Abstract
Objective
To discover genes differentially expressed in the perichondrium of the mandibular condylar cartilage (MCC) that might enhance regenerative medicine or orthopedic therapies directed at the tissues of the temporomandibular joint
Design
We used targeted gene arrays (osteogenesis, stem cell) to identify genes preferentially expressed in the perichondrium (PC) and the cartilaginous (C) portions of the MCC in 2 day-old mice
Results
Genes with higher expression in the PC sample related to growth factor ligand-receptor interactions (FGF-13 (6.4X), FGF-18 (4X), NCAM (2X); PGDF receptors, TGF-β, and IGF-1), the Notch isoforms (especially Notch 3 and 4) and their ligands, or structural proteins/ proteoglycans (collagen XIV (21X), collagen XVIII (4X), decorin (2.5X)). Genes with higher expression in the C sample consisted mostly of known cartilage-specific genes (aggrecan (11X), procollagens X (33X), XI (14X), IX (4.5X), Sox 9 (4.4X), and Indian hedgehog (6.7X)). However, the functional or structural roles of several genes that were expressed at higher levels in the PC sample are unclear (myogenic factor 9 (9X), tooth-related genes such as tuftelin (2.5X) and dentin sialophosphoprotein (1.6X), VEGF–B (2X) and its receptors (3–4X), and sclerostin (1.7X)).
Conclusions
FGF, Notch, and TGF-β signaling may be important regulators of MCC proliferation and differentiation; the relatively high expression of genes such as myogenic factor 6 and VEGF–B and its receptors suggests a degree of unsuspected plasticity in PC cells.
Keywords: mouse, temporomandibular joint, perichondrium, gene expression, gene array
Introduction
The mandibular condylar cartilage (MCC) first appears as a condensation of cells adjacent to the periosteum of the mandible around the seventh or eight week in utero. Over the course of the next four to six weeks, a synovial joint develops that is the only site of articulation between the skull and jaw (except for the dentition) and also a major site of growth for the mandible. This origin of the MCC as a secondary cartilage derived from the periosteum of intramembranous bone has been well-documented in the embryological literature (1–3) and its potential implications for the regulation of mandibular growth have been exhaustively debated in the orthodontic literature (4–7). However, attempts to exploit this peculiar developmental history for therapeutic purposes have been impaired by our relatively limited understanding of MCC cell biology. Of central importance are the cells of the prechondroblastic layer deep within the perichondrium, since they (and not the differentiated chondrocytes as in a growth plate) are the locus of nearly all cell divisions in the MCC (8–10).
One of the earliest investigations of the properties of these cells was performed by Stutzmann and Petrovic (11), who published numerous studies supporting the view that orthopedic appliances that altered the postural position of the mandible could stimulate proliferation in MCC prechondroblastic cells leading to increased growth in mandibular length and height (4, 12–13). They postulated that the prechondroblastic zone contained cells in two stages of differentiation: an elongated ‘stem-cell’ type called a ‘skeletoblast’ which divides infrequently and a ‘true prechondroblast’, a rounded cell that divides more frequently. They further proposed that ‘skeletoblasts’ were bipotent (i.e., they would normally differentiate into preosteoblasts, but could develop into ‘true prechondroblasts’ with appropriate biomechanical/ functional stimulation), whereas ‘true prechondroblasts’ had only chondrogenic potential.
Although Petrovic and associates subsequently published data contrasting intracellular calcium levels and concentrations of fibronectin, transglutaminase and heparin sulfate between skeletoblasts and prechondroblasts (11), their work essentially predated the introduction of molecular biological techniques that might have permitted further investigation of prechondroblastic layer cells. Similarly, their characterization of MCC ‘skeletoblasts’ as “fibroblast-like pluripotential stem-cells [italics mine] derived from the embryonic mesenchymal cell” (13) has lost operationality in the succeeding decades of sophisticated applications of embryonic and adult stem cell populations for regenerative medicine. Therefore, their seminal work left important questions unanswered: Are a subset of the cells of the prechondroblastic layer ‘true’ stem cells or something else? If not, how differentiated are they? Although they have repeatedly been shown to be bipotent, are they pluripotent? What factors are of importance for regulating their proliferation and differentiation?
Although Petrovic and associates subsequently published data contrasting intracellular calcium levels and concentrations of fibronectin, transglutaminase and heparin sulfate between skeletoblasts and prechondroblasts (11), their work essentially predated the introduction of molecular biological techniques that might have permitted further investigation of prechondroblastic layer cells. Similarly, their characterization of MCC ‘skeletoblasts’ as “fibroblast-like pluripotential stem-cells [italics mine] derived from the embryonic mesenchymal cell” (13) has lost operationality in the succeeding decades of sophisticated applications of embryonic and adult stem cell populations for regenerative medicine. Therefore, their seminal work left important questions unanswered: Are a subset of the cells of the prechondroblastic layer ‘true’ stem cells or something else? If not, how differentiated are they? Although they have repeatedly been shown to be bipotent, are they pluripotent? What factors are of importance for regulating their proliferation and differentiation?
Cell culture could be a powerful tool for exploring the potential of prechondroblastic cells from the MCC, but the heterogeneity of cell types in or adjacent to the MCC (fibroblasts, prechondroblasts, non-hypertrophic and hypertrophic chondrocytes, osteoblasts/ osteoclasts) has proven a challenge to obtaining a relatively homogeneous culture of prechondroblastic cells. A recurrent theme in these attempts has been the diversity of cell types in the resulting cultures derived from postnatal rodent, rabbit, or primate MCC (14–16). Moreover, most efforts have first removed the perichondrium by mechanical dissection or enzymatic digestion in order to focus on the chondrocytes. The closest attempt to study the prechondroblastic cells in isolation was an explant culture of the prechondroblastic layer isolated from neonatal mice MCC (17), but this study was structural rather than biochemical or molecular in nature. Numerous studies have employed explant culture of MCC with or without attached mandibles (18–24), but this approach limits the cellular/ molecular techniques that can be utilized.
Despite these impediments, several studies over the last decade using a variety of experimental approaches and transgenic animal strains have begun to better define the lineage of prechondroblastic cells and to illuminate potential regulatory genes. Careful study of the developing MCC in rodents has revealed that the future condyle develops from a condensation of alkaline phosphatase-positive cells that are continuous anteriorly with the alkaline phosphatase-positive periosteum of the mandible (25). This suggests that these cells are not truly mesenchymal in character, but have already differentiated into periosteum-like cells that may still be bipotent between osteogenic and chondrogenic lineages, as proposed by Petrovic and associates (4). In the developing MCC, the bipotentiality of prechondroblastic cells is exemplified by their expression of both mRNA for osteogenic lineage markers such as type I collagen, Runx2, and Osterix, and mRNA for Sox 9, a marker for chondrogenic differentiation (26). Thus, the MCC appears to arise from a periosteum, albeit an ‘immature’ one, and that periosteum can be transformed into a perichondrium under some circumstances. Notch1 and Twist, known as cell fate mediators in a variety of tissues, are both expressed largely in the prechondroblastic layer in the developing MCC (27–28), and expression levels of these factors may also play a role in the differentiation pathway.
