Abstract
This work provides an analysis of across-target bioactivity results in the screening data deposited in PubChem. Two alternative approaches for grouping-related targets are used to examine a compound's across-target bioactivity. This analysis identifies compounds that are selectively active against groups of protein targets that are identical or similar in sequence. This analysis also identifies compounds that are bioactive across unrelated targets. Statistical distributions of compound' across-target selectivity provide a survey to evaluate target specificity of compounds by deriving and analyzing bioactivity profile across a wide range of biological targets for tested small molecules in PubChem. This work enables one to select target specific inhibitors, identify promiscuous compounds and better understand the biological mechanisms of target-small molecule interactions.
Contact: ywang@ncbi.nlm.nih.gov; bryant@ncbi.nlm.nih.gov
1 INTRODUCTION
Bringing a new drug to market is a time consuming and costly process that is also governed by complex regulatory procedures. It is a 100 million dollar challenge typically results in failure (Riggs, 2004). As the drug discovery process gets more expensive as it proceeds with time, it is very important to select drug candidates efficiently and to exclude those with undesired characteristics that are likely to fail at a later development stage. One of the cornerstones of the earlier stages of drug development is hit selection employing High-throughput Screening (HTS) technology. With rapid advances in instrumental automation, combinatorial chemistry and assay technologies, hundreds of thousands of compounds can be tested in a matter of days (Burbaum and Sigal, 1997; Hann and Oprea, 2004). However, the effectiveness of the HTS technology is usually compromised by hits that fail in later stages of drug development, especially in time consuming and costly clinical trials, due to unsatisfactory pharmacokinetic properties, poor pharmacodynamic profiles, limited cell viability, possible toxicity, etc. Many of the false positives are across-target compounds that can act non-selectively on unrelated targets and cause unwanted effects or adverse drug reactions (Azzaoui et al., 2007; Seidler et al., 2003). Therefore, great efforts need to be taken during hit selection to identify compounds with undesired physiochemical properties or unwanted bioactivities towards certain targets, in order to exclude them from the lead optimization process.
One straightforward way to verify the target specificity of a compound is through a data mining process via literature and/or patent searches. However, this is an expensive approach that requires extensive expert knowledge. Another approach is to design a profiling assay against a panel of targets. This is a powerful approach, however it is usually aimed at a limited number of targets and a small number of compounds at the decision-making point when the hit is ready to be progressed to a lead (Azzaoui et al., 2007; Whitebread et al., 2005). Furthermore, an assay panel is often designed to include only pre-selected targets with known biological relationships. While such an approach is useful to identify or verify the expected biological activity against related targets, it is not ideal to discover ‘unexpected’ or off-target effects. Additionally, such screening data are mostly proprietary. Thus, it remains a challenge to obtain a comprehensive biological activity profile of a small molecule for investigating its target selectivity and specificity.
In 2005, the NIH Molecular Library Roadmap Initiative (Zerhouni, 2003) funded a nation-wide screening center network (Austin et al., 2004) to perform industrial scale HTS screening tests for a large collection of compounds. All of the biological activity data produced by this HTS campaign over the past 3 years are now publically available through the PubChem Bioassay database (http://pubchem.ncbi.nlm.nih.gov) at the National Center of Biotechnology Information. As of March 1, 2009, over 42 million biological test results were deposited in PubChem. This huge amount of biological activity information, generated by the unprecedented effort led by the National Institutes of Health, provides quantitative biological annotations for 761 772 unique chemical structures which have been tested in a single or multiple bioassays contained in PubChem. The PubChem resource provides for the first time, the opportunity for researchers to freely access screening data. It also opens great challenges to analyze this complex chemical biology data and to explore drug-target specificity and interaction mechanisms for rapid and efficient discovery of drug leads (Gribbon and Sewing, 2005; Macarron, 2006; Marshall, 1987). The growth rate of biological test results in PubChem has accelerated as the NIH Molecular Library Program (MLP) enters its second phase for developing chemical probes. The need to develop means to analyze and evaluate the rich and complex bioactivity results in PubChem has become imperative.
