Abstract
Purpose
Efforts to validate ovarian cancer early detection biomarkers with immunoassays are challenged by limited specimen volumes available. We sought to develop a specimen-efficient assay to measure CA125 in serum, assess its reproducibility, validity and performance, and test its potential for multiplexing and combining with HE4, a promising novel ovarian cancer marker.
Experimental Design
Four pairs of commercially available anti-CA125 antibodies and one pair of anti-HE4 antibodies were evaluated for accuracy in measuring known concentrations of antigen on a bead-based platform. Two best pairs were further assessed for reproducibility, validity, and ability to discriminate between blinded serum samples obtained from ovarian cancer cases (n=66) and women without ovarian cancer (n=125).
Results
Suitability for use in a bead-based assay varied across CA125 antibody pairs. Two CA125 bead-based assays were highly reproducible (overall correlations between replicates ≥0.95; CV’s below 0.2) and strongly correlated with the research standard CA125II RIA (correlations ≥0.9). Their ability to distinguish ovarian cancer cases from non-cases based on ROC analyses (AUCs of 0.85 and 0.84) was close to that of the CA125II RIA (AUC of 0.87). The HE4 bead-based assay showed lower reproducibility but yielded an AUC of 0.89 in ROC analysis. Multiplexing was not possible but a composite marker including CA125 and HE4 achieved an AUC of 0.91.
Conclusion
Optimization procedures yielded two bead-based assays for CA125 that perform comparably to the standard CA125II RIA, can be combined with an HE4 bead-based assay to improve diagnostic performance, and require only 15μl of sample each.
Keywords: CA125, bead-based ELISA assay, ovarian cancer, early detection, HE4
Introduction
Many candidate markers are being evaluated for their use in an early detection biomarker panel for ovarian cancer [1], but they have not yet been evaluated in pre-clinical samples obtained 1+ years prior to diagnosis because such specimens are very precious. CA125, a high molecular weight glycoprotein recognized by antibodies belonging to only three epitope-groups [2–4], is elevated in most women with ovarian cancer [5]. It has been extensively studied [6–10] and is likely to be included in serum marker panels that are proposed for validation in pre-clinical samples. Quantification of serum CA125 levels is currently based on heterologous assays using two monoclonal antibodies (mAb) directed against the epitope groups M11 and OC125, in contrast with the original homologous assay using only one mAb directed against the OC125-like epitope [11]. The inadequate sensitivity of CA125 for early stage disease and its poor specificity to malignancy limit its use for population screening [12–15]. Adding one or several markers to CA125 for use as a composite marker (CM) would improve performance in a screening program if sensitivity were improved with no loss in specificity [16–18] and stability over time yielded better performance in a longitudinal algorithm [16].
The lead time of a marker is critical, as it measures the marker’s ability to identify disease early in the disease process. Repositories developed by the Carotene and Retinol Efficacy Trial (CARET) [19], the Women’s Health Initiative (WHI) [20], and the Prostate, Lung, Colon and Ovary Cancer Screening Trial (PLCO) [21] have pre-clinical samples for a relatively large number of cases for whom blood samples were collected well in advance of diagnosis, making it possible to estimate the lead time of candidate serum biomarkers. Because of the value and scarcity of these resources, however, use of these specimens must be well-justified and optimized. The research standard CA125 radioimmunoassay (RIA) CA125II from Fujirebio Diagnostics, Inc (FDI, Malvern, PA) requires 0.2 ml of serum sample, limiting the number of other candidates that can be evaluated simultaneously in a typical research sample of 0.5 ml. Some clinical assays require less specimen, but they yield results that vary by type, manufacturer and generation [22]. Our goal was to develop a cost-effective research-quality assay for CA125 that would require only a few microliters of serum and enable us to explore the potential for multiplexing and/or combining CA125 with novel markers such as HE4 [23, 24] for use in a CM and a longitudinal algorithm.
Bead-based ELISA assays require relatively small volumes of sample material [25, 26]. The technology derives from sandwich ELISA assays but uses spectrally discrete polystyrene beads, or microspheres, instead of plastic surfaces to immobilize the capture antibody. Bio-Rad Laboratories, Inc., (Hercules, CA) commercializes carboxy-coated microspheres internally labeled with two fluorescent dyes that produce up to 100 different spectral addresses, and a Multiplex Suspension Array System (Luminex 110S system) whose reading system (Bio-Plex reader) functions on the same principles as a flow cytometer. Similar to a sandwich ELISA assay, each antibody-coupled microsphere captures antigens that are detected with a biotinylated antibody and phycoerythrin-conjugated streptavidin (SA-PE). For each capture antibody-coupled microsphere, the Bio-Plex reader simultaneously measures the fluorescent signals of the microsphere particular spectral address and of the SA-PE. Each mean fluorescence intensity (MFI) reading corresponds to the average of the fluorescent signals from 100 antibody-coupled microspheres of a particular spectral address; duplicate measurements are not required. In the absence of cross-reactivity, each reading can assess the concentration of multiple markers that are detected by spectrally distinct beads, further diminishing the sample volume needed.
We developed CA125 bead-based assays using commercially available antibodies directed against the M11-like or the OC125-like epitopes, and an HE4 bead-based assay using the only available antibody pair for HE4 [13, 24]. Evaluation of the candidate bead-based assays was based on several objectives and criteria. First, laboratory analyses were performed to assess the antibody binding to the beads, the antigen affinity of the capture antibody after immobilization on the beads, and the antibody performance as a sandwich pair against known concentrations of antigen using purified antigen and selected sera. The top two performing CA125 antibody pairs in the bead-based assays were then evaluated for multiplexing with HE4, and the best assays were evaluated in patient sera for their reproducibility, validity, and performance relative to the standard CA125II RIA. As the ultimate objective is to be able to identify ovarian cancer cases early, the bead-based assays were also evaluated for their temporal stability in serial blood samples [27]. Screening decisions based on CA125 most commonly use a single-threshold screening rule that prompts referral for follow up when CA125 values are found above a normal reference range, but performance can be improved by use of a longitudinal algorithm when a marker is stable over time within women [28].
