | 420 | 8 (0.06) |
| BD | 212 | 188 | 27 | 121 | 434 | 0.067 (0.040–0.099) | 73 | 444 | 4 (0.05) |
| Total | 766 | 506 | 228 | 345 | 403 | 0.074 (0.061–0.091) | 211 | 430 | 13 (0.06) |
| Genome | — | 23,049 | — | 14,963 | 399 | 0.128 (0.125-0.131) | 9,071 | 416 | 460 (0.05) |
SV = seminal vesicles, AP = anterior prostate, VP = ventral prostate, DP = dorsolateral prostate, BU = bulbourethral gland, BD = bulbourethral diverticulum. Total = the group of male reproductive proteins identified across all six tissues.
The number of unique proteins identified by MS.
The number of identified proteins that could be mapped to a single region in the genome (see text and supplementary table 1, Supplementary Material online).
The number of proteins unique to that tissue.
Summary statistics from analyses of one-to-one orthologs between mouse and rat. N = number of alignments that passed various quality control measures. Length = median length of the analyzed portion of the protein (= protein length minus the unalignable portions). dN/dS = pairwise estimate of the rate of nonsynonymous substitution that has occurred since rat and mouse last shared a common ancestor. Numbers in parentheses indicate 95% CI derived from bootstrap resampling.
Summary statistics from analyses of one-to-one orthologs aligned from mouse, rat, human, cow, and dog. N = number of alignments that passed various quality control measures. Length = median length of the analyzed portion of the protein (= protein length minus any unalignable portions). Pos. sel. = the number of alignments that produced statistically significant evidence of recurrent positive selection.
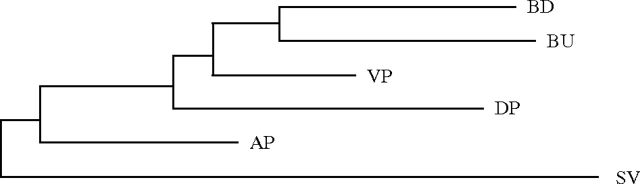
To visualize the degree of specialization across tissues, we constructed Neighbor-Joining phenograms (using Swofford 2002), based on protein presence/absence (fig. 1). There is strong divergence among tissues. For example, the three lobes of the prostate do not form a distinct clade, suggesting they are as divergent in protein complement from each other as they are from other tissues. Other studies have shown distinct patterns of gene expression among prostatic lobes (Abbott et al. 2003; Berquin et al. 2005; Fujimoto et al. 2006).
FIG. 1.—
Neighbor-Joining tree with midpoint rooting inferred from a protein presence/absence matrix (see Materials and Methods). SV = seminal vesicles, AP = anterior prostate, VP = ventral prostate, DP = dorsolateral prostate, BU = bulbourethral gland, and BD = bulbourethral diverticulum.
There is a broad overlap between the proteins identified in mouse male reproductive tissues and orthologous proteins identified in human seminal fluid. Of 506 mouse male reproductive proteins, 367 (73%) have a one-to-one ortholog in human (compared with the genome average of 14,925 of 23,049 = 65%), of which 136 were detected in the human seminal fluid data of Pilch and Mann (2006). The amount of overlap between studies is likely to be an underestimate, because we extracted proteins from dissections whereas Pilch and Mann (2006) isolated ejaculates directly.
Certain models of adaptive evolution predict that male reproductive genes should be favored on the X chromosome (Rice 1984; Vicoso and Charlesworth 2006). The 506 single gene proteins were not significantly more common on the X chromosome (14 of the 506 male reproductive proteins were X-linked compared with 954 of 21,813 genes in the genome, Fisher's Exact Test, P = 0.10).
New Genes in the Mouse Genome
We performed a search against the entire genome translated in all six reading frames with the goal of identifying genes or splice variants that are currently not annotated. Such a search can potentially improve the annotation of the mouse genome with direct evidence of translation. We detected three regions of the genome that contained peptides identified with high confidence, but where genes are currently not annotated (supplementary table 3, Supplementary Material online). Each of these peptides was at least 10 amino acids long and was detected with at least three spectra.
One of these proteins occurs on chromosome 7 in the vicinity of a gene called “Fc fragment of IgG-binding protein” (Fcgbp) (transcript = ENSMUST00000076648, gene = ENSMUSG00000047730). We detected 1,890 spectra matching 41 unique peptides in the annotated region that spans the 3′ end of the Fcgbp gene and the 5′ end of the next gene, Fbl (supplementary fig. 1, Supplementary Material online, Supplementary browser track). The human ortholog of Fcgbp spans the entire region between the annotated mouse Fcgbp and the unannotated regions hit by our six-frame search. If the mouse gene model of Fcgbp were annotated in a similar way, it would contain all of our unique peptides and the predicted transcripts ENSMUST00000082859, ENSMUST00000059886, and ENSMUST00000098633 (supplementary fig. 1, Supplementary Material online). We suggest that all of our hits to this unannotated region are part of one large Fcgbp gene. The size of the gene may make it difficult to sequence the complete mRNA (>15 kb) using a high throughput approach. For most of our unannotated peptides, there are no corresponding expressed sequence tag (EST) data (UCSC Genome Browser, http://genome.ucsc.edu).
In addition to this region of chromosome 7, we found two other peptides matching unannotated regions (supplementary table 3, Supplementary Material online). One on chromosome 1 was found four times in the anterior prostate, and the other on chromosome 13 was found three times in the dorsolateral prostate. Currently there is no EST evidence for transcription at either of these loci. Targeted amplification of cDNA from the regions could reveal the extent of their genes.
Mouse and Human Seminal Proteins Carry Out Similar Functions
To better understand the functions of proteins found in the six male reproductive tissues, we tested for overrepresentation of GO terms (Ashburner et al. 2000) using ONTOLOGIZER version 2.0 (Robinson et al. 2004), with the “Term-For-Term” calculation method and Bonferroni-corrected P < 0.05. We compared the functions inferred for mouse seminal proteins with those inferred from a high-quality human seminal fluid data set (Pilch and Mann 2006).
