Abstract
Food allergy is a potential risk associated with use of transgenic proteins in crops. Currently, safety assessment involves consideration of the source of the introduced protein, in silico amino acid sequence homology comparisons to known allergens, physicochemical properties, protein abundance in the crop, and, when appropriate, specific immunoglobulin E binding studies. Recently conducted research presented at an International Life Sciences Institute/Health and Environmental Sciences Institute–hosted workshop adds to the scientific foundation for safety assessment of transgenic proteins in five areas: structure/activity, serum screening, animal models, quantitative proteomics, and basic mechanisms. A web-based tool is now available that integrates a database of allergenic proteins with a variety of computational tools which could be used to improve our ability to predict allergenicity based on structural analysis. A comprehensive strategy and model protocols have been developed for conducting meaningful serum screening, an extremely challenging process. Several animal models using oral sensitization with adjuvant and one dermal sensitization model have been developed and appear to distinguish allergenic from non-allergenic food extracts. Data presented using a mouse model suggest that pepsin resistance is indicative of allergenicity. Certain questions remain to be addressed before considering animal model validation. Gel-free mass spectrometry is a viable alternative to more labor-intensive approaches to quantitative proteomics. Proteomic data presented on four nontransgenic varieties of soy suggested that if known allergen expression in genetically modified crops falls within the range of natural variability among commercial varieties, there appears to be no need to test further. Finally, basic research continues to elucidate the etiology of food allergy.
Keywords: Food Allergy, biotechnology, genetically modified crops, plant incorporated pesticides, safety assessment
INTRODUCTION
Food allergy is a relatively new concern for toxicologists as a result of the genetic engineering of novel proteins into food crops in order to promote resistance to herbicides, pests or other stresses, improve nutrition, or otherwise modify the plant phenotype. Allergic reactions to food are relatively rare. The incidence of food allergy in the United States and other “westernized” countries ranges from 1 to 2% in adults and 6 to 8% in children (GAO, 2002; Ladics et al. 2003). Relatively few foods are responsible for the vast majority of significant food-induced allergic reactions although what makes them unique is not entirely clear. Food allergy can manifest as inflammation of the skin (hives), gut, and/or lung, and in the most extreme cases (three individuals per 100,000/year) can result in anaphylactic shock and death (Burks and Sampson, 1997). The responses are most commonly associated with production of protein-specific immunoglobulin E (IgE). Thus, although transgenic modification of crops has many advantages over more conventional approaches, there is some concern that introduction of a novel protein into the food supply could increase the risk of food allergy in susceptible individuals. There are three possible scenarios: (1) transferring an existing allergen or cross-reactive protein from one crop to another, (2) creating food allergens de novo, or (3) altering or quantitatively increasing an endogenous (existing) allergen (i.e., increasing exposure to known food allergens). In addition to genetic engineering, conventional breeding approaches, such as chemical and radiation mutation, can also alter levels of existing proteins and may pose a potential risk (Batista et al., 2008).
Over the last 12 years, several guidance documents have been written to provide recommendations for assessing the potential allergenicity of transgenic proteins (Codex Alimentarious Commission, 2003; FAO/WHO, 2001; Metcalfe et al., 1996). Updates occurred with each successive document; however, there is no single, definitive test for determining the allergenic potential of novel proteins. Therefore, the U.S. agencies responsible for the regulation of foods derived from modern biotechnology use a “weight-of-evidence” approach. This approach is consistent with the Annex to the Codex Alimentarius “Guideline for the Conduct of Food Safety Assessment of Foods Derived from Recombinant-DNA Plants” (henceforth, referred to as Codex [Codex Alimentarious Commission, 2003; Ladics, 2008]). The recommended assessments include consideration of the source of the introduced protein (i.e., whether the gene source for the new protein is known to induce allergy), the host crop's propensity to cause allergy, similarity of the introduced protein to known allergens (in silico amino acid sequence similarity comparisons to known human allergens), physicochemical properties (e.g., susceptibility to acid and enzymatic digestion in vitro, heat stability), and protein abundance in the crop. When appropriate (i.e., a positive amino acid sequence match to a known allergen is observed or the transgenic protein is derived from a known allergenic source), specific IgE binding studies are considered. These studies require the use of well-characterized sera from individuals known to be allergic (or skin prick test positive) to the identified source and present ongoing challenges in terms of standardization of test materials, lack of available sera, and validation of procedures. Codex also recognized that certain methods previously recommended (e.g., animal models; targeted serum screening) were not validated but may prove useful in the future in assessing the allergenic potential of transgenic proteins “as scientific knowledge and technology evolves” (Codex Alimentarious Commission, 2003).
Over the past few years, the U.S. Environmental Protection Agency (EPA), Health Canada, and industry have funded research in an effort to increase the scientific knowledge and technology available to assess the potential allergenicity of transgenic proteins, and a 2008 joint EPA and National Institute of Allergy and Infectious Disease (NIAID) exploratory research initiative has recently focused on basic mechanisms of food allergy. On 15–16 October 2008, industry, academic, and government scientists gathered in Washington, DC, at a meeting sponsored by the Health and Environmental Sciences Institute (HESI) of the International Life Sciences Institute, to review the results of this new research and consider its implications and applications as well as to discuss additional research and validation needs. Research in four areas was considered: use of protein structure to predict allergenicity, serum screening, newly developed animal models, and use of proteomics to assess the amount of allergenic protein in plants. The final session described some of the new research projects recently funded by the joint EPA/NIAID initiative.