Materials and Methods
1) Overall strategy to develop, calibrate and evaluate the bead-based assays
In the first phase of the work, we compared the performance of five anti-CA125 antibody couples (three antibody pairs in only one orientation and one antibody pair in both orientations) in a bead-based ELISA platform. All capture antibodies were coupled to the microspheres at five different concentrations and each coupling was validated by direct detection with a goat anti-mouse antibody. After validation, capture antibody-coupled microspheres were incubated with the control antigen diluted in assay buffer or in pooled control sera (CA125 <13 U/ml as measured by CA125II RIA) diluted four, twenty, fifty or two hundred fold. The antigen binding was detected with a biotin-conjugated antibody followed by SA-PE (Bio-Rad Laboratories) and measured in MFI by the Bio-Plex reader. Antibody pairs that performed well were used to test non-blinded positive and negative control sera as identified by CA125II RIA and diluted four or twenty fold. A bead-based assay was similarly optimized for HE4, using the only available pair of capture and detection antibodies. The potential for multiplexing was explored by combining beads for the HE4 assay with beads for each of the 2 best CA125 assays in a single well, and comparing MFI levels to those obtained without multiplexing for both CA125 and HE4.
In the second phase of the work, the two best pairs of CA125 antibodies and the HE4 antibody pair were analyzed in blinded patient sera. To assess reproducibility, for each of the 3 antibody pairs pooled negative serum and CA125/HE4 positive serum were measured repeatedly over 10 days. Serum replicates were used to estimate coefficients of variation (CV). In addition, 204 patient samples from 191 women with and without ovarian cancer were assayed twice over the same 10 days; reproducibility was assessed by the correlation between the 2 experiments. Results from the first experiment for each CA125 and HE4 antibody pair were also used to correlate the findings to the standard CA125II RIA or HE4 ELISA and to generate Receiver Operating Characteristic (ROC) curves assessing diagnostic accuracy.
2) Mouse Monoclonal Antibodies
Anti-CA125 mAb were acquired from FDI (OC-125 and M11), Fitzgerald Industries International, Inc. (FIII, Concord, MA) (M8072320 and M8072321; M002201 and M002203) and Research Diagnostics, Inc. (RDI, Flanders, NJ) (X306 and X52). M8072320, M002201 and X306 epitopes are OC-125-like while M8072321, M002203 and X52 are M11-like. Manufacturers recommend using M11, X306 and M002201 as capture antibodies. For the HE4 assay and to test multiplexing, one complementary pair of anti-HE4 mAb (3D8 and 2H5) were used [24]. Antibodies were dialyzed against Dulbecco’s phosphate buffered saline (PBS) (Invitrogen Corporation, Carlsbad, CA) when needed. Carboxy-coated microspheres (Bio-Rad Laboratories) were coupled with five concentrations (0.2, 1, 5, 10, and 20 μg/mL) of the capture antibodies or with both antibodies of the same pair if no recommendation was given (FIII M8072320 and M8072321). Detection antibodies were biotinylated using the EZ-Link-sulfo-NHS-biotinylation kit (Pierce, Rockford, IL) according to the manufacturer’s instructions and dialyzed against PBS (Pierce Slide-a-Lyzer, 7kDa MWCO).
3) Study population and serum sample collection
Study participants were recruited between 1999 and 2003 to support protocols of the Pacific Ovarian Cancer Research Consortium (POCRC) by physicians at Pacific Gynecology Specialists (PGS), Swedish Medical Center (SMC), the University of Washington/Seattle Cancer Care Alliance (UW/SCCA), and Virginia Mason Medical Center (VMMC). Cases (n=66) were defined as having invasive epithelial carcinoma confirmed by standardized review of medical records and pathologist examination of paraffin-embedded tissue; histologies represented clinical practice and included 33 serous, 7 endometrioid, 3 mucinous, 3 clear cell, 6 undifferentiated and 14 other. FIGO stage distribution similarly reflected clinical practice, including 11 Stage I, 5 Stage II, 37 Stage III, 11 Stage IV tumors and 2 unknown stages. Non-cases, representing women free of ovarian cancer (n=125), consisted of surgical patients (n=72) and women (n=53) enrolled in screening trials who were free of ovarian cancer 2+ years after serum collection. Thirteen of the screening trial participants provided serial blood samples collected one year apart, yielding 66 blood samples for analysis from the screening population. Participants’ characteristics are reported in Table 1. In addition, a pool of sera from nine healthy post-menopausal women collected in a volunteer blood drive was used as negative control. Each patient provided written informed consent and a medical records release form approved by the FHCRC institutional review board.
Table 1. Participant’s characteristics.
The median, minimum, and maximum ages of the participants, and the median, minimum, and maximum CA125 II RIA values by clinical group.
| N | Age Median | Age Min | Age Max | CA125II Median | CA125II Min | CA125II Max | |
|---|---|---|---|---|---|---|---|
| Cases | 66 | 59 | 37 | 89 | 247.26 | 5.16 | 2797.14 |
| Non-cases (total) | 125 | 55 | 35 | 83 | 12.12 | 3.45 | 1790.39 |
| • Surgical | 72 | 59 | 35 | 83 | 13.65 | 3.45 | 1790.39 |
| • Screening | 53 | 53 | 38 | 81 | 10.25 | 3.77 | 190.5 |
All blood samples were collected in Serum Separator Tubes (SST) tubes (BD Vacutainer, Becton, Dickinson and Company, Franklin Lakes, NJ USA) and processed according to the manufacturer’s instructions. Blood was allowed to coagulate at room temperature for at least thirty minutes but no longer than four hours. The serum was aliquoted and stored at −80°C until analysis (three to five years later). Surgical samples were obtained prior to surgery at a clinic visit or in the operating room. All serum samples were characterized for CA125 with the research standard CA125II RIA (FDI) in duplicate using 0.2 ml of serum as recommended by the manufacturer to support POCRC work [16, 24]. A subset of the patient specimens (n=68, including 6 cases and 62 controls) also had CA125 measurements obtained for a different study using a Microparticle Enzyme Immunoassay (MEIA) (AxSYM CA125 assay, Abbott laboratories, Abbott Park, IL). HE4 serum levels were measured in another subset (n=106, including 35 cases and 71 controls) using sandwich ELISA for a POCRC validation study [18].