Among the 506 mouse male reproductive proteins, there were 161 GO terms that were significantly overrepresented (supplementary table 4, Supplementary Material online). Under the biological process partition, many GO terms were related to various aspects of metabolism (alcohol metabolism, carbohydrate metabolism, nucleotide metabolism, protein metabolism, and organic acid metabolism), physiological processes (cell organization, muscle contraction), and response to stress (response to chemical, biotic, heat, and protein stimuli). Under the molecular function partition, nucleotide binding, protein binding, enzyme regulation, and catalytic activity were overrepresented among male reproductive proteins.
Analysis of the cellular component partition of the GO yielded a surprising result. Even though our technique of protein isolation targeted extracellular proteins, functional analyses suggested that intracellular proteins were significantly overrepresented among male reproductive proteins. Reanalysis of the human seminal protein data (Pilch and Mann 2006, supplementary table 4, Supplementary Material online) gave the same result. The human seminal proteins are unlikely to be confined to intracellular space because they were identified from real ejaculates rather than dissections. Therefore, this result probably reflects imperfect knowledge about the complex pathways in which proteins are exported, although it is possible that some cellular sloughing or leakage occurs too.
Many of the 161 significantly overrepresented terms in our data set overlap functional terms identified as overrepresented among human seminal proteins. Specifically, 68 of the 161 terms were also overrepresented in the human data (supplementary table 4, Supplementary Material online). These 68 GO terms represent only identical overlap, ignoring similar functions that are assigned different GO terms. However, the hierarchical nature of GO terms also introduces some degree of nonindependence in these analyses. It was not possible to compare functional overlap to non-reproductive tissues, because human expression data derive largely from cell lines (i.e., Su et al. 2002).
We next focused on mouse male reproductive genes without one-to-one orthologs in the human genome. Among the 63 mouse genes without human orthologs, there were five significantly overrepresented terms, all related to enzyme inhibitor activity (table 2). Of these, three were also significantly overrepresented among the 61 human seminal genes with no mouse ortholog (table 2). Therefore, enzyme inhibitors appear to be particularly prone to gene turnover, either as an increased birth–death process or through rapid evolution that hides true orthology.
Table 2.
GO Functions That Were Significantly Overrepresented among Mouse Male Reproductive Proteins That Have No Form of Orthology to Humans
| Functional Terma | Genomeb (n = 17,075) | Malec (n = 49) | Pd | Humane |
| Molecular function | ||||
| Enzyme inhibitor activity (GO:0004857) | 254 (0.01) | 8 (0.16) | 3.E−04 | X |
| Protease inhibitor activity (GO:0030414) | 164 (0.01) | 8 (0.16) | 9.E−06 | X |
| Endopeptidase inhibitor activity (GO:0004866) | 163 (0.01) | 8 (0.16) | 8.E−06 | X |
| Serine-type endopeptidase inhibitor activity (GO:0004867) | 111 (0.01) | 8 (0.16) | 4.E−07 | |
| […] Protease inhibitor activity (GO:0017114) | 4 (0.00) | 3 (0.06) | 4.E−05 | |
GO terms that are significantly overrepresented among mouse reproductive genes that have no form of orthology to humans. GO terms are hierarchical in nature; indentations are meant to give a rough impression of parent versus daughter terms.
The number of genes in the mouse genome that could be associated with that function (of 17,075 genes in total that were assigned functions), according to the GO database.
The number of mouse male reproductive genes that were associated with that function (of 49 genes in total that were assigned functions).
The Bonferroni-corrected probability of observing this level of overrepresentation by chance alone.
Terms with an “X” were significantly overrepresented among human seminal proteins with no form of orthology to mouse.
Some Reproductive Proteins Evolve Rapidly, but Most Show Functional Constraint
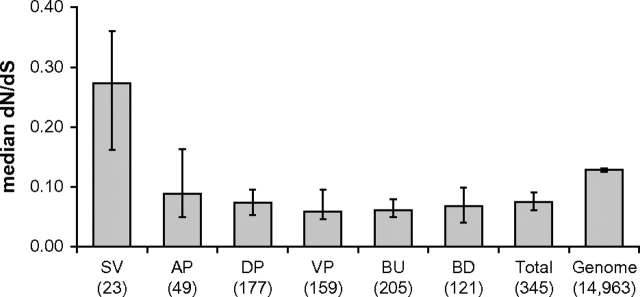
Genes whose proteins were detected in the seminal vesicles showed significantly higher pairwise dN/dS (95% credible interval = 0.162–0.360) compared with the genome average (0.125–0.131) (table 1, figs. 2 and 3). In contrast to seminal vesicle genes, genes whose proteins were detected in the other five male reproductive tissues showed significantly lower dN/dS compared with the genome, suggesting that most male reproductive proteins are evolutionarily constrained (table 1, figs. 2 and 3). There was no significant difference in the length of protein analyzed (the length of the protein, minus regions where mouse and rat do not align) between male reproductive proteins and the genome average (Wilcoxon Rank Sum Test [WRST] = 2,613,762, P = 0.69, table 1), so these patterns are unlikely to be caused by differences in power. There was also no significant difference in the frequency of one-to-one orthology among the six reproductive tissues compared with the genome (table 1). Furthermore, genes that failed quality control measures were distributed equally across the six reproductive tissues (supplementary table 1, Supplementary Material online).
FIG. 2.—
Median pairwise dN/dS estimated from one-to-one orthologs between mouse and rat. Error bars indicate 95% CIs determined from bootstrap resampling. SV = seminal vesicles, AP = anterior prostate, VP = ventral prostate, DP = dorsolateral prostate, BU = bulbourethral gland, BD = bulbourethral diverticulum. Total = the group of male reproductive proteins identified across all six tissues. Numbers in parentheses indicate the number of genes. SV proteins evolve significantly more rapidly than the genomic average. Proteins from all other tissues evolve significantly more slowly than the genomic average.