PROTEIN STRUCTURE
There are currently no known unique motifs that identify a protein as an allergen; however, a better understanding of structural attributes could prove valuable for assessing allergenic potential. Early guidelines proposed linear bioinformatic searches using 8–12 contiguous amino acids in common with a known allergen as an indicator of potential allergenic risk based on sequence similarity (Metcalfe et al., 1996). However, the use of a six-amino acid sliding window was later recommended (FAO/WHO, 2001) but was subsequently found to yield an unacceptably high number of false positives (Hileman et al., 2002; Silvanovich et al. 2006; Stadler and Stadler, 2003). It was concluded that using greater than 35% shared identity over any 80 amino acid section (based on FASTA or other equivalent programs) is a highly conservative estimate (i.e., many false positives) of the potential for cross-reactivity (Ladics et al., 2007; Thomas et al., 2005a). FASTA (Pearson et al., 1988) is a program used for amino acid sequence (or nucleotide) comparisons and database searches. Hence, although these sequence analogy approaches are useful, it is likely that many potentially beneficial protein products are unnecessarily eliminated early in the evaluation process. Additional approaches to sequence/structure analysis would be welcome.
Dr Werner Braun presented information on a structural database of allergenic proteins (SDAPs) (http://fermi.utmb.edu/SDAP/) that is integrated with a variety of computational tools (Table 1). In addition to the above-mentioned FASTA and short amino acid sliding window approaches, several additional types of structural analyses are available at this web site. All allergens in SDAP are classified into their closest protein families as defined by the Pfam database of information about protein domains and families (http://pfam.sanger.ac.uk/). Allergens are found in only a small subset of all known protein families (Radauer et al. 2008). However, protein families that contain allergens also include hundreds of non-allergenic proteins. Using SDAP tools, sequence motifs unique to the allergens in three families (storage proteins, Bet v 1, and tropomyosin) were found to overlap with known IgE epitopes (Ivanciuc et al., 2009a), suggesting that such motifs might be useful is separating non-allergenic from allergenic members within protein families. Another tool available in SDAP is the computation of property distance (PD) values for measuring peptide similarity. A protein sequence similarity search is based on the five-dimensional descriptors E1–E5 of amino acid properties derived from a pool of 237 physicochemical properties. The similarity between two sequences A and B, each one consisting of N residues, is the PD.
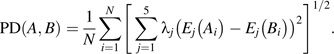 |
TABLE 1.
Tools Available for Assessing Potential Allergenicity Based on Structural Analyses
| Structural characterization | Sourcea | Comments |
| 35% shared identity over any 80 amino acids | http://www.ebi.ac.uk/Tools/webservices/services/fasta | Current approach; highly conservative |
| Protein families | http://pfam.sanger.ac.uk/ | Allergens limited to a small subset of familiesb |
| PD | Ivanciuc et al. (2009b) | Based on physicochemical propertiesb |
| 3D structure | Oezguen et al. (2008) | Conformation of epitope is important; 433 reliable 3D modelsb |
All may be accessed at http://fermi.utmb.edu/SDAP/.
Add value to current approach.
The PD index is a statistically validated method to detect discrete regions of proteins that have a high probability of cross-reacting with IgE from allergic patients (Ivanciuc et al., 2009b). Finally, the SDAP Web site provides the means to examine similarities to allergenic proteins with respect to three-dimensional (3D) structure. 3D structures are available in the Protein Data Bank for only 5% (45/829) of all allergens catalogued in the SDAP. To overcome this limitation, an automated procedure was used to prepare 3D models of all allergens where there was no experimentally determined 3D structure or high identity (95%) to another protein of known 3D structure. After a final selection using quality criteria, 433 reliable 3D models were retained and are now available from the SDAP Web site (Oezguen et al., 2008).
From the preceding, it is clear that several new approaches are available that might be useful in the development of more refined bioinformatics analyses for determining food safety (Table 1). Significant progress has been made since 2005 when application of in silico methods to predict allergenicity was the topic of an HESI workshop (Thomas et al., 2005a). Using the SDAP, it would be possible to use a tiered or weight of evidence approach to protein structure analysis that could provide a more accurate identification of potential allergens than the current, highly conservative, approach. This could include assessing potential allergens according to protein families, PD, and 3D structure in addition to the sliding window analysis of amino acid sequence and FASTA. Further discussion between scientists and the regulatory community are needed to determine the best use of these new in silico tools.
Dr Catherine Schein presented plans for a newly funded grant that will test the hypothesis that computational tools developed as part of the SDAP can detect IgE epitopes responsible for cross-reactions among distantly related nut proteins, according to their common physicochemical properties. She intends to test whether the affinity of the IgE/protein epitope interaction is consistent with clinically relevant cross-reactivity among nuts. Results from this grant should further substantiate the use of structural tools that have been developed.
The greatest research need in this area is to identify additional sequences of allergenic IgE epitopes (Bannon and Ogawa, 2006). Also needed is a comparison of 35 vs. 50% or greater homology over 80 or greater amino acids for cross-reactivity using well-characterized patient allergic serum because, as noted above, the 35% homology is a highly conservative and 50% might be a better indicator.
SERUM SCREENING
A “specific” serum screen involves testing a protein of interest with sera from patients with documented clinical food allergy to a specific allergen to confirm that the tested protein is not cross-reactive with the protein to which the patient produces IgE antibodies. A “targeted” serum screen involves testing the protein of interest with sera from patients sensitive to food or aeroallergens from the same broad group. In either case, the principles associated with the assays used are similar. As noted above, the Codex recommendations include specific, but not “targeted,” serum screening as part of the weight-of-evidence approach to identifying potential allergens. However, practical guidance on how to conduct either of these tests has been lacking. Issues associated with obtaining and validating sera for such tests have been previously described (Thomas et al., 2007a). Dr Richard Goodman presented results from a grant to develop a comprehensive strategy and model protocols to evaluate IgE binding and the range of IgE binding/cross-reactivity.
Serum IgE tests must be reproducible, sensitive, and specific. This begins with documentation that the serum donor is indeed allergic to the allergen of interest. A positive response to oral food challenge is the most definitive test for specificity, but this test is often not performed for practical or ethical reasons. Careful clinical history and positive skin prick tests results are viable alternatives for establishing specificity. If serum from different donors is pooled, it is important to characterize the individual sera to understand the similarities or dissimilarities. Otherwise one serum sample could dominate the results or dilute out IgE that is present in low abundance. Dr Goodman has collected a large set of well-characterized sera specific for a variety of food allergens. One of the most difficult parts of this experimental approach is the accessibility to enough specific donor sera.