4) CA125 and HE4 bead-based immunoassays
Development and evaluation of the bead-based assays were performed in filter plates (Millipore Corporation, Billerica, MA). A vacuum manifold (Millipore) was used to drain reagents and wash steps. All incubations were performed at room temperature, in the dark and on a plate shaker. Carboxy-coated microspheres (Bio-Rad Laboratories) were resuspended at each step by ramping the plate shaker to a speed of 1000 rpm, then reducing to 300 rpm for the incubation. Filter plates were pre-wet with assay buffer prior to adding the microspheres. Capture antibodies were covalently coupled to the microspheres using the Amine Coupling Kit (Bio-Rad Laboratories) according to the manufacturer’s instructions. Briefly, microsphere stock solutions were dispersed by vortexing and water bath sonication (Branson ultrasonic cleaner, Danbury, CT) then checked visually for aggregation. For each coupling, a 100 μl aliquot of 1.25 × 106 monodispersed microspheres was transferred to a coupling reaction tube provided in the kit. The microspheres were washed with the kit bead wash buffer, resuspended in 80 μl of the kit bead activation buffer, vortexed and sonicated. Freshly made solutions of 10 μl of 50 mg/ml 1-ethy-3-(3-dimethylaminopropl)-carbodiimide hydrochloride (EDC, Pierce) and N-hydroxy-sulfosuccinimide (Sulfo-NHS, Pierce) were added sequentially to the coupling tubes to stabilize the reaction and activate the microspheres. After vortexing, the microspheres were incubated rotating for 20 minutes, then washed and resuspended. Various dilutions of capture antibodies were added, the volumes adjusted to 500 μl and the coupling reaction was incubated for two hours. After one wash, the coupled microspheres were blocked with the kit’s blocking buffer for 30 minutes. Finally, microspheres were resuspended in storage buffer, counted using a hemocytometer and stored at 4°C. To test the coupling efficiency, 5,000 microspheres were incubated for 30 minutes with 2μg/ml of biotinylated goat anti-mouse IgG H+L (Invitrogen) followed by three washes and a 10 minute-incubation with SA-PE. Labeled microspheres were washed three times, resuspended in 125μl of assay buffer and analyzed with the Bio-Plex reader. MFI of at least 2,000 was considered an efficient coupling. Our criterion to identify the best pairs of antibodies in the first phase of the work was the greatest ratio between the MFIs measuring 0.1 U/ml and 1000 U/ml of CA125. HE4-Ig fusion recombinant protein was assayed in the same samples on the same plates, independently and multiplexed with each of the CA125 bead-based assays. Criterion for acceptable multiplexing was MFI consistency in the absence and in the presence of reagents for another bead-based assay. All assays of patient samples were run using the same lots of coupled microspheres and biotin-conjugated detection antibodies.
5) Statistical Analyses
All analyses were performed in S-Plus version 6.2 (Insightful, Seattle, WA). Assay reproducibility for the two CA125 bead-based assays and the HE4 assay was assessed by calculating the coefficient of variation (CV) among control replicates and estimating Pearson correlations between repeated experiments. Positive control serum (CA125 level = 231 U/ml as measured by CA125II RIA; HE4 level = 5 ng/ml as measured by ELISA) was measured 19 times in 10 separate experiments, providing 19 replicates for each assay. The pool of negative control serum (CA125 < 7 U/ml by RIA; HE4 undetectable by ELISA) was measured 8 times in triplicate and 11 times alone in 10 separate experiments that included one or two plates, providing 35 replicates for each assay. Among the negative and positive serum controls, an analysis of variance (ANOVA) was used to assess day-to-day variation and plate variations within days. Criteria for inclusion of data were that no errors were reported by BioPlex. CV’s were calculated for positive and negative serum controls, on the raw scale for comparability with published estimates for standard assays. All other analyses were performed after a log transformation was applied to reduce skewness in the distribution. Pearson correlations were estimated using all serum specimens for which repeated results were obtained.
Validity of the CA125 bead-based ELISAs was assessed by correlating their respective CA125 concentrations to those obtained by the standard CA125II RIA in the 204 patient serum samples. Pearson correlation coefficients were calculated overall and within each patient subgroup (cases and non-cases including surgical patients and screening participants). Agreement of the HE4 bead-based assay with traditional ELISA was similarly assessed by Pearson correlation using only the 106 serum samples for which HE4 ELISA results were available [24]. When a sample was dropped due to errors (n=6) it was replaced by its replicate for validity and performance evaluations.
The performance in terms of diagnostic accuracy of the assays was assessed by estimation of ROC curves for cases vs. all non-cases, cases vs. surgical controls, and cases vs. healthy screening controls in 64 cases and 125 noncases (55 screening normals, 70 surgical normals) who reported no history of ovarian cancer and for whom all 4 assay measurements were available, including the three bead-based assays (two CA-125 and HE4) and the CA125II RIA. The ROC curves describe jointly the sensitivity and specificity of the assay as the threshold for a positive test is allowed to vary. The Area Under the Curve (AUC) [29] is reported, where 1.0 represents perfect performance and 0.5 represents what one would expect of chance alone. Equality of the AUC’s was tested using a nonparametric method developed by Delong et al [30]. ROC curves for individual bead-based assays were compared to those for the standard CA125II RIA. In addition, two composite markers (CM) were constructed by combining each of the CA-125 bead-based assays with the HE4 bead-based assay using main effects only obtained by logistic regression [31]; the AUC for each CM was compared to that of CA125II RIA.