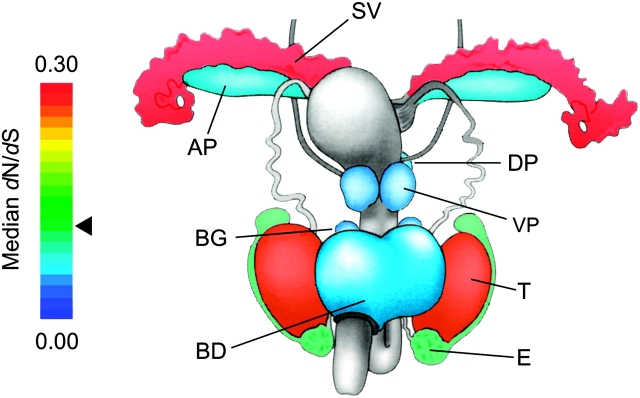
FIG. 3.—
A heat map of median dN/dS identified across the male reproductive tract. Red indicates relatively rapid evolution, and the seminal vesicles and testis show significantly elevated dN/dS compared with the genome median. All other reproductive tissues show significant reduction in dN/dS compared with the genome average. Testis-(dN/dS = 0.261) and epididymis-selective (dN/dS = 0.131) genes were defined according to patterns of gene expression and were taken from Dean et al. (2008). The triangle indicates the median dN/dS = 0.128 across the whole genome. Figure modified from Nagy et al. (2003).
Among the 506 genes detected here, those that were exclusively expressed in male reproductive tissue showed significantly higher dN/dS than genes that are also expressed in nonreproductive tissues. We reanalyzed the expression data of Su et al. (2002), focusing on 21 main tissues (following Dean et al. 2008). Of the 506 genes identified here, 378 could be linked to these expression data (not all genes were represented on the microarray chip of Su et al. 2002), of which 280 yielded pairwise estimates of dN/dS. Genes that were never detected in nonreproductive tissues showed significantly higher dN/dS (N = 34, median dN/dS = 0.17) compared with those genes that were detected in nonreproductive tissues (N = 246, median dN/dS = 0.06) (WRST = 6004, P < 10−5).
Not all proteins are equally detectable using MS, making it difficult to use the number of spectra identified per protein (or the number of peptides identified per protein) to meaningfully quantify relative or absolute abundance. With this caveat in mind, the 143 proteins without a one-to-one ortholog in rat were detected with significantly more total spectra (median = 25 spectra) compared with the 363 proteins with orthologs (median = 12 spectra) (WRST = 20,170, P < 10−4). If the number of spectra at least partly reflects relative protein abundance (Liu et al. 2004), this pattern suggests that more abundant proteins experience greater gene turnover or evolve so rapidly that orthology is obscured.
The proteins identified here were collected from dissected male reproductive glands and are not necessarily ejaculated. To address this issue, we repeated the above analyses of dN/dS including only the 136 genes with one-to-one orthologs in humans that occur in human ejaculates (Pilch and Mann 2006). It is likely that such proteins are also found in mouse ejaculates. Qualitatively, our results do not change: Seminal vesicle proteins still showed higher median dN/dS, and the proteins from the other five tissues still showed lower median dN/dS, compared with the genome (supplementary fig. 2, Supplementary Material online). However, the differences were no longer statistically significant for seminal vesicle and anterior prostate proteins, but this is probably due to the large reduction in sample size (supplementary fig. 2, Supplementary Material online). Although direct confirmation that the proteins identified here are included in the ejaculate awaits further study, the overall patterns of evolution (i.e., fig. 3) are robust. Therefore, differences in evolutionary rate cannot be explained by methodological issues, such as differential ease of dissection across the reproductive tract.
The significantly higher pairwise dN/dS observed for seminal vesicle proteins could have arisen through either recurrent adaptive evolution or relaxed selective constraint. To distinguish between these alternatives, we performed codon-based maximum likelihood estimates of the frequency of positive selection across five diverse mammal species. There were only 12 seminal vesicle genes that had a one-to-one ortholog in all five species analyzed. Of these, one gene (transferrin [ENSMUSG00000032554]) showed significant evidence of positive selection (table 1). There was no significant difference in the length of protein analyzed (the length of the protein, minus regions where there was not full alignment across the five species) between male reproductive proteins and the genome average (WRST = 949,238, P = 0.84, table 1). The small number of seminal vesicle genes that could be analyzed in this framework probably did not yield enough power to detect a higher rate of adaptive evolution. For example, six related genes (SVS2 [ENSMUSG00000040132], SVS3a [ENSMUSG00000017003], SVS3b [ENSMUSG00000050383], SVS4 [ENSMUSG00000016998], SVS5 [ENSMUSG00000017004], and SVS6 [ENSMUSG00000017000]) are located in a ∼100-kb window that has experienced an unusual history of duplication, deletion, and conversion (Hurle et al. 2007). These dynamics make orthology assignment in this region difficult, in turn causing all these genes to “drop out” of the five species analyses. Still, seminal vesicle genes evolve about as rapidly as testis-specialized genes (fig. 3), where adaptive evolution has been repeatedly reported (Good and Nachman 2005; Turner et al. 2008).
Discussion
We have undertaken the first comprehensive study of potentially ejaculated proteins in the house mouse. By targeting six distinct regions of the male reproductive tract, we discovered a previously unappreciated amount of heterogeneity in evolutionary rate experienced by reproductive proteins (fig. 3). Although it is true that many reproductive genes are rapidly evolving in mammals (Waterston et al. 2002; Castillo-Davis et al. 2004; Gibbs et al. 2004; Nielsen et al. 2005), proteins from five of six male accessory glands showed significantly lower rates of evolution compared with the genome average. Therefore, although many rapidly evolving genes have reproductive functions (reviewed in Clark et al. 2006), most proteins found in male accessory glands do not evolve rapidly. Although further work is required to confirm the reproductive roles of most of the proteins identified here, these conclusions remain essentially unchanged if we confine our analyses to those proteins that are also found in human ejaculates.
