Abstract
Gonadotrophin-releasing hormone (GNRH1) regulates pituitary luteinizing hormone (LH). Previous studies have delineated a mechanism for GNRH1-induced LHβ subunit gene (Lhb) transcription, the rate-limiting step in LH production. GNRH1 induces expression of early growth response 1 (EGR1), which interacts with steroidogenic factor 1 (SF1) and paired-like homeodomain transcription factor 1 (PITX1) to regulate Lhb promoter activity. Though the cis-elements for these factors are conserved across species, regulation of human LHB transcription has not been thoroughly investigated. We therefore characterized LHB transcriptional regulation by GNRH1 using promoter-reporter analyses in LβT2 cells. GNRH1 stimulated LHB transcription via an extracellular signal-regulated kinase 1/2 pathway. EGR1 bound to two binding sites on the LHB promoter and this binding was increased by GNRH1. Mutation of either site or knockdown of endogenous EGR1 decreased basal and/or GNRH1-regulated promoter activity. The human LHB promoter also contains low and high affinity SF1 binding sites. Mutation of these elements or depletion of endogenous SF1 impaired basal and ligand-induced transcription. Knockdown of PITX1 or PITX2 isoforms impaired GNRH1 induction, and endogenous PITX1 bound to the candidate PITX binding site on the LHB promoter. Thus, the mechanism described for GNRH1 regulation of Lhb in other species is largely conserved for human LHB. We also uncover a previously unappreciated role for PITX2 isoforms in this process.
Keywords: gonadotrophin, pituitary, EGR1, SF1, PITX
Introduction
Luteinizing hormone (LH) is a dimeric pituitary glycoprotein comprised of the unique LHβ (LHB) subunit and a common α subunit (CGA) which it shares with follicle-stimulating hormone (FSH), thyroid-stimulating hormone (TSH) and, in humans, chorionic gonadotrophin (hCG). LH and FSH are produced and secreted by the same cells in the pituitary gland, gonadotropes, and expression of the β subunits is the rate-limiting step in their synthesis. The primary stimulus for both LH release and LHB transcription is pulsatile gonadotrophin-releasing hormone (GNRH1) secretion from the hypothalamus.
Results from several groups working on the Lhb promoters in rat, cow and horse, as well as data from knockout mouse models, have converged to suggest a general model of Lhb transcriptional regulation by GNRH1 [reviewed in Jorgensen et al. (2004)]. GNRH1 rapidly stimulates early growth response 1 (Egr1) expression within 30 min (Tremblay and Drouin, 1999). Upon translation, the EGR1 protein binds the proximal Lhb promoter via two conserved cis-elements (Halvorson et al., 1998; Wolfe and Call, 1999; Call and Wolfe, 2002), both of which are critical for induction of the Lhb gene in various species (Halvorson et al., 1998, 1999; Dorn et al., 1999; Tremblay and Drouin, 1999; Wolfe and Call, 1999; Kaiser et al., 2000; Weck et al., 2000). The importance of EGR1 in vivo was demonstrated in female Egr1 null mice, which are infertile due to the loss of Lhb expression (Lee et al., 1996; Topilko et al., 1998).
EGR1 acts in concert with the nuclear receptor steroidogenic factor 1 (SF1, NR5A1), which binds to conserved elements occurring in tandem with the two EGR sites in the Lhb promoter from various species. Both SF1 sites are required for maximal induction of Lhb by GNRH1 (Halvorson et al., 1996, 1998, 1999; Keri and Nilson, 1996; Dorn et al., 1999; Tremblay and Drouin, 1999; Kaiser et al., 2000). Targeted deletion of Sf1 in gonadotropes results in significant reduction of LH production in mice (Zhao et al., 2001, 2004), confirming the important role for SF1 in Lhb expression in vivo. Over-expression analyses in heterologous cells show that EGR1 and SF1 act together through their tandem response elements to stimulate Lhb transcription (Halvorson et al., 1998; Dorn et al., 1999; Tremblay and Drouin, 1999; Kaiser et al., 2000).
Several observations suggest that a binding site for Bicoid-related homeodomain transcription factors (hereafter ‘PITX’ element), which occurs between the tandem EGR/SF1 sites, is also important for maximal induction of the Lhb promoter by GNRH1 (Tremblay and Drouin, 1999; Quirk et al., 2001; Jiang et al., 2005). The exact identity of the protein(s) binding this element has not be unequivocally determined (Rosenberg and Mellon, 2002), though evidence from several groups implicates paired-like homeodomain transcription factor 1 (PITX1) or the related PITX2 (Tremblay et al., 1999, 2000; Tremblay and Drouin, 1999; Quirk et al., 2001; Jiang et al., 2005; Lamba et al., 2008a). Mice with homozygous deletion of Pitx1 die after birth, precluding an assessment of PITX1 in LH synthesis in adult animals (Lanctot et al., 1999; Szeto et al., 1999). Mice with gonadotrope-specific deletion of Pitx2 are fertile (Charles et al., 2008), though it is possible that PITX1 can compensate for loss of PITX2 in these animals. Nonetheless, several studies show that PITX1 and PITX2 isoforms can independently and synergistically regulate Lhb transcription with SF1 and EGR1 (Keri and Nilson, 1996; Halvorson et al., 1998, 1999; Dorn et al., 1999; Tremblay and Drouin, 1999; Kaiser et al., 2000; Quirk et al., 2001). Thus, the current model holds that GNRH1 stimulates EGR1 expression, which then acts in concert with SF1 and PITX1 to regulate Lhb transcription through the proximal promoter, which contains a Pitx binding site flanked by tandem Egr/Sf1 elements (Jorgensen et al., 2004).
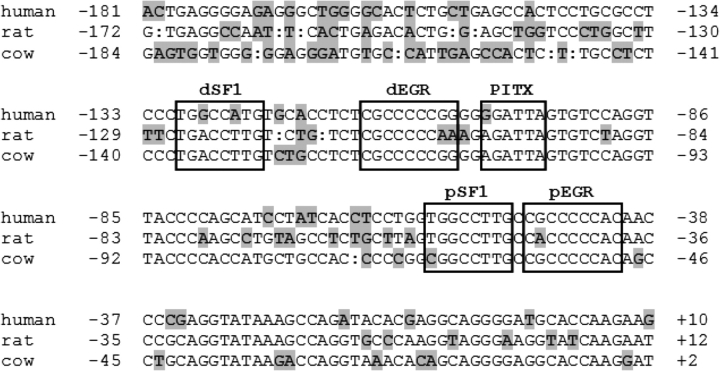
Most investigations on the transcriptional regulation of the Lhb gene have used the bovine or rodent promoters. In contrast, transcriptional regulation of the human LHB promoter has received considerably less attention. One report indicated that both EGR sites and the proximal SF1 site in the human promoter have higher affinity for their respective transcription factors than do the comparable sites in the rat or bovine promoters (Call and Wolfe, 2002). In addition, the distal SF1 element in the human promoter was reported to be of much lower affinity than in other species (Call and Wolfe, 2002). However, the functional relevance of these sites in the context of basal or GNRH1-regulated transcription was not reported. Further, the role of the putative PITX site in the LHB promoter and the identity of the protein(s) binding there are unknown. Sequence alignment of the LHB/Lhb promoters from several species reveals base-pair differences in the EGR, SF1 and PITX elements (Fig. 1), which may be functionally significant. Therefore, we characterized transcriptional regulation of the human LHB promoter by GNRH1. Collectively, the data suggest that the primary mechanisms by which GNRH1 regulates the Lhb/LHB promoter are conserved between humans and other species.
Figure 1.
Aligment of proximal Lhb/LHB promoters from human, rat and cow.
In all cases, +1 refers to the transcription start site. Nucleotides that differ from the consensus are shaded. The conserved SF1, EGR and PITX elements are boxed. ‘d’: distal, ‘p’: proximal.