To evaluate the stability over time of the markers using bead-based assays, the correlation between two measures made in serum samples collected one year apart are reported for each bead-based assay and for the standard CA125II RIA. Temporal stability of a marker is critical for the exploitation of a longitudinal algorithm in a screening program [28].
Results
1) Determining optimal conditions for each antibody pair
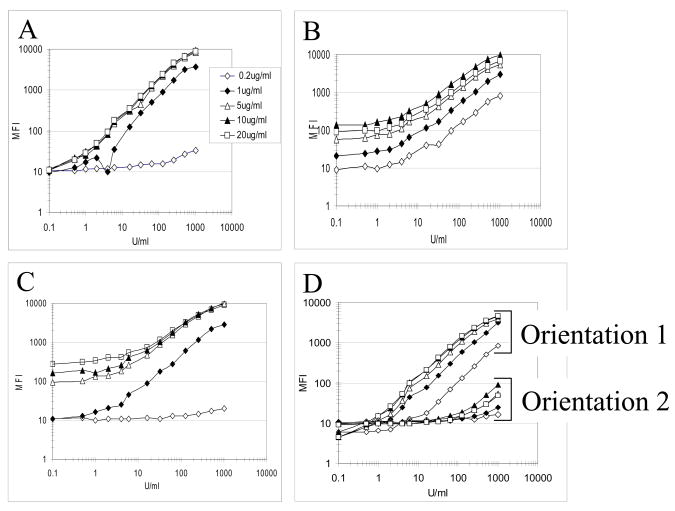
Microspheres coupled to serial dilutions of anti-CA125 and anti-HE4 capture antibodies all surpassed the recommended MFI of 2,000 in validation tests (data not shown). Assays were first performed with 0.1U/ml to 1000U/ml of CA125 antigen purified from human ascites (FIII) and serially diluted in assay buffer or in pooled control sera. Figure 1 shows that the optimal coupling concentration of capture antibodies varied for each antibody and that the orientation of the FIII antibody pair M8072320/21 was critical (compare orientation 1 to 2 in fig. 1D). The addition of serum interfered with the fluorescent signal only when the microspheres were coated with a low concentration of capture antibody (<1 μg/ml or less) (data not shown). Based on a criterion of greatest ratio between the MFIs measuring 0.1 U/ml and 1000 U/ml of CA125 purified antigen, we eliminated the FIII pair with clone M8072321 as capture antibody (fig. 1D orientation 2). Beads coupled with capture antibody concentrations that discriminated best between various antigen concentrations were selected for further tests: RDI X306, 5μg/ml; FDI M11, 1μg/ml; FIII M002201, 1μg/ml; and FIII M8072320, 20μg/ml.
Figure 1. Determination of optimal coupling concentrations with purified CA125 antigen for five pairs of anti-CA125 antibodies.
Five coupling concentrations of capture antibody (0.2, 1, 5, 10 and 20 μg/ml) were incubated with serial dilutions of CA125 antigen in the assay diluent. The captured antigen was detected by 1 μg/ml of biotinylated detection antibody. A: RDI antibodies, X306 capture, X52 detection; B: FDI antibodies, M11 capture, OC-125 detection; C–D: FIII antibodies, C: M002201 capture, M002203 detection; D, orientation 1: M8072320 capture, M8072321 detection; orientation 2: M8072321 capture, M8072320 detection.
For assays with human sera, 15 μl of each serum sample was diluted with 45 μl of human serum diluent (Bio-Rad Laboratories) and capture antibody-coupled microspheres were incubated for 30 minutes with 50 μl of the diluted serum. All four remaining antibody pairs were able to distinguish CA125 positive and two CA125 negative sera (as identified by CA125II RIA) at a concentration of 25% (fig. 2A) or 5% (data not shown). Because the ratios between positive and negative sera were higher with the RDI and FIII M8072320/21 antibody pairs than with the FIII M002201/03 and FDI M11/OC125 pairs, even at the best serum dilution of 25%, we focused further efforts on only two CA125 antibody pairs, RDI and FIII M8072320/21.
Figure 2. Calibration and multiplexing of the bead-based assays.
A- Four sera with known CA125 concentration levels were diluted 4 fold and used to compare four CA125 bead-based assays. All detection antibodies were used at 1 μg/ml. The capture antibodies were immobilized on microspheres at 5 μg/ml for RDI mAb; 20 μg/ml for FIII M8072320 mAb; 1 μg/ml for FDI mAb and 1 μg/ml for FIII M002201 mAb. B- Micropheres were coupled with five concentrations of 2H5 anti-HE4 capture mAb as shown, and incubated with two HE4 positive sera (pos1 and pos 2; HE4 concentrations of 5 ng/ml as determined by ELISA) and two HE4 negative sera (neg 1 and neg 2; HE4 concentrations non detectable by ELISA) diluted 4 fold in assay buffer. HE4 serum levels were detected with 6 μg/ml of 3D8 anti-HE4 biotin-conjugated mAb. C- Optimized HE4 bead-based assay was tested in the absence or in the presence of CA125 bead-based assay reagents and/or in presence of serial dilutions of CA125 purified antigen in assay diluent (as shown: 0; 30; 300; 3000 U/ml). D- CA125 bead-based assay using RDI antibodies was tested in the absence or in the presence of HE4 bead-based assay reagents and/or in presence of serial dilutions of HE4-Ig antigen (as shown: 0; 0.5; 1; 2 μg/ml).
To determine the optimal concentration of detection antibodies X52 (RDI) and M8072321 (FIII), coupled microspheres with the appropriate capture antibody were incubated with four sera characterized by CA125II RIA and two fold serial dilutions of their corresponding detection antibody (1 to 8 μg/ml). The overall MFI were not significantly modified by the different concentrations of detection antibodies (data not shown).