An appropriately validated IgE assay is also essential to the success of any particular serum screening effort. A number of issues must be addressed to ensure the integrity of the assay including appropriate blocking agents to prevent nonspecific binding, overcoming the much larger higher concentrations of IgG relative to IgE that are generally present in sera and can interfere with IgE detection, and demonstrating specificity of the anti-IgE component (secondary antibody) in the ELISA (Holzhauser et al., 2008). A variety of approaches are possible including ELISA and dot and Western blots. It is best to confirm results using more than one approach. Generally, IgE binding to carbohydrate domains is irrelevant to clinical allergy, unless there are multiple carbohydrates on the potentially allergenic protein (Altmann, 2007). However, because posttranslational modifications (such as glycosylation) may contribute to allergenic potential of a protein, more work is needed on cross-reactive carbohydrate determinants in order to establish criteria for when and how to exclude such results. Competitive inhibition tests can be used to establish specificity of serum and to rule out carbohydrate effects on IgE binding. Well-characterized positive and negative controls should be included in all assays, a challenge in itself. More details on key factors of experimental design and methodology for serum IgE tests, especially strategies to minimize false negatives and false positives, are published elsewhere (Goodman, 2008).
Dr Goodman's experience is that these in vitro tests can over predict allergy. A major uncertainty is how much in vitro cross-reactivity is required to trigger allergic reactions. It is difficult to verify biological relevance (allergy) when subjects are not available for challenge tests. For this reason, Dr Goodman proposed to utilize the rat basophilic leukemia (RBL) line 30/25, transfected with the human IgE receptor, to further develop an assay for testing human allergic sera described by Ladics et al. (2008). Because this approach assesses antibody binding to basophils and release of mediators, it may provide the means to demonstrate biologically relevant allergenic activity when food challenge or skin prick testing of human subjects is not possible. Due to the considerable effort associated with obtaining adequate amounts of appropriate sera and validating assays, conducting meaningful serum screening is still extremely challenging. Without rigorous attention to detail, there is great potential for generating data that may not be an accurate prediction of allergenicity.
MOUSE MODELS OF FOOD ALLERGY
The desire for an animal model that could be used to establish the relative allergenicity of a transgenic protein as compared to conventional food proteins has been recognized since the first discussions on safety assessment of transgenic proteins (Metcalfe et al., 1996). As with any toxicity assessment, an appropriate animal model should produce sensitization and/or elicitation of allergic symptoms at a physiologically relevant dose, via the relevant route of exposure (generally thought to be ingestion) in a standard (readily available) mouse strain. However, oral tolerance is a major barrier to developing such an ideal model. In mice, as in most humans, the immune response to an ingested protein (that survives digestion in the stomach) is an active process (oral tolerance) that blocks the development of IgE and delayed-type hypersensitivity responses (Strobel and Mowat, 2006). Five investigators presented work on six new mouse models. (See Table 2 for a summary and comparison of these models.) All but two involve the use of adjuvant at the time of sensitization to circumvent oral tolerance, and all but one used the oral route of sensitization. Earlier attempts to develop a model using ip sensitization have been discussed elsewhere (Dearman et al., 2003; Thomas et al., 2005b) and were only briefly mentioned at the current meeting.
TABLE 2.
Summary of Mouse Models
| Investigator | Mouse strain | Sensitization route (mg/mouse/dose) | Adjuvant | Challenge route (mg/mouse) | End points | Food extracts or allergens |
| Gangura | BALB/c | Dermal (0.05, 0.5, 1), six dose at weekly interval | None | Oral (13) | IgE | Hazelnutb |
| Cashewnutb | ||||||
| Symptoms score | Sesameb | |||||
| Eggb | ||||||
| Milkb | ||||||
| Shellfishb | ||||||
| Temperature | Amaranth seedb | |||||
| Kidney beanc | ||||||
| Pinto beanc | ||||||
| Blueberryc | ||||||
| Bowmand | C3H/HeJ | Oral (1, 2, 5), two doses at weekly interval | CT | None | IgE | Peanutb |
| Brazil nutb | ||||||
| Egg whiteb | ||||||
| Turkeyc | ||||||
| Spinachc | ||||||
| Bowmane | C3H/HeJ | Oral (1, 2), one dose | None | ip (0.1) | IgE | Peanutb |
| Brazil nutb | ||||||
| Egg whiteb | ||||||
| Ovalbuminb | ||||||
| Turkeyc | ||||||
| Spinachc | ||||||
| Lefevbre | C3H/HeJ | Oral (2), two doses at weekly interval | CT | None | IgE | Peanutf |
| Hazelnutb | ||||||
| Potatoc | ||||||
| Spinachc | ||||||
| HoganEsch | A/J | Oral (1), six doses over 3 weeks | CT | None | IgE | Peanutf |
| BALB/cJ | Ovalbuminb | |||||
| C3H/HeJ | Spinachc | |||||
| Potatoc | ||||||
| Brycef | BALB/c or C57BL/6 | Oral (0.1), eight doses at daily interval | SEB | Oral (5) | IgE | Ovalbuminb |
| Symptoms score | ||||||
| Physiologyg | ||||||
| Eosinophils | Peanutb | |||||
| Mast cell degranulation |
Note. CT, cholera toxin.
Known food allergens.
Not thought to be food allergens.
Temperature, blood pressure, and plethysmography.