Materials and Methods
Reagents
Dulbecco's modified Eagle medium (DMEM) with 4.5 g/l glucose, l-glutamine and sodium pyruvate was purchased from Wisent (St Bruno, QC, Canada). DMEM/F-12 Ham's media (1:1) with 2.5 mM l-glutamine and 15 mM HEPES was purchased from HyClone (South Logan, UT, USA). Fetal bovine serum (FBS), Lipofectamine, Lipofectamine 2000 and gentamycin were purchased from Invitrogen (Burlington, ON, Canada). Polyclonal anti-Flag (F7425) and anti-c-myc (M5546) antibodies, aprotinin, leupeptin, pepstatin, PMSF, GNRH1 (LHRH) and SP600125 were from Sigma (St Louis, MO, USA). SB202190 was from Calbiochem (San Diego, CA, USA). Deoxynucleotide triphosphates (dNTPs), T4 DNA ligase, T4 polynucleotide kinase, restriction endonucleases, 5× Passive Lysis Buffer (PLB) and U0126 were from Promega (Madison, WI, USA). DNA polymerases (Pfu Ultra and Turbo) were purchased from Stratagene (La Jolla, CA, USA). [γ-32P] ATP was from PerkinElmer (Boston, MA, USA). Egr1 (D-040286-01), Sf1 (D-051262-01), Pitx1 (D-043250-03), Pitx2 (D-058287-01) and control (D-001210-05) short interfering RNAs (siRNAs) were purchased from Dharmacon (Lafayette, CO, USA). The SF1 rabbit polyclonal antibody (PA1-800) was from Affinity Bioreagents (Golden, CO, USA). PITX1N-15 (sc-18922X) and EGR1 C-19 (sc-189X) rabbit polyclonal antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Normal rabbit IgG (12–370) was from Upstate (Lake Placid, NY, USA). Protease inhibitor tablets (Complete Mini) were purchased from Roche (Indianapolis, IN, USA). Oligonucleotides were synthesized by IDT (Coralville, IA, USA). ECL-plus reagent were from Amersham Biosciences (GE Healthcare, Piscataway, NJ, USA).
Constructs
The LHB luciferase reporters were produced by PCR amplification from genomic DNA (for primers see Table I) as described earlier for the 0.2 kb construct and ligated into pA3-luc (Wang et al., 2008). Mouse EGR1 (NGFIA) in pJDM464 and NR5A1 (SF1) in pCMV5 were generous gifts from Drs Jeffrey Milbrandt (Washington University School of Medicine, St Louis, MO) and Keith Parker (UT Southwestern Medical Center, Dallas, TX), respectively. Murine PITX1, Flag-PITX1, myc-PITX1 and PITX2 expression vector were described earlier (Lamba et al., 2008a, b). To make Flag-tagged EGR1 and SF1, the parental constructs were sub-cloned using strategies described earlier for Flag-PITX1 (Lamba et al., 2008b). Constitutively active MKK6 was a gift from Dr David Engelberg (Hebrew University, Jerusalem, Israel), and Raf-CAAX was from Dr Linda Van Aelst (Cold Spring Harbor Lab). The mutant promoter-reporter and siRNA resistant expression vectors were constructed using the QuikChange protocol (Stratagene) using the primers described in Table I. All constructs were verified by sequencing (Genewiz, South Plainfield, NJ, USA).
Table I.
Primer and probe sequences
| Promoter-reporter cloninga | |
| −197/−178.hLHB.F | GCGGGTACCCTCACCTCTGGCGCTAGACC |
| +8/−12.hLHB.R | CGGAAGCTTCTTGGTGCATCCCCTGCCTC |
| −500/−481.hLHB.F | CGGGGTACCCATCTGGGTCAAGTGGCTTC |
| −1068/−1049.hLHB.F | CGGGGTACCGCCCTGTCTCTGGCTCAGGA |
| Reporter mutagenesisb | |
| hLHB.xdSF1.F | CCTGCGCCTCCCTGGaatTGTGCACCTCTCGCC |
| hLHB.xdEGR.F | CTGCGCCTCCCTGGCCATGTGCACCTCTtagtaCtcGGGGGATTAGTGTCCA |
| hLHB.xPITX.F | CTCTCGCCCCCGGGGGttgTAGTGTCCAGGTTACC |
| hLHB.xpSF1.F | TCACCTCCTGGTGGaaTTcCCGCCCCCACAACC |
| hLHB.xpEGR.F | CTATCACCTCCTGGTGGCCTTGCgGttCttAtAACCCCGAGGTATAAAGCCAGAT |
| Gel shiftb | |
| −134/−103 LHB | TCCCTGGCCATGTGCACCTCTCGCCCCCGGGG |
| −134/−103 xdSF1 LHB | TCCCTGGaatTGTGCACCTCTCGCCCCCGGGG |
| −134/−103 xdEGR LHB | TCCCTGGCCATGTGCACCTCTtagtaCtcGGG |
| −104/−79 LHB | GGGGATTAGTGTCCAGGTTACCCCAG |
| −104/−79 PITX mut LHB | GGGttgTAGTGTCCAGGTTACCCCAG |
| −66/−33 LHB | CTCCTGGTGGCCTTGCCGCCCCCACAACCCCG |
| −66/−33 xpSF1 LHB | CTCCTGGTGGaaTTcCCGCCCCCACAACCCCG |
| −66/−33 xpEGR LHB | CTCCTGGTGGCCTTGCgGttCttAtAACCCCG |
Mutations are in lowercase. aRestriction sites are underlined; bOnly sense strand is shown.
Cell culture, transfections and reporter assays
LβT2 cells were gift from Dr Pamela Mellon (University of California, San Diego, CA). CHO and CV1 cells were provided by Dr Patricia Morris (Population Council, New York, NY). All cells were cultured and transfected as described previously (Lamba et al., 2008b; Wang et al., 2008). Briefly, LβT2 cells were transfected overnight with 450 ng reporter/well and the indicated amounts of plasmid DNA or siRNA. Total DNA transfected was balanced across all conditions. Control siRNAs used in our experiments consistently had non-specific effects on reporter activity, and therefore could not be used as a valid negative control. Indeed, the manufacturer (Dharmacon) cautioned that several of their control siRNAs may have unwanted effects in some contexts (http://www.dharmacon.com/catalog/Item.aspx?Product=31197). The following day, transfection medium was replaced with serum-free DMEM, and cells were starved overnight. Next, cells were treated with GNRH1 and harvested in PLB. Luciferase assays were performed as described previously (Wang et al., 2008). For experiments using pharmacological inhibitors, compounds were applied 30 min before GNRH1 treatment. CV1 cells were transfected with the indicated plasmids using the calcium–phosphate method and harvested the following day for luciferase assays. All reporter experiments were performed a minimum of three times with duplicates or triplicates of all treatments.
Electrophoretic mobility shift, DNA affinity pull-down and immunoblot assays
For EMSA and DNA affinity pull-down (DNAP) experiments, LβT2 cells were grown until confluent in 10-cm plates. Cells were stimulated or not with 10−7 M GNRH1 for 1 h prior to collection of nuclear or whole cell lysates. CHO cells in 10-cm plates were transfected with the indicated plasmid DNA using Lipofectamine, and cells harvested the following day. Nuclear extracts were prepared and gel-shift assays were performed as described previously (Lamba et al., 2006). Briefly, the binding reactions were composed of 10 mM KCl, 25 mM HEPES (pH 7.2), 5 mM dithiothreitol (DTT), 20% glycerol, 500 ng of salmon sperm DNA and equivalent amounts of protein. Where appropriate, cold competitor probe or antibodies were added and reactions were incubated for 10 min at room temperature. Following the addition of 0.05 pmol of 32P-labeled double-stranded probe and incubation for 15 min at room temperature, protein:DNA complexes were resolved on 5% native polyacrylamide gels at 4°C. DNAP assays using streptavidin-coupled Dynabeads® M-280 (Dynal, Invitrogen) were performed as previously described (Lamba et al., 2006, 2008b) using the biotinylated probes. Following elution from the beads, proteins were resolved on 10% SDS–PAGE gels as described previously (Bernard, 2004). Sequences of the probes used for gel-shift and DNAP assays are described in Table I.
Statistical analysis
The data presented are the mean (±SEM) of representative experiments. Differences between means were compared using one-, two- or three-way analyses of variance (ANOVA), followed by pair-wise comparisons using the Tukey post hoc test where appropriate (Systat 10.2, Richmond, CA, USA). In some experiments, data were log transformed when the variances were unequal between groups. Significance was assessed relative to P < 0.05.
Results
The proximal LHB promoter is time- and dose-dependently stimulated by GNRH1 in LβT2 cells
LβT2 cells express both the α and β subunits of LH as well as the GNRH1 receptor, and produce LH in response to GNRH1 stimulation (Turgeon et al., 1996). Because no human gonadotrope cell lines are currently available, we used LβT2 cells as a model system to study the regulation of the human LHB promoter. Cells were transfected with human LHB promoter-reporter constructs of varying lengths and treated with GNRH1 at different doses for different times. The GNRH1 responsive region mapped to the proximal 0.2 kb (Supplementary data, Fig. S1A), with maximal induction observed after 8 h at 10−7 and 10−6 M concentrations of the ligand (Supplementary data, Fig. S1B). In subsequent experiments, we used the 10−7 M GNRH1 concentration and 6 h treatment.