This work is aimed at data mining of the biological test results generated by the NIH MLP initiative by analyzing the target specificity properties of small molecules. We propose two approaches to derive and analyze the across-target bioactivity of a set of compounds. One is based on distinct targets, while the other is based on target cluster to discover their target selectivity and promiscuity. By examining and comparing the biological activity of the small molecules, one may identify compounds with desired selectivity for a given protein target or identify compounds that exhibit promiscuity. The intent of the article is to provide a timely survey on the bioactivities of newly identified bioactive compounds and provide additional insights into some well-studied compounds.
2 METHODS
2.1 BioAssay and compound collection in the across-target compound bioactivity analysis
As of March 1, 2009, there were 660 single target bioassays deposited in PubChem, which contained biological activity outcomes, identified as ‘active’ versus ‘inactive’ status specified by the assay result provider, as well as detailed screening results for the respective protein targets. This analysis utilized the activity outcome as specified by each bioassay data depositor. For screening assays, active and inactive statuses were defined based on the percentage of inhibition from test at a single concentration. For confirmatory assays, active and inactive statuses were defined based on EC50/IC50 values, derived from dose response curves following testing with multiple concentrations. The biological activities of 588 918 small molecules toward 267 unique protein targets were reported in these bioassay records, and results were found for a total of 103 518 compounds. These small molecules and their corresponding bioactivity outcomes were extracted from the PubChem database. In a case when contradicting test results were observed in a bioassay, such results were treated as inconclusive bioactivity outcome and excluded from this analysis. As a result, 10 957 compound-target pairs demonstrating inconclusive results were excluded from a total of 42 349 644 results. In addition, by taking advantage of the property profiling bioassays recently deposited in PubChem, compounds identified as active in the dithiotreitol (DTT) profiling bioassay (AID: 1234) and luciferase inhibition profiling bioassay (AID: 1269, 1379, 411 and 773) were excluded from test results of bioassays employing similar assay protocols.
2.2 Non-redundant target-based compound bioactivity analysis
In this analysis, the across-target compound activities were analyzed by comparing their biological test results across distinct protein targets. As multiple bioassays can be designed for the same protein target, one compound can be tested in different bioassays. In this case, the test results were grouped based on target sequence identity. As a result, a total of 588 918 compounds resulting from 660 bioassays were divided into 267 groups in such a way that each group represents the combined test results for one unique protein target. The number of distinct targets for each compound was summarized, and statistical distribution of compounds showing across-target activities was obtained subsequently.
2.3 Target cluster-based compound bioactivity analysis
Target cluster-based compound bioactivity analysis was performed to provide insights into compound activity across-target families. Protein target clusters were derived using a single linkage clustering method based on sequence similarity. The BLAST (Altschul et al., 1997) program was employed to compare and calculate sequence similarities among all of the 267 distinct protein target sequences under this study. A BLAST E-value threshold of 0.001 was used as sequence similarity cutoff to draw boundaries between target clusters. Following that, the 588 918 compounds resulting from 660 bioassays were divided into target cluster groups so that each group represented the combined test results for one protein target cluster. Test results were grouped and analyzed in a similar way as in the target identity-based analysis.
2.4 Analysis using a subset of biological test results
PubChem bioassays were characterized based on how the activity outcome was derived. Often a series of bioassays were performed for the same biological target system and results were reported as separate bioassay records, whereas analysis of bioactivity outcome was first done based on a single concentration test in the primary screening, and was then confirmed based on a follow up multiple concentration-response test. Usually, only a small fraction, 0–10% of the compounds were confirmed as ‘active’ in the later multiple concentration experiment. As the confirmatory bioassays most likely have a lower false positive rate and suggest a more accurate activity measurement, both target and target cluster-based analysis were carried out based on test results reported through confirmatory bioassays, in addition to the analysis using all of the available screen results for providing general background information. In this study, 341 assays out of a total of 660 were confirmatory assays, whereas a total of 331 702 unique compounds were tested in these confirmatory assays, involving 165 distinct protein targets.