The above results were used to determine the experimental conditions for the rest of the study: the FIII capture antibody M8072320 was coupled at 20 μg/ml and the RDI capture antibody X306 was coupled at 5 μg/ml, while the detection antibodies were used at 1 μg/ml for the FIII antibody M8072321 and at 2 μg/ml for the RDI antibody X52.
Similarly, an HE4 bead-based assay was validated with HE4-Ig fusion recombinant protein and calibrated with four positive and negative control sera characterized by sandwich ELISA test [24]. As previously described, beads coupled to 2H5 anti-HE4 capture antibody were validated with biotinylated goat anti-mouse IgG (data not shown). Figure 2B shows that microspheres coupled with 10μg/ml of 2H5 and detected with 6μg/ml of biotinylated 3D8 discriminated best between the positive and negative control sera.
2) Multiplexing
Multiplexing of CA125 and HE4 bead-based assays was tested with both the FIII and RDI antibody pairs. The HE4 MFI readings were not affected by the presence of the anti-CA125 reagents (fig 2C). However, the MFI of both the RDI (fig 2D) and the FIII (data not shown) CA125 assays increased with the addition of HE4 bead-based assay components, suggesting a cross-reactivity of the anti-HE4 antibodies with the anti-CA125 antibodies. Accordingly, multiplexing was not used for analyses in blinded patient sera.
3) Assay reproducibility
The best CA125 antibody pairs (FIII 8072320/21 and RDI X306/X52) and the HE4 antibody pair were then evaluated in blinded serum from our study population. All the assays were performed with the sera diluted four fold, thus requiring only 0.015 ml of serum per assay per sample. Bead coupling reactions were highly reproducible as demonstrated by validation results with a goat anti-mouse antibody to quantify the mouse antibody bound to the beads (MFI standard deviation <8% for eight couplings performed over 6 months).
Reproducibility was assessed using multiple replicates of negative (n=35) and positive (n=19) serum controls to estimate a CV for each assay. Plate effects assessed by ANOVA included small day-to-day variations found in both sera controls for the CA125 RDI and HE4 assays, and small order effects found both in the positive controls for the CA125 FIII and HE4 assays and in the negative controls for the CA125 RDI and HE4 assays (data not shown). The average CVs for the CA125 RDI and FIII bead-based assays were respectively 18.2 and 15.7 for the pooled control sera and 18.9 and 10.2 for the CA125/HE4 positive sera. The CVs for the HE4 assay were higher, ranging from 26.6 for the positive sera to 32.6 for the pool of negative sera.
For further assessment of reproducibility, blinded samples from191 patients were tested twice in independent experiments conducted identically 1 week apart. Overall, the Pearson correlations for replicates were 0.99, 0.95 and 0.95 for the RDI, FIII and HE4 assays respectively (Table 2). The estimated correlations within subgroups (i.e., cases, surgical controls and screening controls) ranged from 0.64 (FIII screening controls) to 1.00 (RDI cases) providing strong evidence of assay reproducibility, even within the narrower range of antigen levels characteristic of healthy controls. The RDI assay yielded modestly higher correlations between replicates than the FIII assay overall and in all 3 subgroups. Reproducibility for the HE4 bead-based assay was similar, with correlations slightly higher in screening controls (0.86 vs 0.83 and 0.64) but lower in surgical controls (0.90 vs 0.98 and 0.97) (Table 2).
Table 2. Reproducibility and validity of the bead-based assays.
Reproducibility of case and control measurements for the CA125 RDI, CA125 FIII and HE4 bead-based assays was assessed by Pearson correlation (r) between two replicate experiments, calculated on the log scale.
Validity of the CA125 RDI and FIII bead-based assays was assessed by Pearson correlations with the research standard CA125II RIA (CA125 RDI/RIA and CA125 FIII/RIA respectively), calculated on the log scale.
| Reproducibility | Validity | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| CA125 RDI assay | CA125 FIII assay | HE4 assay | CA125 RDI/RIA | CA125 FIII/RIA | |||||
| N | r | N | R | N | r | N | r | r | |
| Overall | 197 | 0.99 | 195 | 0.95 | 194 | 0.95 | 204 | 0.95 | 0.91 |
|
| |||||||||
| Cases | 64 | 1.00 | 64 | 0.93 | 62 | 0.94 | 66 | 0.93 | 0.88 |
|
| |||||||||
| Non-cases (total) | 133 | 0.94 | 131 | 0.89 | 132 | 0.89 | 138 | 0.85 | 0.80 |
| • Surgical | 69 | 0.98 | 68 | 0.97 | 69 | 0.90 | 72 | 0.91 | 0.81 |
| • Screening | 64 | 0.83 | 63 | 0.64 | 63 | 0.86 | 66 | 0.64 | 0.73 |
4) Validity of the bead-based assays: Agreement with the CA125II RIA and HE4 ELISA
The CA125 bead-based assays using either the RDI or the FIII antibody pairs were highly correlated with the standard CA125II RIA (correlations of 0.95 and 0.91 for RDI and FIII respectively) and also highly correlated with each other (correlation = 0.95) (Table 2). Within subgroups, where variability is reduced, agreement remained high, with the RDI assay showing somewhat better agreement with the standard RIA than the FIII assay in cases (0.93 vs 0.88) and surgical controls (0.91 vs 0.81) (Table 2), and the FIII assay showing better agreement with the standard in healthy controls (0.73 vs 0.64). For comparison purposes, the results of the CA125II RIA were also compared to those of the (clinical standard) AxSYM CA125 MEIA assay using the 68 samples (6 cases and 62 controls) for which both measures were available; they yielded an overall correlation of 0.95 but poorer agreement in healthy controls (r=0.57) (data not shown). Correlation between the HE4 bead-based assay and ELISA based on 106 samples (35 cases and 71 controls) was 0.90 overall and in cases, but lower in controls where HE4 variability is very low, ranging from 0.4 (surgical controls) to 0.52 (screening controls) (data not shown).