GNRH1 stimulates transcriptional activity of the LHB promoter through an ERK, but not JNK or P38, mediated pathway
GNRH1 activates the extracellular signal-regulated kinase 1/2 (ERK1/2), mitogen-activated protein kinase 14 (p38) and c-jun N-terminal kinase (JNK) MAPK pathways in LβT2 cells (Naor et al., 2000; Harris et al., 2002; Liu et al., 2002) (data not shown). The ERK1/2 and JNK branches of the MAPK cascades have been implicated in the regulation of the rat Lhb promoter by GNRH1 (Yokoi et al., 2000; Harris et al., 2002). To assess the requirements for GNRH1-mediated LHB promoter activation, we antagonized all three pathways using previously validated inhibitors at validated concentrations (Wang et al., 2008). The p38 (SB202190) and JNK (SP600125) inhibitors did not affect the fold GNRH1 response. In contrast, pretreatment with the MEK1 inhibitor, U0126, markedly suppressed GNRH1-stimulated transcriptional activity by almost 70% (Supplementary data, Fig. S2A). To confirm a role for ERK (MEK1) signaling, we co-transfected the LHB-luc construct with expression vectors for constitutively active (ca-) forms of MKK6 and Raf1 (Raf-CAAX), upstream kinases of p38 and MEK1, respectively. Whereas Raf-CAAX potently stimulated reporter activity, ca-MKK6 had no effect when expressed alone and did not alter the Raf-CAAX effect (Supplementary data, Fig. S2B). Together, these data indicate that GNRH1 stimulates expression of human LHB through an MEK1 (ERK1/2), but not p38 or JNK, dependent pathway in LβT2 cells.
Two EGR binding sites confer GNRH1 responsiveness to the LHB promoter
Having mapped GNRH1 responsiveness of the LHB promoter to within the proximal 0.2 kb, we next sought to identify critical cis-elements. Two conserved EGR response elements, at −111/−103 (‘distal’, d) and −49/−41 (‘proximal’, p), are present in the human promoter (Fig. 1). These two elements, which mediate the GNRH1-induced trans-activation of the Lhb promoter by EGR1 in other species (Dorn et al., 1999; Tremblay and Drouin, 1999; Wolfe and Call, 1999), are perfectly conserved with those in the bovine promoter, but differ from the rat's proximal and distal sites at one and two base-pairs, respectively (Fig. 1). To assess the role of these sites in the human promoter, we mutated each, either alone or in combination. Corresponding mutations have been shown to functionally inactivate the conserved elements in the rat and bovine promoter (Dorn et al., 1999; Tremblay and Drouin, 1999). Mutation of either the distal or proximal site decreased basal reporter activity (Fig. 2A). The proximal, but not distal site mutation also decreased the fold GNRH1 response. The mutations together further decreased the fold GNRH1 induction. These data indicated that the two conserved EGR sites are critical for basal and GNRH1-regulated human LHB promoter activity.
Figure 2.
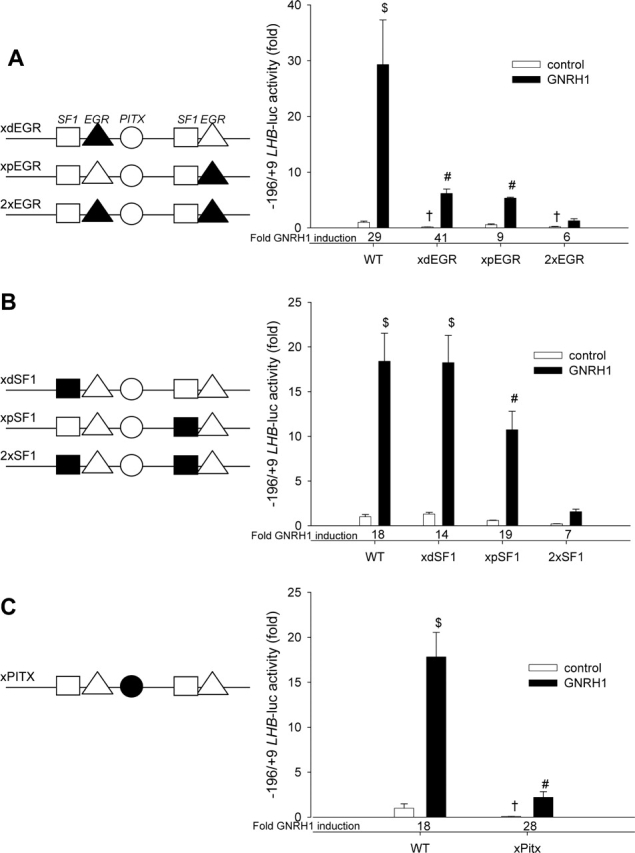
Schematic representations of the proximal LHB promoter are shown at the left of each graph. The SF1, EGR and PITX elements are represented by squares, triangles and a circle, respectively. Black symbols indicate mutated sites. (A) LβT2 cells were transfected with 450 ng/well of the indicated LHB-luc reporters. WT, wild-type; xdEGR, mutated distal EGR site; xpEGR, mutated proximal EGR site; 2xEGR, both EGR elements mutated. Cells were treated or not with 10−7 M GNRH1 for 6 h. (B) LβT2 cells were transfected as above with either WT 0.2 kb LHB-luc reporter or mutant constructs with the inactivated distal (xdSF1), proximal (xpSF1) or both (2xSF1) SF1 sites. Where indicated, GNRH1 treatment was given for 6 h. (C) LβT2 cells were transfected as above with either WT LHB-luc reporter or a construct with a mutated PITX element (xPITX). Differences in reporter activity were measured after 6 h GNRH1 treatment. The fold induction by GNRH1is indicated at the bottom of the graph. Bars with different symbols differ significantly. n = 3 for all treatments.
Two SF1 binding sites and a PITX binding site are required for maximal GNRH1 induction of the LHB promoter
In the rodent and bovine Lhb promoters, two binding sites for SF1 are located 5′ end to the two EGR elements and are important for trans-activation (Halvorson et al., 1996, 1998, 1999; Keri and Nilson, 1996; Dorn et al., 1999; Tremblay and Drouin, 1999; Kaiser et al., 2000). These sites are also present in the human promoter (at −130/−123 and −58/−51), although the distal element differs from those in the bovine or rodent promoters and diverges from the consensus binding sequence for SF1 relative to the other species (Fig. 1). Mutation of the distal SF1 site alone had no effect on either the basal or GNRH1-stimulated LHB promoter activity (Fig. 2B). In contrast, inactivation of the proximal site decreased the basal reporter activity, without altering the fold GNRH1 induction. The two mutations in combination further decreased basal activity and significantly impaired the fold GNRH1 response.
Between the two tandem SF1/EGR elements, at −100/−95, is a binding site for paired-like homeodomain transcription factors, which has been implicated in transcription of the Lhb promoter of various species (Tremblay and Drouin, 1999; Quirk et al., 2001; Rosenberg and Mellon, 2002; Jiang et al., 2005). This site is also present in the human promoter; but, unlike in the other species, perfectly matches the consensus site for PITX proteins [GGATTA (Driever and Nusslein-Volhard, 1989)] (Fig. 1). Introducing a mutation in the element dramatically decreased the basal, but not GNRH1-induced transcriptional activity (Fig. 2C). These results indicate that none of the SF1 or PITX elements alone are required for GNRH1 responsiveness, but all contribute to basal activity and therefore maximal induction of transcription by GNRH1. At the same time, GNRH1 induction of the promoter requires at least one intact SF1 site.