3 RESULTS AND DISCUSSION
3.1 Identification of compounds with specific and across-target bioactivity
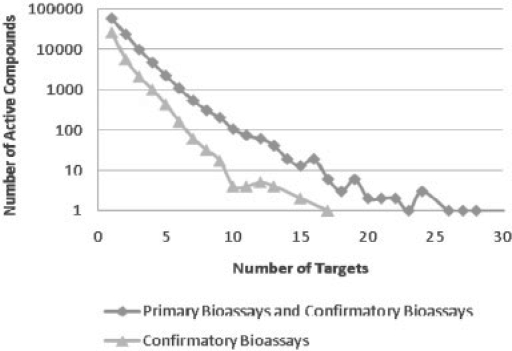
In this study, each compound was analyzed by enumerating their distinct targets over their demonstrated biological activity. Distribution of bioactivity among the compounds and the corresponding targets derived from the test results for the 267 distinct targets was analyzed and is shown in Figure 1. The x-axis indicates the number of targets and the y-axis gives the frequency of compounds that are active across a given number of distinct targets.
Fig. 1.
Distributions of compound activity among 267 targets.
There are two datasets shown in the Figure 1: 1> statistics for active compounds based on all bioassays (including primary, confirmatory); 2> statistics for active compounds based on confirmatory bioassays. For both datasets, a substantial fraction of the compounds (57.7% and 73.5%, respectively) demonstrated single target activity. We also observed a significant fraction of compounds exhibiting across-target activity, with the number of compounds dropping exponentially as the number of targets increases. Comparing the results obtained by confirmatory bioassays and those derived from all bioassays, it suggested that the number of compounds active against multiple targets reduced significantly in the confirmatory assays, by 5–10-fold on average. This may be attributed to the fact that a substantial number of the hits in the primary bioassays turned out to be negative in the confirmatory screenings. It should also be noted that not every hit in the primary screening was necessarily subjected to confirmatory test.
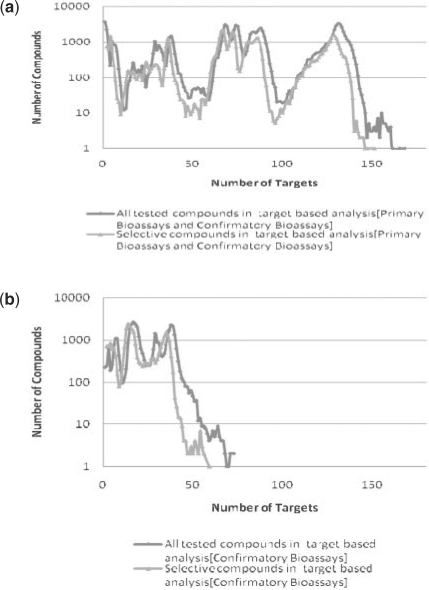
It is essential to examine the data completeness when analyzing compound target selectivity (Mestres et al., 2008). To further evaluate the suggested target selectivity, the profile of tested targets was also analyzed for each bioactive compound as shown in Figure 2a and b, which utilizes all screening (primary + confirmatory) results as well as confirmatory results alone. Figure 2 shows that the majority of the target specific compounds have been tested in many targets. For example, over 50% of the selective compounds have been tested in more than 70 distinct targets (Fig. 2a), which suggests a higher potential of target selectivity for such compounds. This should be of interest for chemical probe analysis or lead compound development, though the target specificity and selectivity of individual compounds needs to be investigated further.
Fig. 2.
(a) Completeness of compound tested among primary and confirmatory targets. (b) Completeness of compound tested among confirmatory targets.
This analysis enables one to identify potential target specific compounds, examine their potency, and evaluate target selectivity by considering all of the screening results provided in PubChem. These analyses used to be time consuming as a laborious search of various bioactivity information was required.