Heterogeneity in evolutionary rate may result from the partitioning of functions across the reproductive tract. Genes whose proteins were detected in the seminal vesicles showed a significantly higher rate of evolution compared with the genomic average. This pattern may be related to a higher degree of specialization for reproductive functions. Seminal vesicle proteins were fewer in number but were identified with significantly more spectra per protein.
To better understand this heterogeneity, we focus on the functions of seminal vesicle proteins. Thirty-seven proteins were identified from seminal vesicles (table 1). Of these, 21 showed the highest number of spectra in seminal vesicles compared with the other five tissues, and all showed >5X the median number of spectra detected in the other five tissues (supplementary table 1, Supplementary material online). These 21 proteins, which appear to be most common in the seminal vesicles, seem to fall into two functional classes: 1) proteins that participate in the formation of the copulatory plug and 2) proteins that function in immune response. It should be stressed, however, that most functional assignments are inferred by computational annotation and not direct assay, and it is probably true that no gene is fully functionally characterized with respect to all its functions.
Copulatory Plug Proteins
Shortly after copulation, a hard coagulum forms in the vaginal–cervical region in many rodent species and may persist for more than 24 h. In related work, we have identified the proteins found in the copulatory plug (unpublished data) using MS. The proteins encoded by seven of the 21 seminal vesicle–specific genes—SVS1 (ENSMUSG00000039215), SVS2 (ENSMUSG00000040132), SVS3A (ENSMUSG00000017003), SVS3B (ENSMUSG00000050383), SVS4 (ENSMUSG00000016998), SVS5 (ENSMUSG00000017004), and SVS6 (ENSMUSG00000017000)—were all detected in the copulatory plug with very high spectral counts. The proteins from SVS1 and SVS2 and SVS3a or SVS3b have been experimentally demonstrated to form transglutaminase-induced cross-linking, a central feature of the copulatory plug (Lundwall et al. 1997; Lin et al. 2002).
Two main hypotheses regarding the role of the copulatory plug in reproductive biology predict rapid evolutionary change in the constituent proteins. First, the copulatory plug may inhibit the sperm of other males. There is a strong correlation between the number of males that a female mates with per estrus cycle and the size and solidification of the copulatory plug in rodents and primates (Dixson and Anderson 2002; Ramm et al. 2005). Furthermore, rodents that do not form a copulatory plug appear to have evolved a different form of mate guarding by which a mechanical lock forms during copulation (Hartung and Dewsbury 1978). In addition, the rate of nonsynonymous substitution in SVS2 is positively correlated with the predicted frequency of multiple mating in rodents (Ramm et al. 2008). Taken together, these patterns suggest that copulatory plugs are an adaptive response to sperm competition. In nature, female mice mate with more than a single male in at least 22% of all estrus cycles (Dean et al. 2006; Firman and Simmons 2008), so sperm competition is a potentially powerful evolutionary force.
Second, the copulatory plug may alter female mating behavior by making her more resistant to subsequent mating (Goldfoot and Goy 1970; Carter and Schein 1971; Hardy and DeBold 1972). Although these behavioral shifts are likely adaptive from the perspective of the male (i.e., he is reducing the chance of future sperm competition), they could be deleterious to females (i.e., her litter is predicted to be less genetically diverse or more likely to carry meiotic drivers, as shown by Price et al. 2008). Sexual antagonism would predict rapid evolution among copulatory plug proteins as they continually change in response to female proteins.
Patterns of evolution are consistent with the predictions of male–male or male–female interactions. Of the three SVS genes that have a rat ortholog and pass quality control measures, all have pairwise dN/dS above 0.40 (more than 3X the genome median, supplementary table 1, Supplementary Material online). Formal tests of positive selection could not be undertaken for any SVS genes because they do not have clear orthologs in the other four species examined here. Nevertheless, some SVS proteins have been shown to experience recurrent positive selection with broader sampling within rodents (Karn et al. 2008; Ramm et al. 2008).
Immunity Proteins
A literature search revealed that 11 of the 21 seminal vesicle-specific proteins appear to function in the context of immunity activation or inhibition. Immunity genes are a classic example of an arms race model, whereby interacting proteins (usually host–pathogen) continually evolve to detect or evade each other. In the context of reproduction, immunity is an especially interesting biological process, as sperm are foreign bodies that must evade the female's immune response in order to successfully fertilize. The female mounts a massive immune response shortly after copulation, potentially posing a strong barrier to male reproductive success.
Two of these 11 genes, nucb2 (ENSMUSG00000030659) and dnase2B (ENSMUSG00000028185), bind or hydrolyze DNA, respectively. Shortly after copulation, the female reproductive tract is inundated with white blood cells. Some of these white blood cells react to foreign bodies by expelling a net of proteases attached to a DNA backbone (Wartha et al. 2007). DNA binding or hydrolysis by seminal vesicle proteins may counteract this response and free entangled sperm (Alghamdi and Foster 2005).
Four additional genes—serpine2 (ENSMUSG00000026249), spink3 (ENSMUSG00000024503), timp1 (ENSMUSG00000001131), and 9530002K18RIK (ENSMUSG00000053729)—all inhibit certain proteolytic enzymes. It is possible that protease inhibitors in seminal fluid protect sperm from proteolytic attack. It is also plausible that protease inhibitors slow down the degradation of the copulatory plug, as some proteases degrade coagulated seminal vesicle secretions (Lilja 1985; Lundwall et al. 2006). Either scenario potentially places male-expressed protease inhibitors in a sexually antagonistic role with female-expressed proteases. Consistent with this model, rapid evolution has been observed in multiple Drosophila proteases and protease inhibitors (Swanson et al. 2004; Mueller et al. 2005; Panhuis and Swanson 2006; Kelleher et al. 2007; Lawniczak and Begun 2007; Findlay et al. 2008; Wong et al. 2008). Interestingly, protease inhibition is a function that was significantly overrepresented among mouse seminal proteins without a human ortholog (table 2), suggesting this class of genes may experience rapid turnover.