5) Diagnostic accuracy
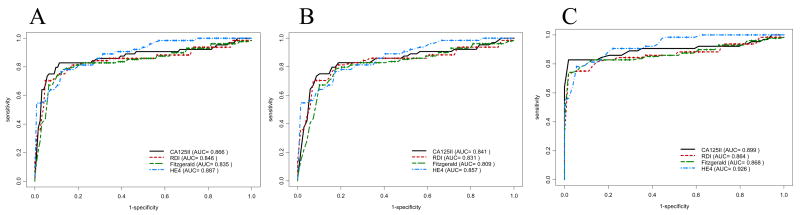
ROC curves describing the ability to distinguish cases from non-cases for each of the two CA125 assays, HE4 and the standard CA125II RIA (fig. 3A–C) suggest that all four assays provide comparable classification ability over the full range of specificity, even in analyses restricting the non-cases to either the surgical controls (fig. 3B) or the screening controls (fig. 3C). AUC statistics ranged from 0.93 for the HE4 assay in cases vs. screening controls to 0.81 for the FIII assay in cases vs. surgical controls, with the HE4 assay consistently providing the highest values followed by the RIA, RDI and FIII assays in that order. Differences in AUC values for the individual markers were not statistically significant.
Figure 3. CA125 and HE4 bead-based assay performance compared to CA125II using ROC.
The ability of the CA125 II RIA to distinguish women with ovarian cancer (cases) from non-cases (A), cases from women who underwent pelvic surgery for reasons unrelated to cancer (B), and cases from a healthy screening population (C) was compared among the two CA125 bead-based assays (RDI and FIII), the HE4 bead-based assay, and the CA125II RIA separately. Total AUC values for each assay are listed in parentheses.
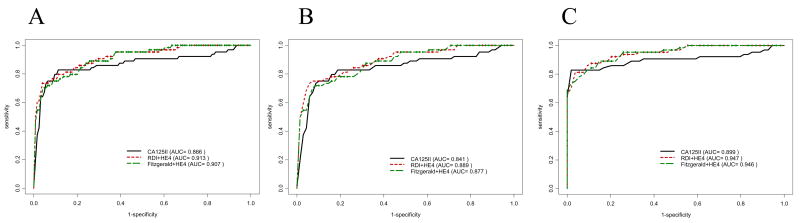
Higher ROC curves were obtained by combining the bead-based HE4 assay with one or the other of the CA125 bead-based assays (fig. 4A–C). Based on cases vs. noncases, logistic regression yielded weights for the RDI/HE4 CM of 0.56 and 1.19 for RDI and HE4 respectively; for the FIII/HE4 CM the weights were 0.42 and 1.27 respectively. The AUC for both FIII/HE4 and RDI/HE4 CM combinations were identical (0.91) and higher than for the standard CA125 RIA (0.87; p=0.032 for RDI, p=0.072 for FIII). Similarly, for cases vs. screening controls (fig. 4C) the AUC for FIII/HE4 and RDI/HE4 CM combinations were identical (0.95) and exceeded that for the CA125II alone (0.90, p = 0.044 for RDI/HE4, p=0.056 for FIII/HE4).
Figure 4. CA125 and HE4 bead-based assay composite marker (CM) performance compared to CA125II using ROC.
Each of the two CA125 bead-based assays (RDI and FIII) was combined with the HE4 bead-based assay to form 2 composite markers (CM). The ability of the CA125II RIA to distinguish women with ovarian cancer (cases) from non-cases (A), cases from women who underwent pelvic surgery for reasons unrelated to cancer (B), and cases from a healthy screening population (C) was compared to each of the CM. Total AUC values for each CM and the RIA are listed in parentheses.
Serial correlations for 13 women with 2 serum samples collected 1 year apart were 0.53 and 0.50 for the RDI and FIII assays respectively, compared to 0.63 for the CA125II RIA and 0.76 for the HE4 assay in the same samples. When an outlier was excluded, serial correlation for the HE4 assay was reduced to 0.51. The outlier is a false positive that is included in the ROC analyses, reducing the performance of HE4.
Discussion
The bead-based ELISA platform was developed to measure multiple markers in small sample volumes and thus is ideally suited for large validation studies. We conducted rigorous analyses to identify CA125 antibody pairs that could be used in a bead-based assay, and evaluated their performance with respect to reproducibility, validity and diagnostic performance.
At least two of the four commercially available antibody pairs (RDI X306/X52 and FIII M8072320/21) are suitable for the bead-based platform. Orientation of capture and detection antibodies was important, and not necessarily consistent with the manufacturer’s reported results in sandwich test ELISA. Prediction programs based on 3-dimensional protein structures as well as new technologies to rapidly isolate numerous antibodies against each candidate marker may be needed to realize fully the potential of the bead-based assay platform. In the absence of better tools to predict the best antibodies for bead-based assays, an empirical approach is needed to compare antibody performance.
To explore the potential for multiplexing, and use of a CM, we have also developed an HE4 [24] bead-based assay and tested it with the two CA125 bead-based assays. The HE4 bead-based assay was robust, but the CA125 MFI increased with the addition of the HE4 bead-based assay reagents suggesting an interaction of the anti-HE4 antibodies with the CA125 assay. Interestingly, the change was unilateral and resulted in an increase of signal that is inconsistent with competition. It might be possible that conformational changes impact the MFI reading. For example, complex formation would yield a larger protein volume, changing the distance between the detection antibody and the beads and perhaps improving the fluorescent signal spatial location for the BioPlex reader. Non-specific binding of the anti-HE4 reagents to CA125, a very large glycoprotein, could produce complex formation. The interaction between CA125 and the c-terminal domain of mesothelin [32], another putative serum marker for ovarian cancer [33], could be an additional source of conformational changes. Alternative antibodies may be needed against markers that are to be multiplexed with CA125.
