Abstract
The evolutionary history of α-satellite DNA, the major component of primate centromeres, is hardly defined because of the difficulty in its sequence assembly and its rapid evolution when compared with most genomic sequences. By using several approaches, we have cloned, sequenced, and characterized α-satellite sequences from two species representing critical nodes in the primate phylogeny: the white-cheeked gibbon, a lesser ape, and marmoset, a New World monkey. Sequence analyses demonstrate that white-cheeked gibbon and marmoset α-satellite sequences are formed by units of ∼171 and ∼342 bp, respectively, and they both lack the high-order structure found in humans and great apes. Fluorescent in situ hybridization characterization shows a broad dispersal of α-satellite in the white-cheeked gibbon genome including centromeric, telomeric, and chromosomal interstitial localizations. On the other hand, centromeres in marmoset appear organized in highly divergent dimers roughly of 342 bp that show a similarity between monomers much lower than previously reported dimers, thus representing an ancient dimeric structure.
All these data shed light on the evolution of the centromeric sequences in Primates. Our results suggest radical differences in the structure, organization, and evolution of α-satellite DNA among different primate species, supporting the notion that 1) all the centromeric sequence in Primates evolved by genomic amplification, unequal crossover, and sequence homogenization using a 171 bp monomer as the basic seeding unit and 2) centromeric function is linked to relatively short repeated elements, more than higher-order structure.
Moreover, our data indicate that complex higher-order repeat structures are a peculiarity of the hominid lineage, showing the more complex organization in humans.
Keywords: evolution, primates, centromere
Introduction
The centromere is a highly specialized region of a chromosome essential for the correct chromosome segregation during mitosis and meiosis in eukaryotic cells. Primate centromeres are mainly composed of repeated DNA, known as α-satellite DNA, made up of a basic 171-bp unit organized as tandemly repeated units (Maio 1971; Manuelidis 1978). Human α-satellite has been classified into two types according to its organization and sequence properties: higher-order α-satellite and monomeric α-satellite.
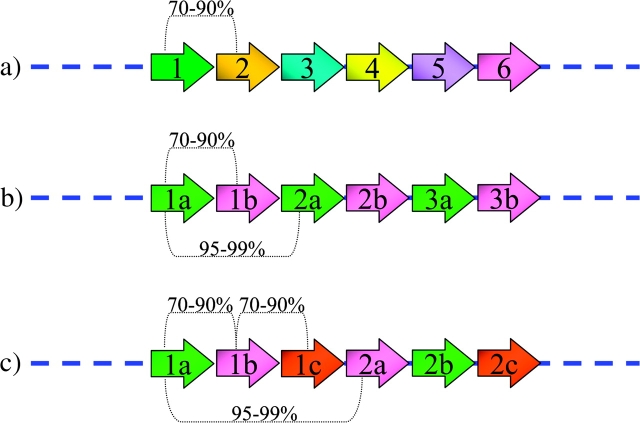
The higher-order structure is based on multiple copies of the 171 bp monomers, assembled into subfamilies at constant unit periodicity. In each subfamily, monomers of the same unit (1a, 1b) differ greatly in primary sequence and are not necessarily any closer in sequence similarity than each is to monomers from different subfamilies. In contrast, monomers within different units at specific periodic distance are virtually identical, sharing high sequence similarity (<2% sequence divergence) (1a–2a) (fig. 1). Higher-order structures are composed of these monomers organized into multimeric repeat units ranging in size from 2 to 5 Mb. Organization and unit periodicity are specific to each human centromere (Willard and Waye 1987b; Lee et al. 1997), or to a small group of chromosomes, identifying the different suprachromosomal families (SFs) (Choo et al. 1988; Jorgensen et al. 1988).
FIG. 1.—
Schematic representation of structure and organization of alphoid sequences (modified from Alexandrov et al. 1993b). Arrows indicate single 171 bp monomers arranged in tandem stretches in (a) monomeric, (b) dimeric, and (c) trimeric structures. The similarity percentage between monomers has been reported on the arrows (see text for details).
The monomeric α-satellite has no detectable higher-order periodicity and its monomers are far less homogeneous than the higher-order repeat (HOR) units (Warburton and Willard 1990; Alexandrov et al. 2001; Rudd and Willard 2004). Phylogenetic analyses suggested that the higher-order α-satellite DNA emerged more recently than the monomeric repeat (Alkan et al. 2004). Recent hypotheses state that the higher-order α-satellite evolved from ancestral arrays of monomeric α-satellite and was subsequently transposed to the centromeric regions of all great-ape chromosomes (Warburton et al. 1996; Alexandrov et al. 2001; Schueler et al. 2001, 2005; Kazakov et al. 2003).
α-Satellite DNA, like other tandemly repeated sequences, undergoes concerted evolution, showing greater similarity within a species than between species (Willard and Waye 1987a). The evolutionary process is known as molecular drive and includes mechanisms such as unequal crossing-over, gene conversion, and transposition (Dover 1982). Thanks to these molecular mechanisms, the structure and genomic organization of centromeric DNA can change very rapidly. Fluorescent in situ hybridization (FISH) studies with human chromosome–specific α-satellite probes against great-ape chromosomes, in fact, have demonstrated that only the organization of the X chromosome α-satellite subset is conserved among closely related species (Baldini et al. 1992; Archidiacono et al. 1995). Furthermore, the rapid α-satellite DNA evolution has been confirmed comparing its organization among Primates. For example, every human α-satellite SF map to nonorthologous chromosomes in chimpanzee, despite the fact that alphoid sequences in human and chimpanzee share high homology. Similarly, comparisons between ape and Old World monkey α-satellite DNA indicate two radically distinct patterns of centromeric organization and chromosome distribution (Haaf and Willard 1998).
Due to this rapid diversification and complex structure, studies on centromeric sequence and organization have been uncoupled from genomewide efforts to sequence genomes. In fact, for each human chromosome assembly, the largest gaps correspond to the centromere gap located between the most proximal p and q arm contigs (Rudd and Willard 2004). For other primate and mammalian genomes, the location and sequence of centromere repeats often remain uncharacterized due to the difficulties in assembling these portions of the genome from whole-genome shotgun sequence (WGSS).
In order to gain an understanding of the evolution, biology, and organization of centromeric sequences, we have cloned and characterized α-satellite sequences from two different primate species: a lesser ape, the white-cheeked gibbon (Nomascus leucogenys, NLE), and a New World monkey (NWM), the common marmoset (Callitrix jacchus, CJA), using two complementary approaches. Our results reveal new features in the organization and evolution of centromeric sequences: 1) Both NLE and CJA alphoid sequences lack HOR or subfamily organization thus supporting the hypothesis that HOR structure arose specifically in the great ape–human lineage of evolution (Alkan et al. 2004); 2) CJA sequence analysis reveals a dimeric structure of ∼342 bp largely different from the previously reported dimeric structure in other Primates; and 3) in NLE alphoid sequences are detected at the centromeres, telomeric, and interstitial regions. We hypothesize that these noncentromeric loci enriched in repetitive alphoid sequences might represent regions of evolutionary genomic instability and partially could explain the high evolutionary rearrangement rates of white-cheeked gibbon karyotype.
Materials and Methods
Cell Line
Metaphase preparations were obtained from a lymphoblastoid cell line of C. jacchus (CJA) and N. leucogenys (NLE), kindly provided by S. Muller (Munchen). Human (HSA) metaphase spreads were prepared from Phytohemagglutinin-stimulated peripheral lymphocytes of normal donors by standard procedures.
FISH and Image Analysis
DNA extraction from BACs and plasmids has already been reported (Ventura et al. 2001). FISH experiments were essentially performed as previously described (Ventura et al. 2003). Briefly, DNA probes were directly labeled with Cy3-dUTP (Perkin–Elmer) or Fluorescein-Deoxicitidinetriphospate (dCTP) (Fermentas) by nick translation. Two hundred nanograms of labeled probe was used for the FISH experiments. Hybridization was performed at 37 °C in 2× sodium chloride, sodium citrate (SSC), 50% (v/v) formamide, 10% (w/v) dextran sulfate, 5 mg of COT1 DNA (Roche), and 3 mg of sonicated salmon sperm DNA in a volume of 10 μl. Posthybridization washing was at 60 °C in 0.1× SSC (three times, high stringency). Washes of FISH experiments were performed at lower stringency: 37 °C in 2× SSC, 50% formamide (X3), followed by washes at 42 °C in 2× SSC (X3).
Digital images were obtained using a Leica DMRXA epifluorescence microscope equipped with a cooled CCD camera (Princeton Instruments). Cy3 (red), fluorescein (green), and 4′,6-diamidino-2-phenylindole (DAPI) (blue) fluorescence signals, detected with specific filters and recorded separately as grayscale images. Pseudocoloring and merging of images were performed using Adobe PhotoShop software.