Five of the 11 immune-related genes may be involved in pathogen defense, as they encode proteins that target pathogens (ceacam10 [ENSMUSG00000054169]), present antigens (b2m [ENSMUSG00000060802]), perform lysis (9530003J23RIK [ENSMUSG00000020177]), degrade proteins (plau [ENSMUSG00000021822]), or alter glycosylated proteins (man2b1 [ENSMUSG00000005142]). It is possible that some of these proteins serve as a kind of paternal investment, whereby the male donates a suite of proteins that protect the female from incoming bacterial or viral infections following copulation. Here, rapid evolution would be predicted not because of sexual antagonism, but because of an arms race between host immunity genes and targeted pathogen molecules. The seven immunity-related genes that have a rat ortholog and pass quality control have a median pairwise dN/dS = 0.304 (∼2.5X the genome median, range = 0.123–0.95, supplementary table 1, Supplementary Material online).
Conclusions
Although it is true that many reproductive proteins evolve rapidly, our work shows that the majority of proteins isolated from the male reproductive tract have been subjected to strong evolutionary constraint. Proteins found in the seminal vesicles are the notable exception, showing patterns of rapid evolution consistent with positive selection driving recurrent nonsynonymous changes. Interestingly, seminal vesicles produce a variety of proteins that form the copulatory plug and apparently suppress the female immune response. These proteins might mediate competitive outcomes among males or sexual conflict between males and females. Furthermore, many seminal vesicle proteins appear to participate in immune response. All of these interactions predict rapid evolution among seminal vesicle proteins due to either sexual or natural selection. Future investigation of ejaculated proteins will shed light on the functional partitioning across the male reproductive tract and how these functions affect male reproductive variance and/or reproductive isolation.
Supplementary Material
Supplementary tables 1–4 and supplementary figures 1 and 2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by NIH postdoctoral fellowship F32GM070246-02 to M.D.D., NIH training grant T32 HG00035 to G.D.F., NICHHD senior postdoctoral fellowship 5F33HD055016-02 to R.C.K., NIH grant P41 RR011823 to M.J.M., and NSF and NIH grants to M.W.N. M. Carneiro read an early version of the manuscript. D. Tautz and two anonymous reviewers improved the manuscript.
References
- Aagaard JE, Yi X, MacCoss MJ, Swanson WJ. Rapidly evolving zona pellucida domain proteins are a major component of the vitelline envelope of abalone eggs. Proc Natl Acad Sci USA. 2006;103:17302–17307. doi: 10.1073/pnas.0603125103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott DE, Pritchard C, Clegg NJ, Ferguson C, Dumpit R, Sikes RA, Nelson PS. Expressed sequence tag profiling identifies developmental and anatomic partitioning of gene expression in the mouse prostate. Genome Biol. 2003;4:R79. doi: 10.1186/gb-2003-4-12-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal Y, Vanha-Perttula T. Effect of secretory particles in bovine seminal vesicle secretion on sperm motility and acrosome reaction. J Reprod Fertil. 1987;79:409–419. doi: 10.1530/jrf.0.0790409. [DOI] [PubMed] [Google Scholar]
- Alghamdi AS, Foster DN. Seminal DNase frees spermatozoa entangled in neutrophil extracellular traps. Biol Reprod. 2005;73:1174–1181. doi: 10.1095/biolreprod.105.045666. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Evol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anderson DJ, Tarter TH. Immunosuppressive effects of mouse seminal plasma components in vivo and in vitro. J Immunol. 1982;128:535–539. [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, et al. (20 co-authors) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines JF, Harr B. Reduced X-linked diversity in derived populations of house mice. Genetics. 2007;175:1911–1921. doi: 10.1534/genetics.106.069419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berquin IM, Min Y, Wu R, Wu H, Chen YQ. Expression signature of the mouse prostate. J Biol Chem. 2005;280:36442–36451. doi: 10.1074/jbc.M504945200. [DOI] [PubMed] [Google Scholar]
- Carballada R, Esponda P. Electrophoretic pattern of rodent seminal vesicle proteins as revealed by silver staining. Int J Androl. 1991;14:52–57. doi: 10.1111/j.1365-2605.1991.tb01062.x. [DOI] [PubMed] [Google Scholar]
- Carballada R, Esponda P. Fate and distribution of seminal plasma proteins in the genital tract of the female rat after natural mating. J Reprod Fertil. 1997;109:325–335. doi: 10.1530/jrf.0.1090325. [DOI] [PubMed] [Google Scholar]
- Carballada R, Esponda P. Binding of seminal vesicle proteins to the plasma membrane of rat spermatozoa in vivo and in vitro. Int J Androl. 1998;21:19–28. doi: 10.1046/j.1365-2605.1998.00082.x. [DOI] [PubMed] [Google Scholar]
- Carballada R, Esponda P. Effect of antibodies against seminal vesicle secretion on fertility in the rat. Zygote. 1999;7:223–231. doi: 10.1017/s096719949900060x. [DOI] [PubMed] [Google Scholar]
- Carter CS, Schein MW. Sexual receptivity and exhaustion in the female golden hamster. Horm Behav. 1971;4:191–200. doi: 10.1016/0018-506x(74)90017-8. [DOI] [PubMed] [Google Scholar]
- Castillo-Davis CI, Kondrashov FA, Hartl DL, Kulathinal RJ. The functional genomic distribution of protein divergence in two animal phyla: coevolution, genomic conflict, and constraint. Genome Res. 2004;14:802–811. doi: 10.1101/gr.2195604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark NL, Aagaard JE, Swanson WJ. Evolution of reproductive proteins from animals and plants. Reprod Fertil Dev. 2006;131:11–22. doi: 10.1530/rep.1.00357. [DOI] [PubMed] [Google Scholar]