Dr Venu Gangur described a model that sensitizes mice to food extracts by transdermal exposure. Although not thought to be the principal route of exposure, data suggest humans can be sensitized through the skin (Lack et al., 2003). Food extract was applied weekly for 6 weeks followed by oral challenge and observation for anaphylactic end points. IgE levels were also assessed (Birmingham et al., 2007; Navuluri et al., 2006). Using this model, it was possible to distinguish between allergenic food extracts and non-allergenic extracts with the exception that two non-allergenic extracts appeared to have sensitizing (IgE inducing) potential, but neither produced an elicitation response following the oral challenge.
Three investigators, Drs Christal Bowman, Harm HoganEsch, and David Lefebvre described variations of a mouse model originally developed to study peanut and milk allergies (Li et al., 1999, 2000) and recently applied (Bowman and Selgrade, 2008a) to assess the relative allergenicity of various food extracts. Mice were sensitized with two doses of food extract (in the 1–5 mg range), orally, 1 week apart, with cholera toxin as adjuvant, and specific IgE was assessed 1 week after the second dose. Two of the three investigators were able to distinguish between allergenic and non-allergenic food extracts based on the IgE response. The A/J mouse appeared to be the most sensitive mouse strain, but the C3H/HeJ mouse produced similar results.
Dr Paul Bryce described an oral exposure model that used a much lower sensitizing dose (0.1 mg) given on eight consecutive days using staphylococcal enterotoxin B (SEB) instead of cholera toxin as the adjuvant (Ganeshan et al., 2009). In addition to measuring antigen-specific IgE, a challenge dose without adjuvant was administered 24 h after the last sensitization, and various physiologic and immunologic end point characteristic of anaphylaxis were assessed. This model gave promising results for peanut extract and ovalbumin; however, as yet, neither non-allergenic food extracts nor non-allergenic proteins have been tested in this model. It should be noted that previous attempts to validate a model that employed the ip route of exposure were thwarted by mixed results in that some investigators observed positive responses to purportedly non-allergenic proteins (Herouet-Guicheney et al., 2009; Thomas et al., 2005b). Also, in the Bowman model, non-allergenic food extracts began to show significant increases in specific serum IgE at high doses when the sensitization regimen was extended to 4 weekly exposures (Bowman and Selgrade, 2008a). This observation suggests that it is important to test non-allergenic food extracts in the Bryce model, which has a longer sensitization time, albeit with lower doses.
The last oral exposure model presented assessed the induction of oral tolerance rather than sensitization because an allergic response requires the ability to sensitize as well as to avoid the induction of oral tolerance. Mice were fed a single dose of vehicle or food extract without adjuvant and challenged 1 week later by the ip route with the food extract of interest. Significantly lower IgE levels in mice that received the oral food extract exposure versus vehicle were considered to be indicative of oral tolerance, which mitigates allergenicity (Bowman and Selgrade, 2008b). Also related to tolerance, Dr HoganEsch presented data (unpublished) using mice deficient in TCR delta gamma–positive T cells, which are thought to be important in the induction of oral tolerance. However, oral administration of allergenic food extracts (without adjuvant) to mice lacking this receptor did not lead to the expected induction of IgE antibodies.
Predictions of allergenicity based on Bowman's mouse model appear to be consistent with predictions based on resistance to pepsin digestibility and lend credence to the use of data from digestibility assays in decision making. Pepsin (stomach digestion enzyme) stability is clearly important for sensitization (Bowman and Selgrade, 2008a), but resistance to both pepsin and trypsin (intestinal digestion enzyme) appears to be required for oral tolerance (Bowman and Selgrade, 2008b). Thus, proteins that escape pepsin digestion in the stomach but undergo trypsin digestion in the gut may pose more of a risk than those that escape both pepsin and trypsin digestion because they can induce sensitization, but not tolerance. Altering digestibility, pH, and/or solubility of the sensitizing food extract can change the results obtained in the oral animal models (Bowman and Selgrade, 2008b), suggesting that the matrix in which proteins are presented can affect results. Others have drawn similar conclusions (Foss et al., 2006; Mills and Mackie, 2008; Thomas et al., 2007b). In fact, it has been suggested that purified peanut allergens possess little intrinsic immune-stimulating capacity in contrast to a whole peanut extract (van Wijk et al., 2005). The role of the food matrix requires further study. From some regulators’ point of view, it would be desirable to test purified, bacterial-expressed, transgenic proteins because the transgenic protein, not the food crop, is regulated. However, this may not adequately mimic real world exposure. Whereas, availability of transgenic crop extracts is limited, for positive and negative controls extracts are more easily obtained than purified proteins.
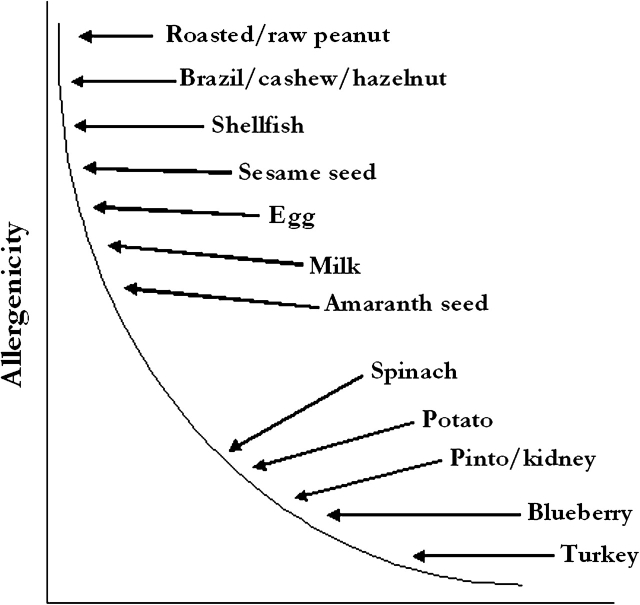
There was general agreement that significant progress has been made toward developing animal models, but that none of the currently available models are ready for validation. More work is needed on identification of appropriate end points, particularly those that reflect anaphylactic activity. It would be especially useful to understand the relationship between IgE levels and some of the manifestations of disease, an ongoing issue for all atopic diseases. It will also be important to resolve the role that matrix plays before designing a validation study. Research is needed to compare the allergenicity of food extracts versus the purified allergens and, if necessary, to devise a representative food matrix. In addition, before validation can be considered, decisions have to be made regarding which mouse strains and adjuvants to include, as well as the ideal doses of test materials. Appropriate test substances that represent a range from highly (commonly) allergenic to poorly (rarely) allergenic need to be selected. The goal of animal testing should be to establish a spectrum of food allergy potencies (conceptualized for food extracts in Fig. 1) or for specific food proteins and then determine where in that spectrum transgenic foods (or novel proteins) fit. Especially when IgE is used as the end point, the goal should not be to show no response but to show a response no greater than that associated with most non-allergenic foods and/or proteins.