Polymerase Chain Reaction (PCR) Labeling
DNA probes were directly labeled with Cy5-dUTP by PCR labeling; 200 ng of labeled probe was used for the FISH experiments. The use of PCR labeling avoids the possible contamination from genomic DNA by nick translation labeling of PCR products.
PCR labeling was carried out in a final volume of 20 μl that contained 100 ng PCR product, 2.5 μl reaction buffer 10×, 2 μl MgCl2 50 mM, 0.5 μl each primer 10 μM, 0.5 μl dACG 2mM, 2.5 Cy5-dUTP 1 mM, 5 μl BSA 2%, and 0.3 μl Taq polymerase 5 U/μl.
Library Screening
Library-hybridization was carried out according to the protocol available at CHORI BACPAC resources (http://bacpac.chori.org/highdensity.htm). The CH271 segment 1 represents a 5.0-fold clone coverage library (http://www.chori.org/bacpac/).
Alpha PCR
Human genomic DNA and common marmoset genomic DNA were obtained from human lymphoblast cell lines by standard methods.
α27 (CATCACAAAGAAGTTTCTGAGAATGCTTC) and α30 (TGCATTCAACTCACAGAGTTGAACCTTCC) primers were used to amplify genomic DNA by Polymerase Chain Reactions. They were obtained from the most conserved regions of human alphoid consensus (Waye and Willard 1986; Choo et al. 1991).
The PCR cycling parameters used were as follows: 2 min initial denaturation at 94 °C, followed by 10 cycles of: 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 1 min; followed by 20 cycles of 94 °C for 15 s, 58 °C for 30 s, and 72 °C for 1 min (20 s more each cycle). Final extension was at 72 °C for 7 min (and then at hold 12 °C).
The reaction mixture consisted of 5 μl dNTPs (10×), 0.5 μl each primer (10 μM), 0.3 μl Platinum Taq DNA polymerase (5 U/μl), 1.5 μl MgCl2 (50 mM), 5 μl reaction buffer (Invitrogen) (10×), 3 μl of DNA template (50 ng/μl), and water up to 25 μl.
PCR products were analyzed by 1% agarose gel electrophoresis. They were labeled and used as a probe for FISH experiments on HSA and CJA metaphase spreads.
Sequence Analysis
CJA alphoid cloned sequences were analyzed using the NCBI Blast 2 Sequences tool (http://blast.ncbi.nlm.nih.gov/bl2seq/wblast2.cgi) (BlastN program), using 1 as reward for a match, −2 as penalty for a mismatch, and 5 and 2 as open and extension gap penalties. We identified a conserved T-rich motif every 171 bp, GTTTTG(A, T or/)GTTTTAGA, and we used this motif to distinguish in CJA alphoid sequences the monomers of 171 bp. The monomers of 342 and 171 bp were multi-aligned using ClustalW algorithm, and consensus sequences were extracted using a modified version of the MaM software (Alkan et al. 2005). MaM returned one character for every column of the ClustalW alignment by “compressing” all the information in a given base position into a single character. For example, if all the bases in a column are G, then the consensus character for that position is G. However, if there are substitutions in a column, then all observed characters are encoded into a single marker: Y for pyrimidines (C or T), R for purines (A or G), N for any (A, C, G, and T), etc.
All-by-All Sequence Comparison of NLE Sequences
All pairwise sequence alignments for NLE sequences were performed with an in-house implementation of the Needleman–Wunsch global alignment algorithm (Needleman and Wunsch 1970). The divergence of two sequences is then computed by calculating the ratio of the Hamming distance of the aligned sequences, and the alignment length (Hamming 1950). For all pairs of sequences, the divergence ratio calculation is also repeated for the reverse complement of the second sequence.
C410 Probe
C410 is a specific NLE alphoid DNA obtained by chromatin immunoprecipitation of NLE lymphoblastoid cells with rabbit polyclonal antibodies directed against human anti-CENP-C centromeric protein (S. Trazzi et al., manuscript in preparation).
Cloning
PCR products were cloned in pCR-XL-TOPO using the standard protocol Topo cloning XL PCR kit (invitrogen).
Southern Blot Analysis
Genomic DNAs from marmoset lymphoblastoid cell lines were prepared by following standard procedures (Maniatis et al. 1982). Endonuclease digestions were performed using a 4-fold excess of enzyme under the conditions suggested by the suppliers. Gel electrophoresis was performed in 1× tris-acetate (TAE) (1× TAE = 40 mM Tris-acetate, 1 mM ethylenediaminetetraacetic acid, EDTA). Genomic DNAs were run in a 0.8% agarose gel for 16–18 h, denatured, and DNA transferred to Hybond membrane (Amersham), using as transfer buffer 20× SSC (1× SSC = 150 mM sodium chloride, 15 mM sodium citrate, pH 7). Clone inserts (50 ng) were labeled with 32P-dCTP (3,000 Ci/mmol; Amersham) by using random oligomer priming. Filters were exposed and developed using storm imaging system.
Consensus Sequences
Consensus sequence obtained from gibbon BAC end sequences: CACTTGCAGTTTCTACAGAAAGAGTGTTTCAAAACTGCTCAATCAAAAGTAAGGTTCAACTCTGTTAGTTGAATGCACAGAACAGAAAGAAGTTTCACAGAATGCTTCTGTGTAGTTTTTATTTGAAGATATTCCTTTTTCCACTATAGGCCTCTTAGCGCTCTGAATGTCCACTTGCAGTTTCTACAGAAAGAGTGTTTCAGAACTGCTCAATCAAAAGTAAGGTTCAACTCTGTTAGTTGAATGCACAGAACAGAAAGAAGTTTCACAGAATGCTTCTGTGTAGTTTTTATTTGAAGATATTCCTTTTTCCACTATAGGCCTCTTAGCGCTCTGAATGTCCACTTGCAGTTTCTACAGAAAGAGTGTTTCAGAACTGCTCAATCAAAAGTAAGGTTCAACTCTGTTAGTTGAATGCACAGAACAGAAAGAAGTTTCACAGAATGCTTCTGTGTAGTTTTTATTTGAAGATATTCCTTTTTCCACT primers designed on gibbon consensus sequence: α NLE_F: TCAACTCTGTTAGTTGAATGCACA and α NLE_R: CTCTTTCTGTAGAAACTGCAAGTG.
Sequences Accession Numbers
Insert sequences from gibbon plasmid clones: pA_12FJ346627, pGAMMA_3FJ346628, pGAMMA_5FJ346629, pGAMMA_6FJ346630, pGAMMA_8FJ346631, pGAMMA_23 FJ346632, pGAMMA_24 FJ346633, pGAMMA_34 FJ346634, pGAMMA_35 FJ346635,p GAMMA_36 FJ346636, pGAMMA_37 FJ346637, pGAMMA_38 FJ346638, pGAMMA_41 FJ346639, pGAMMA_42 FJ346640, pGAMMA_43 FJ346641, pK_64 FJ346642, pK_19 FJ346643, pK_23, FJ346644, pK_25 FJ346645, pK_30 FJ346646, pK_41 FJ346647, pK_43 FJ346648, pK_51 FJ346649, pK_69 FJ346650, pK_1 FJ346651, pK_3 FJ346652, pK_7 FJ346653, pK_20 FJ346654, pK_21 FJ346655, pK_34 FJ346656, pK_63 FJ346657, and pK_68 FJ346658;
Gibbon BAC end sequences: CH271_0010P22_B1 FJ346579, CH271_0010P22_G1 FJ346580, CH271_0046E21_G1 FJ346581,CH271_0046E21_B1 FJ346582, CH271_0002J18_G1 FJ346583,CH271_0002J18_B1 FJ346584, CH271_0029E05_B1 FJ346585, CH271_0029E05_G1 FJ346586, CH271_0032G05_G1 FJ346587, CH271_0032G05_B1 FJ346588, CH271_0036I10_B1 FJ346589, CH271_0036I10_G1 FJ346590, CH271_0023A02_G1 FJ346591, CH271_0023A02_B1 FJ346592, CH271_0059L04_G1 FJ346593, CH271_0059L04_B1 FJ346594, CH271_0083K12_B1 FJ346595, CH271_0083K12_G1 FJ346596, CH271_0042E17_G1 FJ346597, CH271_0042E17_B1 FJ346598, CH271_0005P01_G1 FJ346599, CH271_0005P01_B1 FJ346600, CH271_0015O18_B1 FJ346601, CH271_0015O18_G1 FJ346602, CH271_0039I23_B1 FJ346603, CH271_0039I23_G1 FJ346604, CH271_0054E20_B1 FJ346605, CH271_0054E20_G1 FJ346606, CH271_0096M12_B1 FJ346607, CH271_0096M12_G1 FJ346608, CH271_0015L04_G1 FJ346609, CH271_0015L04_B1 FJ346610, CH271_0024A08_B1 FJ346611, CH271_0024A08_G1 FJ346612, CH271_0048A09_B1 FJ346613, CH271_0048A09_G1 FJ346614, CH271_0084N15_G1 FJ346615, CH271_0084N15_B1 FJ346616, CH271_0072O18_G1 FJ346617, CH271_0072O18_B1 FJ346618, CH271_0047O10_B1 FJ346619, CH271_0047O10_G1 FJ346620, CH271_0027P04_b1 FJ346621, CH271_0027P04_G1 FJ346622, CH271_0091K12_G1 FJ346623, CH271_0091K12_B1 FJ346624, CH271_0007O15_G1 FJ346625, and CH271_0007O15_B1 FJ346626).