Although prechondroblastic cells are bipotent, it is perhaps not surprising that their osteogenic lineage is primary in light of their periosteal derivation. Experiments in secondary cartilage on the intramembranous bones of the chick suggest that movement/ articulation is necessary for diverting the otherwise osteogenic precursors to chondrogenesis (29). This osteogenic bias is further evidenced by the fact that mice genetically altered so as not to express the osteogenic lineage precursor Runx2 do not develop a mandibular condylar cartilage (30). Viewed in this context, prechondroblastic cells of the MCC are clearly not ‘stem cell-like’ in the current usage of this term. They represent pre-osteogenic cells diverted to chondrogenesis in the region of articulation between two bones. However, we know relatively little about differences in gene expression between this periosteum turned perichondrium and the underlying cartilage layers.
The goal of this study was to identify genes that are differentially expressed in the perichondrium or cartilaginous portions of the developing MCC to guide future studies of growth regulation and tissue regeneration. Although limited comparisons of gene expression have been performed contrasting cell layers in the growth plate (31) or intersutural tissue from different sutures (32), to our knowledge no investigation of this sort has been attempted for different zones of the MCC.
Materials and Methods
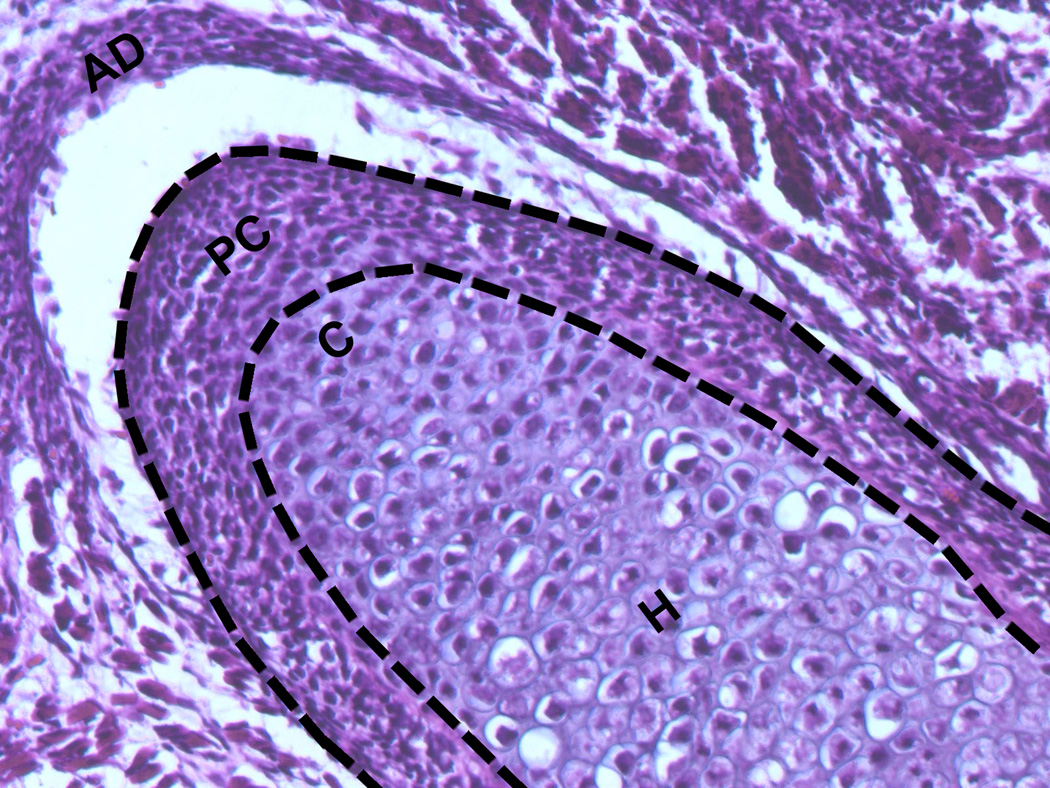
The mandibular condyle and adjacent ramus were dissected from two day-old CD-1 mouse pups. This age was chosen because the MCC was larger than in late embryonic stage pups, but still permitted the perichondrium to be removed with relative ease. Under a dissecting microscope, the perichondrium (PC) was gently teased away from the underlying cartilage (Fig. 1) and the cartilage (C) was separated from the bone. The PC and C samples were then snap frozen in liquid nitrogen. RNA was extracted from pooled samples of around 50 tissues using the RNEasy Micro RNA Isolation Kit (Qiagen, Valencia, CA). The quantity and quality of mRNA were measured by an Agilent 2100 Bioanalyzer.
Figure 1.
Coronal section of embryonic day 18 (E18) mouse mandibular condylar cartilages illustrating perichondrial (PC) and cartilaginous (C) portions of the tissue. Dashed lines delimit tissue removed for the PC sample. AD, articular disc. Hematoxylin and eosin, 20X.
The RNA samples were then analyzed using the Mouse Osteogenesis RT² Profiler™ PCR Array (SuperArray PAMM-026), which profiles the expression of 84 genes related to osteogenic differentiation. In a separate experiment, additional PC and MC samples were analyzed using the Mouse Stem Cell RT² Profiler™ PCR Array (SuperArray PAMM-405), which profiles the expression of 84 genes related to the identification, growth and differentiation of stem cells. Genes were considered to be differentially expressed if they were expressed at least 1.5 times higher in either the PC or C sample.
Results
The Osteogenesis and Stem Cell arrays identified 22 and 26 genes, respectively, that showed higher expression in the PC sample relative to the C sample (Table 1 and Table 2). The highest expression was noted for type XIV collagen (21X), myogenic factor 6 (9X), fibroblast growth factor-13 or FGF-13 (6.4X), followed by several genes in the 3–4X range (collagens IV, VIII, and XVIII; Notch 3 and 4, and cadherins 9, 13, and 15). The Osteogenesis and Stem Cell arrays identified 13 and 20 genes, respectively, that showed higher expression in the C sample relative to the PC sample (Table 3 and Table 4). The highest expression was noted for types XI and X procollagen (14X and 33X), aggrecan (11X), bone morphogenetic protein (BMP) 7 and 8 (8X and 10X), Indian hedgehog (6.7X), matrix metalloproteinase (MMP) 13 (5.9X), and osteopontin (5.3X), followed by several genes in the 3–4X range (procollagen IX, Sox 9, MMP 9, and vitamin D receptor). Most of these genes are characteristic of cartilage as a tissue or typically expressed at high levels in cartilage. Other genes that were over-expressed in the C sample at levels between 3–5X included Wnt inhibitory factor 1 or WIF1, tubulin beta-3, snail 1, frizzled homolog 1, cadherin 2, and bone sialoprotein.