3.2 Identification of compounds with specific and across-target cluster bioactivity
The discovery of compounds across-target activities may stimulate a broad interest to investigate the underlying biological mechanisms. In the case when a compound hits closely related protein targets, it might be due to the significant similarities among the protein target sequences, or due to the highly conserved 3D structures around the binding pockets of the target proteins. On the other hand, if a compound shows multiple activities across unrelated targets, it might be interesting to investigate the possible causes behind its promiscuity. Thus, it is desirable to further explore the biological relationship of the protein targets, and to distinguish compounds responding to biologically related targets from those which interact with unrelated protein targets presumably through different molecular mechanisms. Towards this end, protein targets in the PubChem BioAssay database were compared and clustered. Protein targets can be clustered based on 3D structure comparison to ensure representation of distinct binding pockets within each cluster. This is, however, impractical in the current analysis due to the lack of known experimental structural data for certain targets. Thus, in this study, the target clusters were derived based on the sequence similarity.
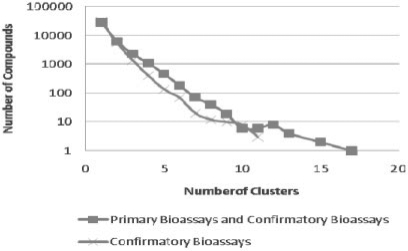
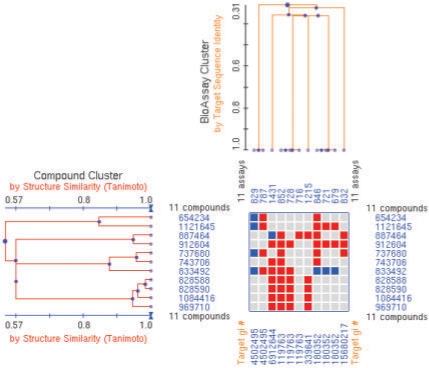
In this analysis, the 267 non-redundant targets were clustered based on their sequence similarity measured by the BLAST (Altschul et al., 1997) algorithm. As a result, 116 target clusters were derived, which include protein families such as kinase, phosphatase, protease and G protein-coupled receptor. Similar to the target identity-based analysis as described above, distributions of compound activity across the derived target clusters were obtained for two datasets as shown in Figure 3. This analysis shows that >50% of the compounds are active only against targets similar in sequences. It also shows that there is a substantial number of compounds revealing activity across non-related or distantly related protein targets. Comparison of the results given in Figures 1 and 3 suggests that the trends of the respective distributions of each dataset resembled each other. Compounds showing selectivity to a single target cluster should include those specific to a single target as well as those which are active to multiple members of the same target family, but otherwise inactive in other target clusters. One example of this would be a group of compounds showing activity across several members of one protease family including Complement factor C1s, Factor XIa, Factor XIIa, Thrombin, Kallikrein-related peptidase and Cathepsin G as shown in Figure 4.
Fig. 3.
Distributions of compound activity among 116 target clusters.
Fig. 4.
A PubChem Heatmap display showing a cluster of compounds together with their biological test results across a group of related protein targets. Clusters of compounds (represented as PubChem Compound identifier ‘CID’) were derived based on 2D structure similarity and shown vertically. Clusters of BioAssays (represented as PubChem BioAssay identifier ‘AID’) were derived based on the sequence similarity of the tested targets and shown horizontally, where the GenBank identifiers of the corresponding protein targets are listed at the bottom of the heatmap view. Each cell in the Heatmap represents an individual activity outcome of a small molecule for the corresponding target, with ‘active’ results denoted by red color, and ‘inactive’ results denoted by blue color.
There were relatively few compounds that showed activity across a wide range of target clusters. While there were 43 753 compounds observed with multiple target activity, only 10 202 compounds were found to be active across multiple target clusters, and no compounds showed activity across > 11 target clusters based on the confirmatory dataset. When looking into such compounds and the corresponding protein clusters, it becomes apparent that one cause for the across-target activity is the structure conservation among distantly related protein families. For instance, it is known that some members of the cysteine protease family share low-sequence similarities. By examining the experimental crystal structures for certain members of this protein family, however, highly conserved 3D structures are observed around the binding pocket. Thus, it is not surprising that a number of compounds showing activity for both Cathepsin B and Cathepsin G based on screening results in PubChem.