Insert sequences from marmoset plasmid clones: C1.1.19 FJ867326, C1.1.74 FJ867327, C2.1.37 FJ867328, C2.1.65 FJ867329, C2.1.73 FJ867330, C3.1.5 FJ867331, C3.1.8 FJ867332, C4.1.13 FJ867333, C4.2.21 FJ867334, C5.1.12 FJ867335, C6.1.1 FJ867336, C6.1.29 FJ867337, C7.1.3 FJ867338, and C7.1.4 FJ867339.
Results
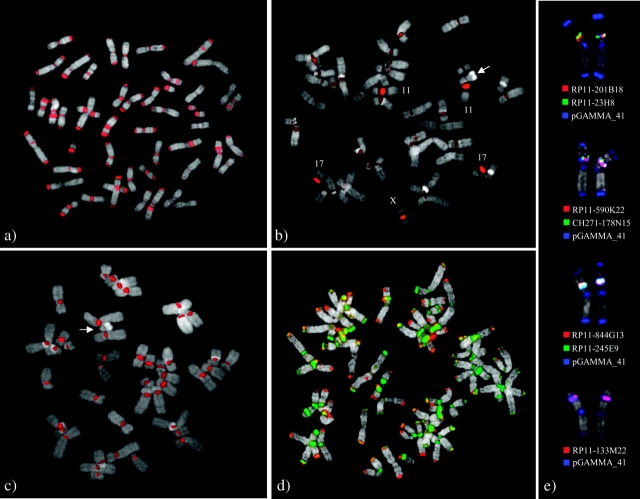
The goal of this work was to characterize the structure and map the location of alphoid sequences in the N. leucogenys (NLE) and C. jacchus (CJA) genomes. A variety of complementary experimental and computational methods was employed. We initially performed α-satellite PCR on genomic DNA from NLE, CJA, and human (HSA) using human alphoid primers (α27/α30). The PCR products, α27/α30-NLE, α27/α30-CJA, and α27/α30-HSA, were used as probes to perform cross-species (NLE, CJA, and HSA) FISH experiments to validate the centromeric localization and find sequence homology among species. The probe α27/α30-NLE tested on NLE metaphases generated consistent signals for both centromeres and telomeres of all white-cheeked gibbon chromosomes as well as interstitial heterochromatic regions on chromosomes 3, 5, 9, and 14 (location defined according to NLE standard karyotype by Rens et al. 2001). In humans, α27/α30-NLE gave strong signals on chromosomes 11, 17, and X; weak signals on the other chromosomes; and no signals on the Y chromosome (fig. 2A and B). No signals were detected on marmoset (CJA) metaphase chromosomes using α27/α30-NLE probe. No differences were observed in signal pattern or distribution under low or high stringency hybridization conditions (table 1). The probe α27/α30-CJA did not show any signal in all the tested species suggesting high sequence divergence between human and CJA. The α27/α30-HSA showed signals only in human, hybridizing to all the centromeres except for the Y chromosome (fig. 2C, table 1).
FIG. 2.—
Examples of hybridization experiments on gibbon and human with α-PCR products. FISH experiments using probe α27/α30-NLE PCR product on NLE (A) and human metaphases (B). (C) Hybridization signals using α27/α30-HSA amplification product on human chromosomes. No signals were detected on chromosome Y indicated by an arrow. (D) Cohybridization using the clone pGamma_36 (green) and a telomeric probe (telomere PNA FISH, kit Cy3, DAKO) (red) showing colocalization of telomeric and centromeric sequences in gibbon chromosomes. (E) The figure shows the colocalization of NLE α satellite clone pGAMMA_41 with human and gibbon BAC clones that cover NLE EBs reported by Roberto et al. (2007) (see text and supplementary table S9, Supplementary Material online, for details).
Table 1.
FISH Pattern Signals of Amplification Centromeric Products in Gibbon and Human Hybridization Results Using the PCR Product α27/α30-NLE and α27/α30-HSA on Gibbon (NLE) and on Human (HSA) at Low and High Stringency Condition
| NLE (Low Stringency) | NLE (High Stringency) | HSA (Low Stringency) | HSA (High Stringency) | |
| α27/α30-NLE | All centromeres and telomeres + interstitial regions | All centromeres and telomeres + interstitial regions | Centromeres (strong signals on 11,17 and X centromeres) | Centromeres (strong signal on 11,17 and X centromeres) |
| α27/α30-HSA | No signal | No signal | Centromeres (except Y centromere) | Centromeres (except Y centromere) |
Because our initial results suggested considerable divergence between human and CJA centromeric sequences, we used CJA-specific α-satellite to identify and characterize centromeres in marmoset.
White-Cheeked Gibbon α-Satellite DNA
To analyze in detail the sequence organization of NLE centromeric DNA, we subcloned and sequenced 32 clones from α27/α30-NLE α-satellite PCR (sequences accession numbers are reported at the end of this paragraph). Analysis of the sequences by RepeatMasker indicated that all of them were composed entirely of α-satellite. We performed comparative FISH experiments using each of the 32 clones as probes on both white-cheeked gibbon and human chromosome metaphase spreads. Three different hybridization patterns were observed for NLE: 1) exclusively centromeric with variable signal intensity (10/32); 2) to the centromeres and telomeres of all the chromosomes and interstitial regions on chromosomes 3, 5, 9, and 14 (12/32); and 3) to the telomeres of all the chromosomes and interstitial regions on chromosomes 3, 5, 9, and 14 (10/32). The chromosome mapping for each clone is shown in supplementary table S1, Supplementary Material online (the clone p_K68 showed Y chromosome specificity). Identical patterns were observed for clones mapping to centromeric regions at low and high stringency conditions. None of the gibbon-derived clones generated signals on HSA metaphases.
The localization of satellite at interstitial regions was previously reported by Chen et al. (2007), but their mapping is inconsistent with our results based on the standard N. leucogenys karyotype (Rens et al. 2001). We located the interstitial signals to chromosomes 3, 5, 9, and 14, whereas Chen et al. reported interstitial signals for chromosomes 3, 5, 8, and 11. To solve this discrepancy in mapping data, we performed cohybridization experiments on NLE using the centromeric probe pGAMMA_41 showing interstitial signals, and human and NLE probes at known mapping (Roberto et al.). Such experiments helped us to establish positively the identities of the NLE chromosomes involved in the rearrangements (fig. 2E).
Due to the mapping of alphoid sequences to telomeric regions, the α-satellite telomeric probe pK_7 was further used in a cohybridization experiment with a telomeric-derived ttaggg probe (telomere PNA FISH, kit Cy3, DAKO) (fig. 2D). At the level of metaphase resolution, the signals completely overlapped even though sequence analysis of the clone pK_7 did not reveal any similarity with telomeric sequences.
In order to explain the different chromosomal locations of the NLE alphoid sequences, we selected two clones for each hybridization pattern (six in total) for restriction analysis (pGamma_35, pGamma_43 as centromeric and telomeric clones; pK_23, pK_7 as telomeric clones; and pK_19, pGamma_8 as centromeric clones). All showed identical HaeIII restriction digest patterns displaying bands in multiples of 171 bp in length, further supporting their alphoid nature. We compared these patterns with those obtained by Southern blot analysis of the NLE genome. We obtained the same identical characteristic HaeIII ladder pattern for three different probes (PCR product α27/α30-NLE, the centromeric clone [pGamma_8] and the centromeric–telomeric clone [pGamma_35]) (data not shown).
Further, we generated a multiple sequence alignment of the 32 cloned sequences. For each alignment, a divergence score ranging from 0 (sequence identity) to 0.882 (highest sequence divergence) was obtained (supplementary table S2, Supplementary Material online). Notably, the degree of sequence identity did not correspond with FISH hybridization pattern. Both high and low sequence similarity clones had the same mapping pattern as well as those showing different hybridization patterns.
To avoid potential biases in the PCR amplification, we screened the large-insert genomic gibbon BAC library CH271 segment 1 (http://www.chori.org/bacpac/) for clones containing centromeric DNA using three different probes: α27/α30-NLE, α27/α30-HSA, and C410. The latter was a specific NLE alphoid DNA obtained by immunoprecipitation with antibody against CENP-C centromeric protein and was used as centromeric positive control in our experiments. These three different hybridization experiments gave the same positive clones (n = 422).