Table 1.
Genes with higher expression in the perichondrium identified by SuperArray™ Osteogenesis Array.
| Gene | Ratio* | Gene | Ratio |
|---|---|---|---|
| Alkaline phosphatase 2 | 1.7 | Growth differentiation factor 10 | 2.0 |
| Bigylcan | 1.5 | Insulin-like growth factor 1 | 1.9 |
| BMP-4 | 1.8 | Matrix metalloproteinase 2 | 1.6 |
| Procollagen XIV | 20.6 | Sclerostin | 1.7 |
| Procollagen XVIII | 3.6 | SPARC | 1.5 |
| Procollagen IV | 4.1 | Tuftelin interacting protein 11 | 1.6 |
| Procollagen VI | 1.5 | TGF-β3 | 1.9 |
| Procollagen VIII | 3.3 | TGF-βr2 | 2.6 |
| Decorin | 2.4 | Tuftelin 1 | 2.5 |
| Dentin sialophosphoprotein | 1.6 | Vascular cell adhesion molecule 1 | 2.1 |
| FMS-like tyrosine kinase | 5.1 | VEGF-B | 2.1 |
Ratio refers to gene expression in the perichondrial sample divided by gene expression in the cartilage sample. Only ratios greater than or equal to 1.5 are included.
Table 2.
Genes with higher expression in the perichondrium identified by SuperArray™ Stem Cell Array.
| Gene | Ratio | Gene | Ratio |
|---|---|---|---|
| ATP-binding cassette | 2.5 | Kinase insert domain protein (VEGF receptor) |
4.3 |
| Actin, alpha, cardiac | 5.7 | Myogenic factor 5 | 1.5 |
| Aldehyde dehydrogenase | 4.2 | Myogenic factor 6 | 9.1 |
| Cadherin 13 (H-cadherin) | 3.4 | MYST histone acetyltransferase 4 | 1.8 |
| Cadherin 15 (M-cadherin) | 4.4 | Neural cell adhesion molecule 1 (NCAM) |
1.8 |
| Cadherin 9 (T-cadherin) | 4.7 | Notch 1 | 1.6 |
| Delta-like 3 | 2.8 | Notch 3 | 3.5 |
| FGF-7 | 1.8 | Notch 4 | 4.1 |
| FGF-13 | 6.4 | Platelet-derived growth factor receptor |
2.4 |
| FGF-18 | 3.8 | Peroxisome proliferator activated receptor-gamma (PPAR-γ) | 2.7 |
| Flt 1 (VEGF receptor) | 2.7 | Pre-T cell antigen receptor | 1.7 |
| Insulin-like growth factor 1 | 1.6 | RAS-related C3 botulinum | 1.7 |
| Jagged 1 | 1.7 |
Table 3.
Genes with higher expression in the cartilage identified by SuperArray™ Osteogenesis Array.
| Gene | Ratio |
|---|---|
| BMP-7 | 6.7 |
| BMP-8 | 5.3 |
| Integrin binding sialoprotein | 4.0 |
| Insulin-like growth factor 1 receptor | 2.2 |
| Matrix metalloproteinase 9 | 3.4 |
| Matrix metalloproteinase 13 | 5.9 |
| Procollagen IX | 4.5 |
| Procollagen X | 33.3 |
| Procollagen XI | 14.3 |
| Scavenger receptor class | 2.9 |
| Sox 9 | 4.3 |
| Osteopontin | 3.4 |
| Vitamin D receptor | 3.7 |
Table 4.
Genes with higher expression in the cartilage identified by SuperArray™ Stem Cell Array.
| Gene | Ratio | Gene | Ratio |
|---|---|---|---|
| Aggrecan | 11.1 | Indian hedgehog | 6.7 |
| BMP-7 | 8.3 | Procollagen X | 25.0 |
| BMP-8 | 10.0 | Ras homolog gene family | 2.1 |
| BMP-binding endothelial | 1.6 | S100 protein, beta polypeptide | 2.0 |
| Cyclin A2 | 1.6 | Snail 1 | 3.3 |
| Cyclin D2 | 2.2 | Snail 2 | 1.6 |
| Cadherin 2 (N-cadherin) | 3.1 | Osteopontin | 5.3 |
| Cadherin 6 | 1.5 | Staufen (RNA1 binding protein) | 2.0 |
| Cyclin-dependent kinase 2b | 4.8 | Tubulin beta 3 | 3.7 |
| Frizzled homolog 1 | 2.6 | Wnt inhibitory factor 1 | 4.8 |
Discussion
In the C sample, the high expression of genes often highly expressed in cartilage can be viewed as a “positive control” for the dissection procedure. In particular, the expression of genes such as collagen X and aggrecan at very high levels (33X and 11X, respectively) in the MC sample suggests that the tissue harvest was fairly accurate in separating cartilage from perichondrium. Evidence that our technique was replicable is provided by the similarity of expression levels in those genes present in both arrays: BMP-7 (6.7X in Osteogenesis Array, 8.3X in Stem Cell Array), BMP-8 (5.3X, 10X), insulin-like growth factor-1 (1.9X, 1.6X), osteopontin (3.4X, 5.3X), and procollagen X (33X, 25X).
Genes with higher expression in the perichondrial (PC) sample
Some of the genes with higher expression in the PC sample have antecedents in the literature or fit with other observations. In other instances, their functional importance requires further investigation, while in still other cases the higher-expressed genes were unexpected. These genes can therefore be discussed in three groups: 1) genes that may be mediators of proliferation and differentiation of prechondroblastic cells; 2) genes for structural and adhesion proteins that are plausibly linked to the architecture and cell communication in the perichondrium; and 3) unexpected genes for which a ready explanation is elusive.
Potential mediators of proliferation and differentiation
This group includes the FGF isoforms and other receptors (platelet-derived growth factor receptor (PDGFr), insulin-like growth factor--1 receptor (IGF-1r), Notch 1, 3, and 4). Three FGF isoforms were enriched in the PC sample: FGF-13 (6.4X), FGF-18 (4X), and FGF-7 (1.8X). In limb bones, FGF-18 has been localized to the periosteum, where it inhibits chondrocyte proliferation and differentiation (33), apparently under the influence of Twist-1 (34). Since Twist-1 has been immunohistochemically localized to the prechondroblastic layer (27), FGF-18 may play a similar role in the MCC, probably signaling via Ffgr2, which is also highly expressed in periosteum and in the prechondroblastic layer of the MCC (24). Neural cell adhesion molecule (NCAM), a cell-surface glycoprotein that mediates cell-cell signaling in the nervous system, was expressed almost 2X greater in the PC sample than in the C sample. A possible explanation may relate to the recent demonstration that NCAM is a major regulator of the interaction of FGF-2 with its receptors in two fibroblast cell lines (35). NCAM, which has been reported to bind to Fgfr2 (the predominant FGF receptor sub-type in the prechondroblastic layer (24), interferes with the binding of the FGF receptor to FGF, thereby inhibiting the cellular response to FGF.