FIG. 1.
Conceptualized spectrum of allergenic potency of food extracts based on perceived allergenicity in humans (adapted from Kimber, unpublished data).
PROTEOMICS
Proteomics is a rapidly progressing technology used for a wide range of applications (Canovas et al., 2004; Jorrin et al., 2007). This session considered the use of proteomics to assess the possibility that biotechnology could alter the amount of existing (endogenous) protein allergens in food crops. Quantitative proteomics is technologically challenging. Therefore, work was presented comparing three analytical methods: antibody-based—ELISA/Western blot, SDS-polyacrylamide gel electrophoresis/2-dimensional gel electrophoresis (2-DGE), and liquid chromatography (LC)/mass spectrometry. The main limitation of the ELISA/Western blot approach is access to validated methods, reagents, and standards, similar to the problems encountered in serum screening. The 2-DGE method has been found to be an effective approach (FAO/WHO, 2001; Ruebelt et al., 2006) as presented by Dr Corrine Herouet-Guicheney. This proteomics profiling approach may detect potential alterations with regards to the amount and/or expression of protein without the use of antibodies or human sera. Data were presented, suggesting that LC/mass spectrometry may be a viable, less labor intensive alternative (http://biochem.missouri.edu/faculty/). Dr Jay Thelen presented data on variation in the profile of major seed allergens in four non-transgenic varieties of soy planted in seven different regions. There was a fair amount of natural variability, probably because many variables including genetic background, climate, nutrient availability, and other factors can affect the amount of protein present in a crop. It was concluded, regardless of the method used to quantify protein, that as long as allergen expression in genetically modified crops falls within the range of natural variability, there should be no exposure concerns or need to test further. Thus, it is important to determine the natural variation range of protein allergen levels in non-genetically modified crop species before any of these methods can be utilized to evaluate protein levels in genetically modified crops. It is currently impossible to correlate protein expression with biological relevance because of limited data on quantitative thresholds for sensitizing individuals to most allergens.
BASIC RESEARCH
Plans for three recently funded grants from the joint NIAID/EPA initiative were described in the final session of the meeting. Dr Cecilia Berin described plans to use a mouse model to understand the role of thymic stromal lymphopoietin (TSLP) in food allergy. TSLP is an epithelial-derived cytokine that has a central role in the development of allergic inflammation in the skin and lung. The hypothesis to be tested is that overexpression of TSLP promotes allergic sensitization to food allergens. In addition to identifying risk factors that may explain individual susceptibility to food allergy, results of this grant could provide additional end points to be assessed in mouse models and could also lead to the development of a transgenic mouse that overexpresses TSLP and, hence, is more susceptible to food allergy.
The goal of the grant presented by Dr Anne Sperling is to determine whether allergen-specific IgG contributes to the cellular and molecular processes involved in generating food allergy. There is growing evidence that IgG may play a role in food allergy and other atopic diseases (Lau et al. 2005; Leung et al. 2003; Sicherer and Sampson, 2007). The anaphylactic reactions elicited in mice following sensitization with cholera toxin differ from those observed in the SEB model (described above) in that the latter exhibits a late-phase airway response that is not seen with cholera toxin–driven sensitization (Ganeshan et al., 2009), as well as greater eosinophilia and plasma histamine. Both models promote equivalent levels of antigen-specific IgE. However, cholera toxin promotes both IgG1 and IgG2a, whereas SEB promotes only IgG1. Hence, the investigators hypothesize that mast cells and macrophage-mediated responses during anaphylaxis are altered by activation via the IgG receptor and that the differences in IgG1 and IgG2a production between cholera toxin and SEB-driven sensitization underlie the differences in anaphylactic responses observed. Others have demonstrated in vitro activation of human mast cells with IgG1 (Woolhiser et al., 2003). Clearly, the results of this work could influence adjuvant selection for mouse models, in addition to increasing our understanding of the role of IgG subclasses in food allergy.
The final presentation, by Dr Fred Finkelman, also focused on the elicitation (anaphylactic) response, specifically to peanut allergens and the roles played by innate immunity and complement (C). The investigators intend to test the following hypotheses: (a) peanuts have components that induce shock primarily by causing the production of C-derived anaphylatoxins that induce macrophages and mast cells to produce platelet-activating factor and histamine; (2) the C3-related anaphylactoid response may act synergistically with peanut-induced IgE-mediated mast cell degranulation to induce the severe anaphylaxis experienced by some peanut-allergic patients; and (c) the inflammatory response stimulated by peanut components acts as an adjuvant that promotes the induction of a Th2 response to the major peanut allergens. The investigators theorize that similar responses may occur with tree nuts, but not milk or egg white. A better understanding of the elicitation response may suggest other end points that may be more predictive of anaphylaxis than IgE and provide a better understanding of the structural components of peanuts and tree nuts that cause them to be potent food allergens. Through the joint exploratory research initiative, NIAID also funded several other grants that were not discussed at this meeting.