Seventy-four BACs, corresponding to the strongest hybridization signals, were selected and used in FISH experiments on NLE chromosomes under high stringency conditions as well as on HSA under low stringency conditions. These BAC probes gave strikingly different hybridization patterns on NLE chromosomes: telomeres, centromeres, both centromeres and telomeres, telomeres and interstitial regions, or centromeres and interstitial regions. Only a small subset of clones showed signals at centromeric regions on human chromosomes (supplementary table S3, Supplementary Material online). We analyzed BAC end sequences generated from a subset of these (n = 140 end sequences; see Materials and Methods) using RepeatMasker (sequence accession numbers are reported at the end of this paragraph): 110/140 detected alphoid sequences, 20/140 detected other repetitive elements (long interspersed nuclear elements, short interspersed nuclear elements, and associated tandem repeats) and 10/140 did not show any similarity with known classes of repetitive sequences (supplementary table S4, Supplementary Material online). These latter sequences were analyzed by Blast versus human genome and showed mapping to the human region 3p12.3 and to the pericentromeric regions 22q11.1 and 20p11.1 that correspond, respectively, to the NLE chromosomes 21, 7, and 13 pericentromeric regions (supplementary table S5, Supplementary Material online). All of them mapped in segmental duplications as reported by UCSC genome Browser.
We specifically searched for NLE HOR patterns by comparing (Blast 2) the α-satellite sequences generated from the 110 bac end sequences and the longest sequences obtained from the genomic DNA subclones (the centromeric and telomeric clones pGamma_35 and pGamma_43, the telomeric clones pK_23 and pK_7, and the centromeric clones pK_19 and pGamma_8). The first 171 bp of each selected sequence was used as query versus the complete sequence from which the 171-bp monomeric unit was derived. High similarity percentage ranging 76–84% was found between the extracted block of 171 bp and each 171-bp monomeric repeats in the complete sequence, but no periodicity was discovered (supplementary table S6, Supplementary Material online), suggesting an alphoid organization characterized by tandemly repeated monomeric units without HOR.
Further support for the lack of HOR within white-cheeked gibbon was obtained by Blast 2 sequence analysis on known HOR structures in macaque and human alphoid sequences. The analysis was carried out on two macaque dimeric sequences ch250-379m3-sp6 (Pike et al. 1986) and ch250-317d4-sp6 (the latter sequence was obtained by a Blast of the clone ch250-379M3-sp6 against the trace archive, http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi?PAGE=Nucleotides&PROGRAM=blastn&BLAST_SPEC=TraceArchive&BLAST_PROGRAMS=megaBlast&PAGE_TYPE=BlastSearch) and, respectively, on pentameric, dimeric, and monomeric human clones pHS53 (Zaitsev and Rogaev 1986), pSE16-2 (Alexandrov et al. 1993a), and Z12013 (Alexandrov et al. 1993b). The analysis performed on the sequences of ch250-317d4-sp6, pHS53, and pSE16-2, which contain HORs, showed a recurrent trend with a periodicity of 2, 5, and 2, respectively, of the similarity percentage, between the first block of ∼171 bp and the other 171-bp blocks inside the complete sequence (supplementary table S7, Supplementary Material online). The Z12013 Blast 2 sequence output, instead, showed a consistent and similar percentage among all the 171-bp units present in the complete sequence (supplementary table S7, Supplementary Material online), the same result obtained for the NLE alphoid sequences. Further sequence comparisons were performed between white-cheeked gibbon consensus and human consensus sequences and revealed sequence identities between 80.1% and 91.2% (supplementary table S8, Supplementary Material online).
Because the use of human primers in our approach could lead to the generation of products that are not entirely of NLE origin, we generated gibbon specific primers from NLE consensus sequences obtained by multi-alignment of all gibbon BES with Megalign software. These primers were tested by PCR on NLE genomic DNA and the amplification product (αBES) was used as probe in FISH experiments on NLE and HSA metaphase. Under low and high stringency conditions, signals for both centromeres and telomeres of all white-cheeked gibbon chromosomes as well as interstitial heterochromatic regions of chromosome 3, 5, 9, and 14 were detected, whereas no signals were detected on human metaphase chromosomes (table 1). The different FISH results obtained with α27/α30-NLE and αBES probes on human metaphases could be explained by different primer origin, human and NLE specific, respectively.
Because the family of the gibbons (Hylobatidae) is divided into four genera: Hylobates, Hoolock, Nomascus, and Symphalangus (Geissmann 2002; Mootnick and Groves 2005), we tested the isolated N. leucogenys centromeric sequences on Hylobates lar. We performed FISH experiments using α27/α30-NLE and αBES: Only signals at centromeric level were observed, thus showing that alphoid centromeric sequences between Hylobates and Nomascus genera are shared, but telomeric and interstitial localizations of alphoid sequences are specific of NLE. The unavailability of samples for the other genera prevented us to define the organization in the other genera.
The gibbon karyotype is known to be extensively rearranged when compared with the human and to the ancestral primate karyotype. Evolutionary breakpoint (EB) refinement of the white-cheeked gibbon (N. leucogenys, NLE) has been performed by Roberto et al. with respect to the human genome. They provided a detailed clone framework map of the gibbon genome and refine the location of 86 EBs to <1-Mb resolution. Comparisons of NLE breakpoints with those of other gibbon species revealed variability in the position, suggesting that chromosomal rearrangement has been a longstanding property of this particular ape lineage. Using the list of refined breakpoints by Roberto et al. (2007), we found that the location of our clones giving signals on interstitial alphoid regions matched to four EBs on chromosomes 3, 5, 9 (evolutionary reciprocal translocations), and 14 (evolutionary inversion) NLE specific. In this regard, two or three color FISH experiments were performed using human and white-cheeked gibbon BAC clones to identify exactly the EB with our alphoid clone. At the cytogenetic level, we found a perfect overlapping between the interstitial signals and the mentioned EBs. No specific NLE clone was reported by Roberto et al. for chromosome 14 in the white-cheeked gibbon; for this reason, only the human clone (RP11-133M22) was used to compare the EB and interstitial α-satellite locations on NLE14 (supplementary table S9, Supplementary Material online, and fig. 2E).
We compared the localizations of interstitial heterochromatic blocks and previously reported ancestral centromeres or neocentromeres (Cardone et al. 2002, 2006, 2007, 2008; Ventura et al. 2004; Misceo et al. 2005; Carbone et al. 2006a; Ventura et al. 2007; Stanyon et al. 2008), but no evidence of colocalization was found.
(Insert sequences from plasmid clones: pA_12FJ346627, pGAMMA_3FJ346628, pGAMMA_5FJ346629, pGAMMA_6FJ346630, pGAMMA_8FJ346631, pGAMMA_23 FJ346632, pGAMMA_24 FJ346633, pGAMMA_34 FJ346634, pGAMMA_35 FJ346635,p GAMMA_36 FJ346636, pGAMMA_37 FJ346637, pGAMMA_38 FJ346638, pGAMMA_41 FJ346639, pGAMMA_42 FJ346640, pGAMMA_43 FJ346641, pK_64 FJ346642, pK_19 FJ346643, pK_23, FJ346644, pK_25 FJ346645, pK_30 FJ346646, pK_41 FJ346647, pK_43 FJ346648, pK_51 FJ346649, pK_69 FJ346650, pK_1 FJ346651, pK_3 FJ346652, pK_7 FJ346653, pK_20 FJ346654, pK_21 FJ346655, pK_34 FJ346656, pK_63 FJ346657, pK_68 FJ346658; BAC end sequences: CH271_0010P22_B1 FJ346579, CH271_0010P22_G1 FJ346580, CH271_0046E21_G1 FJ346581,CH271_0046E21_B1 FJ346582, CH271_0002J18_G1 FJ346583,CH271_0002J18_B1 FJ346584, CH271_0029E05_B1 FJ346585, CH271_0029E05_G1 FJ346586, CH271_0032G05_G1 FJ346587, CH271_0032G05_B1 FJ346588, CH271_0036I10_B1 FJ346589, CH271_0036I10_G1 FJ346590, CH271_0023A02_G1 FJ346591, CH271_0023A02_B1 FJ346592, CH271_0059L04_G1 FJ346593, CH271_0059L04_B1 FJ346594, CH271_0083K12_B1 FJ346595, CH271_0083K12_G1 FJ346596, CH271_0042E17_G1 FJ346597, CH271_0042E17_B1 FJ346598, CH271_0005P01_G1 FJ346599, CH271_0005P01_B1 FJ346600, CH271_0015O18_B1 FJ346601, CH271_0015O18_G1 FJ346602, CH271_0039I23_B1 FJ346603, CH271_0039I23_G1 FJ346604, CH271_0054E20_B1 FJ346605, CH271_0054E20_G1 FJ346606, CH271_0096M12_B1 FJ346607, CH271_0096M12_G1 FJ346608, CH271_0015L04_G1 FJ346609, CH271_0015L04_B1 FJ346610, CH271_0024A08_B1 FJ346611, CH271_0024A08_G1 FJ346612, CH271_0048A09_B1 FJ346613, CH271_0048A09_G1 FJ346614, CH271_0084N15_G1 FJ346615, CH271_0084N15_B1 FJ346616, CH271_0072O18_G1 FJ346617, CH271_0072O18_B1 FJ346618, CH271_0047O10_B1 FJ346619, CH271_0047O10_G1 FJ346620, CH271_0027P04_b1 FJ346621, CH271_0027P04_G1 FJ346622, CH271_0091K12_G1 FJ346623, CH271_0091K12_B1 FJ346624, CH271_0007O15_G1 FJ346625, and CH271_0007O15_B1 FJ346626).