Insulin-like growth factor-1 receptor (IGF-1r), which was more highly expressed in the C sample, has been demonstrated using immunohistochemical techniques to be localized primarily to the chondroblastic and hypertrophic portions of the MCC (24). By contrast, its primary ligand IGF-1, somewhat higher (1.6X) in the PC sample, stimulates proliferation in the perichondrial cells of the MCC (24). Similarly, the receptor for platelet-derived growth factor (PDGF) has been localized to the prechondroblastic layer of the MCC in 1–10 day-old rats (36); in our study it was enriched 2.4 times compared to the MC sample. Finally, transforming growth factor beta receptor 2 (Tgf-βr2) as well as TGF-β3 were increased 2.6 and 1.9 times, respectively, in the perichondrium. This is of great interest since Tgf-βr2 appears to regulate cell proliferation in both osteoprogenitor and chondroprogenitor cells of the developing mandible, where conditional inactivation of Tgf-βr2 also results in major defects in size and organization of the MCC (37).
Members of the Notch family of trans-membrane receptors have been implicated as cell fate mediators in many tissues (38–40). They are expressed in the early stages of chondrogenic differentiation, becoming confined to the perichondrium as differentiation proceeds (41). Of the three isoforms of Notch that were over-expressed in MCC (plus a Notch ligand, Jagged 1(1.7X)), Notch-1 (1.6X) has been localized using immunohistochemistry to the MCC prechondroblastic layer. Moreover, inhibition of Notch reduces proliferation in MCC (28). Our results suggest that Notch-3 (3.5X) and Notch-4 (4.1X), shown to be present in limb articular cartilage (42), may be of greater importance than Notch-1 in the MCC.
Structural and Adhesion Proteins
Some of the other genes that had higher expression in the PC sample were structural proteins or proteoglycans. At least for procollagen XIV (21X higher in the PC sample), this may relate to interactions with type I collagen and/or small proteoglycans. Collagen XIV is distributed preferentially in tissues containing type I collagen fibrils (43) and has been shown to bind to the small proteoglycan decorin (44), which serves to modulate cellular interactions with collagen XIV (45). Since the articular and prechondroblastic layers of the PC are rich in type I collagen (46–47) and decorin (48), the enrichment of the PC sample in mRNA for procollagen XIV and decorin (2.4X) is explicable. Although it might be thought surprising that type I collagen expression did not differ appreciably between the PC and C samples, immunohistochemical studies of the MCC indicate noticeable type I collagen in the deeper (cartilaginous) layers, especially the hypertrophic layer (47).
Still other differential gene expression between the PC and C samples involved various members of the cadherin family, molecules that facilitate cell-cell adhesion and in so doing regulate cellular activities such as differentiation (49). The PC sample was enriched (3–5X) in cadherin 9 (T-cadherin), cadherin 13 (T- or H-cadherin), and cadherin 15 (M-cadherin). The relatively high expression of cadherin 13, which is a modulator of angiogenesis (50–51), may relate to the elevated expression of VEGF–B and its receptors in the PC sample (see below). Similarly, cadherin 15, which facilitates the differentiation of myoblasts by forming a complex with beta catenin (49, 52), may be enriched in concert with the abundant muscle differentiation factor myogenic factor 6 (Myf6) as outlined below.
Unexpected genes
Other matrix proteins with greater expression in the PC sample relative to the C sample are less readily understood. Tuftelin (2.5X), tuftelin interacting protein 11 (1.6X), and dentin sialophosphoprotein or dspp (1.6X) are proteins first identified in the enamel and dentin of the developing tooth (53–55). However, tuftelin and dspp have been reported in bone and other non-dental tissues (53, 56), and dspp has recently been localized immunohistochemically to the prechondroblastic layer of the MCC in very young rats (57). Nevertheless, the role of these proteins in the MCC remains to be elucidated. Similarly, vascular endothelial growth factor-B or VEGF-B, a member of a family of angiogenic agents (58), is expressed at levels twice as high in the PC sample as it is in C sample; the VEGF receptors Flt-1 (2.7X) and kinase insert domain receptor/ Flk-1-KDR (4.3X) are elevated to an even greater extent in the PC sample. Although the role of VEGF-A in endochondral ossification has been well documented (59), current knowledge of VEGF-B does not explain its enrichment, and that of its receptors, in the perichondrium of the MCC. However, chondrocytes secrete all four members of the VEGF family, and chondrogenic stimulation by BMP-2 up-regulates VEGF-B, suggesting that it has a role in growth plate physiology (60). The enrichment of the PC sample (3X) for Peroxisome proliferator activated receptor-gamma (PPAR-γ) is very interesting, since PPAR-γ is known as an adipogenic-specific transcription factor (61, 62).
Sclerostin, enriched 1.7X in the PC sample, is a product of osteocytes which antagonizes Wnt signaling in osteoblasts (61). Perhaps more pertinent to the MCC, it also has been shown to inhibit the differentiation of preosteoblastic cells (64–65). However, perhaps the most puzzling is the 9-fold enrichment of myogenic factor 6 (Myf6) in the PC sample. Myf6 is a transcription factor that is important in the specification and differentiation of skeletal muscle myotubes during embryogenesis (66). Although work on Myf6 has been confined almost completely to muscle, it may be significant that a related gene, Myf5 (which was 1.5X higher in PC), appears to play an important role in rib development (67).
Genes with higher expression in the cartilage (C) sample
As expected, many of the genes that were most highly expressed in the C sample were either characteristic of or specific for cartilage – aggrecan, procollagens IX, X, and XI, Sox9, and Indian hedgehog (68). The greater expression of BMP-7 (6.7X higher) in the C sample is consistent with several reports indicating its activity in promoting chondrogenic differentiation (69–70). Similarly, Cadherin 2 (N-cadherin), the most highly enriched (3X) cadherin in the C sample, is important for chondrogenesis (71).
Although both bone sialoprotein (4X) and osteopontin (5.3X) are important for bone formation (72–73), osteopontin is also expressed by hypertrophic chondrocytes and deep layer articular chondrocytes (74). Both osteopontin and bone sialoprotein have been identified immunohistochemically in the matrix surrounding the hypertrophic chondrocytes of the MCC (57, 75–77), and MMP-13 has likewise been localized to the deepest layer of hypertrophich chondrocytes in 1–10 day-old mouse MCC (78). Snail 1, enriched 3X in the C sample, is also highly expressed in hypertrophic chondrocytes (79), where it is thought to downregulate collagen II and aggregan synthesis, probably via fibroblast growth factor-receptor 3 (fgfr3) signaling (80).