This across-target cluster analysis also revealed compounds that show activity towards non-biologically related proteins. One such compound is myricetin (PubChem CID:5281672), a flavonoid that is commonly found in natural food source. An examination using the PubChem biological test results suggests that this compound is identified as an inhibitor of several proteins such as aldehyde dehydrogenase, Leishmania Mexicana Pyruvate Kinase, Cytochrome P450, Stress-activated protein kinase and human RNase H etc., with a strong potency (IC50 <10 uM). Such observation using PubChem bioactivity data agrees well with reports in the literature where the inhibition activity and possible mechanism of action of this small molecule have been widely discussed (Feng et al., 2008; Lee et al., 2007; Lu et al., 2006; McGovern et al., 2002; Ryan et al., 2003; von Moltke et al., 2004; Wu et al., 2008). The NIH Molecular Libraries-small Molecule Repository (MLSMR) contains a number of compounds that are either drugs on the market, or well-studied small molecules for which biological properties have been reported in the literature. In this case, the biological results in PubChem can be readily compared with previously reported bioactivity data using the PubMed links in PubChem, which are either provided by depositors, authors of articles or through MeSH annotation. However, there is a large portion of compounds in the MLSMR collection that either has not been well characterized, or the biological activity information has not been reported in scientific journals. In this analysis, 42 246 compounds, which account for 96.5% of the total 43 753 compounds identified with across-target activities, do not have any references provided to PubMed articles. The current analysis may provide insights into the biological activities for those compounds identified by the NIH Molecular Library Project.
PubChem is a public and open data repository system. The information content within PubChem was contributed by investigators from many organizations. Biological test results in the BioAssay database are diverse, and the criteria employed when determining bioactivity outcome varies depending on the scientific rationale chosen by each individual investigator. Data analysis using PubChem biological results is conceivably affected by the accuracy of such test results. Our analysis shows that a portion of compounds are observed with single target activity when tested against a wide range of protein targets, which suggests that most of the investigators employed conservative thresholds when assigning bioactivity outcome.
4 CONCLUSION
With the abundance of information on bioactivities of small molecules recently made available through PubChem, it has been a challenging task to mine the biological test results for drug development research. In this study, we proposed a method for target selectivity evaluation and compound promiscuity identification by analyzing the biological test results in PubChem. To the best of our knowledge, this work provides the first analysis on target specificity and promiscuity for a large library of bioactive compounds recently identified by the NIH Molecular Library Program. Statistical distribution of compound target selectivity was obtained and presented as a survey on across-target bioactivities of small molecules. Compounds demonstrating single target selectivity, as well as across-target or across target cluster activity were identified and reported. These data suggest that the proposed approach for across-target activity analysis can be an efficient way for selecting target specific compounds and identifying promiscuous compounds using the biological results in PubChem. The current analysis and survey may provide insights into a compound's bioactivity against previously undiscovered target. Although this method can not address all possible mechanisms, it provides a robust set of candidate compounds for researchers to further investigate their target selectivity and the mechanisms of observed promiscuity, if any. Furthermore, statistical models can be developed based on the accumulated knowledge for in silico analysis and prediction of target specificity for small molecules as well as across-target activity.
As is the norm for the NIH Molecular Library Program, primary HTS screenings for a large compound library are usually performed first. Based on the outcome of this screening, a small subset of compounds, usually no more than a few hundreds, are cherry picked and subjected to confirmatory testing where compounds are analyzed at a series of concentrations. The survey in this study shows that a significant fraction of those selected compounds demonstrated activity against multiple targets or multiple target clusters. Though it is beneficial for the research community to obtain a comprehensive activity profile of a small molecule regardless of previously reported activity, the identification of these compounds as done in the current analysis may provide guidance for compound selection and target specificity evaluation when planning further tests, especially tests at later stages in chemical probe development.
The PubChem BioAssay resource provides unprecedented opportunities for large-scale bioactivity analysis. Such analysis can be used to facilitate hit selection, off-target evaluation and to better understand the biological mechanisms of interactions between small molecules and their targets, thus, to aid the discovery and development of chemical probes and novel drugs.
ACKNOWLEDGEMENTS
We acknowledge the editorial assistance of the NIH Fellows Editorial Board.
Funding: Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Conflict of Interest: none declared.