SUMMARY
The studies presented here provide support for current approaches used to assess the allergenic potential of transgenic proteins (which are highly conservative, but effective) and also provide new tools that could be quite useful for safety assessment. More sophisticated structure-activity tools may prevent the unnecessary elimination of potentially useful products early in the development process. More detailed protocols for serum testing have been developed that should be applied so that the data generated are reliable as there are many pitfalls that can lead to inaccurate prediction of allergenicity using this approach. LC/mass spectrometry may be a viable alternative to more labor-intensive approaches to quantify proteins. Progress has also been made with animal models; data generated using the Bowman mouse model suggest that pepsin digestibility is a good indicator of potential allergenicity. Animal research also suggests that the food matrix may play an important role in the sensitization process. All the above rely heavily on IgE as the indicator for potential allergenicity. Because it is possible for IgE responses to occur in the absence of an adverse reaction in both humans and mice, basic research is currently underway to better understand the cofactors that contribute to anaphylactic/systemic reactions and potentially help to refine our approaches to safety assessment in the future. Likewise research underway to better understand oral tolerance may help to refine future safety assessments. To date, assessments have largely focused on the sensitization process, but work presented here suggests that the ability to sensitize and the ability to evade oral tolerance are not one and the same. Pepsin stability is clearly important for sensitization, but resistance to both pepsin and trypsin appear to be required for oral tolerance. Research on food allergies in general is receiving more attention from the scientific community because of targeted research initiatives, which should result in a better understanding of the disease and all of its causes.
FUNDING
Drs Werner Braun and Catherine Schein, University of Texas Medical Branch, Galveston, TX, Dr Richard Goodman, University of Nebraska, Lincoln, NE; Venu Gangur, Michigan State University, East Lansing, MI, Dr Harm HoganEsch, Purdue University, West Lafayette, IN, Dr Paul Bryce, Northwestern University, Chicago, IL, Dr Cecilia Berin, Mount Sinai School of Medicine, New York, NY; and Dr Anne Sperling, University of Chicago, IL, are supported by EPA Science to Achieve Results grants; Dr Christal Bowman was supported by EPA's intramural program (Research Triangle Park, NC), Dr David Lefebvre by Health Canada (Ottawa), and Dr Fred Finkelstein, University of Cincinnati, OH, by National Institute of Allergy and infectious Disease.
Acknowledgments
Thanks to Drs Michelle Embry (International Life Sciences Institute, HESI), Corinne Herouet-Guicheney (Bayer Cropscience), Linda Meyer (Syngenta), Scott Shore (Syngenta), Scott McClain (Monsanto Company), Nicola Stagg (Dow AgroSciences LLC), and Ms Sue MacIntosh (MacIntosh & Associates, Inc.) and the HESI Protein Allergenicity Committee for financial support of the workshop.
References
- Altmann F. The role of protein glycosylation in allergy. Int. Arch. Allergy Immunol. 2007;142:99–115. doi: 10.1159/000096114. [DOI] [PubMed] [Google Scholar]
- Bannon GA, Ogawa T. Evaluation of available IgE-binding epitope data and its utility in bioinformatics. Mol. Nutr. Food Res. 2006;50:638–644. doi: 10.1002/mnfr.200500276. [DOI] [PubMed] [Google Scholar]
- Batista R, Saibo N, Lourenço T. Microarray analyses reveal that plant mutagenesis may induce more transcriptomic changes than transgene insertion. Proc. Natl. Acad. Sci. U.S.A. 2008;105:3640–3645. doi: 10.1073/pnas.0707881105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmingham NP, Parvataneni S, Hassan HM, Harkema J, Samineni S, Navuluri L, Kelly CJ, Gangur V. An adjuvant-free mouse model of tree nut allergy using hazelnut as a model tree nut. Int. Arch. Allergy Immunol. 2007;144:203–210. doi: 10.1159/000103993. [DOI] [PubMed] [Google Scholar]
- Bowman CC, Selgrade MK. Differences in allergenic potential of food extracts following oral exposure in mice reflect differences in digestibility: Potential approaches to safety assessment. Toxicol. Sci. 2008a;102:100–109. doi: 10.1093/toxsci/kfm288. [DOI] [PubMed] [Google Scholar]
- Bowman CC, Selgrade MK. Failure to induce oral tolerance in mice is predictive of dietary allergenic potency among foods with sensitizing capacity. Toxicol. Sci. 2008b;106:435–443. doi: 10.1093/toxsci/kfn200. [DOI] [PubMed] [Google Scholar]
- Burks AW, Sampson HA. Anaphylaxis and food allergy. In: Metcalfe DD, Sampson HA, Simon RA, editors. Food Allergy: Adverse Reactions to Foods and Food Additives. Cambridge, MA: Blackwell Science; 1997. pp. 25–245. [Google Scholar]
- Canovas FM, Dumas-Gaudot E, Recorbet G, Jorrin J, Mock HP, Rossignol M. Plant proteome analysis. Proteomics. 2004;4:285–298. doi: 10.1002/pmic.200300602. [DOI] [PubMed] [Google Scholar]