- Dean MD, Ardlie KG, Nachman MW. The frequency of multiple paternity suggests that sperm competition is common in house mice (Mus domesticus) Mol Ecol. 2006;15:4141–4151. doi: 10.1111/j.1365-294X.2006.03068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean MD, Good JM, Nachman MW. Adaptive evolution of proteins secreted during sperm maturation: an analysis of the mouse epididymal transcriptome. Mol Biol Evol. 2008;25:383–392. doi: 10.1093/molbev/msm265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean MD, Nachman MW. Faster fertilization rate in conspecific versus heterospecific matings in house mice. Evolution. 2009;63:20–28. doi: 10.1111/j.1558-5646.2008.00499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixson AF, Anderson MJ. Sexual selection, seminal coagulation and copulatory plug formation in primates. Folia Primatol. 2002;73:63–69. doi: 10.1159/000064784. [DOI] [PubMed] [Google Scholar]
- Dlouhy SR, Nichols WC, Karn RC. Production of an antibody to mouse salivary androgen binding protein (ABP) and its use in identifying a prostate protein produced by a gene distinct from Abp. Biochem Genet. 1986;24:743–763. doi: 10.1007/BF00499007. [DOI] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectr. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Findlay GD, MacCoss MJ, Swanson WJ. Proteomic discovery of previously unannotated, rapidly evolving seminal fluid genes in Drosophila. Genome Res. 2009;19:886–896. doi: 10.1101/gr.089391.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay GD, Yi X, MacCoss MJ, Swanson WJ. Proteomics reveals novel Drosophila seminal fluid proteins transferred at mating. PLoS Biol. 2008;6:e178. doi: 10.1371/journal.pbio.0060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firman RC, Simmons LW. The frequency of multiple paternity predicts variation in testes size among island populations of house mice. J Evol Biol. 2008 doi: 10.1111/j.1420-9101.2008.01612.x. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Eskin E, Kang HM, et al. (16 co-authors) A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 2007;448:1050–1053. doi: 10.1038/nature06067. [DOI] [PubMed] [Google Scholar]
- Fujimoto N, Akimoto Y, Suzuki T, Kitamura S, Ohta S. Identification of prostatic-secreted proteins in mice by mass spectrometric analysis and evaluation of lobe-specific and androgen-dependent mRNA expression. J Endocrinol. 2006;190:793–803. doi: 10.1677/joe.1.06733. [DOI] [PubMed] [Google Scholar]
- Geraldes A, Basset P, Gibson B, Smith KL, Harr B, Yu HT, Bulatova N, Ziv Y, Nachman MW. Inferring the history of speciation in house mice from autosomal, X-linked, Y-linked and mitochondrial genes. Mol Ecol. 2008;17:5349–5363. doi: 10.1111/j.1365-294X.2008.04005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, Weinstock GM, Metzker ML, et al. (228 co-authors) Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Goldfoot DA, Goy RW. Abbreviation of behavioral estrus in guinea pigs by coital and vagino-cervical stimulation. J Comp Physiol Psychol. 1970;72:426–434. doi: 10.1037/h0029752. [DOI] [PubMed] [Google Scholar]
- Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences: a maximum likelihood approach. J Mol Evol. 1994;40:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Good JM, Handel MA, Nachman MW. Asymmetry and polymorphism of hybrid male sterility during the early stages of speciation in house mice. Evolution. 2007;62:50–65. doi: 10.1111/j.1558-5646.2007.00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good JM, Nachman MW. Rates of protein evolution are positively correlated with developmental timing of expression during mouse spermatogenesis. Mol Biol Evol. 2005;22:1044–1052. doi: 10.1093/molbev/msi087. [DOI] [PubMed] [Google Scholar]
- Hardy DF, DeBold JF. Effects of coital stimulation upon behavior of the female rat. J Comp Physiol Psychol. 1972;78:400–408. doi: 10.1037/h0032536. [DOI] [PubMed] [Google Scholar]
- Hartung TG, Dewsbury DA. A comparative analysis of copulatory plugs in muroid rodents and their relationship to copulatory behavior. J Mammalogy. 1978;59:717–723. [Google Scholar]
- Hayashi N, Sugimura Y, Kawamura J, Donjacour AA, Cunha GR. Morphological and functional heterogeneity in the rat prostatic gland. Biol Reprod. 1991;45:308–321. doi: 10.1095/biolreprod45.2.308. [DOI] [PubMed] [Google Scholar]
- Henault MA, Killian GJ, Kavanaugh JF, Griel LC., Jr Effect of accessory sex gland fluid from bulls of differing fertilities on the ability of cauda epididymal sperm to penetrate zona-free bovine oocytes. Biol Reprod. 1995;52:390–397. doi: 10.1095/biolreprod52.2.390. [DOI] [PubMed] [Google Scholar]
- Huang YH, Chu ST, Chen YH. A seminal vesicle autoantigen of mouse is able to suppress sperm capacitation-related events stimulated by serum albumin. Biol Reprod. 2000;63:1562–1566. doi: 10.1095/biolreprod63.5.1562. [DOI] [PubMed] [Google Scholar]
- Hurle B, Swanson W, Green ED. Comparative sequence analyses reveal rapid and divergent evolutionary changes of the WFDC locus in the primate lineage. Genome Res. 2007;17:276–286. doi: 10.1101/gr.6004607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin M, Nicholson N, Haywood JT, Poirier GR. Immunofluorescent localization of a murine seminal vesicle proteinase inhibitor. Biol Reprod. 1983;28:1201–1206. doi: 10.1095/biolreprod28.5.1201. [DOI] [PubMed] [Google Scholar]