Marmoset α-Satellite DNA
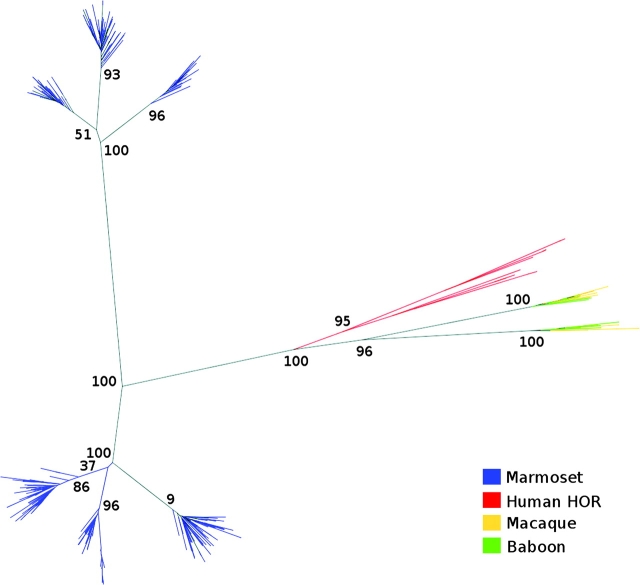
Characterization of marmoset centromeric DNA posed additional challenges due to the considerable sequence divergence of New World monkey α-satellite DNA when compared with human. Although α-satellite DNA is frequently not assembled as part of whole-genome sequencing projects, we previously demonstrated that higher-order α-satellite DNA is well represented in such data sets. We therefore downloaded the marmoset whole-genome shotgun (WGS) sequence library for CJA from NCBI Trace Archive (http://www.ncbi.nlm.nih.gov/Traces/trace.cgi?). We followed the method described in Alkan et al. (2007) to extract and classify alphoid sequences from the WGS data. We first constructed a library of ∼171 bp alphoid monomers using α-satellite sequences from previously characterized New World monkeys: Cebus apella (GenBank: L07926), Cebuella pygmaea (L07928), Chiropotes satanas (L07929), Callicebus moloch (L07930), and C. jacchus. The WGS reads that contain alphoid-like sequences were detected by aligning against the α-satellite set using Blast (parameters: −v 10,000 −b 10,000). CJA α-satellite monomeric repeat units were then extracted from the WGS reads using the RepeatMasker tool and clustered into sets where the pairwise divergence between any pairs of sequences within a set is at most 2% (Alkan et al. 2007). We obtained seven different clusters shared in two main branches by this method and constructed the phylogenetic tree of the clustered sequences using ClustalW (fig. 3).
FIG. 3.—
Phylogenetic analysis of marmoset α-satellite sequences. We used the Neighbor-Joining method (ClustalW) to construct the phylogenetic tree of alphoid monomers extracted from Callithrix jacchus (CJA), Papio anubis (PAN), Macaca mulatta (MMU), and Homo sapiens (HSA). We selected 417 monomeres from CJA (that cluster into 7 sets) and 10 from PAN, MMU, and HSA each. Bootstrap values (n = 100 replicates) are also indicated on the branches that support the evolutionary separation of marmoset alphoids from the human and Old World monkey α-satellite.
We generated marmoset-specific primers (supplementary table S10, Supplementary Material online) from consensus sequences derived from the seven clusters and tested them by PCR on marmoset and human genomic DNAs. No amplification was detected from human DNA. CJA amplification products were tested in FISH experiments on CJA and HSA, respectively. In CJA, signals were detected on different chromosomes with dissimilar signal intensity at low and high stringency conditions (table 2). The same PCR probes did not generate any signals on human metaphase chromosomal spreads.
Table 2.
FISH Results with PCR Product Probes Obtained Using Primers Designed on the Seven Callitrix Cluster Sequences
| CJA Mapping |
||||||||||||||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | X | Y | |
| Cluster 1 | + | + | +++ | +++ | ++ | +++ | + | ++ | ++ | ++ | + | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | ++ | ++ | + | +++ |
| Cluster 2 | + | + | ++ | ++ | + | ++ | + | ++ | ++ | ++ | + | +++ | ++ | +++ | +++ | ++ | ++ | +++ | ++ | ++ | + | + | +++ | |
| Cluster 3 | + | + | + | ++ | ++ | +++ | + | ++ | +++ | + | + | + | ++ | ++ | ++ | ++ | ++ | ++ | ++ | + | + | + | + | + |
| Cluster 4 | * | + | + | + | + | + | +++ | + | + | + | +++ | +++ | + | + | + | + | + | + | + | + | + | + | ||
| Cluster 5 | + | * | + | + | * | * | * | +++ | + | * | * | +++ | +++ | +++ | + | + | + | + | * | + | + | + | * | + |
| Cluster 6 | + | + | + | + | + | + | + | +++ | + | +++ | + | +++ | +++ | + | + | + | + | + | + | + | + | + | + | + |
| Cluster 7 | + | + | +++ | + | + | + | +++ | + | + | + | +++ | + | +++ | +++ | + | +++ | +++ | +++ | +++ | + | + | +++ | ||
Mapping information of the seven cluster amplification products. Plus represents the intensity of the detected signals: + weak, ++ medium and +++ strong. Stars indicate weak signals detected at low stringency conditions.
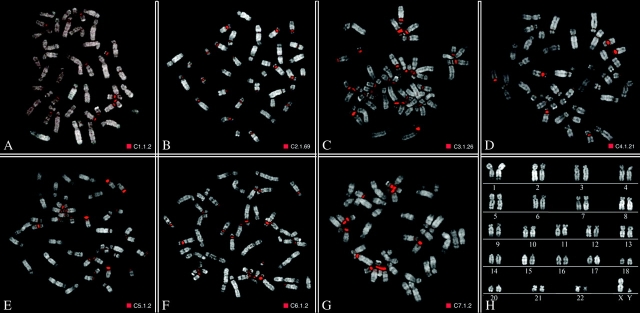
The seven PCR products were subcloned (n = 386 clones) and differential insert sizes were identified by PCR. Accordingly, we further grouped the clones into distinct classes (supplementary table S11, Supplementary Material online). A subset of clones representing each class (59/386) was tested by FISH on CJA metaphases and gave different hybridization patterns (fig. 4, supplementary table S11, Supplementary Material online). Homogeneous hybridization patterns were detected among clones grouped in the same class. Neither SF organization was detected nor correspondence between cluster and map location was observed suggesting a complex heterogeneity of α-satellite DNA.
FIG. 4.—
Examples of FISH pattern signals on CJA metaphases. (A–G) FISH results using clones specific for each of the seven clusters, 1–7, respectively, obtained by bioinformatics approach. Clone names have been reported next to the red square. (H) Marmoset karyotype using standard Q banding according to Sherlock et al. (1996).
To further investigate the sequence structure of the marmoset centromeres, we sequenced 14 clones (accession numbers are reported at the end of this paragraph) selecting the largest inserts for each cluster. No significant sequence similarity was observed between these 14 sequences and representative human alphoid sequences (X07685, Z12013, and Z12009 and M28031, M28032, and M28033). Similar results were obtained by comparison of our sequences with other known primate centromeric sequences including chimpanzee (L08574 and X97003), gorilla (AJ509823 and M62744), and macaque (X04006).
We analyzed the 14 sequences using Blast 2 Sequences (http://blast.ncbi.nlm.nih.gov/bl2seq/wblast2.cgi). The first 171 bp of each sequence was used as a query versus the complete sequence from which the 171 bp were extracted. The query sequence matched multiple sites in the entire sequence of the subject with a periodicity of 342 bp between high-identity matches (79–99%). Further, 34 171 bp monomers were extracted from the 14 clones sequences, and phylogenetic analysis based on a multiple sequence alignment of the 34 monomers showed that 171-bp units grouped into two distinct clades with 50–60% sequence divergence between them (supplementary fig. S1, Supplementary Material online). The similarity between first and second monomers is much lower than previously reported for human and macaque (40–50% in marmoset vs. 75–89% in human and macaque) (supplementary table S7, Supplementary Material online); therefore, marmoset shows a greater divergence rate between monomers in dimeric structures than other Primates. According to these results, we identified in the marmoset repetitive alphoid units with an ancient and more divergent dimeric structure. Moreover, Southern blot analysis confirmed the 342-bp periodicity with a ladder of hybridizing bands of 342-bp spacing (fig. 5). In the light of these results, we conclude that in CJA α-satellite DNA is organized as units of 342 bp, representing the ancient dimeric structure. We found no evidence of HOR structure among the 342 bp, but our data are limited to the insert size of the subcloned sequences we analyzed (the longest consisted of three consecutive 342-bp units). Unlike human and great-ape genomes, analysis of long-range, end-sequence pair data also did not show the presence of any higher-order α-satellite DNA within the marmoset.