REFERENCES
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin CP, et al. NIH Molecular Libraries Initiative. Science. 2004;306:1138–1139. doi: 10.1126/science.1105511. [DOI] [PubMed] [Google Scholar]
- Azzaoui K, et al. Modeling promiscuity based on in vitro safety pharmacology profiling data. Chem. Med. Chem. 2007;2:874–880. doi: 10.1002/cmdc.200700036. [DOI] [PubMed] [Google Scholar]
- Burbaum JJ, Sigal NH. New technologies for high-throughput screening. Curr. Opin. Chem. Biol. 1997;1:72–78. doi: 10.1016/s1367-5931(97)80111-1. [DOI] [PubMed] [Google Scholar]
- Feng BY, et al. Small-molecule aggregates inhibit amyloid polymerization. Nat. Chem. Biol. 2008;4:197–199. doi: 10.1038/nchembio.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribbon P, Sewing A. High-throughput drug discovery: what can we expect from HTS? Drug Discov. Today. 2005;10:17–22. doi: 10.1016/S1359-6446(04)03275-1. [DOI] [PubMed] [Google Scholar]
- Hann MM, Oprea TI. Pursuing the leadlikeness concept in pharmaceutical research. Curr. Opin. Chem. Biol. 2004;8:255–263. doi: 10.1016/j.cbpa.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Lee KW, et al. Myricetin is a novel natural inhibitor of neoplastic cell transformation and MEK1. Carcinogenesis. 2007;28:1918–1927. doi: 10.1093/carcin/bgm110. [DOI] [PubMed] [Google Scholar]
- Lu J, et al. Inhibition of Mammalian thioredoxin reductase by some flavonoids: implications for myricetin and quercetin anticancer activity. Cancer Res. 2006;66:4410–4418. doi: 10.1158/0008-5472.CAN-05-3310. [DOI] [PubMed] [Google Scholar]
- Macarron R. Critical review of the role of HTS in drug discovery. Drug Discov. Today. 2006;11:277–279. doi: 10.1016/j.drudis.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Marshall GR. Computer-aided drug design. Annu. Rev. Pharmacol. Toxicol. 1987;27:193–213. doi: 10.1146/annurev.pa.27.040187.001205. [DOI] [PubMed] [Google Scholar]
- McGovern SL, et al. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J. Med. Chem. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- Mestres J, et al. Data completeness–the Achilles heel of drug-target networks. Nat. Biotechnol. 2008;26:983–984. doi: 10.1038/nbt0908-983. [DOI] [PubMed] [Google Scholar]
- Riggs TL. Research and development costs for drugs. Lancet. 2004;363:184. doi: 10.1016/S0140-6736(03)15370-6. [DOI] [PubMed] [Google Scholar]
- Ryan AJ, et al. Effect of detergent on ‘promiscuous’ inhibitors. J. Med. Chem. 2003;46:3448–3451. doi: 10.1021/jm0340896. [DOI] [PubMed] [Google Scholar]
- Seidler J, et al. Identification and prediction of promiscuous aggregating inhibitors among known drugs. J. Med. Chem. 2003;46:4477–4486. doi: 10.1021/jm030191r. [DOI] [PubMed] [Google Scholar]
- von Moltke LL, et al. Inhibition of human cytochromes P450 by components of Ginkgo biloba. J. Pharm. Pharmacol. 2004;56:1039–1044. doi: 10.1211/0022357044021. [DOI] [PubMed] [Google Scholar]
- Whitebread S, et al. Keynote review: in vitro safety pharmacology profiling: an essential tool for successful drug development. Drug Discov. Today. 2005;10:1421–1433. doi: 10.1016/S1359-6446(05)03632-9. [DOI] [PubMed] [Google Scholar]
- Wu D, et al. D-Alanine:D-alanine ligase as a new target for the flavonoids quercetin and apigenin. Int. J. Antimicrob. Agents. 2008;32:421–426. doi: 10.1016/j.ijantimicag.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Zerhouni E. Medicine. The NIH Roadmap. Science. 2003;302:63–72. doi: 10.1126/science.1091867. [DOI] [PubMed] [Google Scholar]