Conclusions: Implications for orthopedic therapies and regenerative medicine
While the results of this study must be considered preliminary until confirmed by RT-PCR, our findings offer new data regarding how prechondroblastic cells and their surrounding matrix differ in gene expression from the underlying chondrocytes of the mandibular condyle. Our study has confirmed the importance of the members of the FGF and TGF-β family of growth factors for proliferation and differentiation in the MCC, and provided potential insight into specific FGF ligands (e.g, FGF-13 and FGF-18) and other proteins (NCAM) that may be important for FGF signaling in the MCC. Moreover, the relative abundance of three Notch isoforms in the PC sample may be of importance in light of Notch’s growing importance in regenerative medicine efforts (81). Secondly, our results provide information on the characteristics of the matrix of native MCC perichondrium that may be of use in designing replacement tissues for the TMJ. But arguably the most important contribution of our results may derive from the identification of novel, unsuspected genes that are differentially expressed in the PC sample: the tooth-associated genes (tuftelin, tuftelin-interacting protein 11, and dentin sialophosphoprotein), VEGF-B and its receptors and associated cadherin, and myogenic factor 6 and its associated cadherin. Recent evidence has demonstrated that undifferentiated myogenic progenitor cells spontaneously express the osteoblastic-specific genes Runx2 and bone alkaline phosphatase (82). In addition, periosteal cells from adult humans can be made to differentiate into chondrocyte, osteoblast, adipocyte, and skeletal myocyte lineages (83). Therefore, the relatively high expression of genes such as myogenic factor 6 and VEGF-B may indicate a degree of unsuspected plasticity in this bipotent cell population derived from an osteogenic lineage. Unfortunately, it is impossible to discern from our data whether certain of these genes are expressed by a sub-population of cells within the perichondrium. However, our characterization of perichondrial gene expression may serve as a substrate for the burgeoning number of efforts attempting to regenerate the articular disc or MCC (84) or to upregulate growth at the MCC (85).
Clinical Relevance.
With the exception of some basic structural proteins, little is known of the genes that are highly expressed in the dividing cells of the mandibular condylar cartilage. Our study demonstrates differential gene expression in specific growth factor receptors and matrix proteins, as well as in novel, unsuspected genes that hint at an unrecognized plasticity of expression in these cells. Improved understanding of gene expression in native tissue will be essential for regenerative medicine efforts or attempts to upregulate the growth rate at the condylar cartilage for therapeutic purposes.
Acknowledgements
This work was supported by NIH grant DE015401 to RJH.
References
- 1.Hall BK. The fate of adventitious and embryonic articular cartilage in the skull of the common fowl, Gallus Domesticus (Aves: Phasianidae) Aust J Zool. 1968;16:795–805. [Google Scholar]
- 2.Beresford WA. Chondroid Bone Secondary Cartilage, and Metaplasia. Baltimore: Urban and Schwarzenberg; 1981. [Google Scholar]
- 3.Vinkka H. Secondary cartilages in the facial skeleton of the rat. Proc Finn Dent Soc. 1982;78 Suppl VII:1–137. [PubMed] [Google Scholar]
- 4.Petrovic AG, Stutzmann JJ, Oudet CL. Control processes in the postnatal growth of the condylar cartilage of the mandible. In: McNamara JA Jr, editor. Determinants of Mandibular Form and Growth, Monograph Number 4, Craniofacial Growth Series, Center for Human Growth and Development. Ann Arbor: The University of Michigan; 1975. pp. 101–154. [Google Scholar]
- 5.McNamara JA, Bryan FA. Long-term mandibular adaptations to protrusive function: An experimental study in Macaca mulatta. Am J Orthod Dentofac Orthop. 1987;92:98–108. doi: 10.1016/0889-5406(87)90364-7. [DOI] [PubMed] [Google Scholar]
- 6.Johnston LE., Jr . The curious case of the chimerical condyle. In: Graber LW, editor. Orthodontics: State of the Art, Essence of the Science. St. Louis: CV Mosby; 1986. pp. 88–99. [Google Scholar]
- 7.Meikle MC. Remodeling the dentofacial skeleton: the biologic basis of orthodontics and dentofacial orthopedics. J Dent Res. 2007;86:12–24. doi: 10.1177/154405910708600103. [DOI] [PubMed] [Google Scholar]
- 8.Blackwood HJJ. Growth of the mandibular condyle of the ratstudied with tritiated thymidine. Arch Oral Bio. 1966;11:493–500. doi: 10.1016/0003-9969(66)90155-5. [DOI] [PubMed] [Google Scholar]
- 9.Folke LEA, Stallard RE. Cellular kinetics within the mandibular joint. Acta Odontol Scand. 1967;25:437–489. [PubMed] [Google Scholar]
- 10.Luder HU, Leblond CP, von der Mark K. Cellular stages in cartilage formation as revealed by morphometry, radioautography, and type II collagen immunostaining of the mandibular condyle from weanling rats. Am J Anat. 1988;182:197–214. doi: 10.1002/aja.1001820302. [DOI] [PubMed] [Google Scholar]
- 11.Stutzmann JJ, Petrovic AG. Bone cell histogenesis: the skeletoblast as a stem-cell for preosteoblasts and secondary-type prechondroblasts. In: Dixon AD, Sarnat BG, editors. Factors and Mechanisms Influencing Bone Growth. New York: Alan R Liss; 1982. pp. 29–43. [PubMed] [Google Scholar]
- 12.Petrovic A. Mechanisms and regulation of mandibular condylar growth. Acta Morphol Neerl -Scand. 1972;10:25–34. [PubMed] [Google Scholar]
- 13.Petrovic AG. Experimental and cybernetic approaches to the mechanism of action of functional appliances on mandibular growth. In: McNamara JA, Ribbens KA, editors. Malocclusion and the Periodontium. Monograph Number 15, Craniofacial Growth Series, Center for Human Growth and Development. Ann Arbor: The University of Michigan; 1984. pp. 213–268. [Google Scholar]