- Jelinsky SA, Turner TT, Bang HJ, Finger JN, Solarz MK, Wilson E, Brown EL, Kopf GS, Johnston DS. The rat epididymal transcriptome: comparison of segmental gene expression in the rat and mouse epididymides. Biol Reprod. 2007;76:561–570. doi: 10.1095/biolreprod.106.057323. [DOI] [PubMed] [Google Scholar]
- Johnston DS, Jelinsky SA, Bang HJ, Dicandeloro P, Wilson E, Kopf GS, Turner TT. The mouse epididymal transcriptome: transcriptional profiling of segmental gene expression in the epididymis. Biol Reprod. 2005;73:404–413. doi: 10.1095/biolreprod.105.039719. [DOI] [PubMed] [Google Scholar]
- Karn RC, Clark NL, Nguyen ED, Swanson WJ. Adaptive evolution in rodent seminal vesicle secretion proteins. Mol Biol Evol. 2008;25:2301–2310. doi: 10.1093/molbev/msn182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karn RC, Laukaitis CM. Characterization of two forms of mouse salivary androgen-binding protein (ABP): implications for evolutionary relationships and ligand-binding function. Biochemistry. 2003;42:7162–7170. doi: 10.1021/bi027424l. [DOI] [PubMed] [Google Scholar]
- Kawano N, Yoshida M. Semen-coagulating protein, SVS2, in mouse seminal plasma controls sperm fertility. Biol Reprod. 2007;76:353–361. doi: 10.1095/biolreprod.106.056887. [DOI] [PubMed] [Google Scholar]
- Kelleher ES, Swanson WJ, Markow TA. Gene duplication and adaptive evolution of digestive proteases in Drosophila arizonae female reproductive tracts. PLoS Genet. 2007;3:e148. doi: 10.1371/journal.pgen.0030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klammer AA, Wu CC, MacCoss MJ, Noble WS. Peptide charge state determination for low-resolution tandem mass spectra. Proc IEEE Comput Syst Bioinform Conf. 2005:175–185. doi: 10.1109/csb.2005.44. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Laukaitis CM, Dlouhy SR, Emes RD, Ponting CP, Karn RC. Diverse spatial, temporal, and sexual expression of recently duplicated androgen-binding protein genes in Mus musculus. BMC Evol Biol. 2005;5:40. doi: 10.1186/1471-2148-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawniczak MKN, Begun DJ. Molecular population genetics of female-expressed mating-induced serine proteases in Drosophila melanogaster. Mol Biol Evol. 2007;24:1944–1951. doi: 10.1093/molbev/msm122. [DOI] [PubMed] [Google Scholar]
- Lilja H. A kallikrein-like serine protease in prostatic fluid cleaves the predominant seminal vesicle protein. J Clin Invest. 1985;76:1899–1903. doi: 10.1172/JCI112185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H-J, Luo C-W, Chen Y-H. Localization of the transglutaminase cross-linking site in SVS III, a novel glycoprotein secreted from mouse seminal vesicle. J Biol Chem. 2002;277:3632–3639. doi: 10.1074/jbc.M107578200. [DOI] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Lundwall A, Clauss A, Olsson AY. Evolution of kallikrein-related peptidases in mammals and identification of a genetic locus encoding potential regulatory inhibitors. Biol Chem. 2006;387:243–249. doi: 10.1515/BC.2006.032. [DOI] [PubMed] [Google Scholar]
- Lundwall Å, Peter, JLovgren, Lilja H, Malm J. Chemical characterization of the predominant proteins secreted by mouse seminal vesicles. Eur J Biochem. 1997;249:39–44. doi: 10.1111/j.1432-1033.1997.t01-2-00039.x. [DOI] [PubMed] [Google Scholar]
- Mann T, Lutwak-Mann C. Male reproductive function and semen. New York: Springer-Verlag; 1981. [Google Scholar]
- Mueller JL, Ravi Ram K, McGraw LA, Bloch Qazi MC, Siggia ED, Clark AG, Aquadro CF, Wolfner MF. Cross-species comparison of Drosophila male accessory gland protein genes. Genetics. 2005;171:131–143. doi: 10.1534/genetics.105.043844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating the mouse embryo. 2003. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- Nei M, Li W-H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Bustamante C, Clark AG, et al. (13 co-authors) A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O WS, Chen HQ, Chow PH. Effects of male accessory sex gland secretions on early embryonic development in the golden hamster. J Reprod Fertil. 1988;84:341–344. doi: 10.1530/jrf.0.0840341. [DOI] [PubMed] [Google Scholar]
- Pang SF, Chow PH, Wong TM. The role of the seminal vesicles, coagulating glands and prostate glands on the fertility and fecundity of mice. J Reprod Fertil. 1979;56:129–132. doi: 10.1530/jrf.0.0560129. [DOI] [PubMed] [Google Scholar]
- Panhuis TM, Swanson WJ. Molecular evolution and population genetic analysis of candidate female reproductive genes in Drosophila. Genetics. 2006;173:2039–2047. doi: 10.1534/genetics.105.053611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peitz B. Effects of seminal vesicle fluid components on sperm motility in the house mouse. J Reprod Fertil. 1988;83:169–176. doi: 10.1530/jrf.0.0830169. [DOI] [PubMed] [Google Scholar]
- Peitz B, Bennett D. Inhibition of complement-mediated cytotoxicity of antisera by fluid secreted by the seminal vesicle of the house mouse. J Reprod Immunol. 1981;3:109–116. doi: 10.1016/0165-0378(81)90015-2. [DOI] [PubMed] [Google Scholar]
- Peitz B, Olds-Clarke P. Effects of seminal vesicle removal on fertility and uterine sperm motility in the house mouse. Biol Reprod. 1986;35:608–617. doi: 10.1095/biolreprod35.3.608. [DOI] [PubMed] [Google Scholar]
- Pilch B, Mann M. Large-scale and high-confidence proteomic analysis of human seminal plasma. Genome Biol. 2006;7:R40. doi: 10.1186/gb-2006-7-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platz RD, Wolfe HG. Mouse seminal vesicle proteins. The inheritance of electrophoretic variants. J Hered. 1969;60:187–192. doi: 10.1093/oxfordjournals.jhered.a107969. [DOI] [PubMed] [Google Scholar]