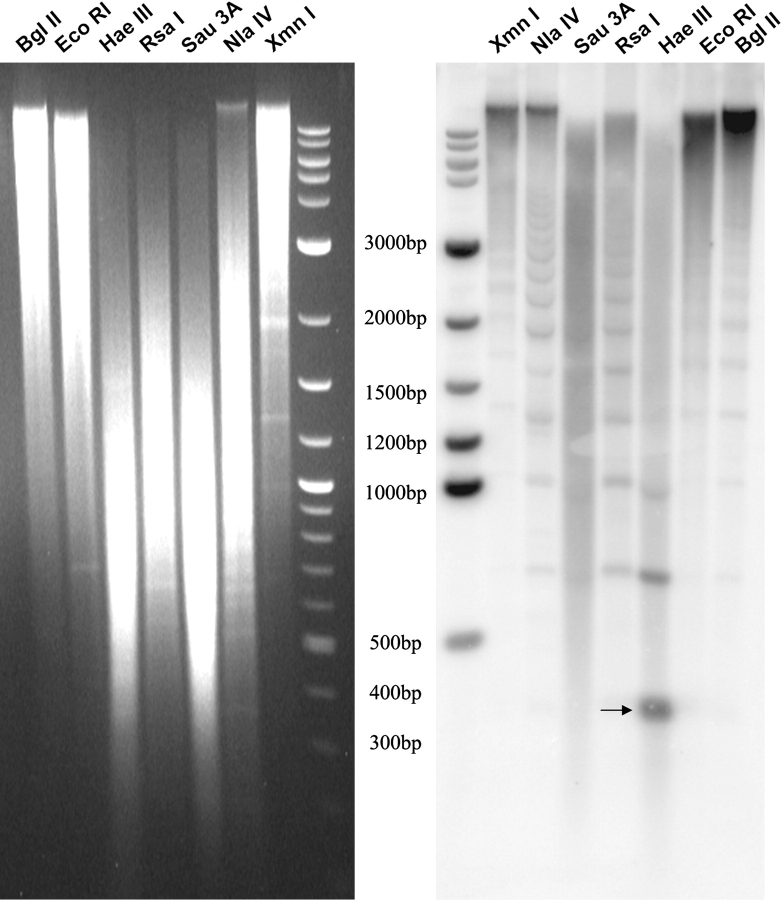
FIG. 5.—
Restriction analysis and southern blot on CJA genomic DNA. Hybridization of clones CJA 3.1.5 to southern blots of CJA genomic DNA digested with the indicated restriction enzymes. The HaeIII lane in the autoradiography on the right, shows hybridizing bands with a periodicity of 342 bp (the arrow shows the 342 bp monomer). We used the 2-log ladder as marker in the last lane of the gel (left) and on the first lane of the southern autoradiography (right). The sizes of the most representative bands of the marker are indicated in the middle.
(Insert sequences from plasmid clones: C1.1.19 FJ867326, C1.1.74 FJ867327, C2.1.37 FJ867328, C2.1.65 FJ867329, C2.1.73 FJ867330, C3.1.5 FJ867331, C3.1.8 FJ867332, C4.1.13 FJ867333, C4.2.21 FJ867334, C5.1.12 FJ867335, C6.1.1 FJ867336, C6.1.29 FJ867337, C7.1.3 FJ867338, and C7.1.4 FJ867339).
Discussion
In the present study, we have analyzed the structure and organization of centromeres in the white-cheeked gibbon (lesser ape) and in marmoset (New World monkey) to gain an insight into centromere satellite organization and evolution. Due to differences in sequence divergence among centromeric sequences in human, gibbon, and marmoset, we used two different approaches to isolate and characterize the centromeric sequences in these species.
We used human sequence–derived degenerate primers to isolate centromeric sequences in NLE. The availability of CENP immunoprecipitation C410 further helped us to prove the ability of our method to identify centromeric alphoid sequences. Comparative FISH analyses moreover confirmed the similarity between NLE α sequences and human centromeres. In particular, the centromeric sequences of human 11, 17 and X chromosomes, grouped in SF 3 (SF3, Jabs and Persico 1987), share the highest degree of similarity with white-cheeked gibbon alphoid sequences as shown by hybridization using the probe α27/α30-NLE. Moreover, our data support the independent evolution of human Y chromosome centromeric sequences: Both α27/α30-NLE and α27/α30-HSA gave signals on all the human chromosomes except Y, showing high divergence between Y and the rest of human centromeric sequences. Therefore, the Y-specific variants of α DNA previously described (Wolfe et al. 1985) likely diverged from the common branch of α DNA before the formation of other chromosome-specific variants (Alexandrov et al. 1988).
We further confirmed by Southern blot and sequence analysis that the 171-bp monomeric unit in NLE, as revealed by HaeIII restriction enzyme digest, lacked any HOR. High-order organization is reduced in complexity in chimpanzee (Alkan et al. 2007) and orangutan (Haaf and Willard 1998), supporting the idea that this highly organized structure reached its most complexity within the human lineage.
The chromosomal localization of alphoid sequences we found in N. leucogenys is quite intriguing. The clones we obtained showed three distinct localizations: centromeres, both centromeres and telomeres, and centromeres/telomeres/interstitial regions. However, no relevant differences were found at the sequence level and no common sequence motif was observed between clones sharing the same map location. These results support the idea that organization and repetition of sequences are crucial to defining the chromosomal localization more than the sequence itself.
Previous studies carried out in Hylobates and Symphalangus showed a quite different pattern on chromosomal distribution of DAPI-positive heterochromatin between these two genera of gibbons. Terminal, interstitial, and paracentric bands have been reported for Hylobates, whereas no interstitial heterochromatin have been detected in Symphalangus (Wijayanto et al. 2005). Our results showed a much more extent of heterochromatin accretion in white-cheeked gibbon compared with Hylobates and Symphalangus genera and disclose the alphoid nature of interstitial and terminal heterochromatin in Nomascus. Taking into consideration this point, the alphoid centromeric/telomeric signals could represent exchange between centromeric and telomeric repetitive elements occurred during the speciation of the N. leucogenys, as reported for duplicons in human (Bailey et al. 2002).
Further detailed mapping comparison between our α-satellite clones with interstitial signals and chromosomal EBs in white-cheeked gibbon (http://www.biologia.uniba.it/gibbon, Roberto et al. 2007) by cohybridization experiments have shown a clear association between them. In particular, NLE chromosomes 3, 5, 9, and 14 showed interstitial signals that overlapped to the EBs specific of NLE previously reported by Roberto et al. (2007) for these chromosomes (fig. 2E). The presence of segmental duplications and various classes of repetitive elements, such as LINE L1, has been recently reported in NLE chromosomal rearrangement breakpoints, suggesting a more complex rearrangement mechanism than simply nonallelic homologous recombination or nonhomologous end joining (Carbone et al. 2006b; Girirajan et al. 2009). According to our findings, the interstitial alphoid regions we detected on chromosomes 3, 5, 9, and 14 could represent a wider accretion of repetitive elements in rearrangement breakpoints thus underpinning a common destiny of the evolution of these regions. In particular, the clustering of repetitive elements in these regions could represent “scars” of evolutionary translocations or inversions occurred during the evolution of N. leucogenys. Even if we found the clustering of alphoid sequences at four EBs, because this is a N. leucogenys specific pattern, it cannot address the general question of the high evolutionary rate of breakpoints in the group of gibbons.
Due to the greater sequence divergence in marmoset, species-specific sequences from WGSSs were obtained and analyzed in detail. Our sequencing data show that the CJA unit is 342 bp in length without any HOR organization, which is in agreement with the satellite organization reported previously for three New World species: C. satanas, Pithecia irrorata, and Cacajo melanocephalus (Alves et al. 1994). Further, in C. satanas and C. melanocephalus, the monomeric repeat unit is 550 bp, whereas in Pithecia, similarly to CJA, the 340 bp monomer accounts for a substantial proportion of the satellite mass. Because the evolutionary distance between CJA and Pithecia (von Dornum and Ruvolo 1999), it can be supposed that in the NWM group, the monomeric ancestral unit was a 340-bp unit that evolved to a 550-bp unit in Chiropotes and Cacajao. However, the absence of HOR in CJA cannot be ruled out, as large contiguous sequences have not yet been generated. Based on the insert sizes of our clones (c2.1.73, 1,142 bp), we could not detect any HOR greater than three monomeric units.