- 14.Engel FE, Khare AG, Boyan BD. Phenotypic changes of rabbit mandibular condylar cartilage cells in culture. J Dent Res. 1990;69:1753–1758. doi: 10.1177/00220345900690110801. [DOI] [PubMed] [Google Scholar]
- 15.Girdler NM. The behaviour of mandibular condylar cartilage in cell culture. Int J Oral Maxillofac Surg. 1993;22:178–184. doi: 10.1016/s0901-5027(05)80248-6. [DOI] [PubMed] [Google Scholar]
- 16.Inoue H, Nebgen D, Veis A. Changes in phenotypic gene expression in rat mandibular condylar cartilage cells during long-term culture. J Bone Min Res. 1995;10:1691–1697. doi: 10.1002/jbmr.5650101111. [DOI] [PubMed] [Google Scholar]
- 17.Weiss A, Livne E, von der Mark K, Heinegard D, Silbermann M. Growth and repair of cartilage: organ culture system utilizing chondroprogenitor cells of condylar cartilage in newborn mice. J Bone Min Res. 1988;3:93–100. doi: 10.1002/jbmr.5650030114. [DOI] [PubMed] [Google Scholar]
- 18.Glasstone S. Differentiation of the mouse embryonic mandible and squamo-mandibular joint in organ culture. Arch Oral Biol. 1971;16:723–729. doi: 10.1016/0003-9969(71)90117-8. [DOI] [PubMed] [Google Scholar]
- 19.Melcher AH. Behaviour of cells of condylar cartilage of foetal mouse mandible maintained in vitro. Archs Oral Biol. 1971;16:1379–1391. doi: 10.1016/0003-9969(71)90075-6. [DOI] [PubMed] [Google Scholar]
- 20.Copray JCVM. Growth of the mandibular condylar cartilage of the rat in serum-free organ culture. Archs Oral Biol. 1983;28:967–974. doi: 10.1016/0003-9969(83)90095-x. [DOI] [PubMed] [Google Scholar]
- 21.Silbermann M, Lewinson D, Gonen H, Lizarbe MA, von der Mark K. In vitro transformation of chondroprogenitor cells into osteoblasts and the formation of new membrane bone. Anat Rec. 1983;206:373–383. doi: 10.1002/ar.1092060404. [DOI] [PubMed] [Google Scholar]
- 22.Garcia AM, Gray ML. Dimensional growth and extracellular matrix accumulation by neonatal rat mandibular condyles in long-term culture. J Orthoped Res. 1995;13:208–219. doi: 10.1002/jor.1100130209. [DOI] [PubMed] [Google Scholar]
- 23.Nakai H, Niimi A, Ueda M. The influence of compressive loading on growth of cartilage of the mandibular condyle in vitro. Archs Oral Biol. 1998;43:505–515. doi: 10.1016/s0003-9969(98)00041-7. [DOI] [PubMed] [Google Scholar]
- 24.Fuentes MA, Opperman LA, Bellinger LL, Carlson DS, Hinton RJ. Regulation of cell proliferation in rat mandibular condylar cartilage in explant culture by insulin-like growth factor-1 and fibroblast growth factor-2. Archs Oral Biol. 2002;47:643–654. doi: 10.1016/s0003-9969(02)00052-3. [DOI] [PubMed] [Google Scholar]
- 25.Shibata S, Fukada K, Suzuki S, Yamashita Y. Immunohistochemistry of collagen types II and X, and enzyme-histochemistry of alkaline phosphatase in the developing condylar cartilage of the fetal mouse mandible. J Anat. 1997;191:561–570. doi: 10.1046/j.1469-7580.1997.19140561.x. 561–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shibata S, Suda N, Suzuki S, Fukuoka H, Yamashita Y. An in situ hybridization study of Runx2, Osterix, and Sox9 at the onset of condylar cartilage formation in fetal mouse mandible. J Anat. 2006;208:169–177. doi: 10.1111/j.1469-7580.2006.00525.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Capps C, So S, Hinton R. Cell fate mediators in mandibular condylar cartilage. J Dent Res. 2006 Spec Iss A, abstract 1233. [Google Scholar]
- 28.So S, Serrano M, Hinton RJ. Notch signaling in mandibular condylar cartilage. J Dent Res. 2007 Spec Iss A, abstract 3010. [Google Scholar]
- 29.Buxton PG, Hall B, Archer CW, Francis-West P. Secondary chondrocyte-derived Ihh stimulates proliferation of periosteal cells during chick development. Development. 2003;130:4729–4739. doi: 10.1242/dev.00610. [DOI] [PubMed] [Google Scholar]
- 30.Shibata S, Suda N, Yoda S, Fukuoka H, Ohyama K, Yamashita Y, et al. Runx2-deficient mice lack mandibular condylar cartilage and have deformed Meckel’s cartilage. Anat Embryol. 2004;208:273–280. doi: 10.1007/s00429-004-0393-2. [DOI] [PubMed] [Google Scholar]
- 31.Nilsson O, Parker EA, Hegde A, Chau M, Barnes K, Baron J. Gradients in bone morphogenetic protein-related gene expression across the growth plate. J Endocrin. 2007;193:75–84. doi: 10.1677/joe.1.07099. [DOI] [PubMed] [Google Scholar]
- 32.Xu Y, Malladi P, Zhou D, Longaker MT. Molecular and cellular characterization of mouse calvarial osteoblasts derived from neural crest and paraxial mesoderm. Plast Reconstr Surg. 2007;120:1783–1795. doi: 10.1097/01.prs.0000279491.48283.51. [DOI] [PubMed] [Google Scholar]
- 33.Liu Z, Xu J, Colvin JS, Ornitz DM. Coordination of chondrogenesis and osteogenesis by fibroblast growth factor 18. Genes Dev. 2002;16:865–869. doi: 10.1101/gad.965602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hinoi E, Bialek P, Chen Y-T, rached M-T, Groner Y, Behringer RR, et al. Runx2 inhibits chondrocytes proliferation and hypertrophy through its expression in the perichondrium. Genes Dev. 2006;20:2937–2942. doi: 10.1101/gad.1482906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Francavilla C, Loeffler S, Piccini D, Kren A, Christofori G, Cavallaro U. Neural cell adhesion molecule regulates the cellular response to fibroblast growth factor. J Cell Sci. 2007;120:4388–4394. doi: 10.1242/jcs.010744. [DOI] [PubMed] [Google Scholar]
- 36.Visnapuu V, Peltomaki T, Ronning O, Vahlberg T, Helenius H. Distribution of fibroblast growth factors (FGFR-1 and −3) and platelet-derived growth factor receptors (PDGFR) in the rat mandibular condyle during growth. Orthod Craniof Res. 2002;5:147–153. doi: 10.1034/j.1600-0544.2002.02205.x. [DOI] [PubMed] [Google Scholar]
- 37.Oka K, Oka S, Sasaki T, Ito Y, Bringas P, Nonaka K, et al. The role of TGf-β signaling in regulating chondrogenesis and osteogenesis during mandibular development. Dev Biol. 2007;303:391–404. doi: 10.1016/j.ydbio.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Curry CL, Reed LL, Golde TE, Nickoloff BJ, Foreman KE. Gamma secretase inhibitor blocks Notch activation and induces apoptosis in Kaposi's sarcoma tumor cells. Oncogene. 2005;24:6333–6344. doi: 10.1038/sj.onc.1208783. [DOI] [PubMed] [Google Scholar]