- Price TAR, Hodgson DJ, Lewis Z, Hurst GDD, Wedell N. Selfish genetic elements promote polyandry in a fly. Science. 2008;322:1241–1243. doi: 10.1126/science.1163766. [DOI] [PubMed] [Google Scholar]
- Prout T, Clark AG. Seminal fluid causes temporarily reduced egg hatch in previously mated females. Proc Roy Soc Lond B Biol Sci. 2000;267:201–203. doi: 10.1098/rspb.2000.0988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queen K, Dhabuwala CB, Pierrepoint CG. The effect of the removal of the various accessory sex glands on the fertility of male rats. J Reprod Fertil. 1981;62:423–426. doi: 10.1530/jrf.0.0620423. [DOI] [PubMed] [Google Scholar]
- Ramm SA, Oliver PL, Ponting CP, Stockley P, Emes RD. Sexual selection and the adaptive evolution of mammalian ejaculate proteins. Mol Biol Evol. 2008;25:207–219. doi: 10.1093/molbev/msm242. [DOI] [PubMed] [Google Scholar]
- Ramm SA, Parker GA, Stockley P. Sperm competition and the evolution of male reproductive anatomy in rodents. Proc Biol Sci. 2005;272:949–955. doi: 10.1098/rspb.2004.3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR. Sex chromosomes and the evolution of sexual dimorphism. Evolution. 1984;38:735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. [DOI] [PubMed] [Google Scholar]
- Robinson PN, Wollstein A, Bohme U, Beattie B. Ontologizing gene-expression microarray data: characterizing clusters with gene ontology. Bioinformatics. 2004;20:979–981. doi: 10.1093/bioinformatics/bth040. [DOI] [PubMed] [Google Scholar]
- Robinson R, Richardson R, Hinds K, Clayton D, Poirier GR. Features of a seminal proteinase inhibitor-zona pellucida binding component on murine spermatozoa. Gamete Res. 1987;16:217–228. doi: 10.1002/mrd.1120160304. [DOI] [PubMed] [Google Scholar]
- Schmidt JA, Oatley JM, Brinster RL. Female mice delay reproductive aging in males. Biol Reprod. 2009;80:1009–1014. doi: 10.1095/biolreprod.108.073619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaji S, Scheit K-H, Bhargava PM. Proteins of seminal plasma. New York: John Wiley & Sons; 1990. [Google Scholar]
- Silver L. Mouse genetics: concepts and applications. New York: Oxford University Press; 1995. [Google Scholar]
- Snyder RL. Collection of mouse semen by electroejaculation. Anat Rec. 1966;155:11–14. doi: 10.1002/ar.1091550103. [DOI] [PubMed] [Google Scholar]
- Snyder RL. Fertility and reproductive performance of grouped male mice. In: Benirschke K, editor. Comparative aspects of reproductive failure. New York: Springer-Verlag; 1967. pp. 458–472. [Google Scholar]
- Su AI, Cooke MP, Ching KA, et al. (14 co-authors) Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Nielsen R, Yang Q. Pervasive adaptive evolution in mammalian fertilization proteins. Mol Biol Evol. 2003;20:18–20. doi: 10.1093/oxfordjournals.molbev.a004233. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Wong A, Wolfner MF, Aquadro CF. Evolutionary expressed sequence tag analysis of Drosophila female reproductive tracts identifies genes subjected to positive selection. Genetics. 2004;168:1457–1465. doi: 10.1534/genetics.104.030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP* Sinaeur Associates; 2002. Sunderland, Massachusetts. [Google Scholar]
- Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tecirlioglu RT, Hayes ES, Trounson AO. Semen collection from mice: electroejaculation. Reprod Fertil Dev. 2002;14:363–371. doi: 10.1071/rd02015. [DOI] [PubMed] [Google Scholar]
- Thaler CJ. Immunological role for seminal plasma in insemination and pregnancy. Am J Reprod Immunol. 1989;21:147–150. doi: 10.1111/j.1600-0897.1989.tb01020.x. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner LM, Chuong EB, Hoekstra HE. Comparative analysis of testis protein evolution in rodents. Genetics. 2008;179:2075–2089. doi: 10.1534/genetics.107.085902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B. Evolution on the X chromosome: unusual patterns and processes. Nat Rev Genet. 2006;7:645–653. doi: 10.1038/nrg1914. [DOI] [PubMed] [Google Scholar]
- Wade CM, Kulbokas EJ, 3rd, Kirby AW, Zody MC, Mullikin JC, Lander ES, Lindblad-Toh K, Daly MJ. The mosaic structure of variation in the laboratory mouse genome. Nature. 2002;420:574–578. doi: 10.1038/nature01252. [DOI] [PubMed] [Google Scholar]
- Wartha F, Beiter K, Normark S, Henriques-Normark B. Neutrophil extracellular traps: casting the NET over pathogenesis. Curr Opin Microbiol. 2007;10:52–56. doi: 10.1016/j.mib.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, et al. (222 co-authors) Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wernersson R, Pedersen AG. RevTrans: multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res. 2003;31:3537–3539. doi: 10.1093/nar/gkg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A, Albright SN, Giebel JD, Ram KR, Ji S, Fiumera AC, Wolfner MF. A role for Acp29AB, a predicted seminal fluid lectin, in female sperm storage in Drosophila melanogaster. Genetics. 2008;180:921–931. doi: 10.1534/genetics.108.092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Bell TA, Churchill GA, Pardo-Manuel de Villena F. On the subspecific origin of the laboratory mouse. Nat Genet. 2007;39:1100–1107. doi: 10.1038/ng2087. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML, a program package for phylogenetic analysis by maximum likelihood. CABIOS. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Pedersen A-MK. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.