EGR1 and SF1 interact with the LHB promoter via two tandem elements
We examined the proteins binding to the putative EGR, SF1 and PITX sites. First, we performed gel-shift assays using two probes containing the distal or proximal tandem SF1/EGR elements and nuclear extracts from LβT2 cells treated or not with GNRH1 for 1 h. We detected four specific complexes (Fig. 3A, lane 1, labeled ‘a’ through ‘d’) binding the proximal SF1/EGR tandem element, which were competed by 100-fold excess cold homologous probe (lane 3). GNRH1 stimulation markedly increased the intensity of complex a (lane 2), which was competed by 100-fold excess wild-type probe (lane 3), but not by a probe containing the inactivating mutation in the presumptive EGR site (lane 5). This complex was super-shifted by an EGR1 antibody (lanes 8 and 9), but not by control IgG (lane 6) or an SF1 antibody (lane 7). A strong complex (complex ‘d’) present under both basal and GNRH1-stimulated conditions (lanes 1 and 2) was competed by 100-fold excess of homologous cold probe (lane 3), but not by a probe containing the inactivating mutation in the putative SF1 element (lane 4). This complex was super-shifted by an SF1 antibody (lane 7), but not by control IgG (lane 6) or the EGR1 antibody (lanes 8 and 9). There was a slight increase in intensity of the SF1-containing complex with GNRH1 treatment (compare lanes 1 and 2). Binding by complexes ‘b’ and ‘c’ was competed by 100-fold excess of probe with a mutant EGR (lane 5), but not SF1 site (lane 4). The intensity of both complexes was mildly decreased by an SF1 antibody (lane 7), but their identities remain to be determined.
Figure 3.
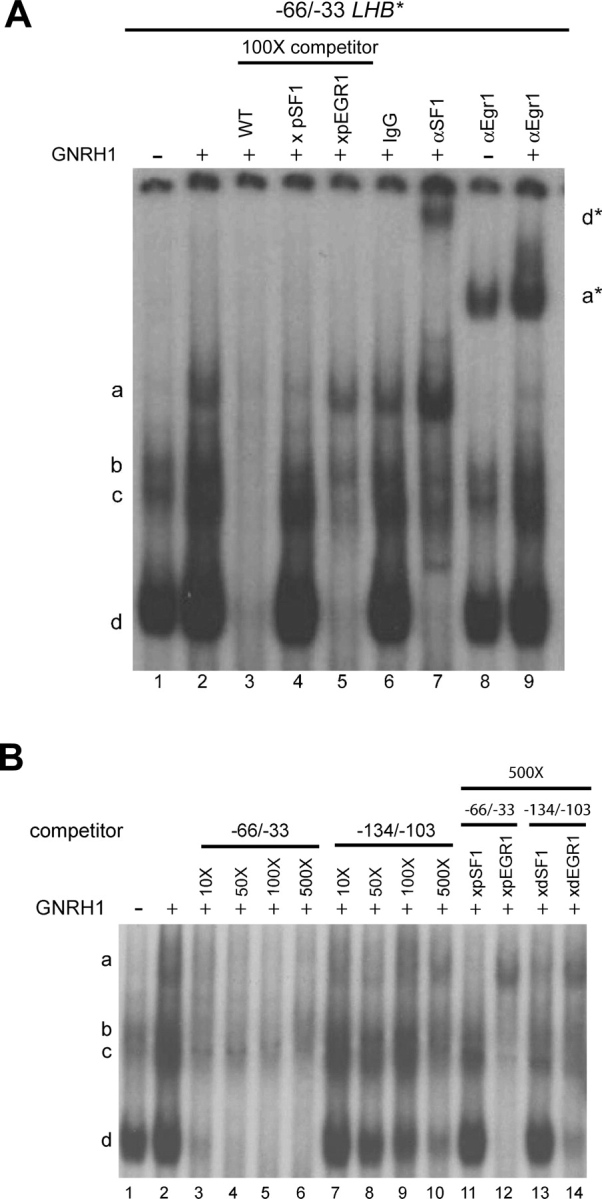
(A) Nuclear extracts from LβT2 cells treated (+) or not (−) with 10−7 M GNRH1 for 1 h were incubated with a radio-labeled probe corresponding to −66/−33 of the LHB promoter. Where indicated, the binding reactions contained 100-fold excess of cold homologous wild-type probe (WT; lane 3) or probes with mutated proximal SF1 (xpSF1; lane 4) or EGR (xpEGR; lane 5) elements. Control IgG (lane 6), or SF1 (lane 7) or EGR1 (lanes 8 and 9) antibodies were added as indicated. Asterisks denote super-shifted complexes. (B) Nuclear extracts from LβT2 cells treated (+) or not (−) with 10−7 M GNRH1 for 1 h were incubated with a radio-labeled probe corresponding to the −66/−33 region of the LHB promoter. Ten, 50, 100 or 500-fold excess homologous cold probe (−66/−33; lanes 3–6), cold probe containing the putative distal SF1 and EGR elements (−134/−103; lanes 7–10), or cold probe with mutated proximal or distal SF1 or EGR elements (500× only; lanes 11–14) were added where indicated.
We next performed a similar analysis with the distal SF1/EGR tandem element. Using nuclear extracts from LβT2 cells stimulated or not with GNRH1, we could not clearly detect any complexes containing SF1 or EGR1 (not shown). To determine whether these observations related to differences in affinities of the proteins for the distal versus proximal sites, we performed competition assays with the radio-labeled proximal probe and varying amounts of cold homologous and distal probes. As little as 10-fold excess cold homologous probe markedly inhibited binding of both SF1 and EGR1 to the proximal SF1 and EGR elements (Fig. 3B, lane 3), complex formation was completely abolished by 50-fold excess cold probe (lane 4). In contrast, although increasing amounts of cold distal probe were able to compete for binding of both SF1 and EGR1 to the proximal elements, complex formation was incompletely abolished even in the presence of 500-fold excess cold probe (lanes 7 through 10). Nonetheless, introduction of the inactivating mutations in the distal elements blocked their abilities to compete for binding to SF1 (compare lanes 10 and 13) and EGR1 (compare lanes 10 and 14). Together, these data indicate that the proximal SF1 and EGR elements are higher affinity binding sites for their respective transcription factors in the LHB promoter than are the more distal sites.
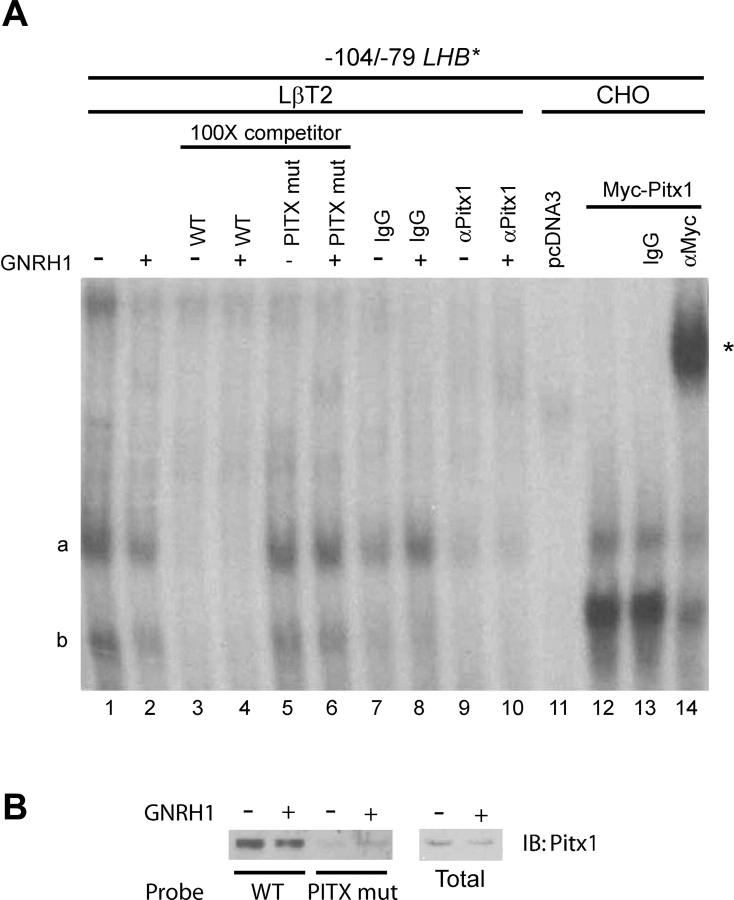
Endogenous PITX1 binds to the LHB promoter
The putative PITX element in the Lhb/LHB promoter could potentially bind several homeodomain transcription factors, and studies in other species have yielded conflicting results regarding the identity of the endogenous proteins occupying this site (Rosenberg and Mellon, 2002). In EMSAs, we detected the formation of two specific complexes (Fig. 4A, lanes 1 and 2, labeled ‘a’ and ‘b’) under both basal and GNRH1-stimulated conditions with a probe containing the PITX element. Complex binding was competed by 100-fold cold homologous probe (lanes 3 and 4), but not by a probe containing the inactivating mutation in the PITX site (lanes 5 and 6). Further, complex formation was disrupted by a PITX1 antibody (lanes 9 and 10), but not by control IgG (lanes 7 and 8). To confirm that these two complexes contained PITX1 proteins, we incubated the probe with nuclear extracts from CHO cells transfected with a myc-tagged PITX1 construct (lanes 12–14). We observed the formation of two complexes co-migrating with the two complexes obtained with the LβT2 nuclear extracts, and both were super-shifted by an anti-myc antibody (lane 14).