- 39.Mitsiadis TA, Regaudiat L, Gridley T. Role of the Notch signalling pathway in tooth morphogenesis. Arch Oral Biol. 2005;50:137–140. doi: 10.1016/j.archoralbio.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 40.Purow BW, Haque RM, Noel MW, Su Q, Burdick MJ, Lee J, et al. Expression of Notch-1 and its ligands, Delta-like-1 and Jagged-1, is crtical for glioma cell survival and proliferation. Canc Res. 2005;65:2353–2363. doi: 10.1158/0008-5472.CAN-04-1890. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe N, Tezuka Y, Matsuno K, Miyatani S, Morimura N, Yasuda M, et al. Suppression of differentiation and proliferation of early chondrogenic cells by Notch. J Bone Miner Metab. 2003;21:344–352. doi: 10.1007/s00774-003-0428-4. [DOI] [PubMed] [Google Scholar]
- 42.Hayes AJ, Dowthwaite GP, Webster SV, Archer CW. The distribution of Notch receptors and their ligands during articular cartilage development. J Anat. 2003;202:495–502. doi: 10.1046/j.1469-7580.2003.00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walchli C, Koch M, Chiquet M, Odermatt BF, Trueb B. Tissue-specific expression of the fibril-associated collagens XII and XIV. J Cell Sci. 1994;107:669–681. doi: 10.1242/jcs.107.2.669. [DOI] [PubMed] [Google Scholar]
- 44.Font B, Aubert-Foucher E, Goldschmidt D, Eichenberger D, van der Rest M. Binding collagen XIV with the dermatan sulfate side chain of decorin. J Biol Chem. 1993;268:25015–25018. [PubMed] [Google Scholar]
- 45.Ehnis T, Walburga D, Bauer M, Schuppan D. Localization of a cell adhesion site on collagen XIV (Undulin) Exp Cell Res. 1998;239:477–480. doi: 10.1006/excr.1997.3895. [DOI] [PubMed] [Google Scholar]
- 46.Silbermann M, Reddi AH, Hand AR, Leapman RD, von der Mark K, Franzen A. Further characterization of the extracellular matrix in the mandibular condyle in neonatal mice. J Anat. 1987;151:169–188. [PMC free article] [PubMed] [Google Scholar]
- 47.Mizoguchi I, Nakamura M, Takahashi I, Kagayama M, Mitani H. An immunohistochemical study of localization of type I and type II collagens in mandibular condylar cartilage compared with tibial growth plate. Histochemistry. 1990;93:593–599. doi: 10.1007/BF00272201. [DOI] [PubMed] [Google Scholar]
- 48.Del Santo M, Jr, Marches F, Ng M, Hinton RJ. Age-associated changes in decorin in rat mandibular condylar cartilage. Archs Oral Biol. 2000;45:485–494. doi: 10.1016/s0003-9969(00)00013-3. [DOI] [PubMed] [Google Scholar]
- 49.Kang J-S, Feinleib JL, Knox S, Ketteringham MA, Krauss RS. Promyogenic members of the Tg and cadherin families associate to positively regulate differentiation. Proc New York Acad Sci. 2003;100:3989–3994. doi: 10.1073/pnas.0736565100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Philippova M, Banfi A, Ivanov D, Gianni-Barrera R, Allenspach R, Erne P, et al. Atypical GPI-anchored T-cadherin stimulates angiogenesis in vitro and in vivo. Arterioscler Thromb Vasc Biol. 2006;26:2222–2230. doi: 10.1161/01.ATV.0000238356.20565.92. [DOI] [PubMed] [Google Scholar]
- 51.Takeuchi T, Adachi Y, Ohtsuki Y, Furihata M. Adiponectin receptors, with special focus on the role of the third receptor, T-cadherin, in vascular disease. Med Mol Morphol. 2007;40:115–120. doi: 10.1007/s00795-007-0364-9. [DOI] [PubMed] [Google Scholar]
- 52.Ishido M, Uda M, Masuhara M, Kami K. Alterations of M-cadherin, neural cell adhesion molecule and β-catenin expression in satellite cells during overload -induced skeletal muscle hypertrophy. Acta Physiol. 2006;187:407–418. doi: 10.1111/j.1748-1716.2006.01577.x. [DOI] [PubMed] [Google Scholar]
- 53.MacDougall M, Simmons D, Dodds A, Knight C, Luan X, Zeichner-David M, et al. Cloning, characterization, and tissue expression pattern of mouse tuftelin cDNA. J Dent Res. 1998;77:1970–1978. doi: 10.1177/00220345980770120401. [DOI] [PubMed] [Google Scholar]
- 54.Wen X, Lei YP, Okamoto CT, Snead ML, Paine ML. Structural organization and cellular organization of tuftelin-interacting protein 11 (TFIP11) Cell Mol Life Sci. 2005;62:1038–1046. doi: 10.1007/s00018-005-4547-z. [DOI] [PubMed] [Google Scholar]
- 55.Mastrangelo F, Scioletti AP, Tranasi M, Tecco S, Sberna MT, Vinci R, et al. Dentin sialophosphoprotein expression during human matrix development. J Biol Regul Homeost Agents. 2007;21:33–39. [PubMed] [Google Scholar]
- 56.Huang B, Sun y, Maciejewska I, Qin D, Peng T, McIntyre, et al. Distribution of SIBLING proteins in the organic and inorganic phases of rat dentin and bone. Eur J Oral Sci. 2008;116:104–112. doi: 10.1111/j.1600-0722.2008.00522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gandhi V, Sun Y, Maciejewska I, Hinton R, Qin C. Distribution of SIBLING proteins in condylar cartilage of rat mandible. J Dent Res. 2008 abstract 433. [Google Scholar]
- 58.Breen EC. VEGF in biological control. J Cell Biochem. 2007;102:1358–1367. doi: 10.1002/jcb.21579. [DOI] [PubMed] [Google Scholar]
- 59.Dai J, Rabie ABM. VEGF: an essential mediator of both angiogenesis and endochondral ossification. J Dent Res. 2007;86:937–950. doi: 10.1177/154405910708601006. [DOI] [PubMed] [Google Scholar]
- 60.Bluteau G, Julien M, Magne D, Mallein-Gerin F, Weiss P, Daculsi G, et al. VEGF and VEGF receptors are differentially expressed in chondrocytes. Bone. 2007;40:568–576. doi: 10.1016/j.bone.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 61.Rich JT, Rosova I, Nolta JA, Myckatyn TM, Sandell LJ, McAlinden A. Upregulation of Runx2 and Osterix during in vitro chondrogenesis of human adipose-derived stromal cells. Biochem Biophys Res Comm. 2008;372:230–235. doi: 10.1016/j.bbrc.2008.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang Y, Mishima H, Sakai S, Liu Y-k, Ohyabu Y, Uuemura T. Gene expression analysis of major lineage-defining factors in human bone marrow cells: effect of aging, gender, and age-related disorders. J Orthop Res. 2008;26:910–917. doi: 10.1002/jor.20623. [DOI] [PubMed] [Google Scholar]