In humans and other studied Primates, the α-satellite unit size has been reported as 171 bp (Rudd et al. 2006; Alkan et al. 2007). This unit has been variously organized during the course of primate evolution, creating human-specific HORs, monomeric gibbon structure, or dimeric structures as in macaque. In Callithrix, it is likely that two of these ancestral monomers fused generating the specific ancient dimer in NWM and no further homogenization occurred between monomers so generating the actual highly divergent dimers.
In the light of our and all published data, we propose a complex model for primate satellite evolution involving genomic amplification, unequal crossover and sequence homogenization. Starting with a 171-bp basic monomeric repeat unit, the centromeric α-satellite evolved by amplification, acquiring increasingly complex genomic structures. In the Platyrrini lineage, two 171-bp units were firstly amplified in dimeric unit and later the two monomers in the same dimer began to acquire differences by the decrease of sequence homogenization, thus forming the specific New World monkeys dimeric repeat unit (∼342 bp). In the Catharrini ancestor, the 171-bp unit continued to amplify and undergo unequal crossover and homogenization thus forming the dimeric structure common to all the centromeres as reported in macaque, baboon, and African green monkey (Musich et al. 1980; Pike et al. 1986). In contrast within the anthropoid lineage, the 171 bp monomer amplified and differentiated in monomeric structure (gibbon and orangutan, present work and Haaf and Willard 1998) or higher-order organization as reported in human (Willard et al. 1989; Arn and Jabs 1990). In any case, the sequence and the detailed organization differ, with the basic 171-bp repeat unit being the only common theme, supporting the notion that centromeric function is linked to relatively short repeated elements, more than sequence specific units (supplementary fig. S2, Supplementary Material online). These data can, moreover, support the idea that also neocentromeres can seed in a repetitive reach DNA domain lacking satellite DNA (Ventura et al. 2004, 2007). The comparison of the evolutionary history of the primate centromeres with other mammalian genomes will likely provide even more insights.
Supplementary Materials
Supplementary figures S1 and S2 and supplementary tables S1–S11 are available
Acknowledgments
This work was supported by MIUR (Ministero Italiano della Universita’ e della Ricerca; Cluster C03, Prog. L.488/92) and European Commission (INPRIMAT, QLRI-CT-2002-01325) are gratefully acknowledged for financial support. This work was also supported in part by R01 GM058815 (to E.E.E.). E.E.E. is an investigator of the Howard Hughes Medical Institute. Mammalian cell lines were kindly provided by the Cambridge Resource Centre.
References
- Alexandrov I, Kazakov A, Tumeneva I, Shepelev V, Yurov Y. Alpha-satellite DNA of primates: old and new families. Chromosoma. 2001;110:253–266. doi: 10.1007/s004120100146. [DOI] [PubMed] [Google Scholar]
- Alexandrov IA, Mashkova TD, Romanova LY, Yurov YB, Kisselev LL. Segment substitutions in alpha satellite DNA. Unusual structure of human chromosome 3-specific alpha satellite repeat unit. J Mol Biol. 1993a;231:516–520. doi: 10.1006/jmbi.1993.1302. [DOI] [PubMed] [Google Scholar]
- Alexandrov IA, Medvedev LI, Mashkova TD, Kisselev LL, Romanova LY, Yurov YB. Definition of a new alpha satellite suprachromosomal family characterized by monomeric organization. Nucleic Acids Res. 1993b;21:2209–2215. doi: 10.1093/nar/21.9.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov IA, Mitkevich SP, Yurov YB. The phylogeny of human chromosome specific alpha satellites. Chromosoma. 1988;96:443–453. doi: 10.1007/BF00303039. [DOI] [PubMed] [Google Scholar]
- Alkan C, Eichler EE, Bailey JA, Sahinalp SC, Tuzun E. The role of unequal crossover in alpha-satellite DNA evolution: a computational analysis. J Comput Biol. 2004;11:933–944. doi: 10.1089/cmb.2004.11.933. [DOI] [PubMed] [Google Scholar]
- Alkan C, Tuzun E, Buard J, Lethiec F, Eichler EE, Bailey JA, Sahinalp SC. Manipulating multiple sequence alignments via MaM and WebMaM. Nucleic Acids Res. 2005;33:W295–W298. doi: 10.1093/nar/gki406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan C, Ventura M, Archidiacono N, Rocchi M, Sahinalp SC, Eichler EE. Organization and evolution of primate centromeric DNA from whole-genome shotgun sequence data. PLoS Comput Biol. 2007;3:1807–1818. doi: 10.1371/journal.pcbi.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves G, Seuanez HN, Fanning T. Alpha satellite DNA in neotropical primates (Platyrrhini) Chromosoma. 1994;103:262–267. doi: 10.1007/BF00352250. [DOI] [PubMed] [Google Scholar]
- Archidiacono N, Antonacci R, Marzella R, Finelli P, Lonoce A, Rocchi M. Comparative mapping of human alphoid sequences in great apes using fluorescence in situ hybridization. Genomics. 1995;25:477–484. doi: 10.1016/0888-7543(95)80048-q. [DOI] [PubMed] [Google Scholar]
- Arn PH, Jabs EW. Characterization of human centromeric regions using restriction enzyme banding, alphoid DNA and structural alterations. Mol Biol Med. 1990;7:371–377. [PubMed] [Google Scholar]
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Baldini A, Archidiacono N, Carbone R, Bolino A, Shridhar V, Miller OJ, Miller DA, Ward DC, Rocchi M. Isolation and comparative mapping of a human chromosome 20-specific alpha-satellite DNA clone. Cytogenet Cell Genet. 1992;59:12–16. doi: 10.1159/000133188. [DOI] [PubMed] [Google Scholar]
- Carbone L, Nergadze SG, Magnani E, et al. (15 co-authors) Evolutionary movement of centromeres in horse, donkey, and zebra. Genomics. 2006a;87:777–782. doi: 10.1016/j.ygeno.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Carbone L, Vessere GM, t. Hallers BFH, et al. (13 co-authors) A high-resolution map of synteny disruptions in gibbon and human genomes. PLoS Genet. 2006b;2:e223. doi: 10.1371/journal.pgen.0020223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone MF, Alonso A, Pazienza M, et al. (13 co-authors) Independent centromere formation in a capricious, gene-free domain of chromosome 13q21 in Old World monkeys and pigs. Genome Biol. 2006;7:R91. doi: 10.1186/gb-2006-7-10-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone MF, Jiang Z, D'Addabbo P, Archidiacono N, Rocchi M, Eichler EE, Ventura M. Hominoid chromosomal rearrangements on 17q map to complex regions of segmental duplication. Genome Biol. 2008;9:R28. doi: 10.1186/gb-2008-9-2-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone MF, Lomiento M, Teti MG, Misceo D, Roberto R, Capozzi O, D'Addabbo P, Ventura M, Rocchi M, Archidiacono N. Evolutionary history of chromosome 11 featuring four distinct centromere repositioning events in Catarrhini. Genomics. 2007;90:35–43. doi: 10.1016/j.ygeno.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Cardone MF, Ventura M, Tempesta S, Rocchi M, Archidiacono N. Analysis of chromosome conservation in Lemur catta studied by chromosome paints and BAC/PAC probes. Chromosoma. 2002;111:348–356. doi: 10.1007/s00412-002-0215-3. [DOI] [PubMed] [Google Scholar]
- Chen L, Ye J, Liu Y, Wang J, Su W, Yang F, Nie W. Construction, characterization, and chromosomal mapping of a fosmid library of the white-cheeked gibbon (Nomascus leucogenys) Genom Proteom Bioinform. 2007;5:207–215. doi: 10.1016/S1672-0229(08)60008-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo KH, Vissel B, Nagy A, Earle E, Kalitsis P. A survey of the genomic distribution of alpha satellite DNA on all the human chromosomes, and a derivation of a new consensus sequence. Nucleic Acids Res. 1991;19:1179–1182. doi: 10.1093/nar/19.6.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo KH, Vissel B, Brown R, Filby RG, Earle E. Homologous alpha satellite sequences on human acrocentric chromosomes with selectivity for chromosomes 13, 14 and 21: implications for recombination between nonhomologues and Robertsonian translocations. Nucleic Acids Res. 1988;16:1273–1284. doi: 10.1093/nar/16.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover G. Molecular drive: a cohesive mode of species evolution. Nature. 1982;299:111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- Geissmann T. Taxonomy and evolution of gibbons. Evol Anthrop. 2002;((Suppl. 1) 1):28–31. [Google Scholar]