Figure 4.
(A) Nuclear extracts from LβT2 cells treated (+) or not (−) with 10−7 M GNRH1 for 1 h (lanes 1–10), or nuclear extracts from CHO cells transfected with empty vector (pcDNA3; lane 11) or Myc-PITX1 (lanes 12–14) were incubated with a radio-labeled probe corresponding to the −104/−79 region of the LHB promoter. Where indicated, the binding reactions contained 100-fold excess of cold homologous WT probe (lanes 3 and 4) or probe with a mutated PITX site (PITX mut; lanes 5 and 6), control IgG (lanes 7, 8 and 13), PITX1 antibody (lanes 9 and 10) or Myc antibody (lane 14). (B) DNAP was performed using the probes described above. Whole cell lysates (total) or proteins interacting with the probes were subjected to immunoblot (IB). Cells were treated (+) or not (−) with 10−7 M GNRH1 for 1 h.
To confirm these results, we performed DNAP experiments using biotinylated probes (Fig. 4B). We pulled down endogenous PITX1 from lysates of control and GNRH1-treated LβT2 cells with a wild-type probe more readily than with a probe containing the inactivating mutation in the PITX site. Together, these results indicate that endogenous PITX1 can bind the LHB promoter.
Because PITX2 proteins bind the same consensus sequence as PITX1 and all known PITX2 isoforms are expressed in LβT2 cells (Lamba et al., 2008a), we next evaluated the possibility that these proteins might be recruited to this element. Using nuclear extracts from transfected CHO cells, we detected binding of all five PITX2 isoforms to the LHB promoter (not shown). However, none of the complexes clearly co-migrated with the endogenous complexes observed using LβT2 nuclear extracts in gel shifts.
EGR1, SF1, PITX1 and PITX2 mediate trans-activation of the LHB promoter
To confirm the roles for EGR1, SF1, PITX1 and PITX2 (isoforms) in the basal and GNRH1-induced LHB transcriptional activity, we first knocked down the expression of the proteins in LβT2 cells using siRNA. siRNAs targeting Egr1 or Sf1 mRNAs markedly decreased both basal reporter activity and fold stimulation by GNRH1 (Fig. 5A). Depletion of PITX1 markedly decreased GNRH1-induced activity and also appeared to inhibit basal reporter activity, but the latter effect was not statistically significant (Fig. 5B).
Figure 5.
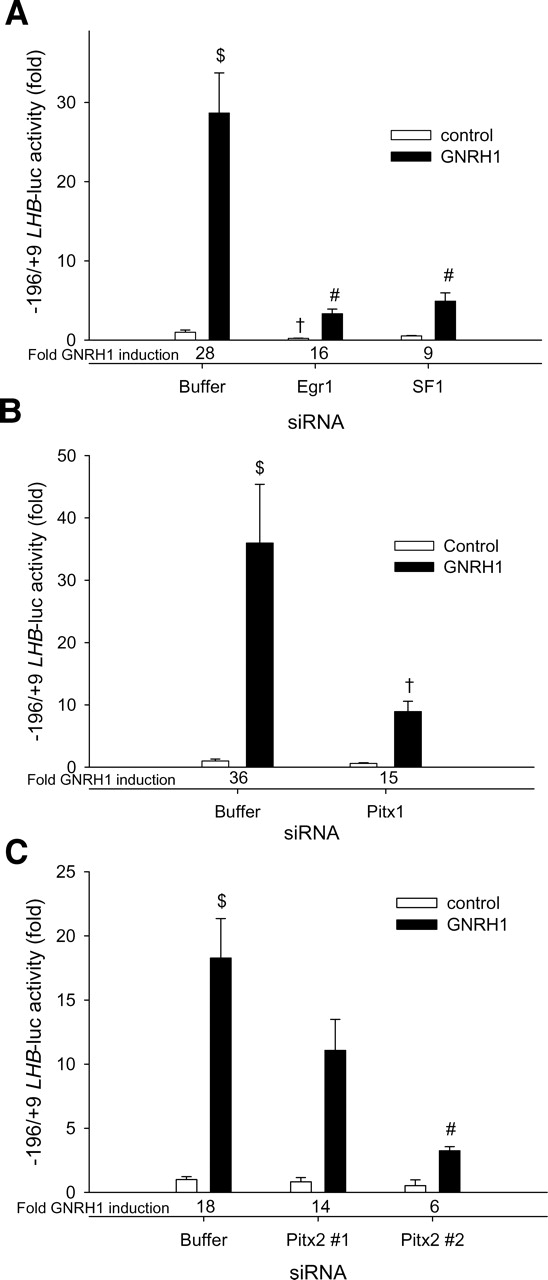
(A) LβT2 cells were co-transfected with 450 ng/well of WT 0.2 kb LHB-luc reporter and 10−8 M of (A) Egr1 or Sf1, (B) Pitx1 or (C) Pitx2 siRNAs. In all cases, 1× siRNA buffer was used as control. Cells were treated or not with 10−7 M GNRH1 for 6 h prior to collection of lysates for luciferase assays. Fold induction by GNRH1 is indicated at the bottom of the graphs. Bars with different symbols differ significantly. n = 3 per treatment.
Notably, knockdown of PITX1 had less effect on reporter activity than did the mutation of the PITX response element (Fig. 2C). This could be attributable to incomplete knockdown and/or to functional compensation by PITX2 proteins. We therefore knocked down PITX2 expression using two siRNAs, one (#1) targeting the first coding exon (exon 2) [expected to affect the PITX2A, B1 and B2 isoforms], and the other (#2) targeting the 3′-end of the coding region (exon 6) (common to all five PITX2 isoforms) (Lamba et al., 2008a). Pitx2 siRNA #2 had a more dramatic effect on LHB promoter activity than Pitx2 siRNA #1 (Fig. 5C). Whereas Pitx2 siRNA #2 consistently decreased basal transcriptional activity, this did not reach statistical significance. The GNRH1-stimulated activity, in contrast, was significantly reduced. Together, these results suggested that endogenous PITX1 and PITX2 proteins in LβT2 cells participate in the trans-activation of the LHB promoter. Control experiments confirmed the efficacy and sequence specificity of the siRNAs (Supplementary data, Fig. S3).
Finally, we used over-expression in heterologous CV-1 cells to examine functional cooperation between EGR1, SF1 and PITX1 at the LHB promoter. Expression of EGR1 or PITX1, but not SF1, by themselves stimulated transcriptional activity of the 0.2 kb promoter-reporter (Fig. 6A and B). Further, PITX1 synergistically induced reporter activity with either EGR1 or SF1. SF1 did not further potentiate the combined effects of PITX1 and EGR1 (data not shown). These results indicate that the transcription factors binding the proximal LHB promoter can co-operate to enhance transcriptional activity.
Figure 6.