- Codex Alimentarius Commission. Alinorm 03/34: Joint FAO/WHO Food Standard Programme, Codex Alimentarius Commission, Twenty-Fifth Session, Rome, Italy, 30 June–5 July, 2003. Appendix III, Guideline for the Conduct of Food Safety Assessment of Foods Derived from Recombinant-DNA Plants, and Appendix IV, Annex on the Assessment of Possible Allergenicity. 2003. pp. 47–60. http://www.codexalimentarius.net/download/report/116/al03_13e.pdf. Accessed April 26, 2009. [Google Scholar]
- Dearman RJ, Skinner RA, Herouet C, Labay K, Debruyne E, Kimber I. Induction of IgE antibody responses by protein allergens: Inter-laboratory comparisons. Food Chem. Toxicol. 2003;41:1509–1516. doi: 10.1016/s0278-6915(03)00167-4. [DOI] [PubMed] [Google Scholar]
- FAO/WHO. Evaluation of Allergenicity of Genetically Modified Foods. 2001. Report of a Joint FAO/WHO Expert Consultation on Allergenicity of Foods Derived from Biotechnology, Rome, Italy. [Google Scholar]
- Foss N, Duranti M, Magni C, Frokiaer H. Assessment of lupin allergenicity in the cholera toxin model: Induction of IgE response depends on the intrinsic properties of the conglutins and matrix effects. Int. Arch. Allergy Immunol. 2006;141:141–150. doi: 10.1159/000094716. [DOI] [PubMed] [Google Scholar]
- Ganeshan K, Neilsen CV, Hadsaitong A, Schleimer RP, Luo X, Bryce PJ. Impairing oral tolerance promotes allergy and anaphylaxis: A new murine food allergy model. J. Allergy Clin. Immunol. 2009;123:231–238. doi: 10.1016/j.jaci.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAO. GAO-02-566, Washington, DC. (U.S.G.A. Office ed.) 2002. Genetically modified foods: Experts view regimen of safety tests as adequate, but FDA's evaluation process could be enhanced. Available at: http://www.gao.gov/new.items/d02566.pdf. Accessed April 26, 2009. [Google Scholar]
- Goodman RE. Performing IgE serum testing due to bioinformatics matches in the allergenicity assessment of GM crops. Food Chem. Toxicol. 2008;46:S24–S34. doi: 10.1016/j.fct.2008.07.023. [DOI] [PubMed] [Google Scholar]
- Herouet-Guicheney C, Aldemir H, Bars R, de Barbeyrac D, Kennel P, Rouquié D, Stahl BU, Kimber I, Dearman RJ. Inter-laboratory comparisons of assessment of the allergenic potential of proteins in mice. J. Appl. Toxicol. 2009;29:141–148. doi: 10.1002/jat.1391. [DOI] [PubMed] [Google Scholar]
- Hileman RE, Silvanovich A, Goodman RE, Rice EA, Holleschak G, Astwood JD, Hefle SL. Bioinformatic methods for allergenicity assessment using a comprehensive allergen database. Int. Arch. Allergy Immunol. 2002;128:280–291. doi: 10.1159/000063861. [DOI] [PubMed] [Google Scholar]
- Holzhauser T, Ree R, Poulsen LK, Bannon GA. Analytical criteria for performance characteristics of IgE binding methods for evaluating safety of biotech food products. Food Chem. Toxicol. 2008;46(Suppl. 10):S15–S19. doi: 10.1016/j.fct.2008.07.026. [DOI] [PubMed] [Google Scholar]
- Ivanciuc O, Garcia T, Torres M, Schein CH, Braun W. Characteristic motifs for families of allergenic proteins. Mol. Immunol. 2009;46:559–568. doi: 10.1016/j.molimm.2008.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanciuc O, Midoro-Horiuti T, Schein CH, Xie L, Hillman GR, Goldblum RM, Braun W. The property distance index PD predicts peptides that cross-react with IgE antibodies. Mol. Immunol. 2009;46:559–568. doi: 10.1016/j.molimm.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorrin JV, Maldonado AM, Castillejo MA. Plant proteome analysis: A 2006 update. Proteomics. 2007;7:2947–2962. doi: 10.1002/pmic.200700135. [DOI] [PubMed] [Google Scholar]
- Lack G, Fox D, Northstone K, Golding J. Factors associated with the development of peanut allergy in childhood. N. Engl. J. Med. 2003;348:977–985. doi: 10.1056/NEJMoa013536. [DOI] [PubMed] [Google Scholar]
- Ladics GS. Current codex guidelines for assessment of potential protein allergenicity. Food Chem. Toxicol. 2008;46(Suppl. 10):S20–S23. doi: 10.1016/j.fct.2008.07.021. [DOI] [PubMed] [Google Scholar]
- Ladics GS, Bannon GA, Silvanovich A, Cressman RF. Comparison of conventional FASTA identity searches with the 80 amino acid sliding window FASTA search for the elucidation of potential identities to known allergens. Mol. Nutr. Food Res. 2007;51:985–998. doi: 10.1002/mnfr.200600231. [DOI] [PubMed] [Google Scholar]
- Ladics GS, Holsapple MP, Astwood JD, Kimber I, Knippels LM, Helm RM, Dong W. Workshop overview: Approaches to the assessment of the allergenic potential of food from genetically modified crops. Toxicol. Sci. 2003;73:8–16. doi: 10.1093/toxsci/kfg055. [DOI] [PubMed] [Google Scholar]
- Ladics GS, van Bilsen JHM, Brouwer HMH, Vogel L, Vieths S, Knippels LMJ. Assessment of three human FcϵRI-transfected RBL cell lines for identifying IgE induced degranulation utilizing peanut-allergic patient sera and peanut protein extract. Regul. Toxicol. Pharmacol. 2008;51:288–294. doi: 10.1016/j.yrtph.2008.04.012. [DOI] [PubMed] [Google Scholar]
- Lau S, Illi S, Platts-Mills TA, Riposo D, Nickel R, Gruber C, Niggemann B, Wahn U. Longitudinal study on the relationship between cat allergen and endotoxin exposure, sensitization, cat-specific IgG and development of asthma in childhood—Report of the German Multicentre Allergy Study (MAS 90) Allergy. 2005;60:766–773. doi: 10.1111/j.1398-9995.2005.00781.x. [DOI] [PubMed] [Google Scholar]