- Girirajan S, Chen L, Graves T, et al. (11 co-authors) Sequencing human–gibbon breakpoints of synteny reveals mosaic new insertions at rearrangement sites. Genome Res. 2009;19:178–190. doi: 10.1101/gr.086041.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T, Willard HF. Orangutan alpha-satellite monomers are closely related to the human consensus sequence. Mamm Genome. 1998;9:440–447. doi: 10.1007/s003359900793. [DOI] [PubMed] [Google Scholar]
- Hamming R. Error detecting and error correcting codes. Bell SystTech J. 1950;29:147–160. [Google Scholar]
- Jabs EW, Persico MG. Characterization of human centromeric regions of specific chromosomes by means of alphoid DNA sequences. Am J Hum Genet. 1987;41:374–390. [PMC free article] [PubMed] [Google Scholar]
- Jorgensen AL, Kolvraa S, Jones C, Bak AL. A subfamily of alphoid repetitive DNA shared by the NOR-bearing human chromosomes 14 and 22. Genomics. 1988;3:100–109. doi: 10.1016/0888-7543(88)90139-5. [DOI] [PubMed] [Google Scholar]
- Kazakov AE, Shepelev VA, Tumeneva IG, Alexandrov AA, Yurov YB, Alexandrov IA. Interspersed repeats are found predominantly in the “old” alpha satellite families. Genomics. 2003;82:619–627. doi: 10.1016/s0888-7543(03)00182-4. [DOI] [PubMed] [Google Scholar]
- Lee C, Wevrick R, Fisher RB, Ferguson-Smith MA, Lin CC. Human centromeric DNAs. Hum Genet. 1997;100:291–304. doi: 10.1007/s004390050508. [DOI] [PubMed] [Google Scholar]
- Maio JJ. DNA strand reassociation and polyribonucleotide binding in the African green monkey, Cercopithecus aethiops. J Mol Biol. 1971;56:579–595. doi: 10.1016/0022-2836(71)90403-7. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning, A laboratory manual. New York (NY): Cold spring harbor laboratory, Cold Spring Harbor. 1982 [Google Scholar]
- Manuelidis L. Chromosomal localization of complex and simple repeated human DNAs. Chromosoma. 1978;66:23–32. doi: 10.1007/BF00285813. [DOI] [PubMed] [Google Scholar]
- Misceo D, Cardone MF, Carbone L, D'Addabbo P, de Jong PJ, Rocchi M, Archidiacono N. Evolutionary history of chromosome 20. Mol Biol Evol. 2005;22:360–366. doi: 10.1093/molbev/msi021. [DOI] [PubMed] [Google Scholar]
- Mootnick A, Groves C. A new generic name for the Hoolock Gibbon (Hylobatidae) Int J Primatol. 2005;26:971–976. [Google Scholar]
- Musich PR, Brown FL, Maio JJ. Highly repetitive component alpha and related alphoid DNAs in man and monkeys. Chromosoma. 1980;80:331–348. doi: 10.1007/BF00292688. [DOI] [PubMed] [Google Scholar]
- Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Pike LM, Carlisle A, Newell C, Hong SB, Musich PR. Sequence and evolution of rhesus monkey alphoid DNA. J Mol Evol. 1986;23:127–137. doi: 10.1007/BF02099907. [DOI] [PubMed] [Google Scholar]
- Rens W, Yang F, O'Brien PC, Solanky N, Ferguson-Smith MA. A classification efficiency test of spectral karyotyping and multiplex fluorescence in situ hybridization: identification of chromosome homologies between Homo sapiens and Hylobates leucogenys. Genes Chromosomes Cancer. 2001;31:65–74. doi: 10.1002/gcc.1119. [DOI] [PubMed] [Google Scholar]
- Roberto R, Capozzi O, Wilson RK, et al. (10 co-authors) Molecular refinement of gibbon genome rearrangements. Genome Res. 2007a;17:249–257. doi: 10.1101/gr.6052507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd MK, Willard HF. Analysis of the centromeric regions of the human genome assembly. Trends Genet. 2004;20:529–533. doi: 10.1016/j.tig.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Rudd MK, Wray GA, Willard HF. The evolutionary dynamics of alpha-satellite. Genome Res. 2006;16:88–96. doi: 10.1101/gr.3810906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueler MG, Dunn JM, Bird CP, Ross MT, Viggiano L, Rocchi M, Willard HF, Green ED. Progressive proximal expansion of the primate X chromosome centromere. Proc Natl Acad Sci USA. 2005;102:10563–10568. doi: 10.1073/pnas.0503346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF. Genomic and genetic definition of a functional human centromere. Science. 2001;294:109–115. doi: 10.1126/science.1065042. [DOI] [PubMed] [Google Scholar]
- Sherlock JK, Griffin DK, Delanthy JDA, Parrington JM. Homologies between human and marmoset (Callithrix Jacchus) chromosomes revealed by comparative chromosome painting. Genomics. 1996;33:214–219. doi: 10.1006/geno.1996.0186. [DOI] [PubMed] [Google Scholar]
- Stanyon R, Rocchi M, Capozzi O, Roberto R, Misceo D, Ventura M, Cardone M, Bigoni F, Archidiacono N. Primate chromosome evolution: ancestral karyotypes, marker order and neocentromeres. Chromosome Res. 2008;16:17–39. doi: 10.1007/s10577-007-1209-z. [DOI] [PubMed] [Google Scholar]
- Ventura M, Antonacci F, Cardone MF, Stanyon R, D'Addabbo P, Cellamare A, Sprague LJ, Eichler EE, Archidiacono N, Rocchi M. Evolutionary formation of new centromeres in macaque. Science. 2007;316:243–246. doi: 10.1126/science.1140615. [DOI] [PubMed] [Google Scholar]
- Ventura M, Archidiacono N, Rocchi M. Centromere emergence in evolution. Genome Res. 2001;11:595–599. doi: 10.1101/gr.152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Mudge JM, Palumbo V, et al. (11 co-authors) Neocentromeres in 15q24-26 map to duplicons which flanked an ancestral centromere in 15q25. Genome Res. 2003;13:2059–2068. doi: 10.1101/gr.1155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Weigl S, Carbone L, et al. (13 co-authors) Recurrent sites for new centromere seeding. Genome Res. 2004;14:1696–1703. doi: 10.1101/gr.2608804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dornum M, Ruvolo M. Phylogenetic relationships of the New World monkeys (Primates, platyrrhini) based on nuclear G6PD DNA sequences. Mol Phylogenet Evol. 1999;11(11):459–476. doi: 10.1006/mpev.1998.0582. [DOI] [PubMed] [Google Scholar]
- Warburton PE, Haaf T, Gosden J, Lawson D, Willard HF. Characterization of a chromosome specific chimpanzee alpha satellite subset: evolutionary relationship to subsets on human chromosomes. Genomics. 1996;33:220–228. doi: 10.1006/geno.1996.0187. [DOI] [PubMed] [Google Scholar]
- Warburton PE, Willard HF. Genomic analysis of sequence variation in tandemly repeated DNA. Evidence for localized homogeneous sequence domains within arrays of alpha-satellite DNA. J Mol Biol. 1990;216:3–16. doi: 10.1016/s0022-2836(05)80056-7. [DOI] [PubMed] [Google Scholar]
- Waye JS, Willard HF. Structure, organization, and sequence of alpha satellite DNA from human chromosome 17: evidence of evolution by unequal crossing-over and an ancestral pentamer repeat shared with the human X chromosome. Mol Cell Biol. 1986;6:3156–3165. doi: 10.1128/mcb.6.9.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijayanto H, Hirai Y, Kamanaka Y, Katho A, Sajuthi D, Hirai H. Patterns of C-heterochromatin and telomeric DNA in two representative groups of small apes, the genera Hylobates and Symphalangus. Chromosome Res. 2005;13:715–722. doi: 10.1007/s10577-005-1007-4. [DOI] [PubMed] [Google Scholar]
- Willard HF, Waye JS. Chromosome-specific subsets of human alpha satellite DNA: analysis of sequence divergence within and between chromosomal subsets and evidence for an ancestral pentameric repeat. J Mol Evol. 1987a;25:207–214. doi: 10.1007/BF02100014. [DOI] [PubMed] [Google Scholar]
- Willard HF, Waye JS. Chromosome-specific subsets of human alpha satellite DNA: analysis of sequence divergence within and between chromosomal subsets and evidence for an ancestral pentameric repeat. J Mol Evol. 1987b;25:207–214. doi: 10.1007/BF02100014. [DOI] [PubMed] [Google Scholar]
- Willard HF, Wevrick R, Warburton PE. Human centromere structure: organization and potential role of alpha satellite DNA. Progr Clin Biol Res. 1989;318:9–18. [PubMed] [Google Scholar]
- Wolfe J, Darling SM, Erickson RP, Craig IW, Buckle VJ, Rigby PWJ, Willard HF, Goodfellow PN. Isolation and characterization of an alphoid centromeric repeat family from the human Y chromosome. J Mol Biol. 1985;182:477–485. doi: 10.1016/0022-2836(85)90234-7. [DOI] [PubMed] [Google Scholar]
- Zaitsev IZ, Rogaev EI. [Structural analysis of alphoid DNA of primates. I. Heterogeneity of nucleotide sequence of alphoid repeats in human DNA] Mol Biol (Mosk) 1986;20:663–673. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.