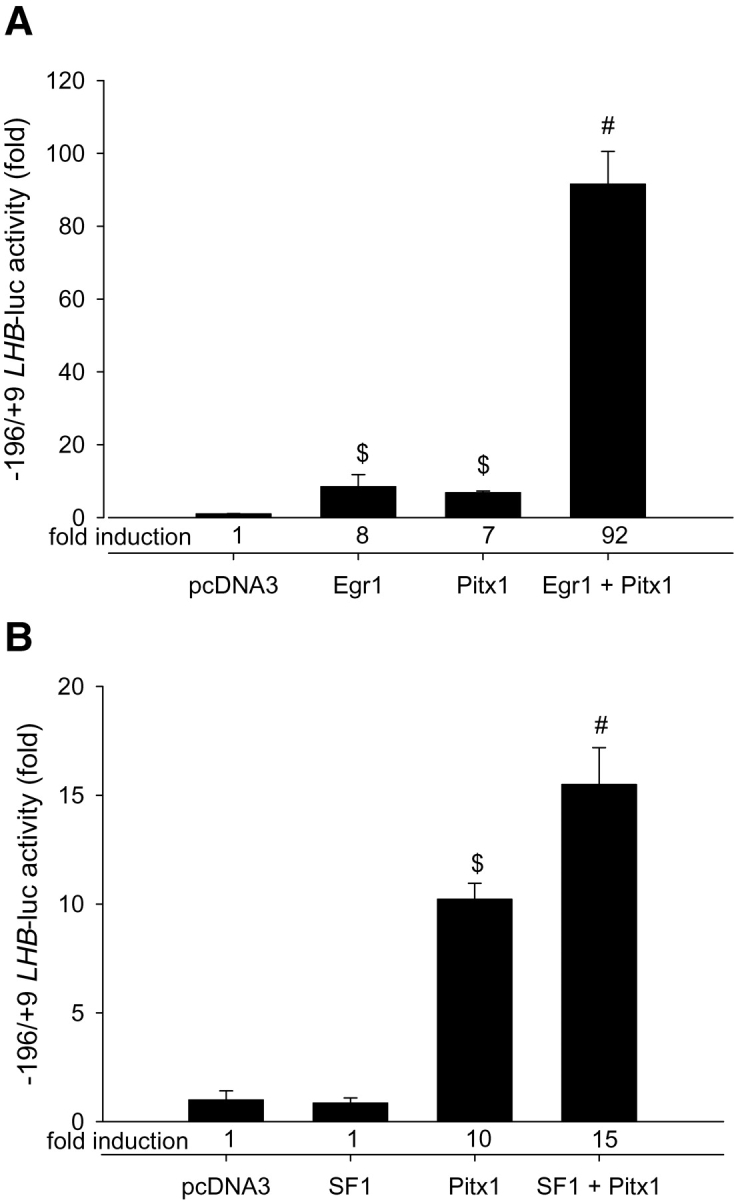
(A) CV-1 cells were transfected with 900 ng/well of the 0.2 kb LHB-luc reporter along with 30 ng/well of EGR1 and/or PITX1 expression vectors or empty vector (pcDNA3).
After overnight recovery, reporter activity was measured. The average fold stimulation, indicated at the bottom of the graph, was normalized to the reporter activity measured in presence of only the empty vector. (B) CV-1 cells were transfected as in panel (A) with 30 ng/well of SF1 and/or PITX1 expression vectors or empty vector (pcDNA3). Reporter activity was measured and normalized as above. Bars with different symbols differ significantly. n = 3 per treatment.
Discussion
Previous studies delineated a mechanism by which GNRH1 signaling induces Lhb transcription. Here, we show that this mechanism is largely conserved in the human LHB promoter. GNRH1 signals through the ERK1/2 MAPK signaling cascade to regulate LHB transcription and does so primarily through the proximal 200 base-pairs. As in rodents, cow, and horse, basal and/or GNRH1-regulated human LHB transcription is dependent upon the coordinated activities of EGR1, SF1 and PITX1 acting through conserved cis-elements within this proximal promoter region. RNA interference experiments confirmed roles for the endogenous proteins in basal and/or GNRH1-regulated promoter activity and further suggest a potential role for PITX2 isoforms.
The data show that GNRH1 induces transcriptional activity of the LHB promoter primarily through an ERK1/2-mediated pathway. Although both the ERK1/2 and JNK MAPK cascades have been implicated in GNRH1 regulation of the Lhb promoter in other species (Yokoi et al., 2000; Harris et al., 2002), a more critical role has been attributed to ERK1/2 (Liu et al., 2002; Kanasaki et al., 2005). GNRH1 stimulates Egr1 expression through the ERK1/2 pathway (Duan et al., 2002; Maudsley et al., 2007), and EGR1 appears to be the primary transducer of the GNRH1 signal to the Lhb promoter (Dorn et al., 1999; Tremblay and Drouin, 1999). Indeed, our data confirm a critical role for EGR1 in regulation of the human LHB promoter through two conserved cis-elements.
In the rat Lhb promoter, a distal region containing at least two Sp1 sites (−450/−441 and −410/−402) contributes significantly to GNRH1 induction (Kaiser et al., 1998a, b, 2000; Weck et al., 2000). Only one of the putative Sp1 sites is partially conserved in the human promoter. Though we noted differences in basal activity between the 0.2, 0.5 and 1 kb LHB promoter-reporters, the fold-induction by GNRH1 was similar among the three (Supplementary data, Fig. S1A). This suggests that distal elements do not significantly contribute to GNRH1 induction of the human LHB gene, at least under the experimental conditions used here.
As in other species, the EGR, SF1 and PITX1 sites are required for maximal induction of the LHB promoter by GNRH1. Mutation of the proximal EGR element or both SF1 sites strongly attenuated the GNRH1 response. We confirmed binding of EGR1 and SF1 binding to their respective sites. Binding to the proximal elements was potentiated following GNRH1 treatment, particularly for EGR1. These results are consistent with the fact that EGR1 levels are markedly increased in gonadotropes upon GNRH1 stimulation (Dorn et al., 1999; Tremblay and Drouin, 1999) (data not shown). Although it has been reported that SF1 levels are unaffected by GNRH1 stimulation in gonadotropes (Dorn et al., 1999; Tremblay and Drouin, 1999), we observed a slight increase in intensity of SF1 binding to the proximal promoter element upon GNRH1 treatment. Therefore, this change in binding might reflect post-translational modifications in SF1 induced by GNRH1 signaling, such as phosphorylation (Hammer et al., 1999; Winnay and Hammer, 2006), and/or potentiation of binding through cooperation with induced EGR1. The data show that the proximal SF1/EGR elements have higher affinity for their respective transcription factors and contribute more than the distal SF1/EGR sites to overall cis-activation of the LHB promoter. Fold GNRH1 induction was decreased when the proximal EGR site was ablated, but was maintained in the presence of a mutated distal EGR element. Inactivation of the distal SF1 site did not affect transcriptional activity either basally on in response to GNRH1. Therefore, this site is likely dispensable for LHB promoter activation. In contrast, inactivation of this element alone prevents normal GNRH1 induction of the bovine Lhb promoter in transgenic mice (Keri and Nilson, 1996), though this was not the case in LβT2 cells (Tremblay and Drouin, 1999). In the rat Lhb promoter, this site contributes significantly to basal activity and shows similar affinity for SF1 compared with the proximal element (Keri and Nilson, 1996; Halvorson et al., 1998; Dorn et al., 1999; Call and Wolfe, 2002). In cow and rat, the distal SF1 element is a perfect match to the consensus site, whereas the human element differs at positions 3 and 6 (Fig. 1). Nevertheless, our data suggest that the distal EGR and SF1 elements can partially compensate for the loss of the proximal sites. Indeed, mutation of the two EGR1 or SF1 elements impairs transcriptional activity to a greater extent than inactivation of the proximal sites alone.
In transgenic mice, there is a clear requirement for the Pitx binding site for activation of the bovine Lhb promoter by GNRH1 (Quirk et al., 2001). Results from mutational analyses reported here similarly indicate a critical role for this site in maximal activity of the human LHB promoter. We also showed binding of endogenous PITX1 to the LHB promoter by gel-shift and DNAP assays, which has not been unequivocally demonstrated in other species (Quirk et al., 2001; Rosenberg and Mellon, 2002; Jiang et al., 2005). This may be explained by the fact that the human PITX binding site conforms perfectly to the consensus site [5′-GGATTA-3′, (Driever and Nusslein-Volhard, 1989)], whereas the corresponding sites in the rodent or bovine promoters do not (5′-AGATTA-3′). Structural analyses indicate that the GG nucleotides are critical for PITX2 binding to the PITX response element (Chaney et al., 2005). Because the homeodomains of PITX2 and PITX1 are 97% identical (Semina et al., 1997), this requirement most likely also applies to PITX1.
Though several studies have implicated PITX1 in the regulation of the Lhb promoter (Tremblay et al., 1999; Tremblay and Drouin, 1999; Quirk et al., 2001; Jiang et al., 2005), possible roles for PITX2 isoforms have been largely overlooked despite the observations that they can trans-activate the bovine Lhb promoter in heterologous cells (Tremblay et al., 2000; Lamba et al., 2008a). Results from RNA interference experiments shown here suggest roles for both PITX1 and PITX2 proteins in basal and GNRH1-regulated LHB promoter activity. However, it was recently reported that targeted deletion of Pitx2 in terminally differentiated gonadotropes had no effect on Lhb expression and fertility in mice (Charles et al., 2008), suggesting either that PITX2 proteins play no role in Lhb regulation in vivo or that PITX1 can compensate for their loss. Additional experiments in which Pitx1 is ablated alone or together with Pitx2 in differentiated gonadotropes will be needed to address these ideas. At the same time, the difference in the PITX binding site between mice and humans leaves open the possibility that different proteins may bind these elements in the two species or that the same proteins may bind with different affinities. As such, results in mice may not be entirely predictive of what occurs in humans. Though the siRNA experiments here suggest a role for PITX2 proteins in regulation of the human LHB gene, we were unable to confirm binding of any endogenous PITX2 protein isoforms in our analyses. Unfortunately, we exhausted the PITX2 antibody we used previously (Lamba et al., 2008a), which precluded super-shift and DNAP analyses of the kind we employed with PITX1.