The CA125 and HE4 bead-based assays are acceptably reproducible and valid, and provide excellent diagnostic accuracy. CV’s for the RDI and FIII CA125 assays were below 0.2; those for the HE4 bead-based assay were about 0.30. These are higher than CV’s for the research standard CA125II RIA (below 0.1). Plate effects were present, suggesting that CV’s could be reduced by the inclusion on each plate of a standard curve, as is done in kits such as the CA125II RIA. Reproducibility of the assays based on repeated experiments in patient sera suggests that the RDI CA125 bead-based assay is as consistent with the CA125II RIA as the AxSYM CA125 MEIA assay that is used routinely in clinical practice (r=0.95 for both). Importantly, the CA125 bead-based assays were equivalent in their disease classification of patient samples as assessed by ROC methods. Correlations of HE4 bead-based assay results with sandwich ELISA test results were above 0.9 overall, and ROC curves for the HE4 bead-based assay were higher than any of the other assays including the research standard CA125II RIA. These results contribute to a growing body of evidence that HE4 is a promising biomarker for early detection of ovarian cancer [34].
Based on tissue arrays, HE4 has been recently reported to complement expression of CA125 in epithelial ovarian cancer [35], but this is the first report that HE4 complements CA125 in serum as well as the first report of a bead-based assay for HE4. Because the CM was estimated using the same data that were used to generate the ROC curves, a subset of which had been used in an earlier report [24], the results reported here must be interpreted as a first report of a panel that includes HE4 and CA125 rather than as a validation study. More research is needed, particularly in preclinical samples, to confirm that HE4 complements CA125 and that a CM including both markers has better performance than CA125 alone.
We conclude from these analyses that bead-based assays can potentially perform as well as traditional sandwich ELISA tests, and specifically that use of the RDI antibody pair to measure CA125 in preclinical samples will yield estimates that are comparable to those obtained from the research standard CA125II RIA. As the RIA kit requires at least 200μl of serum, compared to 15μl for the bead-based assay, the bead-based assay is preferred for its efficient use of specimen in settings where specimen volume is an important consideration. Although multiplexing would further reduce specimen requirements, it is not necessary given the low specimen requirements of individual assays in the bead-based platform and should not be used without rigorous testing to document absence of cross-reactivity.
Acknowledgments
Financial Support. This work was supported by the National Institutes of Health/National Cancer Institute (P50 CA83636) and the Canary Fund.
We are thankful to Irena King for coordinating the CA125 screenings by RIA and MEIA, to Martin McIntosh for helpful comments and discussions, and to Alicia Young for early contribution to the statistical work.
- FIGO
International Federation of Gynecology and Obstetrics
References
- 1.Bast RC, Jr, Urban N, Shridhar V, et al. Early detection of ovarian cancer: promise and reality. Cancer Treat Res. 2002;107:61–97. doi: 10.1007/978-1-4757-3587-1_3. [DOI] [PubMed] [Google Scholar]
- 2.Nap M, Vitali A, Nustad K, et al. Immunohistochemical characterization of 22 monoclonal antibodies against the CA125 antigen: 2nd report from the ISOBM TD-1 Workshop. Tumour Biol. 1996;17:325–31. [PubMed] [Google Scholar]
- 3.Nustad K, Bast RC, Jr, Brien TJ, et al. Specificity and affinity of 26 monoclonal antibodies against the CA 125 antigen: first report from the ISOBM TD-1 workshop. International Society for Oncodevelopmental Biology and Medicine. Tumour Biol. 1996;17:196–219. doi: 10.1159/000217982. [DOI] [PubMed] [Google Scholar]
- 4.Hovig E, Rye PD, Warren DJ, Nustad K. CA 125: the end of the beginning. Tumour Biol. 2001;22:345–7. doi: 10.1159/000050637. [DOI] [PubMed] [Google Scholar]
- 5.Kenemans P, Yedema CA, Bon GG, von Mensdorff-Pouilly S. CA 125 in gynecological pathology--a review. Eur J Obstet Gynecol Reprod Biol. 1993;49:115–24. doi: 10.1016/0028-2243(93)90135-y. [DOI] [PubMed] [Google Scholar]
- 6.Tamakoshi K, Kikkawa F, Hasegawa N, et al. Clinical value of a new serum tumor marker, CA125II, in gynecologic disease: comparison with CA125. Gynecol Obstet Invest. 1995;39:125–9. doi: 10.1159/000292393. [DOI] [PubMed] [Google Scholar]
- 7.Dorum A, Kristensen GB, Abeler VM, Trope CG, Moller P. Early detection of familial ovarian cancer. Eur J Cancer. 1996;32A:1645–51. doi: 10.1016/0959-8049(96)00137-2. [DOI] [PubMed] [Google Scholar]
- 8.Eagle K, Ledermann JA. Tumor Markers in Ovarian Malignancies. Oncologist. 1997;2:324–9. [PubMed] [Google Scholar]
- 9.Fures R, Bukovic D, Hodek B, Klaric B, Herman R, Grubisic G. Preoperative tumor marker CA125 levels in relation to epithelial ovarian cancer stage. Coll Antropol. 1999;23:189–94. [PubMed] [Google Scholar]
- 10.Jacobs IJ, Menon U. Progress and challenges in screening for early detection of ovarian cancer. Mol Cell Proteomics. 2004;3:355–66. doi: 10.1074/mcp.R400006-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Bast RC, Jr, Feeney M, Lazarus H, Nadler LM, Colvin RB, Knapp RC. Reactivity of a monoclonal antibody with human ovarian carcinoma. J Clin Invest. 1981;68:1331–7. doi: 10.1172/JCI110380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bast RC, Jr, Xu FJ, Yu YH, Barnhill S, Zhang Z, Mills GB. CA 125: the past and the future. Int J Biol Markers. 1998;13:179–87. doi: 10.1177/172460089801300402. [DOI] [PubMed] [Google Scholar]