- 63.van Bezooijen RL, Svensson JP, Eefting D, Visser A, van der Horst G, Karperien M, et al. Wnut but not BMP signaling is involved in the inhibitory action of sclerostin on BMP-stimulated bone formation. J Bone Min Res. 2007;21:19–28. doi: 10.1359/jbmr.061002. [DOI] [PubMed] [Google Scholar]
- 64.Winkler DG, Sutherland MK, Geoghegan JC, Yu C, Hayes T, Skonier JE, et al. Osteocyte control of bone formation via sclerostin, a novel BMP antagonist. EMBO J. 2003;23:6267–6276. doi: 10.1093/emboj/cdg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Bezooijen RL, Roelen BAJ, Visser A, van der Wee-Pals L, de Wilt E, Karperien M, et al. Sclerostin is an osteocyte-expressed negative regulator of bone formation, but not a classical BMP antagonist. J Exp Med. 2004;199:805–814. doi: 10.1084/jem.20031454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maak S, Neumann K, Swalve HH. Identification and analysis of putative regulatory sequences for the MYF5/MYF6 locus in different vertebrate species. Gene. 2006;379:141–147. doi: 10.1016/j.gene.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 67.Haldar M, Karan G, Tvrdik P, Capecchi MR. Two cell lineages, myf5 and myf-5 independent, participate in mouse skeletal myogenesis. Dev Cell. 2008;14:437–445. doi: 10.1016/j.devcel.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lefebvre V, Smits P. Transcriptional control of chondrocytes fate and differentiation. Birth Defects Res. 2005;75:200–212. doi: 10.1002/bdrc.20048. [DOI] [PubMed] [Google Scholar]
- 69.Nishihara A, Fujii M, Sampath TK, Miyazono K, Reddi AH. Bone morphogenetic 9 protein signaling in articular chondrocytes differentiation. Biochem Biophys Res Comm. 2003;301:617–622. doi: 10.1016/s0006-291x(02)03068-1. [DOI] [PubMed] [Google Scholar]
- 70.Knippenberg M, Helder MN, Doulabi BZ, Wuisman PIJM, Klein-Nulend J. Osteogenesis versus chondrogenesis by BMP-2 and BMP-7 in adipose stem cells. Biochem Biophys Res Comm. 2006;342:902–908. doi: 10.1016/j.bbrc.2006.02.052. [DOI] [PubMed] [Google Scholar]
- 71.Delise AM, Tuan RS. Analysis of N-cadherin function in limb mesenchymal chondrogenesis in vitro. Dev Dyn. 2002;225:195–204. doi: 10.1002/dvdy.10151. [DOI] [PubMed] [Google Scholar]
- 72.Merry K, Dodds R, Littlewood A, Gowen M. Expression of osteopontin mRNA by osteoclasts and osteoblasts in modeling adult bone. J Cell Sci. 1993;104:1013–1020. doi: 10.1242/jcs.104.4.1013. [DOI] [PubMed] [Google Scholar]
- 73.Malaval L, Wade-Gueye NM, Boudiffa M, Fei J, Zirngibi R, Chen F, et al. Bone sialoprotein plays a functional role in bone formation and osteoclastogenesis. J Exper Med. 2008;205:1145–1153. doi: 10.1084/jem.20071294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schnapper A, Meyer W. Osteopontin distribution in the canine skeleton during growth and structural maturation. Cells Tiss Org. 2004;178:158–167. doi: 10.1159/000082246. [DOI] [PubMed] [Google Scholar]
- 75.Sugiyama H, Imada M, Sasaki A, Ishino Y, Kawata T, Tanne K. The expression of osteopontin with condylar remodeling in growing rats. Clin Orthod Res. 2001;4:194–199. doi: 10.1034/j.1600-0544.2001.40403.x. [DOI] [PubMed] [Google Scholar]
- 76.Shibata S, Fukada K, Suzuki S, Ogawa T, Yamashita Y. In situ hybridization and immunohistochemistry of bone sialoprotein and secreted phosphoprotein 1 (osteopontin) in the developing mouse mandibular condylar cartilage compared with limb bud cartilage. J Anat. 2002;200:309–320. doi: 10.1046/j.1469-7580.2002.00033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hossain KS, Amizuka N, Ikeda N, Nozawa-Inoue K, Suzuki A, Li M, et al. Histochemical evidences on the chronological alterations of the hypertrophic zone of mandibular condylar cartilage. Microsc Res Tech. 2005;67:325–335. doi: 10.1002/jemt.20211. [DOI] [PubMed] [Google Scholar]
- 78.Okhubo K, Shimokawa H, Ogawa T, Suzuki S, Fukada K, Ohya K, et al. Immunohistochemical localization of matrix metalloproteinase 13 (MMP-13) in mouse mandibular condylar cartilage. J Med Dent Sci. 2003;50:203–211. [PubMed] [Google Scholar]
- 79.Seki K, Fujimori T, Savagner P, Hata A, Aikawa T, Ogata N, et al. Mouse Snail family transcription repressors regulate chondrocyte, extracellular matrix, type II collagen, and aggrecan. J Biol Chem. 2003;278:41862–41870. doi: 10.1074/jbc.M308336200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.de Frutos CA, Vega S, Manzanares M, Flores JM, Huertas H, Matrinez-Frias ML, et al. Snail1 is a transcriptional effector of FGFR3 signaling during chondrogenesis and achondroplasia. Dev Cell. 2007;13:872–883. doi: 10.1016/j.devcel.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 81.Carlson ME, Conboy IM. Regulation the Notch pathway in embryonic, adult, and old stem cells. Curr Opin Pharm. 2007;7:303–309. doi: 10.1016/j.coph.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 82.Hasimoto N, Kiyono T, Wada MR, Umeda R, Goto Y, Nonaka I, et al. Osteogenic properties of human myogenic progenitor cells. Mech Dev. 2008;125:257–269. doi: 10.1016/j.mod.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 83.de Bari C, Dell’Acio F, Vanlauwe J, Eyckmans J, Khan IM, Archer CW, et al. Mesenchymal multipotency of adult human periosteal cells demonstrated by single-cell lineage analysis. Arthr Rheum. 2006;54:1209–1221. doi: 10.1002/art.21753. [DOI] [PubMed] [Google Scholar]
- 84.Wang L, Detamore MS. Tissue engineering the mandibular condyle. Tissue Eng. 2007;13:1955–1971. doi: 10.1089/ten.2006.0152. [DOI] [PubMed] [Google Scholar]
- 85.Li QF, Rabie AB. A new approach to control condylar growth by regulating angiogenesis. Arch Oral Biol. 2007;52:1009–1017. doi: 10.1016/j.archoralbio.2007.05.009. [DOI] [PubMed] [Google Scholar]