In summary, our results indicate that the primary mechanisms of GNRH1-induced LHB transcription are conserved between humans and other species. This contrasts with what we have reported for regulation of the FSHB/Fshb in humans and other species (Lamba et al., 2006; Wang et al., 2008). In the latter case, we argued that inter-species differences in transcriptional regulation may relate to observed differences in FSH dynamics in different organisms. When viewed in this light, one might predict conservation of LHB/Lhb transcriptional regulatory mechanisms. That is, in all mammalian species studied to date, GNRH1 pulses are followed faithfully and rapidly by LH pulses. Given the slower kinetics of increases in LHB transcription, one might view this response as a compensatory mechanism to replenish intracellular LH stores in advance of subsequent GNRH1 pulses. This may be particularly important in the context of the LH surge, where GNRH1 pulse frequency and amplitude are elevated, increasing the demand for releasable LH. Given that the dynamics of LH surge generation are common among mammalian species, it is perhaps not surprising that the mechanisms for LHB/Lhb trans-activation would be similarly conserved.
Supplementary data
Supplementary data are available at http://molehr.oxfordjournals.org/.
Funding
This research was supported by grants from the NIH (HD047794) and CIHR (MOP-89991) to D.J.B.
Acknowledgements
The authors thank Drs Pamela Mellon (UCSD) and Patricia Morris (Population Council) for the LβT2 and CHO/CV-1 cells, respectively. The murine EGR1 (NGFIA) expression vector was a generous gift from Dr Jeffrey Milbrandt (Washington University School of Medicine, St Louis, MO). Dr Keith Parker (UT Southwestern Medical Center, Dallas, TX) provided the SF1 (NR5A1) expression vector. Constitutively active MKK6 and Raf-CAAX were gifts from Drs David Engelberg (Hebrew University, Jerusalem, Israel) and Linda Van Aelst (Cold Spring Harbor Lab), respectively.
References
- Bernard DJ. Both SMAD2 and SMAD3 mediate activin-stimulated expression of the follicle-stimulating hormone beta subunit in mouse gonadotrope cells. Mol Endocrinol. 2004;18:606–623. doi: 10.1210/me.2003-0264. [DOI] [PubMed] [Google Scholar]
- Call GB, Wolfe MW. Species differences in GnRH activation of the LHbeta promoter: role of Egr1 and Sp1. Mol Cell Endocrinol. 2002;189:85–96. doi: 10.1016/s0303-7207(01)00744-4. [DOI] [PubMed] [Google Scholar]
- Chaney BA, Clark-Baldwin K, Dave V, Ma J, Rance M. Solution structure of the K50 class homeodomain PITX2 bound to DNA and implications for mutations that cause Rieger syndrome. Biochemistry. 2005;44:7497–7511. doi: 10.1021/bi0473253. [DOI] [PubMed] [Google Scholar]
- Charles MA, Mortensen AH, Potok MA, Camper SA. Pitx2 deletion in pituitary gonadotropes is compatible with gonadal development, puberty, and fertility. Genesis. 2008;46:507–514. doi: 10.1002/dvg.20398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn C, Ou Q, Svaren J, Crawford PA, Sadovsky Y. Activation of luteinizing hormone beta gene by gonadotropin-releasing hormone requires the synergy of early growth response-1 and steroidogenic factor-1. J Biol Chem. 1999;274:13870–13876. doi: 10.1074/jbc.274.20.13870. [DOI] [PubMed] [Google Scholar]
- Driever W, Nusslein-Volhard C. The bicoid protein is a positive regulator of hunchback transcription in the early Drosophila embryo. Nature. 1989;337:138–143. doi: 10.1038/337138a0. [DOI] [PubMed] [Google Scholar]
- Duan WR, Ito M, Park Y, Maizels ET, Hunzicker-Dunn M, Jameson JL. GnRH regulates early growth response protein 1 transcription through multiple promoter elements. Mol Endocrinol. 2002;16:221–233. doi: 10.1210/mend.16.2.0779. [DOI] [PubMed] [Google Scholar]
- Halvorson LM, Kaiser UB, Chin WW. Stimulation of luteinizing hormone beta gene promoter activity by the orphan nuclear receptor, steroidogenic factor-1. J Biol Chem. 1996;271:6645–6650. doi: 10.1074/jbc.271.12.6645. [DOI] [PubMed] [Google Scholar]
- Halvorson LM, Ito M, Jameson JL, Chin WW. Steroidogenic factor-1 and early growth response protein 1 act through two composite DNA binding sites to regulate luteinizing hormone beta-subunit gene expression. J Biol Chem. 1998;273:14712–14720. doi: 10.1074/jbc.273.24.14712. [DOI] [PubMed] [Google Scholar]
- Halvorson LM, Kaiser UB, Chin WW. The protein kinase C system acts through the early growth response protein 1 to increase LHbeta gene expression in synergy with steroidogenic factor-1. Mol Endocrinol. 1999;13:106–116. doi: 10.1210/mend.13.1.0216. [DOI] [PubMed] [Google Scholar]
- Hammer GD, Krylova I, Zhang Y, Darimont BD, Simpson K, Weigel NL, Ingraham HA. Phosphorylation of the nuclear receptor SF-1 modulates cofactor recruitment: integration of hormone signaling in reproduction and stress. Mol Cell. 1999;3:521–526. doi: 10.1016/s1097-2765(00)80480-3. [DOI] [PubMed] [Google Scholar]
- Harris D, Bonfil D, Chuderland D, Kraus S, Seger R, Naor Z. Activation of MAPK cascades by GnRH: ERK and Jun N-terminal kinase are involved in basal and GnRH-stimulated activity of the glycoprotein hormone LHbeta-subunit promoter. Endocrinology. 2002;143:1018–1025. doi: 10.1210/endo.143.3.8675. [DOI] [PubMed] [Google Scholar]
- Jiang Q, Jeong KH, Horton CD, Halvorson LM. Pituitary homeobox 1 (Pitx1) stimulates rat LHbeta gene expression via two functional DNA-regulatory regions. J Mol Endocrinol. 2005;35:145–158. doi: 10.1677/jme.1.01754. [DOI] [PubMed] [Google Scholar]
- Jorgensen JS, Quirk CC, Nilson JH. Multiple and overlapping combinatorial codes orchestrate hormonal responsiveness and dictate cell-specific expression of the genes encoding luteinizing hormone. Endocr Rev. 2004;25:521–542. doi: 10.1210/er.2003-0029. [DOI] [PubMed] [Google Scholar]
- Kaiser UB, Sabbagh E, Chen MT, Chin WW, Saunders BD. Sp1 binds to the rat luteinizing hormone beta (LHbeta) gene promoter and mediates gonadotropin-releasing hormone-stimulated expression of the LHbeta subunit gene. J Biol Chem. 1998;a 273:12943–12951. doi: 10.1074/jbc.273.21.12943. [DOI] [PubMed] [Google Scholar]
- Kaiser UB, Sabbagh E, Saunders BD, Chin WW. Identification of cis-acting deoxyribonucleic acid elements that mediate gonadotropin-releasing hormone stimulation of the rat luteinizing hormone beta-subunit gene. Endocrinology. 1998;b 139:2443–2451. doi: 10.1210/endo.139.5.6003. [DOI] [PubMed] [Google Scholar]
- Kaiser UB, Halvorson LM, Chen MT. Sp1, steroidogenic factor 1 (SF-1), and early growth response protein 1 (egr-1) binding sites form a tripartite gonadotropin-releasing hormone response element in the rat luteinizing hormone-beta gene promoter: an integral role for SF-1. Mol Endocrinol. 2000;14:1235–1245. doi: 10.1210/mend.14.8.0507. [DOI] [PubMed] [Google Scholar]
- Kanasaki H, Bedecarrats GY, Kam KY, Xu S, Kaiser UB. Gonadotropin-releasing hormone pulse frequency-dependent activation of extracellular signal-regulated kinase pathways in perifused LbetaT2 cells. Endocrinology. 2005;146:5503–5513. doi: 10.1210/en.2004-1317. [DOI] [PubMed] [Google Scholar]