- 13.Urban N, McIntosh MW, Andersen M, Karlan BY. Ovarian cancer screening. Hematol Oncol Clin North Am. 2003;17:989–1005. ix. doi: 10.1016/s0889-8588(03)00063-7. [DOI] [PubMed] [Google Scholar]
- 14.Moss EL, Hollingworth J, Reynolds TM. The role of CA125 in clinical practice. J Clin Pathol. 2005;58:308–12. doi: 10.1136/jcp.2004.018077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fung MF, Bryson P, Johnston M, Chambers A. Screening postmenopausal women for ovarian cancer: a systematic review. J Obstet Gynaecol Can. 2004;26:717–28. doi: 10.1016/s1701-2163(16)30643-0. [DOI] [PubMed] [Google Scholar]
- 16.McIntosh MW, Drescher C, Karlan B, et al. Combining CA 125 and SMR serum markers for diagnosis and early detection of ovarian carcinoma. Gynecol Oncol. 2004;95:9–15. doi: 10.1016/j.ygyno.2004.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baron AT, Boardman CH, Lafky JM, et al. Soluble epidermal growth factor receptor (sEGFR) [corrected] and cancer antigen 125 (CA125) as screening and diagnostic tests for epithelial ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:306–18. doi: 10.1158/1055-9965.EPI-04-0423. [DOI] [PubMed] [Google Scholar]
- 18.Gorelik E, Landsittel DP, Marrangoni AM, et al. Multiplexed immunobead-based cytokine profiling for early detection of ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:981–87. doi: 10.1158/1055-9965.EPI-04-0404. [DOI] [PubMed] [Google Scholar]
- 19.Goodman GE, Thornquist MD, Balmes J, et al. The Beta-Carotene and Retinol Efficacy Trial: incidence of lung cancer and cardiovascular disease mortality during 6-year follow-up after stopping beta-carotene and retinol supplements. J Natl Cancer Inst. 2004;96:1743–50. doi: 10.1093/jnci/djh320. [DOI] [PubMed] [Google Scholar]
- 20.Anderson GL. Implementation of the Women’s Health Initiative Study Design. Ann Epidemiol. 2003;13:S5–17. doi: 10.1016/s1047-2797(03)00043-7. [DOI] [PubMed] [Google Scholar]
- 21.Hayes RB, Reding D, Kopp W, et al. Etiologic and early marker studies in the prostate, lung, colorectal and ovarian (PLCO) cancer screening trial. Control Clin Trials. 2000;21:349S–55S. doi: 10.1016/s0197-2456(00)00101-x. [DOI] [PubMed] [Google Scholar]
- 22.Pauler DK, Menon U, McIntosh M, Symecko HL, Skatesn SJ, Jacobs IJ. Factors influencing serum CA125II levels in healthy postmenopausal women. Cancer Epidemiol Biomarkers Prev. 2001;10:489–93. [PubMed] [Google Scholar]
- 23.Schummer M, Ng WV, Bumgarner RE, Nelson PS, et al. Comparative hybridization of an array of 21,500 ovarian cDNAs for the discovery of genes overexpressed in ovarian carcinomas. Gene. 1999;238:375–85. doi: 10.1016/s0378-1119(99)00342-x. [DOI] [PubMed] [Google Scholar]
- 24.Hellstrom I, Raycraft J, Hayden-Ledbetter M, et al. The HE4 (WFDC2) protein is a biomarker for ovarian carcinoma. Cancer Res. 2003;63:3695–700. [PubMed] [Google Scholar]
- 25.Oliver KG, Kettman JR, Fulton RJ. Multiplexed analysis of human cytokines by use of the FlowMetrix system. Clin Chem. 1998;44:2057–60. [PubMed] [Google Scholar]
- 26.Vignali DA. Multiplexed particle-based flow cytometric assays. J Immunol Methods. 2000;243:243–55. doi: 10.1016/s0022-1759(00)00238-6. [DOI] [PubMed] [Google Scholar]
- 27.McIntosh MW, Urban N, Karlan B. Generating longitudinal screening algorithms using novel biomarkers for disease. Cancer Epidemiol Biomarkers Prev. 2002;11:159–66. [PubMed] [Google Scholar]
- 28.McIntosh MW, Urban N. A parametric empirical Bayes method for cancer screening using longitudinal observations of a biomarker. Biostatistics. 2003;4:27–40. doi: 10.1093/biostatistics/4.1.27. [DOI] [PubMed] [Google Scholar]
- 29.Bamber D. The area above the ordinal dominance graph and the area below the receiver operating characteristic graph. Journal of Mathematical Psychology. 1975;12:387–415. [Google Scholar]
- 30.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–45. [PubMed] [Google Scholar]
- 31.McIntosh MW, Pepe MS. Combining several screening tests: optimality of the risk score. Biometrics. 2002;58:657–64. doi: 10.1111/j.0006-341x.2002.00657.x. [DOI] [PubMed] [Google Scholar]
- 32.Rump A, Morikawa Y, Tanaka M, et al. Binding of ovarian cancer antigen CA125/MUC16 to mesothelin mediates cell adhesion. J Biol Chem. 2004;279:9190–8. doi: 10.1074/jbc.M312372200. [DOI] [PubMed] [Google Scholar]
- 33.Scholler N, Fu N, Yang Y, et al. Soluble member(s) of the mesothelin/megakaryocyte potentiating factor family are detectable in sera from patients with ovarian carcinoma. Proc Natl Acad Sci U S A. 1999;96:11531–6. doi: 10.1073/pnas.96.20.11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drapkin R, von Horsten HH, Lin Y, et al. Human epididymis protein 4 (HE4) is a secreted glycoprotein that is overexpressed by serous and endometrioid ovarian carcinomas. Cancer Res. 2005;65:2162–9. doi: 10.1158/0008-5472.CAN-04-3924. [DOI] [PubMed] [Google Scholar]
- 35.Rosen DG, Wang L, Atkinson JN, et al. Potential markers that complement expression of CA125 in epithelial ovarian cancer. Gynecol Oncol. 2005;99:267–77. doi: 10.1016/j.ygyno.2005.06.040. [DOI] [PubMed] [Google Scholar]