- Leung DY, Sampson HA, Yunginger JW, Burks AW, Jr, Schneider LC, Wortel CH, Davis FM, Hyun JD, Shanahan WR., Jr Effect of anti-IgE therapy in patients with peanut allergy. N. Engl. J. Med. 2003;348:986–993. doi: 10.1056/NEJMoa022613. [DOI] [PubMed] [Google Scholar]
- Li XM, Schofield BH, Huang CK, Kleiner GI, Sampson HA. A murine model of IgE-mediated cow's milk hypersensitivity. J. Allergy Clin. Immunol. 1999;103:206–214. doi: 10.1016/s0091-6749(99)70492-6. [DOI] [PubMed] [Google Scholar]
- Li XM, Serebrisky D, Lee SY, Huang CK, Bardina L, Schofield BH, Stanley JS, Burks AW, Bannon GA, Sampson HA. A murine model of peanut anaphylaxis: T- and B-cell responses to a major peanut allergen mimic human responses. J. Allergy Clin. Immunol. 2000;106:150–158. doi: 10.1067/mai.2000.107395. [DOI] [PubMed] [Google Scholar]
- Metcalfe DD, Astwood JD, Townsend R, Sampson HA, Taylor SL, Fuchs RL. Assessment of the allergenic potential of foods derived from genetically engineered crop plants. Crit. Rev. Food Sci. Nutr. 1996;36(Suppl.):S165–S186. doi: 10.1080/10408399609527763. [DOI] [PubMed] [Google Scholar]
- Mills EN, Mackie AR. The impact of processing on allergenicity of food. Curr. Opin. Allergy Clin. Immunol. 2008;8:249–253. doi: 10.1097/ACI.0b013e3282ffb123. [DOI] [PubMed] [Google Scholar]
- Navuluri L, Parvataneni S, Hassan H, Birmingham NP, Kelly C, Gangur V. Allergic and anaphylactic response to sesame seeds in mice: Identification of Ses i 3 and basic subunit of 11s globulins as allergens. Int. Arch. Allergy Immunol. 2006;140:270–276. doi: 10.1159/000093284. [DOI] [PubMed] [Google Scholar]
- Oezguen N, Zhou B, Negi SS, Ivanciuc O, Schein CH, Labesse G, Braun W. Comprehensive 3D-modeling of allergenic proteins and amino acid composition of potential conformational IgE epitopes. Mol. Immunol. 2008;45:3740–3747. doi: 10.1016/j.molimm.2008.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. U.S.A. 1988; 85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radauer C, Bublin M, Wagner S, Mari A, Breiteneder H. Allergens are distributed into few protein families and possess a restricted number of biochemical functions. J. Allergy Clin. Immunol. 2008;121:847–852. doi: 10.1016/j.jaci.2008.01.025. [DOI] [PubMed] [Google Scholar]
- Ruebelt MC, Leimgruber NK, Lipp M, Reynolds TL, Nemeth MA, Astwood JD, Engel KH, Jany KD. Application of two-dimensional gel electrophoresis to interrogate alterations in the proteome of genetically modified crops. 1. Assessing analytical validation. J. Agric. Food Chem. 2006;54:2154–2161. doi: 10.1021/jf0523566. [DOI] [PubMed] [Google Scholar]
- Sicherer SH, Sampson HA. Peanut allergy: Emerging concepts and approaches for an apparent epidemic. J. Allergy Clin. Immunol. 2007;120:491–503. doi: 10.1016/j.jaci.2007.07.015. [DOI] [PubMed] [Google Scholar]
- Silvanovich A, Nemeth MA, Song P, Herman R, Tagliani L, Bannon GA. The value of short amino acid sequence matches for prediction of protein allergenicity. Toxicol. Sci. 2006;90:252–258. doi: 10.1093/toxsci/kfj068. [DOI] [PubMed] [Google Scholar]
- Stadler MB, Stadler BM. Allergenicity prediction by protein sequence. FASEB J. 2003;17:1141–1143. doi: 10.1096/fj.02-1052fje. [DOI] [PubMed] [Google Scholar]
- Strobel S, Mowat AM. Oral tolerance and allergic responses to food proteins. Curr. Opin. Allergy Clin. Immunol. 2006;6:207–213. doi: 10.1097/01.all.0000225162.98391.81. [DOI] [PubMed] [Google Scholar]
- Thomas K, Bannon G, Hefle S, Herouet C, Holsapple M, Ladics G, MacIntosh S, Privalle L. In silico methods for evaluating human allergenicity to novel proteins: International Bioinformatics Workshop Meeting Report, 23–24 February 2005. Toxicol. Sci. 2005a;88:307–310. doi: 10.1093/toxsci/kfi277. [DOI] [PubMed] [Google Scholar]
- Thomas K, Bannon G, Herouet-Guicheney C, Ladics G, Lee L, Lee SI, Privalle L, Ballmer-Weber B, Vieths S. The utility of an international sera bank for use in evaluating the potential human allergenicity of novel proteins. Toxicol. Sci. 2007a;97:27–31. doi: 10.1093/toxsci/kfm020. [DOI] [PubMed] [Google Scholar]
- Thomas K, Herouet-Guicheney C, Ladics G, Bannon G, Cockburn A, Crevel R, Fitzpatrick J, Mills C, Privalle L, Vieths S. Evaluating the effect of food processing on the potential human allergenicity of novel proteins: International workshop report. Food Chem. Toxicol. 2007b;45:1116–1122. doi: 10.1016/j.fct.2006.12.016. [DOI] [PubMed] [Google Scholar]
- Thomas K, Herouet C, Bannon G, Ladics GS, MacIntosh S, Privalle L, Woolhiser M. Evaluation of mouse models for assessing the allergenic potential of proteins. Toxicologist. 2005b;84(Suppl.):1307. [Google Scholar]
- van Wijk F, Nierkens S, Hassing I, Feijen M, Koppelman SJ, de Jong GA, Pieters R, Knippels LM. The effect of the food matrix on in vivo immune responses to purified peanut allergens. Toxicol. Sci. 2005;86:333–341. doi: 10.1093/toxsci/kfi187. [DOI] [PubMed] [Google Scholar]
- Woolhiser MR, Brockow K, Metcalfe DD. Activation of human mast cells by aggregated IgG through FcgammaRI: Additive effects of C3a. Clin. Immunol. 2003;110:172–180. doi: 10.1016/j.clim.2003.11.007. [DOI] [PubMed] [Google Scholar]