- Keri RA, Nilson JH. A steroidogenic factor-1 binding site is required for activity of the luteinizing hormone beta subunit promoter in gonadotropes of transgenic mice. J Biol Chem. 1996;271:10782–10785. doi: 10.1074/jbc.271.18.10782. [DOI] [PubMed] [Google Scholar]
- Lamba P, Santos MM, Philips DP, Bernard DJ. Acute regulation of murine follicle-stimulating hormone beta subunit transcription by activin A. J Mol Endocrinol. 2006;36:201–220. doi: 10.1677/jme.1.01961. [DOI] [PubMed] [Google Scholar]
- Lamba P, Hjalt TA, Bernard DJ. Novel forms of paired-like homeodomain transcription factor 2 (PITX2): generation by alternative translation initiation and mRNA splicing. BMC Mol Biol. 2008;a 9:31. doi: 10.1186/1471-2199-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba P, Khivansara V, D'Alessio AC, Santos MM, Bernard DJ. Paired-like homeodomain transcription factors 1 and 2 regulate follicle-stimulating hormone beta-subunit transcription through a conserved cis-element. Endocrinology. 2008;b 149:3095–3108. doi: 10.1210/en.2007-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctot C, Moreau A, Chamberland M, Tremblay ML, Drouin J. Hindlimb patterning and mandible development require the Ptx1 gene. Development. 1999;126:1805–1810. doi: 10.1242/dev.126.9.1805. [DOI] [PubMed] [Google Scholar]
- Lee SL, Sadovsky Y, Swirnoff AH, Polish JA, Goda P, Gavrilina G, Milbrandt J. Luteinizing hormone deficiency and female infertility in mice lacking the transcription factor NGFI-A (Egr-1) Science. 1996;273:1219–1221. doi: 10.1126/science.273.5279.1219. [DOI] [PubMed] [Google Scholar]
- Liu F, Austin DA, Mellon PL, Olefsky JM, Webster NJ. GnRH activates ERK1/2 leading to the induction of c-fos and LHbeta protein expression in LbetaT2 cells. Mol Endocrinol. 2002;16:419–434. doi: 10.1210/mend.16.3.0791. [DOI] [PubMed] [Google Scholar]
- Maudsley S, Naor Z, Bonfil D, Davidson L, Karali D, Pawson AJ, Larder R, Pope C, Nelson N, Millar RP, et al. Proline-rich tyrosine kinase 2 mediates gonadotropin-releasing hormone signaling to a specific extracellularly regulated kinase-sensitive transcriptional locus in the luteinizing hormone beta-subunit gene. Mol Endocrinol. 2007;21:1216–1233. doi: 10.1210/me.2006-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naor Z, Benard O, Seger R. Activation of MAPK cascades by G-protein-coupled receptors: the case of gonadotropin-releasing hormone receptor. Trends Endocrinol Metab. 2000;11:91–99. doi: 10.1016/s1043-2760(99)00232-5. [DOI] [PubMed] [Google Scholar]
- Quirk CC, Lozada KL, Keri RA, Nilson JH. A single Pitx1 binding site is essential for activity of the LHbeta promoter in transgenic mice. Mol Endocrinol. 2001;15:734–746. doi: 10.1210/mend.15.5.0628. [DOI] [PubMed] [Google Scholar]
- Rosenberg SB, Mellon PL. An Otx-related homeodomain protein binds an LHbeta promoter element important for activation during gonadotrope maturation. Mol Endocrinol. 2002;16:1280–1298. doi: 10.1210/mend.16.6.0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semina EV, Reiter RS, Murray JC. Isolation of a new homeobox gene belonging to the Pitx/Rieg family: expression during lens development and mapping to the aphakia region on mouse chromosome 19. Hum Mol Genet. 1997;6:2109–2116. doi: 10.1093/hmg/6.12.2109. [DOI] [PubMed] [Google Scholar]
- Szeto DP, Rodriguez-Esteban C, Ryan AK, O'Connell SM, Liu F, Kioussi C, Gleiberman AS, Izpisua-Belmonte JC, Rosenfeld MG. Role of the Bicoid-related homeodomain factor Pitx1 in specifying hindlimb morphogenesis and pituitary development. Genes Dev. 1999;13:484–494. doi: 10.1101/gad.13.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topilko P, Schneider-Maunoury S, Levi G, Trembleau A, Gourdji D, Driancourt MA, Rao CV, Charnay P. Multiple pituitary and ovarian defects in Krox-24 (NGFI-A, Egr-1)-targeted mice. Mol Endocrinol. 1998;12:107–122. doi: 10.1210/mend.12.1.0049. [DOI] [PubMed] [Google Scholar]
- Tremblay JJ, Drouin J. Egr-1 is a downstream effector of GnRH and synergizes by direct interaction with Ptx1 and SF-1 to enhance luteinizing hormone beta gene transcription. Mol Cell Biol. 1999;19:2567–2576. doi: 10.1128/mcb.19.4.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay JJ, Marcil A, Gauthier Y, Drouin J. Ptx1 regulates SF-1 activity by an interaction that mimics the role of the ligand-binding domain. EMBO J. 1999;18:3431–3441. doi: 10.1093/emboj/18.12.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay JJ, Goodyer CG, Drouin J. Transcriptional properties of Ptx1 and Ptx2 isoforms. Neuroendocrinology. 2000;71:277–286. doi: 10.1159/000054547. [DOI] [PubMed] [Google Scholar]
- Turgeon JL, Kimura Y, Waring DW, Mellon PL. Steroid and pulsatile gonadotropin-releasing hormone (GnRH) regulation of luteinizing hormone and GnRH receptor in a novel gonadotrope cell line. Mol Endocrinol. 1996;10:439–450. doi: 10.1210/mend.10.4.8721988. [DOI] [PubMed] [Google Scholar]
- Wang Y, Fortin J, Lamba P, Bonomi M, Persani L, Roberson MS, Bernard DJ. AP-1 and Smad proteins synergistically regulate human follicle-stimulating hormone {beta} promoter activity. Endocrinology. 2008;149:5577–5591. doi: 10.1210/en.2008-0220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weck J, Anderson AC, Jenkins S, Fallest PC, Shupnik MA. Divergent and composite gonadotropin-releasing hormone-responsive elements in the rat luteinizing hormone subunit genes. Mol Endocrinol. 2000;14:472–485. doi: 10.1210/mend.14.4.0453. [DOI] [PubMed] [Google Scholar]
- Winnay JN, Hammer GD. Adrenocorticotropic hormone-mediated signaling cascades coordinate a cyclic pattern of steroidogenic factor 1-dependent transcriptional activation. Mol Endocrinol. 2006;20:147–166. doi: 10.1210/me.2005-0215. [DOI] [PubMed] [Google Scholar]
- Wolfe MW, Call GB. Early growth response protein 1 binds to the luteinizing hormone-beta promoter and mediates gonadotropin-releasing hormone-stimulated gene expression. Mol Endocrinol. 1999;13:752–763. doi: 10.1210/mend.13.5.0276. [DOI] [PubMed] [Google Scholar]
- Yokoi T, Ohmichi M, Tasaka K, Kimura A, Kanda Y, Hayakawa J, Tahara M, Hisamoto K, Kurachi H, Murata Y. Activation of the luteinizing hormone beta promoter by gonadotropin-releasing hormone requires c-Jun NH2-terminal protein kinase. J Biol Chem. 2000;275:21639–21647. doi: 10.1074/jbc.M910252199. [DOI] [PubMed] [Google Scholar]
- Zhao L, Bakke M, Krimkevich Y, Cushman LJ, Parlow AF, Camper SA, Parker KL. Steroidogenic factor 1 (SF1) is essential for pituitary gonadotrope function. Development. 2001;128:147–154. doi: 10.1242/dev.128.2.147. [DOI] [PubMed] [Google Scholar]
- Zhao L, Bakke M, Hanley NA, Majdic G, Stallings NR, Jeyasuria P, Parker KL. Tissue-specific knockouts of steroidogenic factor 1. Mol Cell Endocrinol. 2004;215:89–94. doi: 10.1016/j.mce.2003.11.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.